The Diaphanous formins mDia1, mDia2, and mDia3 are involved in the capture of cortical microtubules and ErbB2-dependent directed migration. These functions are independent of actin. They are mediated by mDia FH2 domains, which associate with distinct sets of proteins. Rab6IP2 is a novel interactor of mDia1 that contributes to microtubule tethering.
Abstract
Formins constitute a large family of proteins that regulate the dynamics and organization of both the actin and microtubule cytoskeletons. Previously we showed that the formin mDia1 helps tether microtubules at the cell cortex, acting downstream of the ErbB2 receptor tyrosine kinase. Here we further study the contributions of mDia1 and its two most closely related formins, mDia2 and mDia3, to cortical microtubule capture and ErbB2-dependent breast carcinoma cell migration. We find that depletion of each of these three formins strongly disrupts chemotaxis without significantly affecting actin-based structures. Further, all three formins are required for formation of cortical microtubules in a nonredundant manner, and formin proteins defective in actin polymerization remain active for microtubule capture. Using affinity purification and mass spectrometry analysis, we identify differential binding partners of the formin-homology domain 2 (FH2) of mDia1, mDia2, and mDia3, which may explain their nonredundant roles in microtubule capture. The FH2 domain of mDia1 specifically interacts with Rab6-interacting protein 2 (Rab6IP2). Further, mDia1 is required for cortical localization of Rab6IP2, and concomitant depletion of Rab6IP2 and IQGAP1 severely disrupts cortical capture of microtubules, demonstrating the coinvolvement of mDia1, IQGAP1, and Rab6IP2 in microtubule tethering at the leading edge.
INTRODUCTION
Cell motility depends on the coordinated reorganization of the actin and microtubule cytoskeletons (Waterman-Storer and Salmon, 1999; Wittmann and Waterman-Storer, 2001). In response to a promigratory stimulus, cells assemble actin-based protrusions, lamellipodia and filipodia, at the leading edge. After formation of adhesion sites, thick actomyosin cables called stress fibers connect the adhesion sites and facilitate contraction of the cell body and, upon detachment of the cell rear, cell movement in the forward direction (Le Clainche and Carlier, 2008). In migrating cells, microtubules are nucleated from microtubule-organizing centers and their dynamic plus ends explore the cytoplasmic area through alternating phases of growth and shrinkage until stabilized near the cell cortex at the leading edge. Microtubules contribute to directional motility in several ways, including the establishment of cell polarity and the regulation of adhesion site turnover (Kaverina and Straube, 2011). The molecular mechanisms underlying actin–microtubule cross-talk remain largely unknown.
Formins are a family of ubiquitous and conserved actin filament assembly factors that nucleate linear, unbranched filaments and stay processively attached to the growing barbed ends of filaments, protecting them from capping proteins (Breitsprecher and Goode, 2013). Accordingly, formins have diverse roles in forming primarily unbranched actin structures, such as those found in filopodia, cytokinetic rings, cables, and stress fibers (Pellegrin and Mellor, 2005; Schirenbeck et al., 2005; Hotulainen and Lappalainen, 2006). Formins are large, multidomain proteins defined primarily by the presence of an actin filament end-binding formin homology 2 (FH2) domain. Mammalian cells have 15 different formin genes, which fall into seven phylogenetic subfamilies (Higgs and Peterson, 2005). Most of our knowledge of mammalian formins stems from studies on Diaphanous-related formins (DRFs), which include eight of the 15 formins: three mammalian Diaphanous formins (mDia1/DRF1, mDia2/DRF3, and mDia3/DRF2), two disheveled-associated activators of morphogenesis (DAAM1 and DAAM2), and three formin-related proteins identified in leucocytes (FRL1, FRL2, and FRL3; Higgs, 2005). The FH2 domain of formins directly nucleates actin nucleation and remains bound to the growing barbed end during elongation of the filament, a phenomenon referred to as processive capping (Pruyne et al., 2002; Sagot et al., 2002; Zigmond et al., 2003; Kovar and Pollard, 2004; Moseley et al., 2004). DRFs also contain a proline-rich FH1 domain adjacent to the FH2, which greatly accelerates actin assembly by allowing formins to efficiently recruit profilin-bound actin monomers (Kovar et al., 2006). Finally, DRFs are also characterized by the presence of a C-terminal Diaphanous-autoregulatory domain (DAD), which interacts with an N-terminal Diaphanous inhibitory domain (DID; Alberts, 2001) and contributes to actin nucleation (Gould et al., 2011). Multiple lines of evidence indicate that autoinhibitory DID-DAD interactions in DRFs are released by binding of RhoGTPases to an N-terminal GTPase-binding domain (GBD), and therefore N-terminally truncated DRFs are constitutively active in vitro and in vivo (Watanabe et al., 1999; Sagot et al., 2002).
A growing body of evidence shows that formins, and DRFs in particular, are critical for directly regulating microtubule dynamics in migrating cells, suggesting that formins are potential mediators of actin–microtubule interplay during cell motility. In HeLa cells mDia1 induces the coalignment of stress fiber and microtubule arrays (Ishizaki et al., 2001). In NIH3T3 cells, active mDia2 induces the formation of oriented stable (detyrosinated) microtubules through its interaction with microtubule plus end–tracking proteins EB1 and adenomatous polyposis coli (APC; Palazzo et al., 2001; Wen et al., 2004). Our work showed that in migrating breast carcinoma cells, mDia1 facilitates capture of microtubules at the cell cortex downstream of ErbB2 receptor tyrosine kinase signaling. ErbB2 activation leads to recruitment of mDia1 via the Memo adaptor and RhoA (Zaoui et al., 2008). In turn, this complex controls cortical localization of a microtubule capture complex that includes APC and the spectraplakin ACF7 (Zaoui et al., 2010). mDia1 also controls microtubule dynamics downstream of integrin-linked kinase and IQGAP1 in keratinocytes (Wickstrom et al., 2010) and downstream of Gα12/13 and the RhoGEF LARG in fibroblasts (Goulimari et al., 2008), but downstream effectors were not described.
