Abstract
The FOXO family represents a group of transcription factors that is required for a number of stress related transcriptional programs including antioxidant response, gluconeogenesis, cell cycle control, apoptosis and autophagy. The liver utilizes several FOXO-dependent pathways to adapt to its routine cycles of feeding and fasting and to respond to the stresses induced by disease. FOXO1 is a direct transcriptional regulator of gluconeogenesis, is reciprocally regulated by insulin and has profound effects on hepatic lipid metabolism. FOXO3 is required for antioxidant responses and autophagy and is altered in Hepatitis C infection and fatty liver. Emerging evidence suggests dysregulation of FOXO3 in some hepatocellular carcinomas. FOXOs are notable for the extensive number of functionally significant post-translational modifications that they undergo. Recent advances in our understanding how FOXOs are regulated are providing a more detailed picture of how specific combinations of posttranslational modifications alter both nuclear translocation as well as transcriptional specificity under different conditions. This review summarizes emerging knowledge of FOXO function in the liver, FOXO changes in liver disease, and the posttranslational modifications responsible for these effects.
Keywords: FOXO1, FOXO3, FOXO4, gluconeogenesis, protein methylation, Hepatitis C
1. Introduction
The liver plays a central role in adaptation to stress. It is anatomically situated as the buffer between the gut and the systemic circulation and is required to buffer large transient fluxes of nutrients, exogenous toxins and gut-derived bacterial products. It must optimally utilize or dispose of these products originating from the portal circulation without disturbing the much more stable environment of the systemic circulation. For this reason the liver engages a number of stress response pathways that regulate metabolism, immune response, organic ion transport and cell proliferation. The ability to engage these stress response pathways allows the liver to respond to the changing input environment.
FOXO transcription factors are part of one important stress response pathway that is responsible for many of these regulatory events. They are necessary for plasticity of the organ, adaptation to fasting, response to stress, and regulation of cell proliferation. This article will review the role of the FOXO family of transcription factors in the hepatic homeostatic response and discuss how regulation of this pathway is altered in liver disease.
2. FOXO Transcription Factors
The O branch of the large forkhead family of transcription factors1 is ubiquitously expressed and highly conserved evolutionarily2. The prototype of the FOXO family was first described in C. elegans as daf16, a factor that is required for formation of a long-lived dormant form of the organism called the dauer larval stage. Subsequently, FOXO factors were shown to play a similar role in higher organisms and function to prevent cellular proliferation, induce antioxidant and stress response genes, and modify insulin sensitivity2, 3. In mammals there are 4 FOXO proteins, FOXO1, FOXO3a (sometimes called just FOXO3), FOXO4 and FOXO6. While FOXO6 is largely specific to neurons, the other 3 factors are widely distributed and are present in most tissues. There appears to be considerable overlap in the transcriptional targets of the three, but the consequences of knock outs in mice are very different with FOXO1 knock out being embryonically lethal due to failure of angiogenesis, FOXO3 knock out producing premature ovarian failure, and FOXO4 knock out having no obvious phenotype4. There is also evidence that each of these can compensate to some degree for loss of the others as triple conditional knockouts resulted in lymphomas, hemagiomas and angiosarcomas which did not occur with double knock out combinations4. Some specificity though clearly occurs as FOXO1 plays the major role of regulation of insulin sensitivity (see below) and FOXO3, an important longevity factor in invertebrates, mice and humans5-9, is more prominently associated with the antioxidant stress response and tumor suppressor activity1, 4-6, 10-12. FOXO transcriptional activity is regulated by a complex array of posttranslational modifications (PTMs). In many circumstances, the primary regulatory event is Akt mediated phosphorylation of three conserved amino acids, 2 serines and 1 threonine, that results in binding to 14-3-3 and nuclear export of the protein.
