The mitotic apparatus protein NuMA has an intrinsic membrane-targeting mechanism that is regulated by CDK1-mediated phosphorylation, underlies anaphase-specific cortical accumulation of dynein, and contributes to chromosome separation.
Abstract
Accurate and efficient separation of sister chromatids during anaphase is critical for faithful cell division. It has been proposed that cortical dynein–generated pulling forces on astral microtubules contribute to anaphase spindle elongation and chromosome separation. In mammalian cells, however, definitive evidence for the involvement of cortical dynein in chromosome separation is missing. It is believed that dynein is recruited and anchored at the cell cortex during mitosis by the α subunit of heterotrimeric G protein (Gα)/mammalian homologue of Drosophila Partner of Inscuteable/nuclear mitotic apparatus (NuMA) ternary complex. Here we uncover a Gα/LGN-independent lipid- and membrane-binding domain at the C-terminus of NuMA. We show that the membrane binding of NuMA is cell cycle regulated—it is inhibited during prophase and metaphase by cyclin-dependent kinase 1 (CDK1)–mediated phosphorylation and only occurs after anaphase onset when CDK1 activity is down-regulated. Further studies indicate that cell cycle–regulated membrane association of NuMA underlies anaphase-specific enhancement of cortical NuMA and dynein. By replacing endogenous NuMA with membrane-binding-deficient NuMA, we can specifically reduce the cortical accumulation of NuMA and dynein during anaphase and demonstrate that cortical NuMA and dynein contribute to efficient chromosome separation in mammalian cells.
INTRODUCTION
Faithful separation of duplicated genetic materials into daughter cells is critical for animal development. It occurs at a specific stage of mitosis known as anaphase. Chromosome separation is driven by microtubules (MTs), which organize into spindle-shaped structure during mitosis (Scholey et al., 2003). Many studies from different systems reveal that the driving forces for chromosome separation include kinetochore motors, microtubule flux, and interpolar microtubule sliding (Mitchison, 1989; Desai et al., 1998; Maddox et al., 2002, 2003; Cleveland et al., 2003; Rogers et al., 2005; Ganem and Compton, 2006; Maiato and Lince-Faria, 2010; Roostalu et al., 2010). In addition, cortical dynein–mediated pulling forces on astral MTs emanating from the spindle poles have also been proposed to contribute to anaphase spindle elongation and chromosome separation (Dujardin and Vallee, 2002; Maiato and Lince-Faria, 2010; Roostalu et al., 2010). This is mostly based on laser-mediated microtubule surgery experiments and dynein inhibition in lower organisms such as fungi, Caenorhabditis elegans, and Drosophila (Aist and Berns, 1981; Aist et al., 1991, 1993; Nicklas, 1989; Waters et al., 1993; Grill et al., 2001, 2003; Fink et al., 2006; Pecreaux et al., 2006; Nguyen-Ngoc et al., 2007). In the mammalian system, however, no specific function of cortical dynein in chromosome separation has been found, primarily due to the pleiotropic consequences of dynein inhibition.
The cortical targeting mechanism for dynein was elusive until recently. An evolutionarily conserved α subunit of heterotrimeric G protein (Gα)/mammalian homologue of Drosophila Partner of Inscuteable (LGN)/nuclear mitotic apparatus (NuMA) ternary complex was shown to recruit and anchor dynein at the cell cortex to direct spindle positioning during mitosis (Nguyen-Ngoc et al., 2007; Woodard et al., 2010; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012; Zheng et al., 2013). NuMA is a large nuclear protein that contains globular head and tail domains separated by a huge (1500 amino acids [aa]) coiled-coil domain (Figure 1A; Yang et al., 1992; Radulescu and Cleveland, 2010). During mitosis, NuMA translocates to the spindle poles and the cell cortex (Van Ness and Pettijohn, 1983; Yang and Snyder, 1992; Compton and Cleveland, 1993; Du and Macara, 2004). NuMA can bind to MTs through a C-terminal microtubule-binding domain (Figure 1A) and functions to tether the minus ends of MTs at the spindle poles, which is critical for the establishment and maintenance of mammalian spindle poles (Gaglio et al., 1995, 1996; Merdes et al., 2000; Du et al., 2002; Haren and Merdes, 2002; Silk et al., 2009). LGN is an adaptor protein that can link NuMA and Gα—the N-terminal tetratricopeptide repeats (TPRs) of LGN interact with NuMA and the C-terminal GoLoco motifs of LGN bind to Gα (Mochizuki et al., 1996; Du et al., 2001; Du and Macara, 2004; Blumer et al., 2003; Willard et al., 2008; Zhu et al., 2011). Thus the cortical targeting of NuMA during mitosis is believed to be mediated through LGN and membrane-bound Gα (Du and Macara, 2004; Woodard et al., 2010; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012). Recently live-cell time-lapse analysis of green fluorescent protein (GFP)–dynein revealed that its cortical localization is cell cycle regulated—the cortical accumulation of dynein is significantly enhanced when the cells enter anaphase (Collins et al., 2012). The underlying mechanism, however, is not clear.
FIGURE 1:
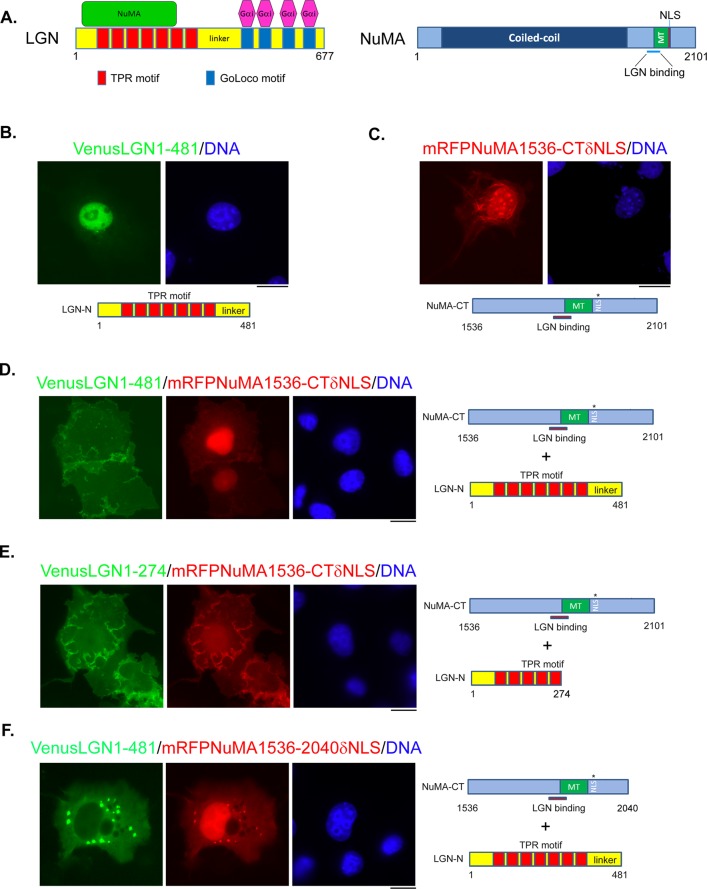
Gα-independent membrane association of LGN-N/NuMA-CT complex. (A) Schematics of LGN and NuMA proteins. Typical domains, binding sites, and binding proteins. MT, microtubule-binding domain; NLS, nuclear localization signal. (B, C) Representative images of Cos 7 cells expressing Venus-LGN1-481 (green) or mRFP-NuMA1536-CTδNLS (red). Cells were transfected with plasmids expressing Venus-LGN1-481 or mRFP-NuMA1536-CTδNLS, respectively. At 24 h later, cells were fixed and stained with DNA dye (Hoechst 33342, blue). (D–F) Representative images of Cos 7 cells coexpressing Venus-LGN1-481 (green, D) and mRFP-NuMA1536-CTδNLS (red, D); Venus-LGN1-274 (green, E) and mRFP-NuMA1536-CTδNLS (red, E); or Venus-LGN-1-481 (green, F) and mRFP-NuMA1536-2040δNLS (red, F). Cells were cotransfected with the indicated combinations of plasmids. At 24 h later, cells were fixed and stained with DNA dye (blue). Schematics of expressed fragments are shown underneath (B, C) or on the right (D–F) of the images. Bars, 10 μm.
Here we show that NuMA has an intrinsic membrane-targeting mechanism, which is regulated by cyclin-dependent kinase 1 (CDK1)–mediated phosphorylation, underlies anaphase-specific cortical accumulation of dynein and contributes to chromosome separation.