In the present study, we investigate the contribution of mDia1, mDia2, and mDia3 to breast carcinoma cell motility and microtubule capture, map the functional domains, identify critical amino acid residues, and characterize a novel mDia1 ligand involved in cortical capture of microtubules.
RESULTS
mDia1, mDia2, and mDia3 contribute to ErbB2-dependent chemotaxis
Heregulin β1 (HRG), a ligand for the ErbB3 and ErbB4 receptor tyrosine kinases, induces breast carcinoma cell motility via transactivation of ErbB2 (Spencer et al., 2000; Marone et al., 2004). When applied to cells in a gradient, HRG triggers a chemotactic response that requires a functional Memo/ACF7 pathway (Benseddik et al., 2013). mDia1 is a critical component of the Memo/ACF7 pathway (Zaoui et al., 2008, 2010), but the specific functional roles of mDia2 and mDia3 in this pathway are unknown.
We designed small interfering RNAs (siRNAs) that efficiently and specifically inhibit the expression of each mDia, as verified by quantitative reverse transcription (RT)-PCR and Western blotting (Figure 1A and Supplemental Figure S1). Then we evaluated their roles in HRG-dependent chemotaxis by tracking SKBr3 breast carcinoma cells as they migrated in response to a HRG gradient in Dunn chambers. In contrast to control cells, cells depleted of mDia1, mDia2, or mDia3 failed to detect the source of the promigratory ligand and therefore migrated randomly (Figure 1B), demonstrating that all three proteins are required nonredundantly for directed cell migration.
FIGURE 1:
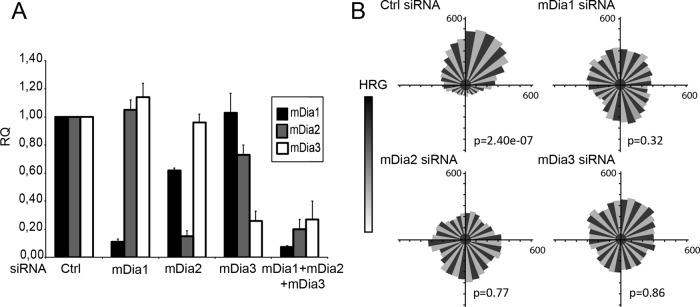
Inhibition of individual mDia formins disturbs directed migration. SKBr3 cells were transfected with the indicated siRNA for 48 h. (A) Levels of mDia1, mDia2. and mDia3 mRNA (RQ) expression relative to control (Ctrl) were evaluated by quantitative PCR. (B) SKBr3 cells were plated in Dunn chambers in the presence of a HRG gradient and tracked by time-lapse microscopy over 8 h. Distribution of migration angles was plotted as Rose diagrams. The Rayleigh test evaluates unimodal distribution of cell directions at end points; p > 0.05 is considered uniform distribution. A total of 150 cells were tracked in three independent experiments.
Roles of mDia proteins in the formation of actin cellular structures
Formins are regulators of actin assembly, with critical roles in the formation of filopodia, lamellipodia, and stress fibers (Schirenbeck et al., 2005; Hotulainen and Lappalainen, 2006; Yang et al., 2007). We evaluated whether depletion of mDia proteins affected the organization of the actin cytoskeleton of migrating SKBr3 cells. Depletion of mDia1, mDia2, or mDia3 individually (unpublished data) or together (Supplemental Figure S2) had no effect on HRG-induced 1) cell protrusion, 2) lamellipodial actin network, 3) filopodia-like structures (microspikes), or 4) stress fiber formation. Thus silencing of the three mDia formins does not alter the general organization of the actin cytoskeleton in SKBr3 cells.
Role of mDia formins in ErbB2-induced microtubule capture
We recently observed that defective microtubule capture in cells impairs the chemotactic response (Benseddik et al., 2013). Thus we investigated the role of mDia formins in microtubule capture.
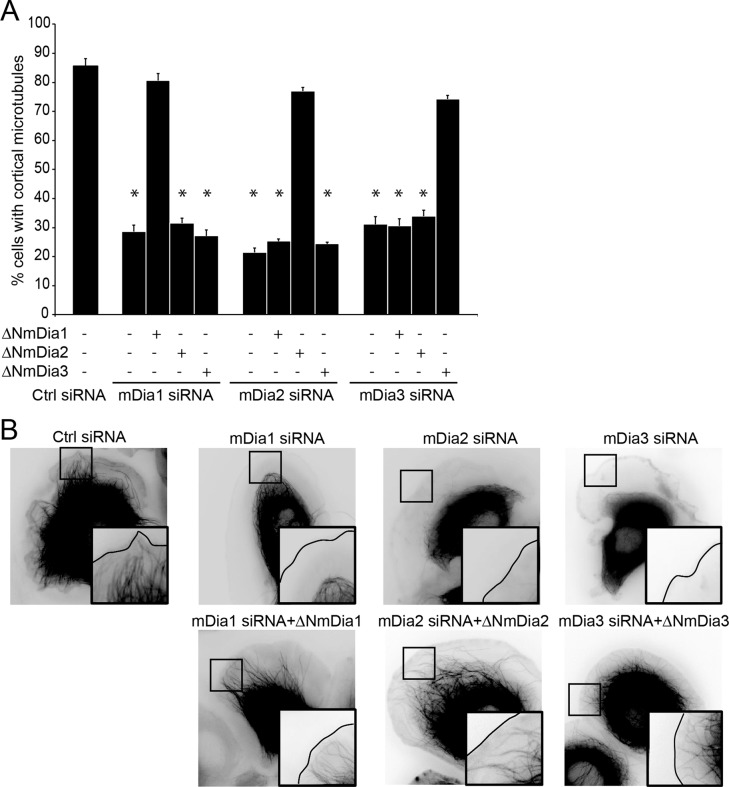
Activation of ErbB2 led to the formation of cell protrusions that contain microtubules extending toward and stabilized at the cell cortex. Silencing of mDia1 expression led to the expected defect in cortical microtubules, as previously reported (Zaoui et al., 2008). Inhibition of mDia2 and mDia3 led to a similar absence of cortical microtubules (Figure 2, A and B). Expression of an N-terminally truncated, constitutively active form of mDia1 (ΔNmDia1) not targeted by the siRNA restored cortical microtubules. ΔNmDia2 and ΔN-mDia3 also restored cortical microtubules in mDia2-depleted and mDia3-depleted cells, respectively, confirming that the observed effect is specifically caused by decreased mDia expression (Figure 2, A and B). To ensure that restoration of function was not simply due to unregulated mDia activity (caused by the ∆N truncation), we verified in similar experiments that full-length wild-type mDia proteins can restore microtubule capture, using a set of siRNAs targeting the 3′ untranslated region (Supplemental Figure S3).