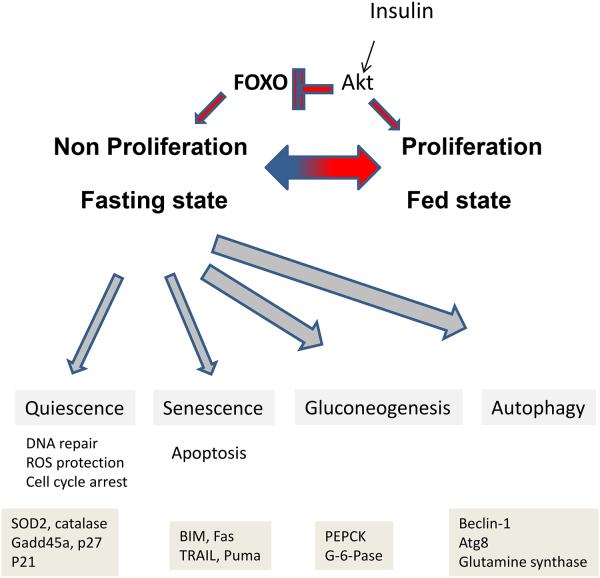
A conceptual theme that has emerged from the study of multiple FOXOs is that they are a major part of the mechanism that allows cells to transition between a fed/unstressed state where cell proliferation is favored and a fasting/stressed state which initially favors cell cycle arrest, DNA repair and antioxidant enzyme induction, but can proceed toward apoptosis and cell death (see Fig. 1). The precise program initiated, the particular FOXO proteins that predominate, and the nature of the PTMs that control the response varies between cell types and the particular stress circumstances that initiate the response.
Figure 1. FOXO functions.
The FOXO transcription factors serve as a counterpoint to Akt in the control of cell proliferation and gluconeogenesis. When active, as occurs when Akt activity is suppressed, FOXOs translocate to the nucleus where they initiate transcriptional programs for cell cycle arrest, oxidative stress protection, gluconeogenesis and autophagy. A brief list of a small subset of relevant target genes is listed for each function.
3. FOXO in normal liver function
Glucose metabolism
FOXO1 plays a major role in regulating the insulin response and the liver is one of its critical sites of action. The liver adapts to feeding through several insulin mediated events including increasing glucose uptake into hepatocytes, suppressing gluconeogenesis and glycogenolysis, and upregulating glycogen synthesis. In fasting, the withdrawal of insulin stimulation results in gluconeogensis through an upregulation of PEPCK and G-6-Pase, and induction of autophagy. This response is largely dependent on the interplay between Akt and FOXO1.
The role of FOXO1 in the adaptation to fasting has been largely documented by animal studies of overexpression and heterozygous null expression leading to increased or decreased FOXO1 expression. When FOXO1 is constitutively expressed in the liver, fasting blood glucose rises 13. Conversely, liver specific FOXO1 knock-out mice develop fasting hypoglycemia14. The mechanism behind these phenomena appears to be relatively straightforward. FOXO1 is active in the fasted state where it is dephosphorylated at the Akt sites, and localized in the nucleus. This results in the transcriptional induction of two gluconeogenic enzymes, G6Pc (Glucose-6-phosphatase catalytic subunit) and PEPCK (phosphoenolpyruvate carboxykinase)15 and increased hepatic glucose production. In the fed state, insulin signaling activates PI3kinase and the subsequent production of PIP3 activates Akt. Akt phosphorylates FoxO1 at Thr24, Ser253 and Ser316 leading to its nuclear exportation and inactivation16 with subsequent suppression of gluconeogenesis.
The importance of FOXO1 as a counter of Akt in the glycogen synthesis-gluconeogensis balance has been recently demonstrated using liver specific knock-out mice for both Akt and FOXO116. Hepatic deletion of Akt resulted in a constitutive hyperglycemia that was completely corrected by concomitant hepatic deletion of FOXO1. In the absence of both Akt and FOXO1 the mice were able to maintain glucose homeostasis through fasting and feeding. This demonstrates that FOXO1 is intrinsically glucogenic and in its absence, glucose homeostasis can be maintained without Akt activation. The primary function of insulin-induced Akt activation is to counteract FOXO1 and thus reduce glucose production during the fed state. This study also demonstrated that FOXO1 does not inhibit the insulin mediated upregulation of anabolic processes such as glycogen and lipid synthesis16.
The activity of FOXO1 as a regulator of blood glucose is also modulated by processes other than Akt phosphorylation. The balance between acetylation and deacetylation is a second order of regulation. Deacetylation by Sirt1under conditions of cellular stress, such as that induced by oxygen free-radicals activates transcription, overriding the nuclear exclusion effect of Akt and causing nuclear translocation/retention and expression of FOXO1 target genes including those involved in gluconeogenesis17. Other deacetylases contribute to FOXO1 activation as well. Class IIa HDACs have been shown to be positive regulators of hepatic FOXO1 in response to glucagon signaling during fasting. They are phosphorylated by AMPK and translocated to the nucleus where they deacetylate and activate FOXOs, inducing transcription of gluconeogenic genes18.
