CHES1/FOXN3 inhibits cell proliferation and protein biosynthesis in tumor cell lines but not in normal fibroblasts. CHES1 directly represses the expression of the gene coding for the protein kinase PIM2, and PIM2 or eIF4E counteracts the antiproliferative effect of CHES1. The levels of CHES1 and PIM2 are inversely correlated in several human cancers.
Abstract
The expression of the forkhead transcription factor checkpoint suppressor 1 (CHES1), also known as FOXN3, is reduced in many types of cancers. We show here that CHES1 decreases protein synthesis and cell proliferation in tumor cell lines but not in normal fibroblasts. Conversely, short hairpin RNA–mediated depletion of CHES1 increases tumor cell proliferation. Growth suppression depends on the CHES1 forkhead DNA-binding domain and correlates with the nuclear localization of CHES1. CHES1 represses the expression of multiple genes, including the kinases PIM2 and DYRK3, which regulate protein biosynthesis, and a number of genes in cilium biogenesis. CHES1 binds directly to the promoter of PIM2, and in cells expressing CHES1 the levels of PIM2 are reduced, as well as the phosphorylation of the PIM2 target 4EBP1. Overexpression of PIM2 or eIF4E partially reverses the antiproliferative effect of CHES1, indicating that PIM2 and protein biosynthesis are important targets of the antiproliferative effect of CHES1. In several human hematopoietic cancers, CHES1 and PIM2 expressions are inversely correlated, suggesting that repression of PIM2 by CHES1 is clinically relevant.
INTRODUCTION
Checkpoint suppressor 1 (CHES1) is a human forkhead transcription factor identified as a suppressor of checkpoint defects in yeast (Pati et al., 1997). CHES1 interacts with the transcription regulator SKIP, also known as NCoA2 (Scott and Plon, 2005), and with the corepressors MEN1, HDAC1, HDAC2, and SAP130/mSin3a (Busygina et al., 2006). CHES1 expression is down-regulated in human cancers, including oral squamous cell carcinoma (Chang et al., 2005), laryngeal cancer (Markowski et al., 2009a, b), and diffuse large B-cell lymphoma (Basso et al., 2005). Furthermore, the distal part of chromosome 14, where the CHES1 locus is located, hosts a tumor suppressor gene (Pehlivan et al., 2008). Knockdown of CHES1 in Xenopus laevis revealed a role for this transcription factor in craniofacial and eye development, and immunoprecipitation analysis confirmed its interaction with several corepressors (Schuff et al., 2007). Similarly, genetic inactivation of CHES1 in mice led to craniofacial defects and in some cases lethality (Samaan et al., 2010).
The paradigm of transcriptional repressors as tumor suppressor genes was based on the discovery of the retinoblastoma tumor suppressor, which represses the expression of E2F target genes required for cell proliferation (Weinberg, 1995). However, other repressors have powerful oncogenic activities (Peinado et al., 2007), indicating that the specificity of gene repression determines the role of this class of transcriptional regulators in human cancers. Hence, for CHES1 to act as a tumor suppressor, it must negatively regulate genes required for cell proliferation. Here we investigate the effects of CHES1 on cell proliferation and gene expression and provide evidence for a role of CHES1 as a repressor of the expression of PIM2 and other genes required for protein biosynthesis and cilium biogenesis.
RESULTS
The forkhead transcription factor CHES1 inhibits cell proliferation in tumor cells but not in normal fibroblasts
To investigate the role of CHES1 in the control of cell proliferation, we cloned CHES1 (NM_001085471.1) into the retroviral expression vector pLPC under the control of the CMV promoter. Introduction of this vector in the human tumor cell line H1299 (lung cancer) or U2OS (osteosarcoma) dramatically inhibited their ability to form colonies (Figure 1, A and B). Conversely, short hairpin RNA (shRNA)–mediated inactivation of CHES1 in U2OS cells moderately increased their growth, and this effect was reversed by expressing a mutated allele of CHES1 (rCHES1) resistant to the effects of the shRNA (Figure 1, C and D). In contrast, expression of CHES1 in human normal fibroblasts IMR90, BJ, or human primary prostate fibroblasts did not significantly affect cell growth (Figure 1, E and F). Consistent with previous work showing that CHES1 is a transcription factor of the forkhead family, we found that the protein localized to the nucleus when expressed in IMR90 cells (Figure 1G).
FIGURE 1:
CHES1 inhibits proliferation of cancer cells. (A) Western blots showing the overexpression of CHES1 in H1299 lung carcinoma cells and U2OS osteosarcoma cells. (B) Colony formation assays of H1299 and U2OS cells transfected with a control vector or CHES1. (C) Growth curves of U2OS cells infected with a vector expressing a shRNA against CHES1 (shCHES1) or a nontargeting control shRNA (shNTC) and vectors expressing a CHES1 mutant resistant to the shRNA (rCHES1). (D) Quantitative PCR (qPCR) of CHES1 mRNA levels from cells described in C. (Note that the scale is cut to better observe both down-regulation and overexpression.) (E) Growth curves of normal human fibroblasts IMR90, BJ, or HprF infected with a control vector or CHES1. (F) Western blot showing the overexpression of CHES1 in fibroblasts cells as in E. (G) Immunofluorescence analysis showing nuclear localization of CHES1 in IMR90.