FIGURE 2:
mDia1, mDia2, and mDia3 have nonredundant functions in ErbB2-induced microtubule capture. SKBr3 cells were transfected with the indicated siRNA and active mDia cDNA (ΔNmDia) as indicated. EGFP- or mCherry-tubulin–expressing cells were visualized by time-lapse microscopy 20 min after addition of HRG. (A) The percentage of cells with cortical microtubules was evaluated; 90–150 cells were counted per condition in three independent experiments; mean ± SEM; *p < 0.01. (B) Still images. Insets, zooms showing the presence or absence of microtubules in cell protrusions.
Of importance, when the three mDia formins were concomitantly depleted (Figure 1A), reexpression of active mDia1, mDia2, or mDia3 individually was not sufficient to restore cortical microtubules (Supplemental Figure S4). Moreover, expression of active mDia1 only restored cortical microtubules in mDia1- but not in mDia2- or mDia3-depleted cells. In the same manner, expression of active mDia2 and mDia3 compensated for loss of mDia2 and mDia3, respectively, but not the loss of other mDia proteins (Figure 2, A and B). Thus each of mDia1, mDia2, and mDia3 is required for cortical microtubules in a nonredundant manner.
Contribution of mDia formins to the Memo/ACF7 pathway
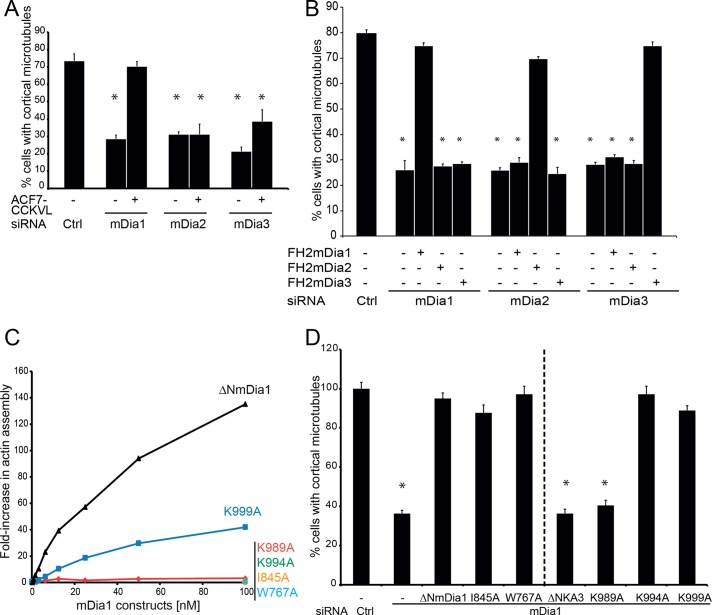
We previously showed that ErbB2-induced cortical microtubules depended on the Memo/ACF7 signaling pathway and that inactivation of the Memo pathway at any level can be compensated for by expression of a shortened, internally deleted ACF7 construct that is constitutively targeted to the cell membrane (called ACF7-CCKVL; Zaoui et al., 2010). Accordingly, we observed that expression of ACF7-CCKVL restored microtubule capture in cells lacking mDia1. In contrast, ACF7-CCKVL failed to restore microtubule capture in the absence of mDia2 or mDia3 (Figure 3A and Supplemental Figure S5A), suggesting that mDia2 and mDia3 function via different effectors.
FIGURE 3:
Characterization of mDia molecular mechanisms of action. (A, B, D) Cells were transfected with the indicated mDia siRNA and with cDNA coding for a membrane-targeted form of ACF7 (ACF-CCKVL) or mDia mutants, as indicated. The percentage of cells with peripheral microtubules was evaluated as described in Figure 1. Mean ± SEM; *p < 0.01. (A) Expression of ACF7-CCKVL compensates for loss of mDia1 but not of mDia2 or mDia3. (B) Expression of the FH2 domain is sufficient to restore peripheral microtubules. Expression of the FH2 domain of an mDia can only compensate for loss of the corresponding mDia. (C) Activity of recombinant ΔNmDia1 constructs was evaluated in a pyrene-actin assembly assay. K989A, K994A, I845A, and W767A mutations, and to a lesser extent the K999A mutation, decrease mDia1-induced actin assembly. (D) Only K989A mutation affects the activity of ΔNmDia1 in peripheral microtubule formation.
FH2 domain of mDia formins is sufficient for microtubule capture
Next we asked what domains in mDia proteins are required for microtubule capture. We already showed that ΔNmDia1, which contains FH1 and FH2 domains, is functional (Figure 2A). Gradual truncations of the FH1 domain revealed that the FH1 domain of mDia1 is not required to mediate microtubule capture (Supplemental Figure S6, A and B). Similarly, we found that the FH2 domains of mDia2 and mDia3 were sufficient to restore cortical microtubules in mDia2- and mDia3-depleted cells, respectively (Figure 3B and Supplemental Figure S5B). Of interest, expression of the FH2 domain of an mDia did not compensate the loss of the other mDia formins (Figure 3B), demonstrating the specificity of the microtubule-regulatory functions contained in each separate mDia FH2 domain.