RESULTS
Gα-independent membrane association of LGN-N/NuMA-CT complex
The N-terminus of LGN physically interacts with a small fragment at the C-terminus of NuMA (Figure 1A; Du et al., 2001). When an N-terminal fragment of LGN (LGN1-481, LGN-N) was ectopically expressed in Cos 7 cells, it localized predominantly in the nucleus (Figure 1B). On the other hand, a C-terminal fragment of NuMA that contains the LGN-binding domain, the microtubule-binding domain, and a mutation in the putative nuclear localization signal (NLS; NuMA1536-CTδNLS, NuMA-CT; Du and Macara, 2004) is distributed mainly in the nucleus (probably due to an additional NLS at the very C-terminal of NuMA; our unpublished data) and on the bundled MTs (Figure 1C). Of interest, the distribution of the two proteins was significantly different when they were coexpressed in Cos 7 cells (Figure 1D). Both proteins exhibited smooth and even distribution all over the cells, which is reminiscent of the characteristics of the membrane-binding proteins. Similar results were obtained using HeLa and Madin–Darby canine kidney (MDCK) cells, for which the membrane localization of the complex was even more evident (Supplemental Figure S1). This was surprising, since LGN1-481 does not contain the Gα-binding GoLoco motifs (Figure 1A), and the membrane association of the LGN-N/NuMA-CT complex should not be mediated through membrane-bound Gα. To test which part(s) of LGN-N or NuMA-CT is or are required for the membrane association of the complex, we generated a series of shortened LGN-N and NuMA-CT fragments and tested their localization in cotransfected Cos 7 cells. The results indicated that the linker region of LGN is dispensable (Figure 1E), whereas the very C-terminus of NuMA (aa 2041–2101) is required (Figure 1F).
The C-terminus of NuMA contains a membrane-binding domain
If NuMA is mediating the membrane binding of the LGN-N/NuMA-CT complex, why did not NuMA-CT itself show obvious membrane association? We showed previously that LGN binding could inhibit the association of NuMA with MTs (Du et al., 2002). It is possible that ectopically expressed NuMA-CT preferentially binds to MTs and its membrane association is masked. When LGN-N was coexpressed, it prevented the binding of NuMA to MTs and allowed NuMA-CT to associate with plasma membrane. To test this hypothesis, we partially deleted the microtubule-binding domain of NuMA-CT and expressed it in Cos 7 cells. Indeed, the microtubule-binding and NLS double-deletion mutant of NuMA-CT exhibited membrane distribution in the absence of coexpressed LGN-N (Figure 2A), indicating that NuMA-CT may contain a membrane-binding domain. Through further deletion and transfection experiments, we identified a small region at the C-terminal of NuMA (aa 1981–2060)—right downstream of the putative NLS—that shows robust plasma membrane localization when expressed in Cos 7 or HeLa cells (Figure 2B). We termed this domain the NuMA membrane-binding domain (MBD). Consistent with our cotransfection results (Figure 1F), further deletion from the C-terminal of NuMA MBD (deleting aa 2041–2060) abolished its association with the plasma membrane (Figure 2C).
FIGURE 2:
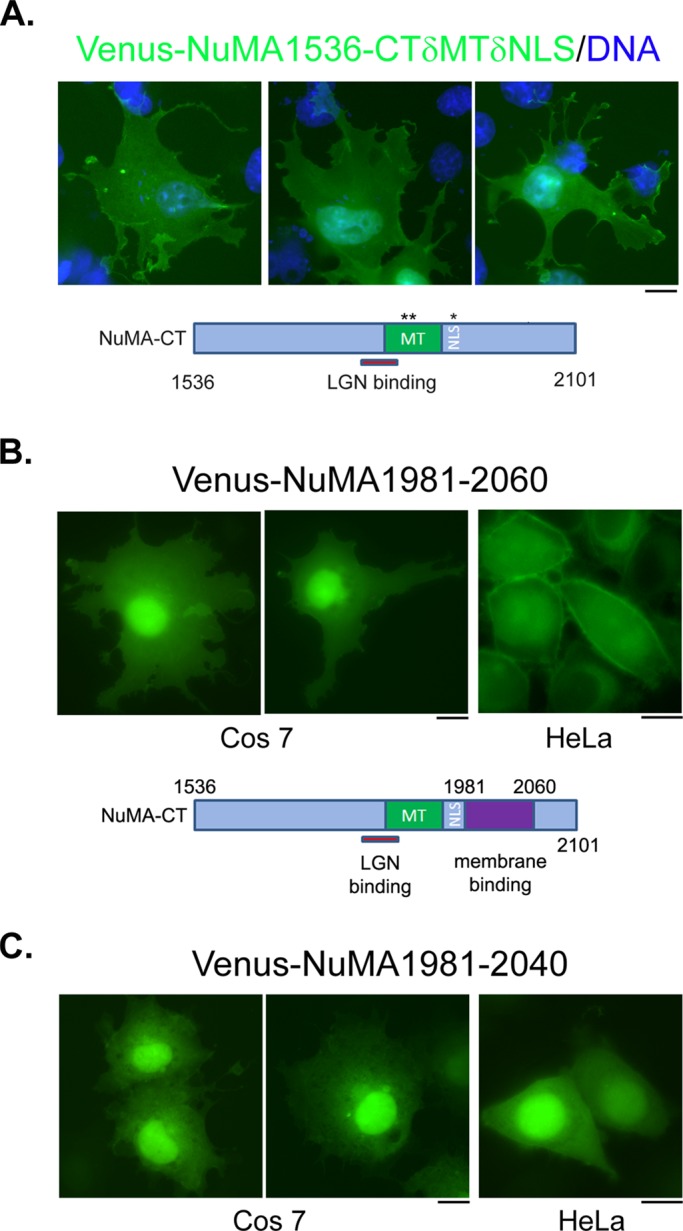
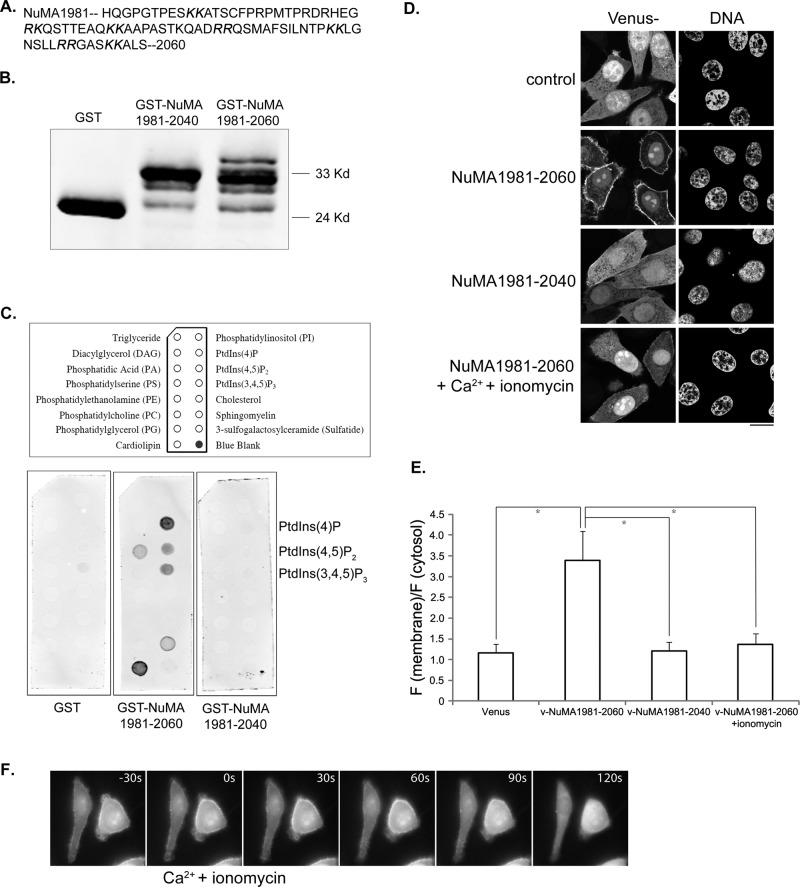
The C-terminus of NuMA contains a membrane association domain. (A) When microtubule binding and nuclear localization are compromised, the C-terminus of NuMA associates with plasma membrane. Representative images of Cos 7 cells expressing a mutant NuMA C-terminal fragment. Cells were transfected with plasmids expressing Venus-NuMA1536-CTδMTδNLS (green). At 24 h later, cells were fixed and stained with DNA dye (blue). A schematic of the expressed fragment is shown underneath the images. (B) A small fragment at the C-terminus of NuMA associates with plasma membrane. Representative images of Cos 7 (left, middle) or HeLa (right) cells expressing Venus-NuMA1981-2060 (green). Cells were transfected with plasmids expressing Venus-NuMA1981-2060. At 24 h later, cells were fixed and imaged. A schematic indicating the membrane-binding domain is shown underneath the images. (C) Further deletion of NuMA1981-2060 impaired its association with plasma membrane. Representative images of Cos 7 (left, middle) or HeLa (right) cells expressing Venus-NuMA1981-2040 (green). Bars, 10 μm.