Next we studied the domain requirements for CHES1-mediated growth suppression in U2OS cells. We generated multiple deletion mutants of CHES1 lacking the N- or C-terminal domains in combination or not with the forkhead DNA-binding domain (DBD) located at the center of the protein (Figure 2A). According to the software PredictNLS (https://rostlab.org/owiki/index.php/PredictNLS), CHES1 possesses a nuclear localization signal (NLS) in the C-terminal domain between residues 418 and 445 (Figure 2A). Because the C-terminal-truncated proteins may not localize to the nucleus to the same extent as the N-terminal-truncated mutants or the wild-type CHES1, we introduced the SV40 NLS sequence at the C-terminus of these mutants to ensure that the subcellular localization would not influence our functional characterization (Figure 2A). We introduced these constructs in U2OS cells (Figure 2, B and C) and measured cell proliferation in a colony formation assay (Figure 2D). We found that deletions including the forkhead DBD abolished CHES1’s growth inhibitory effect (Figure 2, D and E). In addition, mutation of the conserved histidine residue at position 164 into alanine (CHES1H164A) also impaired the growth suppression ability of CHES1 (Supplemental Figure S1). This residue is in the forkhead DNA-binding domain and is equivalent to histidine 169 in HNF-3 (FOXM1), which directly contacts the DNA (Clark et al., 1993). Of interest, deletion of either the N- or the C-terminal domain alone had little or no effect on CHES1’s ability to reduce colony formation, suggesting that CHES1’s DBD is necessary and sufficient to mediate its effects and that other domains are dispensable (Figure 2, D and E). Of interest, CHES1ΔN was as effective at reducing colony formation as wild-type CHES1, although it was expressed at much lower levels (Figure 2, B and C). This effect was not due to differences in transfection efficiency, as levels of cotransfected GFP were similar for all mutants. In agreement, treatment of cells with the proteasome inhibitor MG132 at concentrations that stabilized the proteasome target p53 allowed us to detect the presence of CHES1ΔN (Figure 2C). It is then plausible that, like other transcription factors, CHES1 activity is coupled to proteasome-dependent degradation (Geng et al., 2012). Accordingly, CHES1ΔN may have increased activity, explaining its increased proteasome-dependent turnover. Taken together, the data obtained by expressing different deletion mutants of CHES1 indicate that the DNA-binding domain is necessary for growth suppression and under conditions of overexpression is sufficient to reduce cell growth.
FIGURE 2:
The DNA-binding domain of CHES1 is necessary and sufficient to inhibit cell proliferation. (A) Schematic representation of CHES1 and its derived mutants: ΔN, ΔFN, ΔC, ΔFC, DBD-NLS, ΔC-NLS, and ΔFC-NLS. The DBD, ΔC, and ΔFC mutants were fused to the SV40 NLS and termed DBD-NLS, ΔC-NLS, and ΔFC-NLS respectively. (B) Western blots showing the expression of all the mutants. (C) Western blots showing the expression of CHES1 and the ΔN mutant in cells treated with the proteasome inhibitor MG132 at a concentration of 50 μM for 5 h. Blot for p53 indicates the efficiency of the MG132 treatment. (D) Colony formation assay in U2OS cells transiently transfected with empty vector (V), CHES1, or its derived mutants. (E) Quantification of the growth data presented in D.
Next we developed a fusion protein between CHES1 and the ligand-binding domain of the estrogen receptor (ER) to create a chimeric protein in which CHES1’s functions can be conditionally activated. ER fusions are inactive because ER keeps the fusion protein in an inactive complex in the cytoplasm. However, treatment with 4-hydroxytamoxifen (4-OHT) releases the fusion protein from the inactive conformation. This strategy allows us to generate sufficient amounts of CHES1-expressing cells for biochemical analysis. We introduced CHES1-ER in the human lung cancer cell line H1299 by retroviral gene transfer and obtained a moderately higher expression than endogenous levels (Figure 3A). Adding 4-OHT induced the nuclear localization of CHES1-ER (Figure 3B) and dramatically inhibited cell growth (Figure 3C). We did not notice a significant accumulation of floating (dead cells) in our cultures, suggesting that CHES1 inhibits cell proliferation without causing cell death.
FIGURE 3:
Characterization of the activity of an inducible CHES1. (A) Western blots showing the expression of CHES1 in H1299 lung carcinoma cells infected with a control vector (ER, estrogen receptor ligand-binding domain) or with a vector expressing CHES1-ER fusion protein. Cells were treated with 300 nM 4-hydroxytamoxifen (4-OHT; inductor) for 24 h. (B) Observation of the nuclear localization of the fusion protein upon 24-h treatment with EtOH (negative control) or 300 nM 4-OHT. (C) Growth curves of H1299 cells expressing the control vector (ER) or the fusion protein (CHES1-ER) and treated with EtOH or 300 nM 4-OHT every 2–3 d for a total period of 9 d.
CHES1 as a transcriptional repressor
To identify genes regulated by CHES1, we infected H1299 cells with retroviral vectors expressing CHES1 or a control empty vector. RNA was obtained 3 d after puromycin selection of cell populations expressing the vectors. We found mostly down-regulated genes (Figure 4A), suggesting that CHES1 may regulate cell proliferation by acting predominantly as a transcriptional repressor, as previously suggested (Scott and Plon, 2005; Busygina et al., 2006). Among the down-regulated genes that could potentially explain the proliferation defects of CHES1-expressing cells, we noticed the protein kinases PIM2 and DYRK3, which regulate protein biosynthesis, as well as the tRNA splicing enzyme TSEN2. Gene Ontology analysis revealed the down-regulation of many genes implicated in cilium biogenesis, the flagellum, the cytoskeleton, and the smoothened signaling pathway (Figure 4B, Table 1, and Supplemental Figure S2). We confirmed by quantitative PCR (qPCR) the down-regulation of PIM2, DYRK3, and TSEN2, which are involved in protein biosynthesis, as well as three more genes found to be highly down-regulated: IFTT81, CCDC104, and IQCK (Figure 4C).