The FH2 domain dimerizes; each functional half of an FH2 dimer contains two distinct actin-binding surfaces, marked by conserved Ile and Lys residues, respectively, that are important for actin nucleation and elongation (Xu et al., 2004). In murine mDia1, the first actin-binding site is centered on Ile-845 and the second on Lys-994 (Shimada et al., 2004; Xu et al., 2004; Otomo et al., 2005). We introduced the I845A and K994A mutations in mDia1. These mutations abolished the activity of recombinant mDia1 FH2 domains in bulk pyrene-actin assembly assay (Figure 3C), but the same mutations did not affect microtubule function in vivo when endogenous mDia1 was silenced and cells were rescued with mutant versus wild-type constructs (Figure 3D and Supplemental Figure S6C). These results show that functions of the FH2 domain in microtubule capture are independent of its functions in actin assembly. Moreover, a mutation of the conserved Trp-767 that greatly weakens FH2 dimerization and abolish actin assembly activity (Moseley et al., 2004) did not alter mDia1 function in cortical microtubule capture (Figure 3D), indicating that the mDia1 FH2 domain can perform its microtubule function, but not its actin function, as a monomer.
A previous study identified a lysine cluster in a conserved surface patch of the FH2 domain (Shimada et al., 2004), and it has been shown that a triple mutation of mDia1 Lys-989/994/999 disrupts in vivo actin bundling and alignment of microtubules along actin filaments (Ishizaki et al., 2001). We tested the potential involvement of these lysine residues in microtubule capture. We found that the K989/994/999A mutant failed to support microtubule capture (Figure 3D and Supplemental Figure S6C). Next we mutated the lysine residues individually, which revealed that only Lys-989 is critically required for microtubule capture in vivo (Figure 3D and Supplemental Figure S6C). In vitro, we found that Lys-989, Lys-994, and to a lesser extent Lys-999 are important for actin assembly (Figure 3C).
Identification of FH2-interacting proteins contributing to microtubule capture
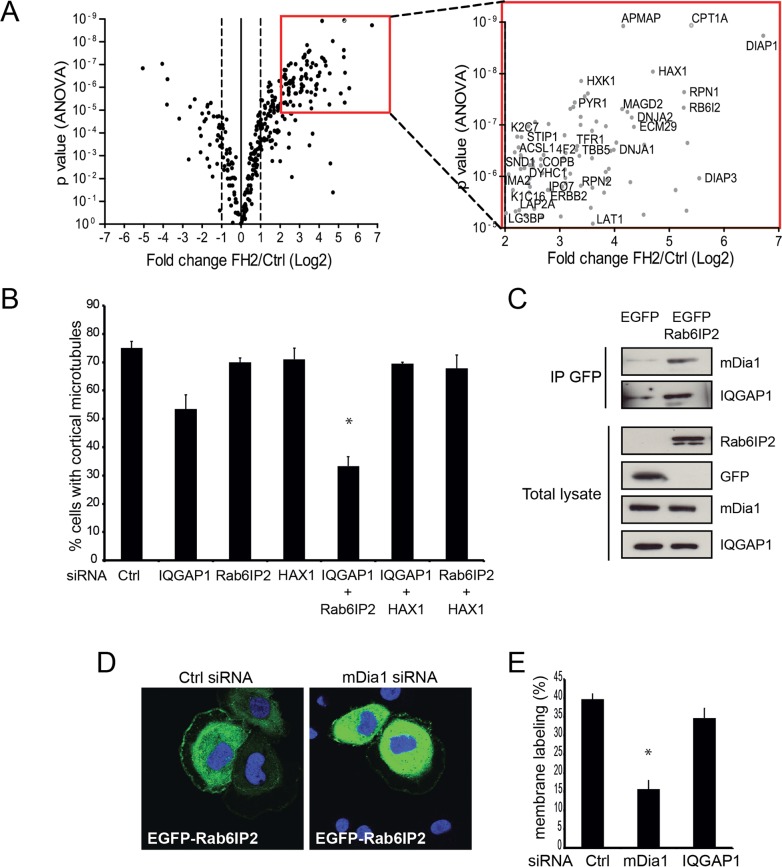
Most of the identified mDia1-interacting proteins are involved in actin regulation (Aspenstrom, 2010). To better understand the cellular mechanism by which mDia1 functions in microtubule capture, we developed a strategy to identify new binding partners of the mDia1 FH2 domain. The FH2 domain, fused to enhanced GFP (EGFP), was stably expressed in SKBr3 cells and used as bait in GFP-trap pull-down experiments. Because the EGFP-FH2 construct is likely to dimerize with endogenous mDia1, which might bring down interactors associating with other domains of mDia1, we depleted endogenous mDia1 by siRNA before collecting the cells. The pulled-down proteins were analyzed by label-free mass spectrometry. The abundance of proteins relative to the control EGFP pull down and statistical significance were calculated (Figure 4A, Table 1, and Supplemental Table S1). By this approach, we identified a limited number of proteins that associate reproducibly with EGFP-FH2. Among them, we clearly identified mDia2, which suggests that the two FH2 domains can heterodimerize. Of note, although tubulin bound to the control beads and thus could be considered background, it was often more strongly associated with GFP-FH2. None of the other proteins identified in our screen were described previously as mDia-interacting proteins (Figure 4A, Table 1, and Supplemental Table S1).