NuMA MBD binds to lipids
The membrane binding of NuMA MBD could be direct or indirect. We analyzed the amino acid composition of NuMA MBD and could not find a typical membrane-binding motif; instead, we found that it contains multiple pairs of positively charged lysine or arginine residues (Figure 3A). The interaction between polybasic clusters with negatively charged lipids contributes to the membrane association of some proteins (Heo et al., 2006; Su et al., 2011). We tested whether NuMA MBD binds to lipids by using purified recombinant proteins and a strip lipid-binding assay. As shown in Figure 3, B and C, although glutathione S-transferase (GST) itself does not bind to any lipid, GST-NuMA1981-2060 showed robust lipid binding, preferentially with phosphorylated forms of phosphatidylinositol (PIPs). Consistent with the inability of Venus-NuMA1981-2040 to localize to the plasma membrane, GST-NuMA1981-2040 also failed to bind to any lipid (Figure 3C). Given that recombinant NuMA MBD showed robust binding to PIPs, we tested whether PIPs mediate the membrane association of NuMA MBD. Activation of phospholipase C with ionomycin leads to depletion of membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate (Varnai and Balla, 1998; Hammond et al., 2012). We treated Venus-NuMA1981-2060–expressing cells with ionomycin and tested its effect on the membrane association of Venus-NuMA1981-2060. Two minutes of ionomycin treatment resulted in significant dissociation of Venus-NuMA1981-2060 from the plasma membrane, as revealed by quantitative analysis of fixed cells (Figure 3, D and E). Live-cell time-lapse analysis also confirmed rapid membrane dissociation of Venus-NuMA1981-2060 upon ionomycin treatment (Figure 3F and Supplemental Movie S1). These results indicate that the membrane association of the identified NuMA MBD is mediated through its direct binding to PIPs.
FIGURE 3:
The membrane association domain of NuMA binds to lipids. (A) Amino acid sequence of the membrane-binding domain of NuMA (aa 1981–2060). The paired, positively charged lysine and arginine residues are highlighted in italic. (B) Expression of recombinant proteins. Purified recombinant GST, GST-NuMA1981-2040, and GST-NuMA1981-2060 (0.5 μg of each) were separated by SDS–PAGE and blotted with anti-GST antibody. (C) Lipid-binding assay. Lipid membrane strips were incubated with GST, GST-NuMA1981-2040, and GST-NuMA1981-2060, respectively, and bound proteins were detected with anti-GST antibody. (D) Ionomycin treatment disrupts the membrane association of NuMA. Representative single-layer confocal images of HeLa cells expressing Venus, Venus-NuMA1981-2040, or Venus-NuMA1981-2060. Transfected cells were either directly fixed (top three) or treated with ionomycin plus calcium for 2 min and then fixed (bottom). Bar, 10 μm. (E) Quantification of membrane-to-cytosol fluorescence intensity ratio from images in D. Results are from three independent experiments. Error bars, SD. n = 50 from each group of cells. *p < 0.01. (F) Fluorescence images from time-lapse analysis of HeLa cells expressing Venus-NuMA1981-2060. Time points are shown as seconds. Time point zero indicates the time when calcium (1 mM final concentration) and ionomycin (10 μM final concentration) were added to the medium.
The membrane association of NuMA MBD is cell cycle regulated during mitosis
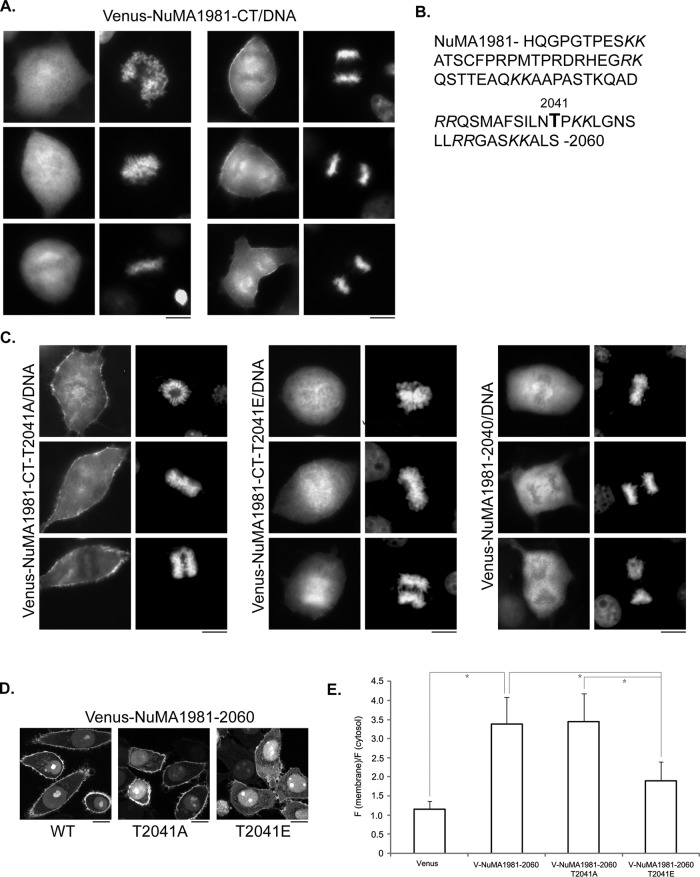
NuMA is a nuclear protein (Van Ness and Pettijohn, 1983). The identified membrane association of NuMA could happen only during mitosis when the nuclear envelope is broken down. We observed mitotic Cos 7 cells expressing Venus-NuMA1981-CT, which contains the membrane-binding domain. Surprisingly, unlike in interphase cells, the expressed protein did not exhibit obvious membrane association in prophase and metaphase cells (Figure 4A; similar results were obtained in 100% of cells observed, n > 50). The membrane association of Venus-NuMA 1981-CT, however, was evident when cells entered anaphase (Figure 4A; similar results were obtained in 100% of cells observed, n > 50), suggesting that the membrane association of NuMA MBD could be cell cycle regulated.
FIGURE 4:
Cell cycle–regulated membrane association of NuMA MBD. (A) The membrane association of NuMA1981-CT is evident only in anaphase cells. Representative images of mitotic Cos 7 cells expressing Venus-NuMA1981-CT. Cells were transfected with plasmids expressing Venus-NuMA1981-CT. At 24 h later, cells were fixed and stained with DNA dye. Left, prophase and metaphase cells; right, anaphase cells. (B) Amino acid sequence of the membrane-binding domain of NuMA (aa 1981–2060). The paired, positively charged lysine and arginine residues are highlighted in italic; Thr-2041 is highlighted in bold. (C) Phosphorylation of Thr-2041 regulates membrane association of NuMA during mitosis. Representative images of mitotic Cos 7 cells expressing Venus-NuMA1981-CT-T2041A (left), Venus-NuMA1981-CT-T2041E (middle), or Venus-NuMA1981-2040 (right). Cells were transfected with plasmids expressing the indicated proteins, fixed, and stained as in A. (D) Phosphorylation of Thr-2041 affects membrane association of NuMA MBD in interphase cells. Representative single-layer confocal images of HeLa cells expressing Venus-NuMA1981-2060 (WT; left), Venus-NuMA1981-2060-T2041A (middle), or Venus-NuMA1981-2060-T2041E (right). (E) Quantification of membrane-to-cytosol fluorescence intensity ratio from images in D. Results are from three independent experiments. Error bars, SD. n = 50 for each group of cells. *p < 0.01. Bars, 10 μm.
The phosphorylation state of T2041 is critical for the membrane association of NuMA MBD
What could be preventing the membrane association of NuMA MBD during prophase and metaphase? A clue came from a previous study from the Compton lab showing that when several putative CDK1 phosphorylation sites at the C-terminal of NuMA were mutated, the mutant proteins mislocalized during mitosis (Compton and Luo, 1995). In particular, when Thr-2041 (T2041, Figure 4B; originally described as T2040 in the Compton article and as T2055 when another human NuMA cDNA [NP_006176] was used) was mutated to alanine, the mutant protein appeared to localize to the plasma membrane during mitosis (Compton and Luo, 1995). The underlying mechanism for this altered localization is not known. T2041 locates within the identified NuMA MBD (Figure 4B). It was confirmed to be a CDK1 substrate (Blethrow et al., 2008). CDK1 is a cell cycle–regulated protein kinase—it is active during prophase and metaphase and is inactivated before anaphase initiation (Rhind and Russell, 2012). It is tempting to speculate that CDK1 phosphorylation of T2041 may inhibit the membrane association of NuMA MBD during prophase and metaphase, and the inhibitory effect is released at the onset of anaphase when CDK1 activity is down-regulated. To test this hypothesis, we mutated Thr-2041 to alanine and tested its effect on the membrane binding of NuMA MBD through mitosis. Unlike the wild-type protein, Venus-NuMA1981-CT(T2041A) was observed to localize at the plasma membrane even in prophase and metaphase cells (Figure 4C, left; similar results were obtained in 100% of cells observed, n > 50). Of importance, when Thr-2041 was mutated to glutamic acid, which mimics the phosphorylated state of the residue, the mutant protein (Venus-NuMA1981-CT(T2041E)) did not show obvious membrane association even in anaphase cells (Figure 4C, middle; similar results were obtained in 32 of 40 anaphase cells). Not surprisingly, Venus-NuMA-1981-2040 did not show membrane localization throughout mitosis (Figure 4C, right; similar results were obtained in 100% of cells observed, n > 50), consistent with its impaired lipid and membrane binding. It is noteworthy that, in interphase cells, whereas the T2041A mutant of NuMA MBD showed robust membrane binding, the T2041E mutant of NuMA MBD exhibited significantly reduced membrane association (Figure 4, D and E). These results indicate that membrane binding of NuMA MBD is regulated by phosphorylation of Thr-2041 in a cell cycle–dependent manner, mostly likely through CDK1.