FIGURE 4:
Microarray analysis reveals that CHES1 acts as a transcriptional repressor of PIM2 and other genes involved in protein biosynthesis and cilium biogenesis. (A) Volcano plot representing the proportion of genes modulated by CHES1 in comparison with cells expressing an empty vector using transcripts with a fold change ≥1.5 and p < 0.05 according to a two-sample Student's t test. We found 214 genes down-regulated (green) and 64 upregulated (red). (B) Pie charts of the most important biological functions and cellular components affected by the expression of CHES1 in H1299 cells. A FatiGO gene enrichment analysis was performed using transcripts down-regulated with a fold change ≥1.5 and p < 0.05 according to a two-sample Student's t test. (C) qPCR analysis of the mRNA levels for the indicated genes (a–c, three independent experiments with RNA obtained in the same way as for the microarray analysis).
TABLE 1:
Gene Ontology categories of CHES1-regulated genes.
| Biological process category | Genes | p |
|---|---|---|
| Determination of bilateral symmetry | KIF3B, KIF3A, ARL6, IFT88, DYNC2H1, DYNC2LI1, RPGRIP1L | 7.92273E-5 |
| Smoothened signaling pathway | KIF3A, TULP3, TCTN1, IFT88, RPGRIP1L | 1.78363E-2 |
| Neural tube patterning | KIF3A, TULP3, TCTN1, RPGRIP1L | 1.97025E-2 |
| Cilium assembly | KIF3A, ARL6, DYNC2H1, DYNC2LI1, BBS1, RPGRIP1L | 7.92273E-5 |
| Cilium morphogenesis | KIF3A, IFT88, BBS1 | 1.97283E-2 |
| Protein translation | PIM2, TSEN2, DYRK3 |
PIM2 is a powerful oncogene in transgenic mice (Allen et al., 1997), and expression of its three isoforms (long, medium, and short) correlates with malignancy in human prostate cancers (Dai et al., 2005). PIM2 is overexpressed in several B-cell cancers, including chronic lymphocytic leukemia, diffuse large B-cell lymphoma, mantle cell lymphoma, and myeloma (Cohen et al., 2004; Huttmann et al., 2006). PIM kinases are under investigation as targets for pharmacological inhibition in hematological cancers (Schatz et al., 2011). To study the mechanism of PIM2 repression by CHES1, we mapped potential forkhead binding sites along the PIM2 promoter from −2.5 to +0.25 of the transcription start site with the Transcriptional Regulatory Element Database (http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=home), using the HNF3-β/FOXA2 matrix (Figure 5A and Supplemental Table S1). We also analyzed the PIM2 locus for CHES1 binding sites using chromatin immunoprecipitation (ChIP) with primers covering the same region (Figure 5B). We found binding of CHES1 to many sites along this region but not to the control HMBS promoter. In addition, the binding at two sites close to the transcription start site at −250 was stronger. We thus cloned the PIM2 promoter region containing the two strong CHES1 binding sites in a luciferase reporter vector. Coexpression of this reporter with increased amounts of CHES1 revealed a dose-dependent repression of luciferase activity. Of note, the maximum concentration of CHES1 used was not able to repress luciferase in a control reporter without the CHES1 binding sites (Figure 5C). In addition, deletion of the forkhead binding sites in the PIM2 proximal promoter fragment abolished the ability of CHES1 to repress reporter gene expression (Figure 5D). These results suggest that CHES1 can directly regulate PIM2 gene expression.
FIGURE 5:
CHES1 represses the PIM2 promoter. (A) Identification of potential forkhead binding sites along the PIM2 promoter according to the Transcriptional Regulatory Element Database (http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=home). The analysis was performed with the matrix search tool by selecting the HNF3-beta/FOXA2 matrix from the JASPAR database as template. Sequence logo of the matrix of HNF-3beta/FOXA2, score, and position of each identified potential CHES1-binding site. (B) Chromatin immunoprecipitation of H1299 cells 48 h after transfection with a control vector (flag) or flag-CHES1, showing the enrichment of CHES1 on the PIM2 promoter (left) but not on the HMBS promoter (right). (C) Luciferase assay performed by transfecting pGL3-PIM2 (left) or pGL3-promoter (right) with different concentrations of a plasmid expressing CHES1 as indicated. (D) Luciferase assay performed by transfecting different mutants of the pGL3-PIM2 reporter plasmid containing both (pGL3-PIM2), only one (pGL3-PIM2 mut1; pGL3-PIM2 mut2), or none (pGL3-PIM2 mut3) of the two forkhead binding sites (FBS1 and 2) identified in the CHES1-binding region of the PIM2 promoter (see B).