FIGURE 4:
Rab6IP2 interacts with FH2 and contributes to microtubule capture. (A) Identification of proteins interacting with the FH2 domain of mDia1. Volcano plot showing the proteins associating with EGFP-FH2 relative to the control bait, identified by mass spectrometry. Fold change vs. significance (ANOVA) are plotted. Right, zoom on the proteins of highest interest. A typical experiment is shown. Proteins that are reproducibly found in biological replicates (see Table 1) are labeled (UniProt nomenclature). DIAP1, DIAP3, and RB6I2 correspond to mDia1, mDia2, and Rab6IP2, respectively. (B) Volcano plot showing proteins whose interaction is diminished in the WIK2 FH2 mutant vs. wild-type FH2. Only proteins that showed significant binding to FH2 in the previous plot (fold change >2) were included. (C) Role of Rab6IP2, HAX1, and IQGAP1 in microtubule capture. Cells were transfected with the indicated siRNAs. The percentage of cells showing peripheral microtubules was evaluated as described in Figure 1. Mean ± SEM; *p < 0.01. (D) Rab6IP2 binds to full-length mDia1 and IQGAP1. An immunoprecipitation using GFP-Trap beads was performed on EGFP-Rab6IP2– or EGFP-expressing cell lysates; binding of ectopically expressed mDia1 and IQGAP1 was visualized by Western blotting. (E) Recruitment of Rab6IP2 to cell membranes and ruffles depends on mDia1. SKBr3 cells were transfected with mDia1 siRNA for 48 h, before addition of HRG for 20 min. EGFP-Rab6IP2 immunofluorescence was visualized by confocal microscopy. The percentage of cells showing membrane labeling was evaluated. A total of 300 cells were counted for each condition in three independent experiments. Mean ± SEM; *p < 0.01.
TABLE 1:
Proteins associating with the FH2 domain of mDia1, identified by label free mass spectrometry.
| UniProt ID | Protein name | Fold change, experiment 1–experiment 2 |
|---|---|---|
| DIAP1a | Protein Diaphanous homologue 1 (bait) | 63.9–105.4 |
| DIAP3 | Protein Diaphanous homologue 3 | 29–47 |
| RB6I2a | ELKS/Rab6-interacting/CAST family member 1 | 17.7–38.4 |
| HAX1 | HCLS1-associated protein X-1 | 28.2–26 |
| APMAP | Adipocyte plasma membrane–associated protein | 27.4–17.9 |
| CPT1A | Carnitine O-palmitoyltransferase 1 | 2.3–42 |
| RPN1a | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 | 3.7–38.7 |
| RPN2 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2 | 19.5–10.4 |
| DNJA2 | DnaJ homologue subfamily A member 2 | 7.8–20 |
| ECM29 | Proteasome-associated protein ECM29 homologue | 3.9–20.5 |
| MAGD2 | Melanoma-associated antigen D2 | 6.4–17.6 |
| DNJA1a | DnaJ homologue subfamily A member 1 | 3.8–16 |
| LAT1 | Large neutral amino acid transporter small subunit 1 (CD98LC) | 5.7–12.2 |
| HXK1 | Hexokinase-1 | 4.7–10.5 |
| TBB5a | Tubulin β chain | 3.7–9.9 |
| DUS23 | Dual-specificity protein phosphatase 23 | 2.2–11.3 |
| 4F2a | 4F2 cell-surface antigen heavy chain (CD98HC) | 3.9–8.4 |
| PYR1a | CAD protein | 2.6–9.7 |
| ACSL1 | Long-chain-fatty-acid–CoA ligase 1 | 7.3–4.5 |
| ERBB2a | Receptor tyrosine-protein kinase erbB-2 | 3.5–7.1 |
| SND1a | Staphylococcal nuclease domain–containing protein 1 | 5–4.9 |
| STIP1 | Stress-induced phosphoprotein 1 | 4.2–4.9 |
| PSMD3 | 26S proteasome non-ATPase regulatory subunit 3 | 2.1–6.9 |
| HS90Ba | Heat shock protein 90-β | 4.8–3 |
| COPB | Coatomer subunit β | 3.4–4.2 |
| IMA2 | Importin subunit α-2 | 2.1–5.5 |
| DYHC1a | Cytoplasmic dynein 1 heavy chain 1 | 2.3–4.9 |
| LG3BP | Galectin-3–binding protein | 3–4 |
| LAP2A | Lamina-associated polypeptide 2, isoform α | 2.1–4.8 |
| HS90Aa | Heat shock protein 90-α | 3.8–2.9 |
| SERA | D-3-Phosphoglycerate dehydrogenase | 2.7–3.7 |
| HSPB1 | Heat shock protein β-1 | 3.4–2.5 |
Fold change relative to control in two independent experiments.
aProtein also identified in the preliminary small-scale experiment. More information is provided in Table S1.
Next we sought to identify proteins specifically involved in microtubule capture. Although the Lys-989 mutant is not functional for microtubule capture, it is also impaired in its actin assembly activities (Figure 3C), precluding its use as an effective tool for identifying interaction partners specific to microtubule functions. Instead, we performed comparative pull-down experiments with one construct specifically defective in actin functions (called FH2-WIK2 [Supplemental Figure S7, A and B], which contains mutations at Trp-767, Ile-845, Lys-994, and Lys-999) and another construct defective in both actin binding/polymerization and microtubule capture (called FH2-WIK3 [Supplemental Figure S7A], which contains the same mutations plus a Lys-989 mutation). We reasoned that proteins diminished in the FH2-WIK3 pull down relative to FH2-WIK2 pull down would likely be involved in microtubule capture. Using this strategy, we found that only mDia2, as expected from the mutation of the dimerization site, Rab6-interacting protein 2 (Rab6IP2), and, to a lesser extent, SND1 were lost in the FH2-WIK2 pull down (Supplemental Figure S7C). No additional proteins were lost specifically in the FH2-WIK3 pull down (Supplemental Figure S7D), and thus this strategy did not identify proteins whose interaction with FH2 is solely dependent on Lys-989.