CDK1-regulated membrane association of NuMA underlies cell cycle–dependent cortical accumulation of NuMA
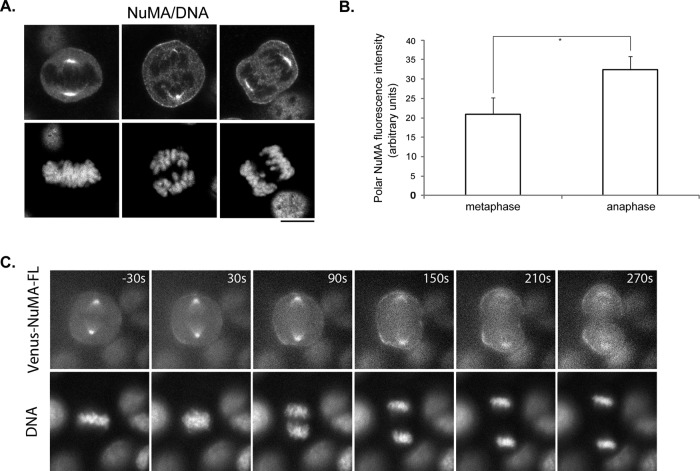
Our finding of the cell cycle–regulated membrane-binding domain of NuMA led us to ask whether it contributes to the localization of full-length NuMA. We carefully compared the localization of endogenous NuMA throughout the cell cycle using antibody staining and found that the immunofluorescence intensity of polar cortical NuMA is significantly stronger in anaphase than metaphase cells (Figure 5, A and B), suggesting that the cortical localization of NuMA could be cell cycle regulated. To directly monitor the dynamic changes of NuMA localization, we established stable MDCK cell lines expressing low levels of Venus-tagged, full-length human NuMA (Venus-NuMA-FL). It is critical to keep the expression of exogenous NuMA at a low level because moderate or high level overexpression of NuMA would result in spindle organization defects and mitotic arrest (Quintyne et al., 2005). Time-lapse analysis of Venus-NuMA–expressing cells through mitosis captured an obvious enhancement of cortical Venus-NuMA right after the cells entered anaphase (Figure 5C and Supplemental Movie S2; similar results were obtained in 18 of 18 cells), indicating that the cortical accumulation of full-length NuMA is indeed cell cycle regulated.
FIGURE 5:
Cortical localization of NuMA is cell cycle regulated. (A) Cortical localization of endogenous NuMA is enhanced during anaphase. Representative images of metaphase (left) and anaphase (middle and right) HeLa cells. Cells were fixed and stained with anti-NuMA antibody (top) and DNA dye (bottom). Bar, 10 μm. (B) Quantification of polar cortical NuMA fluorescence intensity from images in A. Results are from three independent experiments. Error bars, SD. n = 50 for each group of cells. *p < 0.01. (C) Fluorescence images from time-lapse analysis of MDCK cells expressing Venus-tagged full-length NuMA (Venus-NuMA-FL). MDCK cells stably express Venus-NuMA-FL were seeded on glass-bottomed dishes. At 16 h later, cells were briefly stained with Hoechst 33342, washed, and imaged. Fluorescence signals of Venus-NuMA (top) and Hoechst 33342 (bottom) were captured simultaneously every 10 s. Time points are shown as seconds. Time zero was defined as when the congressed metaphase chromosome became loose (the initiation of chromosome segregation). Negative number indicates time point before time zero.
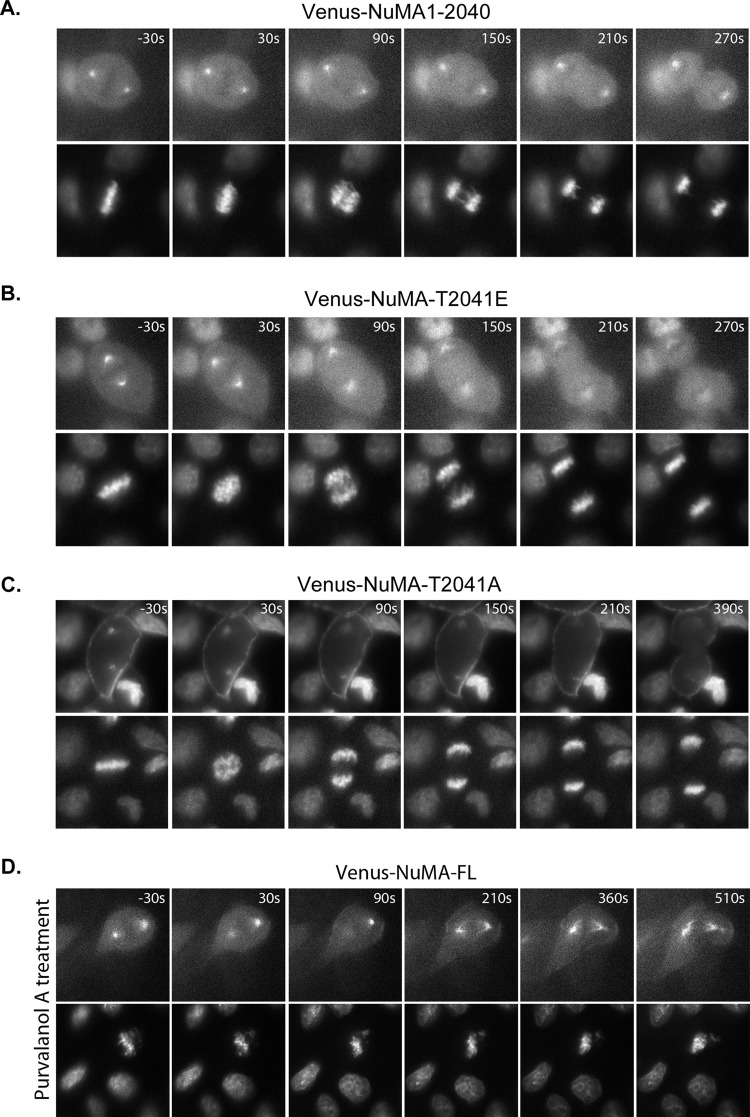
Next we tested whether the identified, cell cycle–regulated membrane association of NuMA is responsible for the observed anaphase cortical enhancement of NuMA. We generated stable MDCK cell lines expressing low levels of Venus-NuMA1-2040, which should have impaired membrane association. Live-cell imaging analysis revealed that unlike full-length NuMA, NuMA1-2040 did not show obvious anaphase cortical enhancement (Figure 6A and Supplemental Movie S3; similar results were obtained in 12 of 12 cells). Similarly, we generated cell lines expressing Venus-NuMAT2041E and did not observe cell cycle–regulated membrane accumulation of the mutant protein (Figure 6B and Supplemental Movie S4; similar results were obtained in 10 of 13 cells). On the contrary, Venus-NuMA-T2041A showed robust and consistent membrane association throughout different stages of mitosis (Figure 6C and Supplemental Movie S5; similar results were obtained in 15 of 15 cells). These results indicate that the membrane association of NuMA is required for the enhanced anaphase cortical accumulation of NuMA. Of importance, the phosphorylation status of Thr-2041 is a critical determinant for the cortical targeting of full-length NuMA.
FIGURE 6:
CDK1-regulated membrane association underlies cell cycle–dependent cortical localization of NuMA. Fluorescence images from time-lapse analyses of MDCK cells. Time-lapse analyses were performed as described in Figure 5C. (A) MDCK cells expressing Venus-tagged NuMA1-2040 (Venus-NuMA1-2040). (B) MDCK cells expressing Venus-tagged NuMA-T2041E mutant (Venus-NuMA-T2041E). (C) MDCK cells expressing Venus-tagged NuMA-T2041A mutant (Venus-NuMA-T2041A). (D) MDCK cells expressing Venus-tagged full-length NuMA (Venus-NuMA-FL) imaged before and after 30 μM purvalanol A treatment. Time points are shown as seconds. For A–C, time zero was defined as when the congressed metaphase chromosome became loose (the initiation of chromosome segregation). For D, time zero was defined as when purvalanol A was added in the medium. Negative numbers indicate time points before time zero.
If CDK1 phosphorylation prevents membrane association of NuMA, we should expect that inhibition of CDK1 should lead to membrane accumulation of NuMA. We treated metaphase Venus-NuMA–expressing cells with the CDK1 inhibitor purvalanol A and observed immediate enhancement of cortical Venus-NuMA (Figure 6D and Supplemental Movie S6; similar results were obtained in five of five cells), indicating that CDK1 is responsible for limiting the cortical accumulation of NuMA before anaphase onset.