CHES1 reduces protein biosynthesis
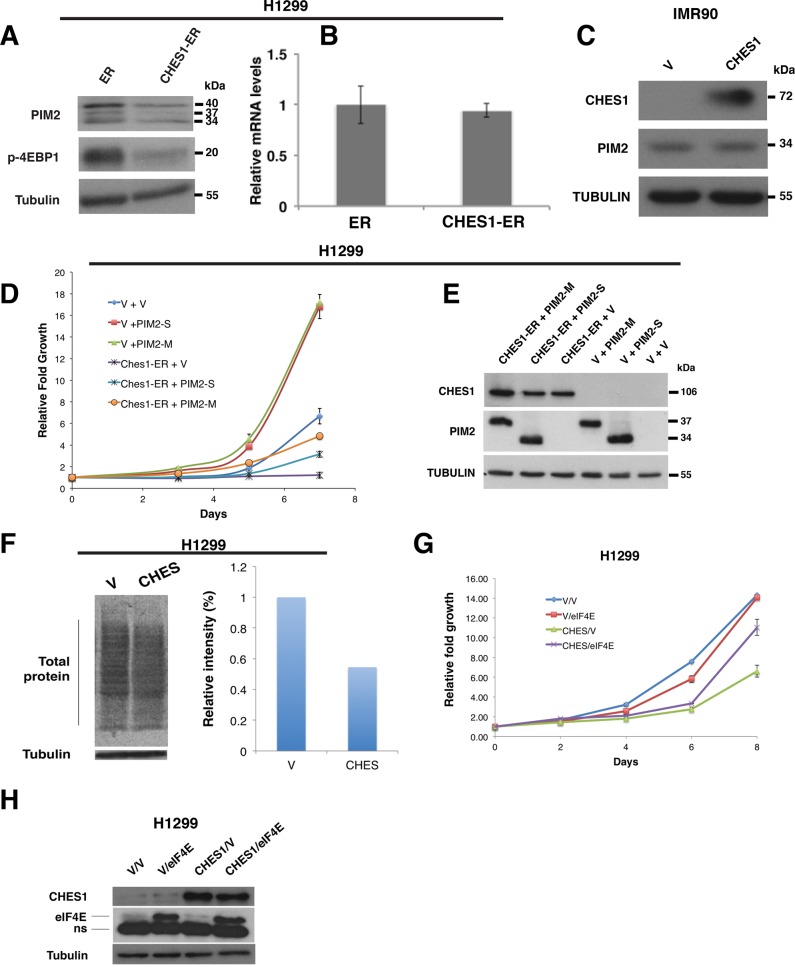
Next we used the conditional CHES1-ER fusion construct to investigate the regulation of PIM2 by CHES1. We found that all protein isoforms of PIM2 were repressed in CHES1-ER–expressing cells treated with 4-OHT (Figure 6A). Of importance, phosphorylation of 4EBP1, a target of the PIM2 kinase, was highly reduced in CHES1-expressing cells (Figure 6A), whereas its transcription did not seem affected (Figure 6B). Of note, total 4EBP1 levels were also reduced (unpublished data) since hypophosphorylated 4EBP1 is degraded rapidly by the proteasome (Yanagiya et al., 2012). In agreement with this result, it has been reported that a PIM kinase inhibitor blocks the growth of lymphoma cells concomitant with a reduction in both phosphorylated and total levels of 4EBP1 (Lin et al., 2010). Finally, expression of CHES1 in normal human fibroblasts IMR90 did not reduce PIM2 expression, although CHES1 was well expressed (Figure 6C), consistent with the lack of growth inhibition previously observed. To investigate whether the reduction of PIM2 in CHES1-expressing tumor cells was important for its antiproliferative effects, we expressed the short or medium isoforms of PIM2 (PIM2-S and PIM2-M, respectively) in H1299 cells expressing CHES1-ER. Treatment with 4-OHT to induce CHES1-ER activity inhibited the growth of cells coexpressing a control vector, but cells coexpressing PIM2-S or M were more resistant to CHES1 induction (Figure 6, D and E). CHES1 represses other proteins—some required for translation—suggesting an explanation for this partial effect. In agreement with a global translational effect of CHES1, cells expressing CHES1 had a reduced incorporation of 35S-labeled methionine into total proteins (Figure 6F). Overexpression of eIF4E, an oncogenic translation factor (Wendel et al., 2004), also inhibited the antiproliferative effect of CHES1 in H1299 cells (Figure 6, G and H).
FIGURE 6:
PIM2 down-regulation and reduced translation are critical for CHES1-induced proliferation inhibition. (A) Western blot of H1299 cells infected with a control vector (ER) or CHES1-ER upon 24-h treatment with 300 nM 4-OHT, showing decreased PIM2 protein levels and phospho-4EBP1. (B) qPCR analysis of EIF4EBP1 mRNA levels in H1299 infected with a control vector (ER) or CHES1-ER upon 24-h treatment with 300 nM 4-OHT. (C) Western blot for IMR90 cells infected with CHES1 or a control vector (V), showing the unchanged levels of PIM2. (D) Growth curves of H1299 cells infected with CHES1-ER and PIM2-S (short isoform) or PIM2-M (medium isoform) and treated with 300 nM 4-hydroxytamoxifen (4-OHT) for a period of 7 d. (E) Western blots of cells as described in D. (F) [35S]methionine incorporation in H1299 cells expressing CHES1 or a control vector (V). The intensity of the signal on the film (left) was measured with ImageJ and plotted (right). (G) Growth curve of H1299 cells infected with CHES1 and eIF4E. (H) Western blot of cells as described in G (ns, nonspecific).
To further investigate whether the antiproliferative effects of CHES1 were due to activation of a checkpoint pathway that arrests cells in a particular stage of the cell cycle, we analyzed CHES1-ER–expressing H1299 cells treated with 4-OHT or vehicle for DNA content using fluorescence-activated cell sorting. We did not find accumulation of cells at any specific cell cycle stage or cells with DNA content lower than G1 (indicative of apoptosis; Figure 7A). Because we found that CHES1 inhibits translation, we next treated H1299 cells with 50 μg/ml protein synthesis inhibitor cycloheximide. We did not found major changes in the cell cycle profile of cells treated with this drug (Figure 7B). This result is consistent with a disabled translational control in CHES1-expressing cells, which likely slows the cell cycle at multiple steps. Moreover, measurement of the levels of cyclin A in cells expressing CHES1-ER treated with 4-OHT did not reveal a significant difference from untreated cells or control cells with an empty vector (Figure 7C). Cyclin A is preferentially expressed in cells during S and G2 (Pines and Hunter, 1990), and its levels would have been high if cells had been arrested in S or G2 and very low if cells had been arrested in G1. We also studied the cell cycle profile of U2OS cells, whose proliferation is also inhibited by CHES1 and, unlike H1299, have a wild-type p53 gene. We introduced CHES1-ER in U2OS cells and treated them with 4-OHT or vehicle. Again, we did not observe a significant accumulation of cells in any stage of the cell cycle upon induction of CHES1-ER activity (Figure 7D). Together these results support the idea that CHES1-expressing cells have a reduced proliferation potential due to reduced protein biosynthesis, which slows transit through all stages of the cell cycle, as recently observed in cells depleted for the ribosomal proteins RPL5/11 (Teng et al., 2013).