Several other more novel mechanisms have also been observed to play a role in FOXO1 regulation and hepatic glucose metabolism. XBP-1, a transcription factor involved in the unfolded protein response that induces expression of genes involved in ER membrane folding, has been shown to increase insulin sensitivity. This activity is independent of its transcriptional effects but can be accounted for by its direct binding to FOXO1, acting as a chaperone to direct it to proteosomal degradation 19. Another mechanism that appears to play a specific role in regulation of the glucuneogensis function of FOXO1 is O-GlcNAc modification 20, 21. This glylcosylation event activates transcriptional activity of FOXOs independently of nuclear translocation and results in upregulation of G6Pase and other gluconeogenic genes. Paradoxically, it is induced by hyperglycemia and appears to result from PGC-1α binding to O-GlcNAc transferase and targeting it to nuclear FOXO122.
Lipid metabolism
The second area of liver metabolic function regulated by FOXO is lipid metabolism. FOXO1 has an important role in the insulin-dependent regulation of hepatic VLDL production and persistence of VLDL in the circulation. This is achieved largely via transcriptional regulation of two important proteins, ApoC-III and microsomal triglyceride transfer protein (MTP) 23 and these play a major role in the regulation of circulating triglycerides during fasting. As discussed, in the absence of insulin, Akt activity is suppressed and FOXO1 is transcriptionally active. This effect results in an increase in MTP, the rate-limiting enzyme in hepatic VLDL production, increasing VLDL secretion. In addition, FOXO1 also results in increased transcriptional activity and hepatic secretion of ApoC-III. In the circulation, this apolipoprotein inhibits the activity of lipoprotein lipase, responsible for hydrolysis and uptake of the triglyceride component of VLDL and chylomicrons, thus prolonging the persistence of VLDL24. In response to feeding, FOXO1 is inactivated, shutting down both these mechanisms and preventing post-prandial hyperglycemia. In states of insulin resistance, this suppression of FOXO1 activity may fail to occur resulting in both hyperglycemia and hypertriglyceridemia. 25
Additional factors appear to be involved in the lipid effects of FOXO1 as well. Early attempts to understand the effects of FOXO on hepatic lipid metabolism involved expression of various mutated forms of FOXO1 that were felt to represent constitutively active forms of the protein. These studies seemed to imply both positive and negative effects of FOXO on lipid production and accumulation. One model for expression of constitutively active FOXO1 using a single S-253 mutated phosphorylation site led to increased hepatic triglyceride levels but lower levels in the circulation26. Another model for expression of constitutively active FOXO1 using alanine substitution at all three Akt phosphorylation sites had normal hepatic triglyceride levels15 but showed that increased FOXO1 activity led to suppression of a number of proteins required for lipid synthesis including SREBP-1c, acetyl-CoA carboxylase-α (ACC), and fatty acid synthase (FAS)15. These data are difficult to interpret unambiguously because the mutated forms of FOXO may behave differently in unanticipated ways.
Perhaps the best systems in which to study the net effects of FOXO proteins on hepatic and serum lipid homeostasis is in liver specific multiple FOXO knockouts. Zhang et al27 showed that ablation of FOXO1 caused a decrease in plasma glucose without a significant effect on lipid metabolism, but simultaneous knock out of FOXO1 and FOXO3 caused hepatic steatosis, increased hepatic lipid secretion and increased serum triglycerides27. While the precise mechanism for these effects could not be determined, these authors showed a negative transcriptional effect of FOXO3 and particularly the FOXO1/FOXO3 combination on two important genes of lipid synthesis, fatty acid synthase and HMG CoA reductase. A similar phenomenon was also observed by Tao et al28 who produced a hepatic-specific knockout of the combination of FoxO1, FoxO3 and FoxO4 in mice. This also resulted in lipid accumulation in the liver and an increase in expression of fatty acid synthase28. The mechanism of the lipid accumulation in this latter model, however, appeared to be primarily a result of the decrease in the FOXO-dependent expression of the enzyme nicotinamide phosphoribosyltransferase (Nampt) which is the rate limiting enzyme in the salvage pathway for NAD+. The FOXO triple knock out resulted in decreased levels of Nampt, a decrease in NAD+ levels and NAD+/NADH ratio, and a subsequent inhibition of NAD+-dependent deacetylases, particularly Sirt1. Direct manipulation of Nampt expression, both positive and negative, confirmed the centrality of this enzyme to regulation of lipid synthesis. The ultimate lipid accumulation could be secondary to SIRT inhibition resulting in increased acetylation of several proteins involved in in lipid synthesis and fatty acid oxidation such as SREBP-1b and PGC-1α 28. Together these results clearly show a lipid modulatory effect of FOXOs. FOXO1 activity by itself promotes hypertriglyceridemia, and FOXO3, in synergy with FOXO1, is able to suppress hepatic lipid accumulation by an indirect process.