LGN appears to be dispensable for anaphase cortical targeting of NuMA
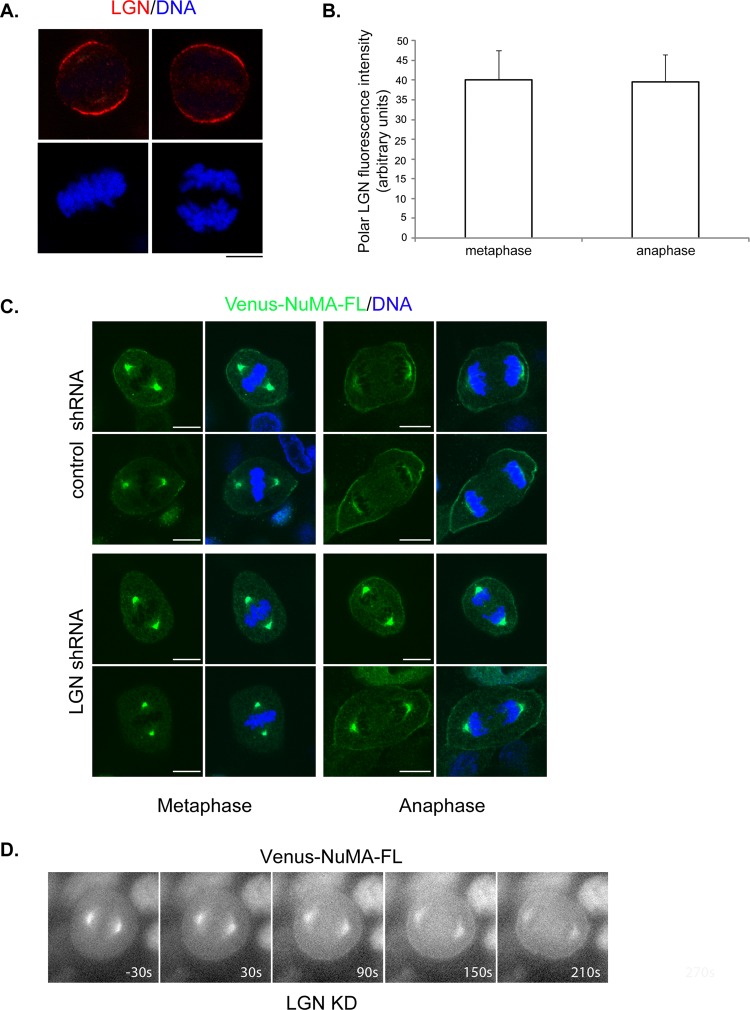
Previous studies suggested that cortical targeting of NuMA is mediated through its interaction with LGN and subsequent binding of LGN to membrane-bound Gα; however, most of the work was done in metaphase cells (Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012; Zheng et al., 2013). Unlike that of NuMA, the intensity of cortical LGN appeared to be unchanged from metaphase to anaphase (Figure 7, A and B). To determine whether LGN is needed for the observed anaphase cortical accumulation of NuMA, we knocked down LGN in Venus-NuMA–expressing MDCK cells, in which NuMA signal is more robust when detected using our anti-NuMA antibody. In agreement with previous reports, knocking down LGN led to complete depletion of cortical NuMA in metaphase cells (Figure 7C). In anaphase cells, however, substantial amounts of NuMA were still observed at the cell cortex, although its intensity appeared to be reduced compared with control short hairpin RNA (shRNA)–transfected cells (Figure 7C). We also performed time-lapse analyses of Venus-NuMA in LGN-depleted cells and observed anaphase-specific cortical enhancement of Venus-NuMA (Figure 7D and Supplemental Movie S7; similar results were obtained in six of six cells). These results suggest that anaphase cortical targeting of NuMA may not necessarily require LGN.
FIGURE 7:
LGN appears to be dispensable for anaphase cortical accumulation of NuMA. (A) Cortical intensity of endogenous LGN does not change from metaphase to anaphase. Representative images of metaphase (left) and anaphase (right) HeLa cells. Cells were fixed and stained with anti-LGN antibody (top) and DNA dye (bottom). Bar, 10 μm. (B) Quantification of polar cortical LGN fluorescence intensity from images in A. Results are from three independent experiments. Error bars, SD. n = 50 for each group of cells. (C, D) Knockdown of LGN did not prevent anaphase cortical accumulation of NuMA. MDCK cells stably expressing low level of Venus-NuMA were transfected with plasmids expressing control shRNA (top) or LGN-specific shRNA (bottom). At 72 h later, cells were either fixed and stained with anti-NuMA antibody (green) and DNA dye (blue; C) or subjected to time-lapse analysis (D) as described in Figure 5C. Bars, 10 μm.
Membrane association of NuMA is critical for anaphase cortical accumulation of dynein
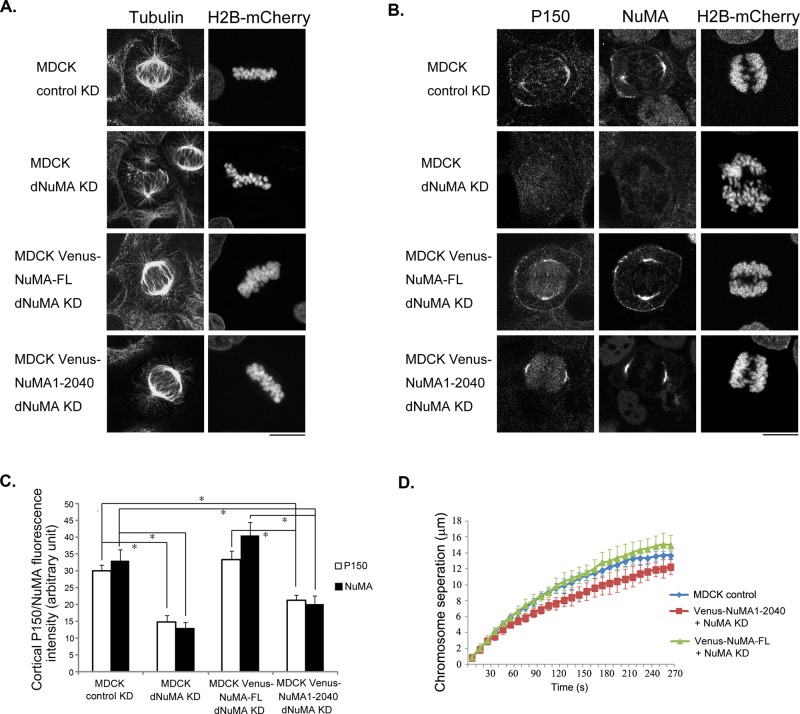
NuMA mediates cortical targeting of cytoplasmic dynein during mitosis (Kotak et al., 2012). Our finding of cell cycle–regulated membrane association of NuMA coincides with recently described anaphase enhancement of cortical dynein (Collins et al., 2012). It is possible that LGN-independent membrane association of NuMA during anaphase underlies enhanced cortical accumulation of dynein. We reasoned that if we could replace endogenous NuMA with a mutant NuMA that cannot associate with plasma membrane, a reduction of cortical dynein during anaphase would support our hypothesis. Given that we established stable MDCK cell lines expressing low levels of Venus-tagged NuMA1-2040 that are deficient in membrane association, knocking down endogenous NuMA in these cells would fulfill our purpose. To efficiently knock down endogenous NuMA in MDCK cells, we used the lentivirus-mediated stable knockdown approach. We engineered lentivirus vectors to express H2B-mCherry, as well as control or canine NuMA–specific shRNA, and tested them in control MDCK cells. As shown in Supplemental Figure S2, transduction of MDCK cells with lentivirus expressing shRNA against NuMA led to efficient knockdown of endogenous NuMA. Virus-transduced cells exhibited typical NuMA loss-of-function phenotypes, including chromosome alignment defects during mitosis (Supplemental Figure S2, bottom and second from bottom, arrows) and formation of multiple micronuclei in postmitotic cells (Yang and Snyder, 1992; Du and Macara, 2004; Haren et al., 2009; Supplemental Figure S2, bottom and second from bottom, arrowheads). Similar results were observed when two different shRNAs against distinct regions of canine NuMA were stably expressed (unpublished data). Previous studies of NuMA function indicated that NuMA is critical for mitotic spindle organization. It functions to tether the minus ends of spindle MTs at the spindle poles (Yang and Snyder, 1992; Compton and Cleveland, 1993). Indeed, stable knockdown of NuMA in MDCK cells led to severe spindle organization defects—the spindle MTs appeared to be less compact and failed to be focused properly at the centrosomes (Figure 8A, second from top). Of interest, such defects were not observed when the same virus was used to transduce MDCK cells expressing low levels of shRNA-resistant Venus-NuMA-FL or Venus-NuMA1-2040 (Figure 8A, bottom and next to bottom), suggesting that they could rescue the function of endogenous NuMA in organizing the mitotic spindles. Consistent with the role of NuMA in cortical targeting of dynein, knocking down NuMA in control cells resulted in complete loss of cortical dynactin subunit P150Glued and dynein heavy chain (DYNC1H1) in a few cells that eventually entered anaphase (Figure 8, B and C, and Supplemental Figure S3, A and B). In Venus-NuMA-FL–expressing cells, depletion of endogenous NuMA did not reduce the cortical accumulation of P150Glued and DYNC1H1 during anaphase (Figure 8, B and C, and Supplemental Figure S3, A and B). Knocking down endogenous NuMA in Venus-NuMA1-2040–expressing cells, however, led to significant reduction of cortical P150Glued as well as DYNC1H1 during anaphase (Figure 8, B and C, and Supplemental Figure S3, A and B). Of importance, the effect is specific to cortically localized dynein, since spindle pole accumulation of DYNC1H1 was completely rescued in Venus-NuMA-FL– and Venus-NuMA1-2040– expressing metaphase and anaphase cells (Supplemental Figure S3C). These results suggest that membrane binding of NuMA is critical for anaphase cortical accumulation of dynein/dynactin.