FIGURE 7:
CHES1 effects on cell cycle profiles. (A) Cell cycle profile of H1299 cells expressing the control vector (ER) or the fusion protein (CHES1-ER). Cells were first put on low serum (0.1% FBS) and then treated with serum and either EtOH or 300 nM 4-OHT for 24 h. (B) Cell cycle profile of H1299 cells treated with 50 μg/ml cycloheximide for the indicated times. (C) Immunoblot showing the levels of cyclin A in cells expressing the control vector (ER) or CHES1-ER. H1299 cells were treated with either EtOH or 300 nM 4-OHT for 24 h. (D) Cell cycle profile of U2OS cells expressing the control vector (ER) or CHES1-ER. Cells were treated with EtOH or 300 nM 4-OHT for 8 d.
To validate whether repression of PIM2 by CHES1 occurs in human cancers, we searched Oncomine for tumors in which CHES1 is reduced. In primary effusion lymphomas, diffuse large B-cell lymphoma, and hairy cell leukemia we found a reduction in CHES1 and a concomitant increase in PIM2 (Supplemental Figure S3).
DISCUSSION
A conserved DNA-binding domain known as the forkhead characterizes the forkhead family of transcription factors. In humans, this family contains 39 members divided into subgroups from A to S (Greer and Brunet, 2005). In human cancers, several members of the FOXO subgroup are inhibited after phosphorylation by oncogenic kinases, such as AKT (Bouchard et al., 2004) and IKKβ (Hu et al., 2004). FOXOs can repress tumor formation by several mechanisms, including the repression of MYC target genes (Bouchard et al., 2004) and induction of several cyclin kinase inhibitors such as p27, p15INK4b, and p19INK4d (Katayama et al., 2008). CHES1 is a forkhead DNA-binding transcription factor of the N subgroup (its alternative name is FOXN3), which is also down-regulated in several human cancers (Basso et al., 2005; Chang et al., 2005; Markowski et al., 2009a, b). CHES1 is unique among forkhead family members because it does not contain a transcriptional activation domain. Instead, CHES1 C-terminus binds several corepressors (Scott and Plon, 2005; Busygina et al., 2006), suggesting that it regulates transcription mostly by inhibiting gene expression. This is in agreement with our microarray analysis showing predominantly down-regulated genes in CHES1-expressing cells.
Unlike the FOXO subgroup, CHES1 seems to inhibit a unique set of genes, including the kinases PIM2 (Song and Kraft, 2012) and DYRK3, which regulate TORC1 signaling and translation (Wippich et al., 2013), and many genes implicated in cilium biogenesis. The role of cilium biogenesis as a target of the antiproliferative functions of CHES1 requires further study, but it is intriguing that the tumor suppressors TSC1/2 also inhibit protein synthesis and cilium biogenesis (Yuan et al., 2012). The FOXO family of tumor suppressors antagonize growth-promoting signals, in part by up-regulating the translational repressor 4EBP1 (Harvey et al., 2008). CHES1 may cooperate in the same pathway by repressing PIM2, which phosphorylates and inactivates 4EBP1 (Hammerman et al., 2005), and by repressing DYRK3, which inhibits TORC1 (Wippich et al., 2013). Intriguingly, PIM2 can also phosphorylate and inactivate the FOXO family (Morishita et al., 2008), so its repression by CHES1 could activate other tumor suppressors of the FOXO family. Of interest, like CHES1, the tumor suppressor p53 can also target protein biosynthesis (Horton et al., 2002), suggesting that this could be a general mechanism of action of multiple tumor suppressors.
In three different lymphomas, the expression of CHES1 negatively correlated with the expression of PIM2 (Basso et al., 2005). Those studies suggest that loss of CHES1 in human tumors leads to an increase in PIM2 kinase levels. It is also intriguing that CHES1 inhibited the growth of tumor cell lines but had no effect in normal human fibroblasts. In Drosophila, the FOXO transcription factor only restricts the growth of cells with hyperactive TOR signaling (Harvey et al., 2008). Although further studies in additional normal cell types are required to explain tumor selectivity of CHES1’s antiproliferative effects, it is likely that oncogenic signals may modify CHES1 properties in a way that increases its ability to repress genes required for cell proliferation. Consistent with this idea, CHES1 was unable to repress PIM2 expression in normal human fibroblasts IMR90, although, as seen with the CHES1-ER fusion protein, it was well expressed and localized to the nucleus. We thus define a novel tumor suppressor pathway by a forkhead transcription factor via repression of PIM2 and the reduction of protein biosynthesis.
MATERIALS AND METHODS
Cell culture
U2OS, H1299, and normal human diploid fibroblasts IMR90 and BJ were purchased from the American Type Culture Collection, (Manassas, VA) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS; Wisent, Montréal, QC, Canada) and 1% penicillin G/streptomycin sulfate. U2OS and H1299 were also supplemented with 2 mM l-glutamine. Human primary prostate fibroblasts (HprFs; Science Cell Research Laboratories, Carlsbad, CA) were grown of FM medium (Science Cell Research Laboratories) supplemented with 2% FBS, 1% fibroblast growth supplement, and 1% penicillin G/streptomycin sulfate.