Autophagy and adaptation to starvation
Based on the above discussion, it is clear that FOXOs are critical for adaptation of the liver to low nutrient states. They are activated by AMPK, increase glucose production, and prevent lipid accumulation seen in insulin resistant states. Another well described mechanism by which FOXOs promote adaptation to starvation is their promotion of autophagy29. There are likely several mechanisms by which FOXOs promote autophagy. Several of the proteins that make up critical parts of the autophagy machinery, including Beclin-1 and Atg8 are direct FOXO transcriptional targets. Recently van der Vos and colleagues30 demonstrated that FOXOs, in particular FOXO3, plays another, more indirect role in autophagy through influencing amino acid metabolism. They determined that glutamine synthase is a target gene of FOXO3 and as a consequence, cellular glutamine levels increase when FOXO3 is active. The increased glutamine inhibits mTORC1 signaling activity, decreasing its negative regulation on autophagy.
In addition to the role of FOXO proteins in autophagy via induction of gene expression, and modulation of glutamine levels, cytosolic FOXO1 has been shown to have a transcriptionally-independent role in autophagy as well. When subjected to stress such as nutrient deprivation, cytosolic FOXO1 dissociates from SIRT2 which leads to its acetylation. Acetylated FOXO1 was then shown to directly interact with Atg7, a key regulator of the formation of the autophagosome31. Overall, these multiple mechanisms show that FOXOs stimulate autophagy and promote adaptation to starvation and fasting. Autophagy stimulation also promotes lipid degredation32 and is thus another mechanism by which active FOXO prevents hepatic steatosis.
4. FOXO and liver disease
In spite of the well documented importance of FOXOs to liver function and the stress response, there is relatively little is known about FOXOs in liver disease. The most data is available for HCV infection where FOXO1 activity appears to be directly increased by the virus and this contributes to HCV-induced insulin resistance33, 34. The mechanisms of these effects are not entirely clear. Banerjee et al33 observed that HCV-induced FOXO1 activation resulted from an HCV core protein dependent process that suppressed the ability of Akt to phosphorylate FOXO133. Similar results were obtained by Deng et al34 although they showed that the HCV simulation of FOXO1 was dependent upon NS5a-induced ROS production and subsequent JNK activation.
A second FOXO-dependent HCV effect has been observed with FOXO3. FOXO3 has been observed to play a role in regulating the innate immune signaling pathway, directly suppressing TLR signaling35. It also is a transcriptional activator of SOCS3, an inhibitor of interferon-mediated signaling and it is itself inactivated by IKK-ε, one of the upstream activators of interferon production. FOXO3 activity was increased by starvation/malnutrition in HCV infection and this effect caused an increased expression of SOCS3 and a consequent suppression of the interferon signaling pathway36. In this case direct viral FOXO activation contributes to both insulin resistance and infection persistence.
FOXOs have been implicated in several other liver diseases as well, but the evidence supporting this is limited. Enhancement of FOXO1 expression and nuclear localization was seen in NASH patients37 and this was felt to be a possible contributor to insulin resistance. Due to their well-documented function as tumor suppressors, there has also been some interest in the role of FOXO in hepatocellular carcinoma. Little is known in this regard although one report observed longer survival in HCC patients with high levels of FOXO3 in their tumors38.