FIGURE 8:
Cell cycle–regulated membrane association of NuMA underlies cell cycle–dependent cortical accumulation of dynein and contributes to anaphase chromosome segregation. (A) Stable knockdown of NuMA in MDCK cells led to spindle organization defects, which could be rescued by expressing low levels of shRNA-resistant full-length NuMA or NuMA1-2040. Normal MDCK cells were transduced with lentivirus expressing control shRNA (top) or NuMA specific shRNA (second from top). MDCK cells expressing Venus-NuMA-FL (second from bottom) or Venus-NuMA1-2040 (bottom) were also transduced with lentivirus expressing NuMA-specific shRNA. Stable, virus-transduced cells were fixed and stained with anti–α-tubulin antibody. Note that both lentiviruses also express H2B-mCherry. A nontransduced, H2B-mCherry–negative cell exhibited normal spindle organization (second from top). Bar, 10 μm. (B) Membrane association of NuMA is required for anaphase cortical accumulation of dynein/dynactin. Cells were infected with lentiviruses as in A. Transduced cells were fixed and stained with anti-P150Glued (P150) and anti-NuMA antibodies. Bar, 10 μm. (C) Quantification of cortical P150 and NuMA fluorescence intensity from images in B. Results are from three independent experiments. Error bars, SD. n = 10 for NuMA KD control cells and n = 30 for other group of cells. *p < 0.01. (D) NuMA-mediated cortical targeting of dynein contributes to anaphase chromosome segregation. Quantitative analysis of chromosome separation. MDCK control, control MDCK cells expressing H2B-mCherry and control shRNA (n = 15); Venus-NuMA-FL + NuMA KD, Venus-NuMA-FL cells transduced with lentivirus expressing H2B-mCherry and NuMA shRNA (n = 12); Venus-NuMA1-2040 + NuMA KD, Venus-NuMA1-2040 cells transduced with lentivirus expressing H2B-mCherry and NuMA shRNA (n = 13). Virus-transduced cells were subjected to time-lapse analyses to record the movement of H2B-mCherry–labeled chromosomes. Distances between separated chromosomes were plotted against time points (seconds). Results are from three independent experiments. Error bars, SD.
NuMA-mediated cortical accumulation of dynein contributes to anaphase chromosome separation
Cortical dynein has been proposed to function in generating pulling forces on astral MTs and contributes to anaphase chromosome segregation (Roostalu et al., 2010). However, direct evidence for the specific involvement of cortical dynein in mammalian cells is missing. By replacing endogenous NuMA with a membrane-binding-impaired NuMA, we can now specifically eliminate anaphase cortical accumulation of dynein while maintaining dynein function in other cellular compartments. Time-lapse analyses of chromosome segregation were performed in control MDCK cells and endogenous NuMA-depleted, Venus-NuMA-FL– or Venus-NuMA1-2040–expressing cells. Quantitative analysis indicated that the chromosome segregation speed was substantially reduced in endogenous NuMA-depleted, Venus-NuMA1-2040–expressing cells (Figure 8D). It is still possible, however, that the observed slower chromosome separation in Venus-NuMA1–2040–expressing cells is not due to reduced cortical localization of NuMA; instead, it is caused by alteration of astral MTs or incomplete rescue of chromosome alignment defects. We did not observe obvious reduction or shortening of astral MTs in endogenous NuMA-depleted, Venus-NuMA1-2040–expressing cells (Figure 8A, bottom). Of importance, careful time-lapse analyses from nuclear envelope breakdown (NEB) throughout mitosis indicated that there is no delay of chromosome alignment or anaphase initiation in Venus-NuMA1-2040–rescuing cells compared with control cells (time spent from NEB to anaphase onset: control cells, 34.6 ± 2.1 min, n = 8; endogenous NuMA-depleted, Venus-NuMA1-2040–expressing cells, 34.4 ± 2.8 min, n = 8). Taking the results together, we conclude that cell cycle–regulated, NuMA-mediated cortical accumulation of dynein contributes to efficient anaphase chromosome segregation.
DISCUSSION
Cortical dynein has been proposed to contribute to anaphase spindle elongation and chromosome segregation by exerting pulling forces on astral MTs (Dujardin and Vallee, 2002; Maiato and Lince-Faria, 2010; Roostalu et al., 2010). Elegant in vitro reconstitution analyses demonstrated that artificially “anchored” dynein can indeed exert forces on astral MTs by modulating microtubule dynamics (Hendricks et al., 2012; Laan et al., 2012). As the major microtubule-based, minus ends–directed motor protein, dynein plays multiple roles during mitosis. It functions not only at the cell cortex, but also at kinetochores and spindle poles (Kardon and Vale, 2009). In mammalian cells, global inhibition of dynein function leads to early-stage mitotic defects, which makes it difficult to dissect the specific contribution of cortical dynein during anaphase. Understanding the detailed cortical targeting mechanisms for dynein will help us to elucidate the specific function of dynein at the cell cortex.
How is dynein anchored at the cell cortex? Recent studies suggest that dynein is recruited and anchored at the cell cortex by the Gα/LGN/NuMA ternary complex (Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012; Zheng et al., 2013). This Gα-mediated membrane-anchoring mechanism is plausible and has been proven to be important in regulating metaphase spindle positioning in multiple systems (Morin and Bellaiche, 2011; McNally, 2013). In addition, cortical actin filaments help to maintain dynein at the cell cortex (Zheng et al., 2013). Of interest, the cortical localization of dynein appears to be cell cycle regulated. Concomitant with its potential role in chromosome separation, a significant rise of cortical dynein at anaphase onset was observed (Collins et al., 2012). What underlies the cell cyclexregulated cortical accumulation of dynein is not clear. We have now shown that it is mediated by NuMA, which has an intrinsic, cell cycle–regulated membrane-binding domain. The direct membrane binding of NuMA provides another interesting cell cycle–regulated cortical anchoring mechanism for dynein. The understanding of such a mechanism allowed us to design experiments to specifically reduce the cortical localization of NuMA and dynein during anaphase without compromising their functions at kinetochores and spindle poles and demonstrate that cortical NuMA and dynein are indeed required for efficient chromosome separation.
It is interesting to observe that when the isolated NuMA-CT was expressed alone, it localized predominantly to bundled MTs and did not show obvious membrane association. Only when LGN-N was coexpressed did the LGN-N/NuMA-CT complex localize to the plasma membrane (Figure 1), suggesting that inhibition of the microtubule binding of NuMA by LGN is required for the membrane association of NuMA-CT. However, in LGN-knockdown cells, substantial amounts of NuMA could be detected at the cell cortex of anaphase cells (Figure 7), indicating that the anaphase cortical localization of NuMA is at least partially independent of LGN. This is consistent with the fact that unlike NuMA, the cortical localization of LGN is not cell cycle regulated (Figure 7). We hypothesize that during mitosis, there is a small amount of NuMA that is not associated with either LGN or MTs, and it is this pool of NuMA that undergoes cell cycle–regulated membrane association.
Recently an LGN-independent cortical targeting mechanism for NuMA and dynein was attributed to the interaction between NuMA and protein 4.1 (Kiyomitsu and Cheeseman, 2013). The protein 4.1–binding domain lies ∼200 aa from the NuMA MBD. The fact that the isolated NuMA MBD shows robust membrane association suggests that the membrane binding of NuMA is protein 4.1 independent. It is possible that the interaction between protein 4.1 and NuMA may further reinforce the membrane association of NuMA.
Our lipid-binding assays suggest that the membrane-binding domain of NuMA preferentially binds to PIPs, which are signaling molecules that also play important roles during mitosis. It is noteworthy that integrin and phosphoinositide 3-kinase (PI3K) signaling appears to be involved in regulating spindle orientation in human cells (Toyoshima et al., 2007; Toyoshima and Nishida, 2007). It would be interesting to study whether integrin and PI3K are involved in regulating the membrane association of NuMA.