Plasmids
CHES1 cDNA was cloned from reverse-transcribed RNA (protocol described later) from human diploid fibroblasts IMR90 and PCR amplified using an EcoRI-tagged sense primer and an XhoI-tagged CHES1 antisense primer. The gene was then cloned into the EcoRI and XhoI restriction sites of the retroviral vector pLPC. From there, all CHES1 constructs were generated by PCR with pLPC-CHES1 as a template and cloned into the EcoRI and XhoI restriction sites of pLPC with an N-terminal Flag tag. Full-length Flag-tagged CHES1 was generated with CHES1 sense flag primer and the CHES1 antisense primer (see Supplemental Table S2 for primer sequences). The ΔFC mutant was generated with CHES1 sense flag and ΔFork-C antisense. The ΔFork-C NLS mutant was generated with CHES1 sense flag and ΔFork-C NLS antisense. The ΔC mutant was generated with CHES1 sense flag sense and ΔC antisense. The ΔC-NLS mutant was generated with CHES1 sense flag sense and ΔC-NLS antisense. The ΔN mutant was generated with ΔN sense and CHES1 antisense. The ΔFN mutant was generated with ΔFN sense and CHES1 antisense. CHES1-DBD was generated with ΔN sense and ΔC-NLS antisense. All sense primers contained a minimal Kozak sequence before the start codon. The pBabe puro CHES1-ER construct was generated by in frame insertion of full-length human CHES1 cDNA (nucleotides 138–1610) into the XhoI/EcoRI sites of pBabe puro-ER.
The rCHES1 (shRNA-resistant CHES1) was generated by mutating the nucleotide sequence targeted by the shRNA but not the protein sequence. We use a two-step PCR protocol. In the first step we prepared two fragments of CHES1 with the mutated sequence using as primers rCHES1-sense and CHES1-antisense for the first fragment and rCHES1 antisense and CHES1 sense for the second fragment. The fragments were generated by PCR with pLPC-CHES1 as a template. The second PCR step was done by combining both fragments and the CHES1 sense and antisense primers. Product of the second PCR was then subcloned into the EcoRI/NdeI sites of pWZL vector. The shRNA against CHES1 was designed using the method described by Paddison et al. (2004). We targeted the sequence CTCTTGAAGAAGGTACTGCCCG. The isoforms (medium and mhort) of PIM2 in pLXSN were kindly provided by Michael Lilly, Loma Linda University School of Medicine, Loma Linda, CA.
The pGL3-PIM2 plasmid was generated as follows: the promoter region (−550;0) of PIM2 was PCR amplified using a SacI-tagged sense primer and a XhoI-tagged antisense primer and ultimately cloned into the SacI and XhoI restriction sites upstream from the SV40 promoter of the pGL3 promoter plasmid. pGL3-PIM2 mutants 1 and 2 were made by PCR mutagenesis from pGL3-PIM2. pGL3-PIM2 mutant 3 was obtained by digesting the pGL3-PIM2 plasmid with AgeI and XhoI. The MSCV plasmid expressing eIF4E was kindly provided by Katherine Borden, IRIC, Université de Montreal, QC, Canada. All primers are shown in Supplemental Table S2.
Retroviral-mediated gene transfer
Retroviral-mediated gene transfer was done as described previously (Ferbeyre et al., 2000). Puromycin and G418/geneticin were used at a concentration of 2.5 μg/ml for 3 d and 400 μg/ml for 7 d, respectively.
Growth curves
We plated IMR90 fibroblasts (2 × 104/well), HprFs (1 × 104/well), and BJs (1 × 104/well) in 12-well plates and U2OS and H1299 (1 × 104/well) in six-well plates. At the indicated times, cells were washed once with 1× phosphate-buffered saline (PBS) and fixed for 15 min at room temperature in 1% glutaraldehyde, and relative cell numbers were estimated at various times using a crystal violet retention assay as previously described (Ferbeyre et al., 2000).
Colony assay
U2OS and H1299 cells were plated at a density of 5 × 105 per 10-cm dish and incubated overnight at 37°C. Cells were transfected using the calcium phosphate precipitation method with 15 μg of the indicated plasmid constructs. Fresh medium supplemented with 2.5 μg/ml of puromycin was added 48 h after transfection, and cells were kept in selection for 7 d until formation of colonies. Afterward, cells were fixed and stained with crystal violet.
Immunoblotting and chromatin immunoprecipitation
ChIP and immunoblotting assays were performed as described before (Calabrese et al., 2009). The following primary antibodies were used: anti-Flag (M2 mouse monoclonal, F1804, 1:1000; Sigma-Aldrich), anti-tubulin (1:2000; Sigma-Aldrich, Oakville, ON, Canada), anti-CHES1 (Ab50756, 1:2000; Abcam, Cambridge, MA), anti-CHES1 (ARP32841_T100; 1:1000; Aviva Systems Biology, San Diego, CA), anti–green fluorescent protein (11814460001, 1:1000; Roche, Laval, QC, Canada), anti–cyclin A (C-19, rabbit, SC-596, 1:200; Santa Cruz Biotechnology), anti-PIM2 (Ab97475, 1:1000; Abcam), anti-PIM2 (ID-12, SC-13514, 1:200; Santa Cruz Biotechnology), anti–phospho-4EBP1 (Thr-37/46) (rabbit, 9459, 1:1000; Cell Signaling), and anti-eIF4E (610269, 1:500; BD Transduction Laboratories). Signals were revealed after incubation with anti-mouse (1:5000) or anti-rabbit (1:5000) secondary antibodies coupled to peroxidase (Dako) by using enhanced chemiluminescence (Amersham, United Kingdom), or Lumi-Light (Roche). Primers used in the ChIP protocol for the PIM2 and HMBS promoters are indicated in Supplemental Table S2.