One final area of FOXO involvement in liver disease is its potential role in fibrosis. FOXOs are known to be survival factors that are required for the quiescent state of long living cells. One area in where this has been well documented is in survival of hematopoetic stem cells39. Adachi et al40 thus examined whether FOXOs play a role in the quiescence of hepatic stellate cells as the transdifferentiation and proliferation of stellate cells is required for nearly all forms of hepatic fibrosis. This study observed that the proliferation of stellate cells in vitro was enhanced by dominant negative forms of FOXO1 and suppressed by constitutively active forms of the protein. Furthermore, FOXO1(+/−) mice were more susceptible to fibrosis. This intriguing result suggests a possible role of FOXOs in hepatic fibrosis.
5. FOXO regulation in liver by post-translational modifications (PTMs)
The above considerations show the importance of multilevel FOXO regulation for adaptation of the liver to stresses. A complete understanding of the role of FOXOs in liver disease requires a more detailed understanding of the many upstream events that affect the functions of these proteins. Multiple post translational modifications of FOXO have been described including phosphorylations, acetylation, and ubiquitination5. These PTMs can be either activating or inactivating. They alter nuclear import and export steps, modify the DNA binding affinity, and alter the pattern of transcriptional activity for specific target genes2, 41.
The first layer of regulation of FOXOs is a series of modifications that controls the translocation between nucleus and cytosol. These FOXO PTMs can be divided into two groups. The first group promotes nuclear export, polyubiquitination and proteosomal degradation. These includes phosphorylation by Akt (the main pathway of FOXO degradation) 9, ERK 42, IKKβ 10 and CDK2 43. Sites for all those modifications have been described and activation of these kinases normally correlates with loss of nuclear FOXOs. Deubiquitination by USP7 is also known to cause the nuclear export of FOXO4 44. Similar mechanisms exist for regulation of other members of FOXO family. The second group of PTMs that control the nuclear-cytosolic distribution are those that promote nuclear localization and are associated with an increase in transcriptional activity. These include phosphorylation by JNK 34, p38 45, 46, AMPK 47, 48, CDK1 49, and MST1 50, as well as monoubiquitination by unknown enzymes 44, and arginine methylation by PRMT1 51. There is an interesting interaction within this group. All FOXO proteins contain numerous phosphorylation ‘SP’ motifs shared by JNK, p38 and ERK. Phosphorylation on these sites has been detected following oxidative stress and other stimuli. FOXO3, for example, contains p38 phosphorylation sites on Ser7, Ser12, Ser294, Ser344, and Ser425 that can be also targeted by JNK (Ser294 and Ser425) and ERK (Ser294, Ser344, and Ser425) 45. While p38 and JNK are known to promote nuclear localization, ERK modification has an opposite effect42. One can speculate that these modifications can happen consecutively by different enzymes and various combinations throughout the FOXO sequence create unique protein conformations that define its localization. Another mechanism of preventing FOXO nuclear export is a direct inhibition of AKT phosphorylation by methylation of closely located arginine residues 51. The balance between these two groups of modifications in the liver creates an environment that defines the amount of FOXOs in the nucleus. Complete loss of nuclear FOXO undoubtedly leads to deregulation of above mentioned pathways controlled by FOXO transcriptional activity.
The second layer of regulation includes a series of modifications that regulate FOXO transcriptional activity by changing DNA binding and promoter binding specificity. This group includes acetylation by the redox activated acetyl transferase, p30052-54, deacetylation by SIRT155-57, SIRT258, 59 and SIRT360, lysine methylation61, 62 and glycosylation20-22. Lysine methylation at K270 of FOXO3 promotes loss of DNA binding and reduces FOXO-mediated apoptosis. Deacetylation by SIRT1 has been shown to differentially alter DNA binding affinity, so that more highly acetylated forms of FOXO3 favor expression of pro-apoptotic genes, (Bim, TRAIL and FasL), while the more deacetylated forms favor expression of antioxidant and cytoprotective genes55. SIRT2 also deacetylates FOXOs and increases their DNA-binding activity58, 59. The binding of CBP/p300 to FOXOs is essential for transactivation of target genes52-54. However, the acetylation itself attenuates FOXO transcriptional activity.