Down-regulation of CDK and rapid dephosphorylation of the phosphorylated proteins upon metaphase-to-anaphase transition has proven to be a general mechanism for mitotic progression (Roostalu et al., 2010). We have now identified NuMA as another mitotic protein that undergoes such regulation. We hypothesize that mammalian cells have developed a strategy to modulate cortical pulling forces during mitosis. The membrane binding of NuMA is inhibited by CDK1-mediated phosphorylation of T2041 during prophase and metaphase, such that the cortical targeting of NuMA and dynein can only be achieved through LGN and Gα. Limited cortical NuMA and dynein are sufficient for directing spindle positioning. At anaphase onset, down-regulation of CDK1 and rapid dephosphorylation of T2041 allow NuMA to associate with the plasma membrane by itself and recruit more dynein to produce robust pulling forces for efficient chromosome separation.
MATERIALS AND METHODS
Cell lines and reagents
Cos 7 and MDCK cells were maintained in DMEM (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) and antibiotics at 37°C in a humidified 5% CO2 atmosphere.
Rabbit anti-LGN antibodies were described previously (Du and Macara, 2004). Rabbit anti-NuMA antibody was a kind gift from Duane Compton (Dartmouth Medical School, Hanover, NH). The following antibodies were also used: primary, monoclonal anti-GST (Santa Cruz Biotechnology, Santa Cruz, CA), anti–α-tubulin (Sigma-Aldrich, St. Louis, MO), anti-p150Glued (Cell Signaling Technology, Danvers, MA), and polyclonal anti-DYNC1H1 (Santa Cruz Biotechnology); secondary, Alexa 488–, 594–, 660–, and 680–conjugated (Invitrogen, Carlsbad, CA) and IRDye800-conjugated (Rockland Immunochemicals, Gilbertsville, PA) goat anti-mouse or rabbit antibodies. Purvalanol A was from Sigma-Aldrich, and ionomycin was from Invitrogen.
Plasmids, cell transfection, and stable cell lines
Human NuMA cDNA was kindly provided by Duane Compton (Dartmouth Medical School). Compared with NuMA1 (Gene Bank NP_006176), the cDNA we used has a 14-aa deletion between aa 1535 and 1536. Plasmids expressing LGN or NuMA fragments were PCR amplified and cloned in pK-Venus or pK–monomeric red fluorescent protein (mRFP) as previously described (Du and Macara, 2004; Zhu et al., 2011). To delete the microtubule-binding domain of NuMA, we deleted sequences encoding aa 1925–1933 by PCR, restriction enzyme digestion, and subsequent cloning. The T2041A and T2041E mutant NuMAs were generated using a QuikChange site-directed mutagenesis protocol (Agilent Technologies, Santa Clara, CA). For transient knockdown of endogenous LGN, the coding sequence of neo from pRNAi-neo (Biosettia, San Diego, CA) was replaced with mCherry to generate pRNAi-mCherry. The target sequence against canine LGN was described previously (Zheng et al., 2010).
Cells were electroporated with mammalian expression vectors using Amaxa Nucleofection device (Lonza, Basel, Switzerland) following the manufacturer's instructions.
Stable Tet-off inducible MDCK cell lines were generated as previously described (Du et al., 2001). Briefly, an enhanced yellow fluorescent protein (Venus) was cloned into pTRE2Hyg vector (Clontech, Mountain View, CA). cDNAs encoding human NuMA-FL (aa 1–2101) and NuMA1-2040 were inserted downstream of and in-frame with Venus, respectively. Again, the T2041A and T2041E mutant full-length NuMAs were generated by site-directed mutagenesis. These plasmids were transfected into MDCK T23 cells, which express the tetracycline-repressible transactivator. Cells were passaged 24 h posttransfection onto P-100 plates in medium containing 200 μg/ml hygromycin B (Mediatech, Manassas, VA) and 20 ng/ml doxycycline (ThermoFisher, Waltham, MA). After selection for 7–10 d, individual colonies were isolated using cloning rings (ThermoFisher), and the expression of Venus-fusion proteins was assessed by immunofluorescence microscopy and Western blotting after removal of doxycycline.
Lentivirus-mediated stable knockdown
The GFP coding sequence from pLV-mU6-EF1-GFP (Biosettia) vector was replaced with H2B-mCherry to generate pLV-mU6-EF1-H2B-mCherry. Lentivirus-mediated stable knockdown of NuMA in MDCK cells was carried out as previously described (Zheng et al., 2010; Wan et al., 2012). Briefly, long oligos containing control and target sequences were cloned downstream of the U6 promoter in pLV-mU6-EF1-H2B-mCherry to generate specific shRNA vectors. Once sequence was verified and knockdown efficiency tested by transient transfection, the shRNA vectors were cotransfected with the lentiviral packaging mix (Invitrogen) into HEK293FT cells, and the pseudovirus containing supernatant was collected 48 h posttransfection. Virus supernatant was used to infect MDCK cells cultured in 12-well plates. At 24 h after infection, the cells were passaged onto P-100 plates, and transduced clones (based on virus-mediated expression of H2B-mCherry) were marked and isolated using cloning rings 1 wk later. The knockdown efficiency was analyzed by Western blot and immunostaining. Target sequences for canine NuMA were 5′-GCTTTCAGCATCCTCAATACA-3′ and 5′-GCTTGCGGATGAGAGAAATAA-3′.
Immunofluorescence microscopy
Cells were grown on polylysine-coated coverglass and fixed using either 4% paraformaldehyde (PFA) or 4% PFA/0.25% Triton X-100 in phosphate-buffered saline (PBS) as indicated. Fixed cells were blocked with 1% bovine serum albumin (BSA)/10% normal goat serum in PBS for 1 h and incubated in primary antibodies for 1 h at room temperature or overnight at 4°C. Cells were then washed and incubated for 1 h with the DNA dye Hoechst 33342 and secondary antibodies coupled with Alexa 488 or Alexa 594 (Invitrogen). A SlowFade Gold AntiFade kit (Invitrogen) was used to reduce photobleaching. Cells were imaged using either a 60×/numerical aperture (NA) 1.2 oil-immersion objective on a Nikon TE2000 inverted microscope (Nikon Instruments, Melville, NY) or a 63×/NA 1.4 objective on a Zeiss 510 LSM confocal microscope (Carl Zeiss, Oberkochen, Germany).
Lipid-binding assay
We expressed GST, GST-NuMA1981-2040, and GST-NuMA1981-2060 in Escherichia coli (BL21DE3) and purified them using Glutathione Sepharose 4 Fast Flow beads (GE Healthcare, Piscataway, NJ) as described (Du et al., 2001).
For lipid-binding assays, membrane lipid arrays (Echelon Biosciences, Salt Lake City, UT) were incubated with 100 ng/ml of each recombinant protein in PBS containing 0.1% Tween-20 and 3% BSA for 1 h at 25°C. After washing, bound proteins were detected using anti-GST antibodies.
Measurement of relative fluorescence intensity of membrane-bound proteins and cortical proteins
Measurement of the relative fluorescence intensity of membrane-bound proteins was performed as described (Zheng et al., 2010). Fluorescence images of transfected cells were taken on the Zeiss 510 LSM confocal microscope using identical microscopic settings. To compare the relative fluorescence intensity of membrane-bound protein, a 30-pixel line was drawn across the cell border using MetaMorph software (Molecular Devices, Sunnyvale, CA). The Line Scan function of MetaMorph was used to reveal the relative fluorescence intensity across the line. Fluorescence intensities at the cell border and 10 pixels away at the cytosol were referred to as F(membrane) and F(cytosol), respectively. The ratio F(membrane)/F(cytosol) was collected for each group of cells and analyzed.
Measurements of the relative fluorescence intensity of cortical LGN, NuMA, P150Glued, and DYNC1H1 were performed as described (Zheng et al., 2013). Briefly, cells were stained by identical procedures, and images were taken with identical microscopic settings. Thirty metaphase or anaphase cells in each group were randomly selected, and the mean fluorescence intensity of cortical LGN, NuMA, or P150Glued was measured using ImageJ software (National Institutes of Health, Bethesda, MD). Standard deviation was calculated, and statistical significance was determined by the Student's t test.
Quantitation of chromosome separation
To quantitate the speed of anaphase chromosome separation, cells expressing H2B-mCherry were imaged using a 60×/NA 1.2 oil-immersion objective on a Nikon TE-2000 inverted microscope. The distance from the center of the mass of separating chromosomes was calculated for each time point using ImageJ.
Live-cell time-lapse analysis
Live-cell time-lapse analyses were performed as described (Du and Macara, 2004). Cells were grown on Delta T dishes (Bioptechs, Butler, PA) in F10 medium supplemented with 10% FBS and antibiotics. For cells that do not express H2B-mCherry, 2 μg/ml Hoechst 33342 was added to the medium, and the cells were incubated for 5 min. After several washes, the dish was filled with F10 medium and sealed with a 40-mm coverslip. The dish was then placed in a temperature control system (Bioptechs) that maintained a temperature of 37°C. An objective heating control system (Bioptechs) was also applied to keep the temperature of the objective lens. Time-lapse sequences were collected on a Nikon TE2000 microscope using a CFI PLAN FLUOR 40×/NA 1.3 oil-immersion objective, a CoolSnap charge-coupled device camera (Photometrics, Tucson, AZ), and MetaMorph software.
Note added in proof.