We further investigated three of the novel binding partners of mDia1 with suggested links to cell motility or microtubules. First, HCLS1-associated protein X-1 (HAX1) is believed to function with RhoA in neutrophil motility and with cortactin in fibroblast motility (Radhika et al., 2004; Cavnar et al., 2011). Second, Rab6IP2 binds to Rab6, facilitates docking of exocytotic vesicles (Monier et al., 2002; Grigoriev et al., 2007), and functions with LL5β in cortical capture of CLASP2-decorated microtubules (Lansbergen et al., 2006). Third, IQGAP, which was found less reproducibly in our FH2 pull downs, functions in microtubule stabilization with the microtubule plus end–tracking protein CLIP170 (Fukata et al., 2002). Depletion of HAX1 or Rab6IP2 had no effect on cortical microtubules, and inhibition of IQGAP1 only slightly decreased the percentage of cells showing cortical microtubules (Supplemental Figure S1 and Figure 4C). Previous work suggested that, whereas Rab6IP2 was part of the LL5β/CLASP2 complex that contributed to microtubule cortical attachment, it had only an ancillary function (Xu et al., 2004; Lansbergen et al., 2006). We wondered whether Rab6IP2 might similarly have an auxiliary role in the IQGAP1-associated complex. Therefore we codepleted IQGAP1 and Rab6IP2, which led to much stronger defects in microtubule capture than depletion of either alone (Figure 4C). Thus Rab6IP2 and IQGAP1 make complementary contributions to this function.
To confirm the physical associations of these proteins, we immunoprecipitated Rab6IP2 and found that it coprecipitated mDia1 and IQGAP1 (Figure 4D). We also observed that, upon addition of HRG, Rab6IP2 associated with the membrane and ruffling areas of SKBr3 cells, similar to mDia1 (Zaoui et al., 2008). Finally, we found that membrane/ruffling recruitment of Rab6IP2 was dependent on mDia1 expression (Figure 4E).
Our data show that the specificity of mDia1, mDia2, and mDia3 functions in microtubule capture lies within their FH2 domains, suggesting that each FH2 domain may have unique activities and/or interactions with distinct cellular ligands. To obtain additional clues about the respective mechanisms of action of each different mDia formin, we compared the in vivo binding partners of each FH2 domain. Using cell lines that separately and stably express the FH2 domains of mDia1, mDia2, and mDia3 fused to EGFP, we isolated and identified the associated proteins as before by pull down and mass spectrometry. The results showed that each mDia formin associates with a distinct set of proteins (Table 2), many of which were not previously reported as mDia-binding partners. For example, LTBP came down as a major interaction partner of mDia3, and mDia2 interacted with NudC and several nuclear proteins, histones, and regulators of transcription.
TABLE 2:
Proteins binding to the FH2 domains of mDia2 and mDia3 identified by pull-down and mass spectrometry analysis.
| Uniprot ID | Protein name |
|---|---|
| mDia2-FH2–binding proteins | |
| DIAP1 | Protein diaphanous homologue 1 (mDia1) |
| NUDC | Nuclear migration protein nudC |
| H15 | Histone H1.5 |
| H2A2B | Histone H2A type 2-B |
| H2B1K | Histone H2B type 1-K |
| H2AZ | Histone H2A.Z |
| H33 | Histone H3.3 |
| H31 | Histone H3.1 |
| H32 | Histone H3.2 |
| H4 | Histone H4 |
| HDA10 | Histone deacetylase 10 |
| DDX21 | Nucleolar RNA helicase 2 |
| TOP2A | DNA topoisomerase 2-alpha |
| TOP2B | DNA topoisomerase 2-beta |
| DDB1 | DNA damage-binding protein 1 |
| SP16H | FACT complex subunit SPT16 |
| SSRP1 | FACT complex subunit SSRP1 |
| LMNA | Prelamin-A/C |
| NUCL | Nucleolin |
| BAF | Barrier-to-autointegration factor |
| PABP1 | Polyadenylate-binding protein 1 |
| LRRF1 | Leucine-rich repeat flightless-interacting protein 1 |
| ROA2 | Heterogeneous nuclear ribonucleoproteins A2/B1 |
| HNRPU | Heterogeneous nuclear ribonucleoprotein U |
| STIP1a | Stress-induced phosphoprotein 1 |
| mDia3-FH2–binding proteins | |
| DIAP1 | Protein diaphanous homologue 1 (mDia1) |
| LTBP3 | Latent-transforming growth factor beta-binding protein 3 |
| UBE2O | Ubiquitin-conjugating enzyme E2 O |
| LMNA | Prelamin-A/C |
| SYK | Lysine–tRNA ligase |
| PYR1a | CAD protein |
| STIP1a | Stress-induced phosphoprotein 1 |
Proteins strongly increased in the EGFP-FH2 pull downs relative to the control pull down, in at least two of three independent experiments, are shown.
aProteins also identified in the mDia1-FH2 pull down.
Our results provide new insights into the molecular and cellular basis of mDia1-mediated capture of cortical microtubule and open up new avenues of research concerning the discrete functions of Diaphanous formins.
DISCUSSION
We found that mDia1, mDia2, and mDia3 are involved in ErbB2-dependent capture of microtubules at the cell leading edge and ErbB2-driven guided migration. The functional domain that mediates microtubule capture was determined to be the FH2 domain. FH2 contributes to microtubule capture independent of its ability to assemble actin filaments. The FH2 domains of mDia1, mDia2, and mDia3 interacted with distinct sets of proteins. We identified Rab6IP2/ELKS as a protein that interacts with the FH2 domain of mDia1 and contributes, in cooperation with IQGAP1, to microtubule tethering.