Several lysines were reported to be acetylated in FOXOs. Brunet et al found that FOXO3 is acetylated at K242, K259, K271, K290 and K569 in the presence of stress stimuli55. Acetylation at K222, K245, K248, K262, K265, K274, K294 of FOXO1 was also reported to regulate its DNA binding affinity and sensitivity to AKT phosphorylation63-65. Acetylation at K242, K245, and K262 of FOXO1 is sufficient to attenuate its transcriptional activity64. Fukuoka et al reported the importance of K186, K189, and K408 deacetylation by HDAC in regulating FOXO4 transciptional activity 66. O-glycosylation is another modification that does not affect the nuclear/cytosolic distribution of FOXOs, but results in the up-regulation of specific gene expression such as G6Pase21 and other gluconeogenic genes20. Recent studies show that some of these effects involve the ability of specific PTMs, such as GlcNAcylation to produce differential binding of FOXOs to cofactors such as PGC-1α with a subsequent increase in specific transcriptional activities22.
This second layer of modifications gives an idea of how FOXO transcriptional activity can be regulated. However, the question of how FOXOs decide which transcriptional program is activated in any given condition is still unclear. Since all FOXO proteins recognize a conserved consensus motif TTGTTTAC67, 68 present in multiple genes, the promoter binding patterns may be defined more by differential binding to various cofactors. FOXOs have been shown to interact with a large number of binding partners resulting in changes in transcriptional activity of both proteins. The list includes a number of nuclear hormone receptors, other transcription factors such as β-catenin, RUNX3, SMADs and histone modifying enzymes such as acetylases and methyltranferases (summarized by 69). In addition to being binding partners, these modifying enzymes can directly affect the PTMs of FOXO itself as well as histone modifications providing an additional level of complexity to the activation of FOXO target genes.
This emerging understanding of the role of FOXO PTMs in cofactor binding can explain the so-called ‘FOXO code’, .i.e. very specific PTM regulated transcriptional programs2. PGC-1α and p300 are two examples of close linkages between FOXO PTM status and transcriptional cofactors interaction. PGC-1α promotes FOXO GlcNacylation. GlcNacylation in turn directs FOXOs towards gluconeogenic genes through interaction with additional cofactors or target gene promoter sequences. The interaction can be disrupted by insulin signaling. This way the balance between two different upstream modifying enzymes regulates the activity of FOXO in the gluconeogenesis pathway.
The interaction with p300, on the other hand, is necessary for FOXO activity, but the direct FOXO acetylation that may result can lead to loss of DNA binding and nuclear export. The amount of active FOXO is constantly replenished by deacetylation enzymes such as the SIRTs. The presence of multiple acetylation sites (7 lysines in FOXO1) provides the potential for considerable promoter specificity by this mechanism. This system creates a dynamic activation of FOXOs, important for quick changes in transcriptional program.
6. Conclusions and future directions
FOXO transcription factors are essential to liver function and liver stress response and their alteration in disease are only now being recognized. In addition to their critical role in carbohydrate metabolism, lipid metabolism and oxidative stress response, the FOXOs are tumor suppressors that promote both cell cycle arrest and apoptosis. Pharmacological manipulation of FOXOs in the liver thus has potential benefit for metabolic liver disease, inflammatory liver disease, and prevention of hepatocellular carcinoma.
The existence of a set of PTMs that regulate transcriptional programs of the FOXO factors is important in that it opens the potential for selective modulation of FOXO function. Studies on sites that alter FOXOs DNA-binding activity and their interaction with transcription-regulatory proteins, as well as their stability and subcellular localization may represent a target for pharmacological manipulation of FOXO activity. The existence of unique acetylation sites for different members of the FOXO family potentially can also provide insight into the non-redundant roles of each of the FOXO proteins in transcriptional regulation of hepatic target genes.