While this paper was under revision, two papers (Kotak et al., 2013; Seldin et al., 2013) describing CDK1-regulated cortical targeting of NuMA were published.
Supplementary Material
Acknowledgments
We thank Duane Compton (Dartmouth Medical School) for providing the anti-NuMA antibody. This work was supported by Grant GM079506 from the National Institutes of Health to Q.D.
Abbreviations used:
- CDK1
cyclin-dependent kinase 1
- Gα
α subunit of heterotrimeric G protein
- LGN
mammalian homologue of Drosophila Partner of Inscuteable
- MBD
membrane-binding domain
- MT
microtubule
- NuMA
nuclear mitotic apparatus
- PIP
phosphorylated phosphatidylinositol
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0474) on December 26, 2013.
*These authors contributed equally to this work.
REFERENCES
- Aist JR, Bayles CJ, Tao W, Berns MW. Direct experimental evidence for the existence, structural basis and function of astral forces during anaphase B in vivo. J Cell Sci. 1991;100:279–288. doi: 10.1242/jcs.100.2.279. [DOI] [PubMed] [Google Scholar]
- Aist JR, Berns MW. Mechanics of chromosome separation during mitosis in Fusarium (Fungi imperfecti): new evidence from ultrastructural and laser microbeam experiments. J Cell Biol. 1981;91:446–458. doi: 10.1083/jcb.91.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aist JR, Liang H, Berns MW. Astral and spindle forces in PtK2 cells during anaphase B: a laser microbeam study. J Cell Sci. 1993;104:1207–1216. doi: 10.1242/jcs.104.4.1207. [DOI] [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci USA. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer JB, Bernard ML, Peterson YK, Nezu J, Chung P, Dunican DJ, Knoblich JA, Lanier SM. Interaction of activator of G-protein signaling 3 (AGS3) with LKB1, a serine/threonine kinase involved in cell polarity and cell cycle progression: phosphorylation of the G-protein regulatory (GPR) motif as a regulatory mechanism for the interaction of GPR motifs with Gi alpha. J Biol Chem. 2003;278:23217–23220. doi: 10.1074/jbc.C200686200. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol Biol Cell. 2012;23:3380–3390. doi: 10.1091/mbc.E12-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA, Cleveland DW. NuMA is required for the proper completion of mitosis. J Cell Biol. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA, Luo C. Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. J Cell Sci. 1995;108:621–633. doi: 10.1242/jcs.108.2.621. [DOI] [PubMed] [Google Scholar]
- Desai A, Maddox PS, Mitchison TJ, Salmon ED. Anaphase A chromosome movement and poleward spindle microtubule flux occur at similar rates in Xenopus extract spindles. J Cell Biol. 1998;141:703–713. doi: 10.1083/jcb.141.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Du Q, Stukenberg PT, Macara IG. A mammalian Partner of Inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- Du Q, Taylor L, Compton DA, Macara IG. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr Biol. 2002;12:1928–1933. doi: 10.1016/s0960-9822(02)01298-8. [DOI] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB. Dynein at the cortex. Curr Opin Cell Biol. 2002;14:44–49. doi: 10.1016/s0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- Fink G, Schuchardt I, Colombelli J, Stelzer E, Steinberg G. Dynein-mediated pulling forces drive rapid mitotic spindle elongation in Ustilago maydis. EMBO J. 2006;25:4897–4908. doi: 10.1038/sj.emboj.7601354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Compton DA. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Compton DA. Functional roles of poleward microtubule flux during mitosis. Cell Cycle. 2006;5:481–485. doi: 10.4161/cc.5.5.2519. [DOI] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- Hammond GR, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Gnadt N, Wright M, Merdes A. NuMA is required for proper spindle assembly and chromosome alignment in prometaphase. BMC Res Notes. 2009;2:64. doi: 10.1186/1756-0500-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Merdes A. Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J Cell Sci. 2002;115:1815–1824. doi: 10.1242/jcs.115.9.1815. [DOI] [PubMed] [Google Scholar]
- Hendricks AG, Lazarus JE, Perlson E, Gardner MK, Odde DJ, Goldman YE, Holzbaur ELF. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr Biol. 2012;22:632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 2013;154:391–402. doi: 10.1016/j.cell.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gonczy P. Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol. 2012;199:97–110. doi: 10.1083/jcb.201203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gonczy P. NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 2013;32:2517–2529. doi: 10.1038/emboj.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P, Desai A, Oegema K, Mitchison TJ, Salmon ED. Poleward microtubule flux is a major component of spindle dynamics and anaphase a in mitotic Drosophila embryos. Curr Biol. 2002;12:1670–1674. doi: 10.1016/s0960-9822(02)01183-1. [DOI] [PubMed] [Google Scholar]
- Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol. 2003;162:377–382. doi: 10.1083/jcb.200301088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Lince-Faria M. The perpetual movements of anaphase. Cell Mol Life Sci. 2010;67:2251–2269. doi: 10.1007/s00018-010-0327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Cho G, Wen B, Insel PA. Identification and cDNA cloning of a novel human mosaic protein, LGN, based on interaction with G alpha i2. Gene. 1996;181:39–43. doi: 10.1016/s0378-1119(96)00456-8. [DOI] [PubMed] [Google Scholar]
- Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gonczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. The motor for poleward chromosome movement in anaphase is in or near the kinetochore. J Cell Biol. 1989;109:2245–2255. doi: 10.1083/jcb.109.5.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecreaux J, Roper JC, Kruse K, Julicher F, Hyman AA, Grill SW, Howard J. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- Radulescu AE, Cleveland DW. NuMA after 30 years: the matrix revisited. Trends Cell Biol. 2010;20:214–222. doi: 10.1016/j.tcb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Russell P. Signaling pathways that regulate cell division. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rogers SL, Sharp DJ. Spindle microtubules in flux. J Cell Sci. 2005;118:1105–1116. doi: 10.1242/jcs.02284. [DOI] [PubMed] [Google Scholar]
- Roostalu J, Schiebel E, Khmelinskii A. Cell cycle control of spindle elongation. Cell Cycle. 2010;9:1084–1090. doi: 10.4161/cc.9.6.11017. [DOI] [PubMed] [Google Scholar]
- Scholey JM, Brust-Mascher I, Mogilner A. Cell division. Nature. 2003;422:746–752. doi: 10.1038/nature01599. [DOI] [PubMed] [Google Scholar]
- Seldin L, Poulson ND, Foote HP, Lechler T. NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol Biol Cell. 2013;24:3651–3662. doi: 10.1091/mbc.E13-05-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk AD, Holland AJ, Cleveland DW. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J Cell Biol. 2009;184:677–690. doi: 10.1083/jcb.200810091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su KC, Takaki T, Petronczki M. Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev Cell. 2011;21:1104–1115. doi: 10.1016/j.devcel.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Matsumura S, Morimoto H, Mitsushima M, Nishida E. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev Cell. 2007;13:796–811. doi: 10.1016/j.devcel.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness J, Pettijohn DE. Specific attachment of nuclear-mitotic apparatus protein to metaphase chromosomes and mitotic spindle poles: possible function in nuclear reassembly. J Mol Biol. 1983;171:175–205. doi: 10.1016/s0022-2836(83)80352-0. [DOI] [PubMed] [Google Scholar]
- Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan QW, Liu J, Zheng Z, Zhu HB, Chu XG, Dong Z, Huang S, Du QS. Regulation of myosin activation during cell-cell contact formation by Par3-Lgl antagonism: entosis without matrix detachment. Mol Biol Cell. 2012;23:2076–2091. doi: 10.1091/mbc.E11-11-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Cole RW, Rieder CL. The force-producing mechanism for centrosome separation during spindle formation in vertebrates is intrinsic to each aster. J Cell Biol. 1993;122:361–372. doi: 10.1083/jcb.122.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard FS, et al. A point mutation to Galphai selectively blocks GoLoco motif binding: direct evidence for Galpha.GoLoco complexes in mitotic spindle dynamics. J Biol Chem. 2008;283:36698–36710. doi: 10.1074/jbc.M804936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol Cell Biol. 2010;30:3519–3530. doi: 10.1128/MCB.00394-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Lambie EJ, Snyder M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J Cell Biol. 1992;116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Snyder M. The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol Biol Cell. 1992;3:1259–1267. doi: 10.1091/mbc.3.11.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Wan Q, Liu J, Zhu H, Chu X, Du Q. Evidence for dynein and astral microtubule-mediated cortical release and transport of Gai/LGN/NuMA complex in mitotic cells. Mol Biol Cell. 2013;24:901–913. doi: 10.1091/mbc.E12-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Zhu HB, Wan QW, Liu J, Xiao ZN, Siderovski DP, Du QS. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J Cell Biol. 2010;189:275–288. doi: 10.1083/jcb.200910021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JW, Wen WY, Zheng Z, Shang Y, Wei ZY, Xiao ZN, Pan Z, Du QS, Wang WN, Zhang MJ. LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and Gai/LGN/NuMA pathways. Mol Cell. 2011;43:418–431. doi: 10.1016/j.molcel.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.