Strict side-by-side comparison of the endogenous activity of the three mDia formins has rarely been performed. However, because the FH2 domains of mDia1, mDia2, and mDia3 share high sequence homology, which phylogenetically categorizes them into a formin subfamily (Higgs and Peterson, 2005), they are generally expected to carry out similar or even redundant functions. Indeed, each of the active forms of the three mDia formins (i.e., FH1-FH2 constructs) stimulates actin nucleation and elongation in vitro, although with variable efficiencies (Higgs, 2005). Similarly, a study in mouse fibroblasts showed that each of the active mDia1, mDia2, and mDia3 induces formation of actin stress fibers, alignment of microtubules along actin filaments, and acetylation of microtubules (Thurston et al., 2012). The observation that knocking down mDia1 and mDia3 together is necessary to cause brain developmental defects in mice (Thumkeo et al., 2011; Shinohara et al., 2012) further suggests that in some capacities mDia formins have overlapping in vivo functions. On the other hand, there is also evidence for isoform-specific functions within the mDia subfamily. For instance, only mDia3 associates with RhoD to control endosome trafficking (Gasman et al., 2003); mDia1 fails to rescue lamellipodia formation in mDia2-depleted melanoma cells (Yang et al., 2007); and all three mDia formins make independent contributions to gelatin degradation and invadopodia formation (Colucci-Guyon et al., 2005). We clearly show that the three mDia formins also have unique roles in chemotaxis and microtubule capture. This is consistent with our observation that each mDia has distinct binding partners. It has been suggested that specificity of function might by dictated by the N-terminal regulatory domains; our data clearly demonstrate that the FH2 domain of each mDia is also endowed with specific properties.
Besides actin, a limited number of mDia interactors have been identified (Aspenstrom, 2010). Most of these proteins interact with the regulatory GBD or proline-rich FH1 domains (Breitsprecher and Goode, 2013). Interactors of the FH2 domain include CLIP170 (Lewkowicz et al., 2008), Dia-interacting protein (Eisenmann et al., 2007), and the transcription factor Pax6 (Tominaga et al., 2002). We identified many new ligands of the FH2 domains of mDia1, mDia2, and mDia3. The FH2 domain of mDia1 specifically associated with several proteins involved in both cell motility and Rho GTPase signaling. For instance, mDia1-FH2 coprecipitated the light and heavy chains of CD98, which is involved in integrin signaling and supports RhoA-driven contractility (Cantor and Ginsberg, 2012). Another mDia1 FH2 ligand is HAX1, which has been linked to cell motility via its stimulation of Rac1 and/or RhoA (Radhika et al., 2004; Cavnar et al., 2011). Directly relevant to the function of microtubules in cell migration, the FH2 domain of mDia1 pulled down dynein 1 heavy chain, an important component of the dynein/dynactin complex; this complex is involved in the retrograde transport of vesicles along microtubules and, when tethered at the cortex, generates pulling forces on microtubules required for centrosome repositioning (Kardon and Vale, 2009).
Of great interest, mDia2-FH2 coprecipitated with NudC, a protein that functions together with Lis1 in dynein-mediated nuclear migration and plus end–directed transport of cytoplasmic dynein and dynactins (Yamada et al., 2010; Cappello et al., 2011). mDia2-FH2 interacted with NudC but did not associate with dynein, whereas mDia1-FH2 did associate with dynein heavy chain but not with NudC. These differences again underscore the specificity of the interactions and functions of each mDia in microtubule regulation. It is tempting to speculate that mDia1 and mDia2 have distinct yet complementary roles in microtubule capture and directed cell migration, in which mDia2 helps to promote plus end–directed transport of dynein via NudC and mDia1 facilitates tethering dynein at the cell cortex. mDia3-FH2 is unique in its interaction with latent transforming growth factor β–binding protein-3 (LTBP3). LTBP3 has been described as a secreted protein that is part of the extracellular matrix; mDia3 might therefore interact with a specific pool of LTBP3 that enters or remains in cells. One must keep in mind, however, that these proteins interacted with the isolated FH2 domain, and that interactions in the context of the full-length protein will need be investigated in the future.
Whereas the molecular basis of mDia processive elongation on actin filament ends is well understood (Chesarone et al., 2010), comparatively less is known about how mDia formins interact with and control microtubules. However, this is likely to involve direct interactions of mDia formins with microtubules (Bartolini et al., 2008; Gaillard et al., 2011) and regulatory molecules that target microtubule plus ends (Wen et al., 2004; Yamana et al., 2006; Zaoui et al., 2010) or alter tubulin posttranslational state (Palazzo et al., 2001; Thurston et al., 2012). One must note that even though mDia1 and mDia2 bind microtubules with submicromolar affinity in vitro (Bartolini et al., 2008; Gaillard et al., 2011), we did not observe any localization of endogenous or ectopically expressed mDia formins along microtubules. Our data indicate that the FH2 domains of mDia formins are fully functional for microtubule capture in vivo. In contrast, expression of truncated FH1 in starved fibroblasts led to reduced or absent FH2-mediated microtubule detyrosination (Wen et al., 2004) and acetylation (Thurston et al., 2012), two posttranslational modifications associated with microtubule stability. This suggests different modes of action by mDia formins for microtubule tethering at the cell front and microtubule stabilization.
We identified Rab6IP2 as an mDia1-associated protein involved in microtubule capture. Rab6IP2 was previously identified in several contexts. It was identified as the active zone protein ELKS/CAST, which plays a role in neurotransmitter release (Ohtsuka et al., 2002; Deguchi-Tawarada et al., 2004) and was shown to interact with GTP-bound Rab6 and contribute to vesicle trafficking and exocytosis (Monier et al., 2002; Grigoriev et al., 2007). Most relevant to our results, Rab6IP2 was recently proposed to form with LL5β a cortical platform that allows attachment of CLASP2-decorated microtubule ends (Lansbergen et al., 2006). Depletion of Rab6IP2 had milder effects on peripheral microtubule density than depletion of CLASP2 or LL5β, suggesting that Rab6IP2 is an accessory protein. Similarly, in our model system Rab6IP2 has an auxiliary function, as silencing of Rab6IP2 alone had no effect, whereas simultaneous knockdown of Rab6IP2 and IQGAP1 caused synergistic defects in microtubule capture. We show here that Rab6IP2 recruitment to the cortical area is dependent on mDia1. IQGAP1 was previously found to interact with the N-terminal DID/FH3 domain of mDia (Brandt et al., 2007). Thus, in the context of ErbB2 signaling, the mDia1/Rab6IP2/IQGAP1 module appears to have a function similar to the LL5β/Rab6IP2 platform in microtubule tethering and could also contribute to the targeted delivery of Rab6-decorated vesicles to cortical areas. The functional relationship between these two modules and their relationship with previously identified microtubule capture complexes (Zaoui et al., 2010; Etienne-Manneville, 2013) clearly deserves further investigation. Defective Rab6-dependent docking and fusion of exocytotic carriers might actually contribute to disturbed directed motility upon disturbance of mDia1 cortical recruitment (Benseddik et al., 2013).