References
- 1.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–87. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 2.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–88. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 3.Burgering BM. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258–62. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- 4.Paik JH, Kollipara R, Chu G, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–23. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–50. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 6.Sedding DG. FoxO transcription factors in oxidative stress response and ageing–a new fork on the way to longevity? Biol Chem. 2008;389:279–83. doi: 10.1515/BC.2008.033. [DOI] [PubMed] [Google Scholar]
- 7.Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–92. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flachsbart F, Caliebe A, Kleindorp R, et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–5. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunet A, Bonni A, Zigmond MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 10.Hu MC, Lee DF, Xia W, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–37. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 11.Arden KC. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 2008;27:2345–50. doi: 10.1038/onc.2008.27. [DOI] [PubMed] [Google Scholar]
- 12.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–59. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto M, Han S, Kitamura T, Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest. 2006;116:2464–72. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto M, Pocai A, Rossetti L, DePinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor foxo1 in liver. Cell Metab. 2007;6:208–16. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Patil S, Chauhan B, et al. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–17. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 16.Lu M, Wan M, Leavens KF, et al. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat Med. 2012;18:388–95. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–95. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 18.Mihaylova MM, Vasquez DS, Ravnskjaer K, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–21. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, Lee J, Reno CM, et al. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat Med. 2011;17:356–65. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Housley MP, Rodgers JT, Udeshi ND, et al. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283:16283–92. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS Lett. 2008;582:829–34. doi: 10.1016/j.febslet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Housley MP, Udeshi ND, Rodgers JT, et al. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem. 2009;284:5148–57. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamagate A, Qu S, Perdomo G, et al. FoxO1 mediates insulin-dependent regulation of hepatic VLDL production in mice. J Clin Invest. 2008;118:2347–64. doi: 10.1172/JCI32914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altomonte J, Cong L, Harbaran S, et al. Foxo1 mediates insulin action on apoC-III and triglyceride metabolism. J Clin Invest. 2004;114:1493–503. doi: 10.1172/JCI19992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim DH, Zhang T, Ringquist S, Dong HH. Targeting FoxO1 for hypertriglyceridemia. Current drug targets. 2011;12:1245–55. doi: 10.2174/138945011796150262. [DOI] [PubMed] [Google Scholar]
- 26.Qu S, Altomonte J, Perdomo G, et al. Aberrant Forkhead box O1 function is associated with impaired hepatic metabolism. Endocrinology. 2006;147:5641–52. doi: 10.1210/en.2006-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K, Li L, Qi Y, et al. Hepatic suppression of Foxo1 and Foxo3 causes hypoglycemia and hyperlipidemia in mice. Endocrinology. 2012;153:631–46. doi: 10.1210/en.2011-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao R, Wei D, Gao H, Liu Y, DePinho RA, Dong XC. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. J Biol Chem. 2011;286:14681–90. doi: 10.1074/jbc.M110.201061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferdous A, Battiprolu PK, Ni YG, Rothermel BA, Hill JA. FoxO, autophagy, and cardiac remodeling. Journal of cardiovascular translational research. 2010;3:355–64. doi: 10.1007/s12265-010-9200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Vos KE, Eliasson P, Proikas-Cezanne T, et al. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat Cell Biol. 2012;14:829–37. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, Yang J, Liao W, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–75. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 32.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee A, Meyer K, Mazumdar B, Ray RB, Ray R. Hepatitis C virus differentially modulates activation of forkhead transcription factors and insulin-induced metabolic gene expression. J Virol. 2010;84:5936–46. doi: 10.1128/JVI.02344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng L, Shoji I, Ogawa W, et al. Hepatitis C Virus Infection Promotes Hepatic Gluconeogenesis through an NS5A-Mediated, FoxO1-Dependent Pathway. J Virol. 2011;85:8556–68. doi: 10.1128/JVI.00146-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luron L, Saliba D, Blazek K, Lanfrancotti A, Udalova IA. FOXO3 as a new IKK-epsilon-controlled check-point of regulation of IFN-beta expression. Eur J Immunol. 2012;42:1030–7. doi: 10.1002/eji.201141969. [DOI] [PubMed] [Google Scholar]
- 36.Honda M, Takehana K, Sakai A, et al. Malnutrition impairs interferon signaling through mTOR and FoxO pathways in patients with chronic hepatitis C. Gastroenterology. 2011;141:128–40. doi: 10.1053/j.gastro.2011.03.051. 40 e1-2. [DOI] [PubMed] [Google Scholar]