Quantitative PCR
qPCR with Syber Green technology was performed as previously described (Vernier et al., 2011). The relative quantification of target genes over housekeeping genes (HMBS, TBP) was determined by using the ΔΔCT method. Primers used in the qPCR protocol are indicated in Supplemental Table S2.
Microarray analysis
RNA was collected from H1299 cells 5 d after infection with pLPC-flag-CHES1 or control vector pLPC-flag. Total RNA samples were sent to the Genome Quebec facility at McGill University for cRNA amplification and subsequent hybridization on GeneChIP Human Gene 2.0 ST Array Affymetrix DNA Chip. Data were analyzed using Affymetrix Expression Console Software and Transcriptome Analysis Console (www.affymetrix.com). Data are available at www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49422.
Fluorescence microscopy
For fluorescence microscopy, 4 × 104 cells were plated on coverslips in six-well plates. At 24 h after plating, the cells were fixed with 4% paraformaldehyde for 15 min at room temperature. Then the cells were washed in 1× PBS and permeabilized using ice-cold 0.2% Triton X-100 in PBS/BSA 3% solution for 5 min. Then cells were washed three times with PBS/BSA and incubated for 1 h at room temperature with anti-CHES1 CHESC9H4 mouse monoclonal antibody (1:200; Abcam). The cells were washed and incubated for 1 h with the appropriate conjugated secondary antibody (1:4000, Alexa Fluor 488 goat anti-mouse; Molecular Probes-Invitrogen). Finally, the cells were washed three times with 1× PBS, incubated in 300 nM 4′,6-diamidino-2-phenylindole for 10 min and mounted on microscope slides. Images were obtained using a Nikon Eclipse TE2000-U microscope and MetaMorph software (Universal Imaging).
Luciferase assay
We plated 7.5 × 104 H1299 cells in 12-well plates and cotransfected 0.1 μg of the pGL3-PIM2 reporter plasmid, in which the firefly luciferase is under the control of the PIM2 promoter followed by the SV40 promoter, with different concentrations of pLPC-CHES1 as indicated. The total quantity of plasmid was kept constant by adding the empty vector pLPC. The pGL3-promoter reporter plasmid, in which the firefly luciferase is under the control of the SV40 promoter only, was used as control. To normalize the data for possible transfection variations, we cotransfected cells with 0.1 mg of a pLPC plasmid expressing LacZ. Cells were transfected with Lipofectamine 2000 (Invitrogen), according to manufacturer's instructions. At 24 h posttransfection, cells were lysed in 200 ml of lysate buffer (Dual Luciferase Assay System; Promega), and luciferase assays were performed using the Dual Luciferase Reporter Assay System according to the manufacturer's instructions. The activity of LacZ was measured by incubating 50 ml of each lysate with 50 ml of 2Xb-gal buffer (200 mM sodium phosphate buffer, pH 7.3, 2 mM magnesium chloride, 100 mM β-mercaptoethanol, 1.33 mg/ml ortho-nitrophenyl-β-galactoside) at 37°C for 1 h. The absorbance was measured at 420 nm. Firefly counts were obtained using Fusion a-FP (Perkin Elmer).
[35S]methionine in vitro labeling
Nine days after infection with a plasmid expressing CHES1 or an empty vector, 1 × 106 H1299 cells were plated in 6-cm plates. At 24 h after plating, the cells were incubated for 2 h in methionine/cysteine-free DMEM (Wisent). Then the cells were incubated in methionine/cysteine-free DMEM complemented with 0.2 mCi for 1½ h before collection by trypsinization. Afterward, the cells were resuspended in Laemmli buffer. Total protein extract was subjected to 10% PAGE and transferred to Immobilon-P membranes (Millipore). The membranes were exposed to x-ray film for 24 h and developed.
Supplementary Material
Acknowledgments
We thank M. Lilly and K. Borden for reagents and members of the Ferbeyre lab for critical reading of the manuscript. This work was funded by a grant from the Cancer Research Society. G.F. is a Chercheur National of the Fonds de Recherche du Québec-Santé.
Abbreviations used:
- CHES1
checkpoint suppressor 1
- ER
ligand-binding domain of the estrogen receptor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-02-0110) on January 8, 2014.
*These authors contributed equally.