- 37.Valenti L, Rametta R, Dongiovanni P, et al. Increased expression and activity of the transcription factor FOXO1 in nonalcoholic steatohepatitis. Diabetes. 2008;57:1355–62. doi: 10.2337/db07-0714. [DOI] [PubMed] [Google Scholar]
- 38.Lu M, Ma J, Xue W, et al. The expression and prognosis of FOXO3a and Skp2 in human hepatocellular carcinoma. Pathol Oncol Res. 2009;15:679–87. doi: 10.1007/s12253-009-9171-z. [DOI] [PubMed] [Google Scholar]
- 39.Tothova Z, Gilliland DG. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell. 2007;1:140–52. doi: 10.1016/j.stem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Adachi M, Osawa Y, Uchinami H, Kitamura T, Accili D, Brenner DA. The forkhead transcription factor FoxO1 regulates proliferation and transdifferentiation of hepatic stellate cells. Gastroenterology. 2007;132:1434–46. doi: 10.1053/j.gastro.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 41.Daitoku H, Sakamaki JI, Fukamizu A. Biochim Biophys Acta. 2011. Regulation of FoxO transcription factors by acetylation and protein-protein interactions. [DOI] [PubMed] [Google Scholar]
- 42.Yang JY, Zong CS, Xia W, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–48. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–7. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 44.van der Horst A, de Vries-Smits AM, Brenkman AB, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–73. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 45.Ho KK, McGuire VA, Koo CY, et al. Phosphorylation of FOXO3a on Ser-7 by p38 promotes its nuclear localization in response to doxorubicin. J Biol Chem. 2012;287:1545–55. doi: 10.1074/jbc.M111.284224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asada S, Daitoku H, Matsuzaki H, et al. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell Signal. 2007;19:519–27. doi: 10.1016/j.cellsig.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 47.Chiacchiera F, Matrone A, Ferrari E, et al. p38alpha blockade inhibits colorectal cancer growth in vivo by inducing a switch from HIF1alpha- to FoxO-dependent transcription. Cell Death Differ. 2009;16:1203–14. doi: 10.1038/cdd.2009.36. [DOI] [PubMed] [Google Scholar]
- 48.Li XN, Song J, Zhang L, et al. Activation of the AMPK-FOXO3 pathway reduces fatty acid-induced increase in intracellular reactive oxygen species by upregulating thioredoxin. Diabetes. 2009;58:2246–57. doi: 10.2337/db08-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan Z, Becker EB, Merlo P, et al. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science. 2008;319:1665–8. doi: 10.1126/science.1152337. [DOI] [PubMed] [Google Scholar]
- 50.Lehtinen MK, Yuan Z, Boag PR, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 51.Yamagata K, Daitoku H, Takahashi Y, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–31. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 52.Nasrin N, Ogg S, Cahill CM, et al. DAF-16 recruits the CREB-binding protein coactivator complex to the insulin-like growth factor binding protein 1 promoter in HepG2 cells. Proc Natl Acad Sci U S A. 2000;97:10412–7. doi: 10.1073/pnas.190326997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perrot V, Rechler MM. The coactivator p300 directly acetylates the forkhead transcription factor Foxo1 and stimulates Foxo1-induced transcription. Mol Endocrinol. 2005;19:2283–98. doi: 10.1210/me.2004-0292. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, Zhao Y, Liao W, et al. Acetylation of FoxO1 activates Bim expression to induce apoptosis in response to histone deacetylase inhibitor depsipeptide treatment. Neoplasia. 2009;11:313–24. doi: 10.1593/neo.81358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 56.Motta MC, Divecha N, Lemieux M, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–63. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 57.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–9. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 58.Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–14. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 59.Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol Biol Cell. 2009;20:801–8. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–71. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calnan DR, Webb AE, White JL, et al. Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging. 2012;4:462–79. doi: 10.18632/aging.100471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie Q, Hao Y, Tao L, et al. Lysine methylation of FOXO3 regulates oxidative stress-induced neuronal cell death. EMBO reports. 2012;13:371–7. doi: 10.1038/embor.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiang L, Banks AS, Accili D. Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J Biol Chem. 2010;285:27396–401. doi: 10.1074/jbc.M110.140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102:11278–83. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daitoku H, Hatta M, Matsuzaki H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–7. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fukuoka M, Daitoku H, Hatta M, Matsuzaki H, Umemura S, Fukamizu A. Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int J Mol Med. 2003;12:503–8. [PubMed] [Google Scholar]
- 67.Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–34. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xuan Z, Zhang MQ. From worm to human: bioinformatics approaches to identify FOXO target genes. Mechanisms of ageing and development. 2005;126:209–15. doi: 10.1016/j.mad.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 69.van der Vos KE, Coffer PJ. FOXO-binding partners: it takes two to tango. Oncogene. 2008;27:2289–99. doi: 10.1038/onc.2008.22. [DOI] [PubMed] [Google Scholar]