REFERENCES
- Allen JD, Verhoeven E, Domen J, van der Valk M, Berns A. Pim-2 transgene induces lymphoid tumors, exhibiting potent synergy with c-myc. Oncogene. 1997;15:1133–1141. doi: 10.1038/sj.onc.1201288. [DOI] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Marquardt J, Bras A, Medema RH, Eilers M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 2004;23:2830–2840. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busygina V, Kottemann MC, Scott KL, Plon SE, Bale AE. Multiple endocrine neoplasia type 1 interacts with forkhead transcription factor CHES1 in DNA damage response. Cancer Res. 2006;66:8397–8403. doi: 10.1158/0008-5472.CAN-06-0061. [DOI] [PubMed] [Google Scholar]
- Calabrese V, Mallette FA, Deschenes-Simard X, Ramanathan S, Gagnon J, Moores A, Ilangumaran S, Ferbeyre G. SOCS1 links cytokine signaling to p53 and senescence. Mol Cell. 2009;36:754–767. doi: 10.1016/j.molcel.2009.09.044. [DOI] [PubMed] [Google Scholar]
- Chang JT, et al. Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int J Cancer. 2005;114:942–949. doi: 10.1002/ijc.20663. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Cohen AM, Grinblat B, Bessler H, Kristt D, Kremer A, Schwartz A, Halperin M, Shalom S, Merkel D, Don J. Increased expression of the hPim-2 gene in human chronic lymphocytic leukemia and non-Hodgkin lymphoma. Leuk Lymphoma. 2004;45:951–955. doi: 10.1080/10428190310001641251. [DOI] [PubMed] [Google Scholar]
- Dai H, Li R, Wheeler T, Diaz de Vivar A, Frolov A, Tahir S, Agoulnik I, Thompson T, Rowley D, Ayala G. Pim-2 upregulation: biological implications associated with disease progression and perinueral invasion in prostate cancer. Prostate. 2005;65:276–286. doi: 10.1002/pros.20294. [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14:2015–2027. [PMC free article] [PubMed] [Google Scholar]
- Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annu Rev Biochem. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Hammerman PS, Fox CJ, Birnbaum MJ, Thompson CB. Pim and Akt oncogenes are independent regulators of hematopoietic cell growth and survival. Blood. 2005;105:4477–4483. doi: 10.1182/blood-2004-09-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Mattila J, Sofer A, Bennett FC, Ramsey MR, Ellisen LW, Puig O, Hariharan IK. FOXO-regulated transcription restricts overgrowth of Tsc mutant organs. J Cell Biol. 2008;180:691–696. doi: 10.1083/jcb.200710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton LE, Bushell M, Barth-Baus D, Tilleray VJ, Clemens MJ, Hensold JO. p53 activation results in rapid dephosphorylation of the eIF4E-binding protein 4E-BP1, inhibition of ribosomal protein S6 kinase and inhibition of translation initiation. Oncogene. 2002;21:5325–5334. doi: 10.1038/sj.onc.1205662. [DOI] [PubMed] [Google Scholar]
- Hu MC, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- Huttmann A, et al. Gene expression signatures separate B-cell chronic lymphocytic leukaemia prognostic subgroups defined by ZAP-70 and CD38 expression status. Leukemia. 2006;20:1774–1782. doi: 10.1038/sj.leu.2404363. [DOI] [PubMed] [Google Scholar]
- Katayama K, Nakamura A, Sugimoto Y, Tsuruo T, Fujita N. FOXO transcription factor-dependent p15(INK4b) and p19(INK4d) expression. Oncogene. 2008;27:1677–1686. doi: 10.1038/sj.onc.1210813. [DOI] [PubMed] [Google Scholar]
- Lin YW, et al. A small molecule inhibitor of Pim protein kinases blocks the growth of precursor T-cell lymphoblastic leukemia/lymphoma. Blood. 2010;115:824–833. doi: 10.1182/blood-2009-07-233445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowski J, et al. Gene expression profile analysis in laryngeal cancer by high-density oligonucleotide microarrays. J Physiol Pharmacol. 2009a;60(Suppl 1):57–63. [PubMed] [Google Scholar]
- Markowski J, et al. Metal-proteinase ADAM12, kinesin 14 and checkpoint suppressor 1 as new molecular markers of laryngeal carcinoma. Eur Arch Otorhinolaryngol. 2009b;266:1501–1507. doi: 10.1007/s00405-009-1019-3. [DOI] [PubMed] [Google Scholar]
- Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68:5076–5085. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Cleary M, Silva JM, Chang K, Sheth N, Sachidanandam R, Hannon GJ. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat Methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- Pati D, Keller C, Groudine M, Plon SE. Reconstitution of a MEC1-independent checkpoint in yeast by expression of a novel human fork head cDNA. Mol Cell Biol. 1997;17:3037–3046. doi: 10.1128/mcb.17.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehlivan D, et al. Loss of heterozygosity at chromosome 14q is associated with poor prognosis in head and neck squamous cell carcinomas. J Cancer Res Clin Oncol. 2008;134:1267–1276. doi: 10.1007/s00432-008-0423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype. Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature. 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- Samaan G, Yugo D, Rajagopalan S, Wall J, Donnell R, Goldowitz D, Gopalakrishnan R, Venkatachalam S. Foxn3 is essential for craniofacial development in mice and a putative candidate involved in human congenital craniofacial defects. Biochem Biophys Res Commun. 2010;400:60–65. doi: 10.1016/j.bbrc.2010.07.142. [DOI] [PubMed] [Google Scholar]
- Schatz JH, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med. 2011;208:1799–1807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff M, Rossner A, Wacker SA, Donow C, Gessert S, Knochel W. FoxN3 is required for craniofacial and eye development of Xenopus laevis. Dev Dyn. 2007;236:226–239. doi: 10.1002/dvdy.21007. [DOI] [PubMed] [Google Scholar]
- Scott KL, Plon SE. CHES1/FOXN3 interacts with Ski-interacting protein and acts as a transcriptional repressor. Gene. 2005;359:119–126. doi: 10.1016/j.gene.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Song JH, Kraft AS. Pim kinase inhibitors sensitize prostate cancer cells to apoptosis triggered by Bcl-2 family inhibitor ABT-737. Cancer Res. 2012;72:294–303. doi: 10.1158/0008-5472.CAN-11-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T, Mercer CA, Hexley P, Thomas G, Fumagalli S. Loss of tumor suppressor RPL5/RPL11 does not induce cell cycle arrest but impedes proliferation due to reduced ribosome content and translation capacity. Mol Cell Biol. 2013;33:4660–4671. doi: 10.1128/MCB.01174-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Begin V, Saad F, Mes-Masson AM, Ferbeyre G. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev. 2011;25:41–50. doi: 10.1101/gad.1975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, Imataka H, Mikami S, Martineau Y, Ronai ZA, Sonenberg N. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell. 2012;46:847–858. doi: 10.1016/j.molcel.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Li J, Diener DR, Choma MA, Rosenbaum JL, Sun Z. Target-of-rapamycin complex 1 (Torc1) signaling modulates cilia size and function through protein synthesis regulation. Proc Natl Acad Sci USA. 2012;109:2021–2026. doi: 10.1073/pnas.1112834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.