FH2 interactions with actin are known to depend on two highly conserved surface residues, Ile-845 and Lys-994, in mDia1. Our results indicate that microtubule capture does not require either of the two critical residues for actin polymerization, showing that microtubule functions of the FH2 are independent of actin assembly effects. On the other hand, we found that all three residues of the conserved lysine cluster (Lys-989, Lys-994, and Lys-999) were involved in actin polymerization but only Lys-989 was required for microtubule capture, indicating that the FH2 surface requirements for actin and microtubule functions are distinct but intersecting. The observation that mDia-mediated actin assembly and microtubule capture are mediated by overlapping surfaces on the FH2 also suggests that mDia formins may not be capable of performing their microtubule and actin functions simultaneously, and indeed this is consistent with a report showing that release of mDia from actin filament ends in vivo liberates them to perform their microtubule functions more robustly (Bartolini et al., 2012).
Given their links with cortically located actin regulatory molecules such as IQGAP1, ACF7, and APC, mDia formins should be ideally positioned during cell motility to coordinate actin assembly, microtubule tethering and stability, and even intermediate filament organization (Kodama et al., 2003; Brandt et al., 2007; Zaoui et al., 2010; Breitsprecher et al., 2012; Sakamoto et al., 2013). Although our work offers new insights into the mechanisms by which mDia formins control microtubule capture, further investigation is needed to decipher the coordinated regulation of the various components of the cytoskeleton during cell migration.
MATERIALS AND METHODS
Transfection of siRNA and plasmid constructs
SKBr3 breast carcinoma cells were obtained from ATCC-LGC Standards (Molsheim, France). Cells grown in DMEM and 10% fetal calf serum were transfected by nucleofection (Lonza, Basel, Switzerland) with 21-mer siRNAs (mDia1 [AF051782-132], mDia2 [AB244756-961], mDia3 [Y15909-1077], Rab6IP2 [AB053471.1-5100], IQGAP1 [L33075.1-4615], HAX1 [U68566.1-1139] or LacZ [M55068-4277] used as negative control [target identifier-first nucleotide of the targeted sequence are indicated]) or cDNAs: GFP-ΔGBDDia2c, Dia2c-Myc, ΔGBDDia2c-Myc (S. Gasman, Institut des Neurosciences Cellulaires et Intégratives, Strasbourg, France), pEGFP/C1-mDia2, YFPDGBDmDia2 (T. Svitkina, University of Pennsylvania, Philadelphia, PA), and pEGFP-Rab6IP2B (B. Goud, Institut Curie, Paris, France). Other plasmid constructs were described previously (Zaoui et al., 2008, 2010). ΔΔmDia1, EGFP-mDia1-FH2, EGFP-mDia2-FH2, and EGFP-mDia3-FH2 constructs were generated by Gateway cloning. Point mutations were generated by the QuikChange Site-Directed Mutagenesis kit (Agilent, Les Ulis, France). All constructs were sequence verified.
Western blotting
Cells were collected in lysis buffer as described (Zaoui et al., 2008). Western blotting was performed using antibodies directed against mDia1, IQGAP1, HAX1 (BD Biosciences, Le Pont de Claix, France), Dia2c (Santa Cruz Biotechnology, Dallas, TX), mDia2/DRF3 (a kind gift from H. N. Higgs, Dartmouth Medical School, Hanover, NH), α-tubulin, ELKS (Sigma-Aldrich, Lyon, France), and GFP (Roche, Meylan, France).
Chemotaxis assay
Chemotaxis was measured by tracking cells on collagen-coated glass coverslips in a stable HRG gradient using Dunn chemotaxis chambers (Hawksley Technology, Lancing, United Kingdom) as described (Benseddik et al., 2013). Images were acquired using the 10× plan apochromat (numerical aperture [NA], 1.4) objective of a Zeiss Axiovert 200 microscope with a time interval of 8 min for 8 h using a digital camera (CoolSNAP HQ; Roper Scientific, Evry, France) driven by MetaMorph 6.3 software (Molecular Devices, Sunnyvale, CA). Tracks were analyzed using the Chemotaxis and Migration Tool of ImageJ (National Institutes of Health, Bethesda, MD). Cell orientation is shown as Rose plots. The Rayleigh test for unimodal clustering of directions was used to test directionality in the chemotactic gradient.
Live and confocal fluorescence microscopy
Cells transfected with EGFP-tubulin were grown on collagen-coated glass coverslips for 48 h and observed upon addition of 5 nM HRG using the 63× objective (plan apochromat, NA 1.4) of a Zeiss Axiovert 200 microscope. Images were acquired using a CoolSNAP HQ camera (Roper Scientific). Cells transfected with pEGFP-Rab6IP2 were grown on collagen-coated coverslips and fixed in PBS, 4% paraformaldehyde, 3% sucrose. DNA was counterstained with Hoechst dye (Sigma-Aldrich). Images were recorded with a Z1confocal Imager (Zeiss), using a 100× plan objective (1.4 NA) coupled to an AxioCam MRm camera. The percentage of EGFP-positive cells displaying cortical labeling was quantified. Results were analyzed with Prism (GraphPad, La Jolla, CA).
Protein purification
Rabbit skeletal muscle actin was purified as previously described (Spudich and Watt, 1971) and fluorescently labeled with pyrenyl-iodoacetamide on cysteine 374. Wild-type and mutant mDia1 C-terminal polypeptides (amino acids 543–1192) were subcloned in pGEX-6P-3 (GE Healthcare, Piscataway, NJ) and purified from Escherichia coli Rosetta-2 cells (Merck Millipore, Billerica, MA) by glutathione affinity chromatography essentially as described (Harris and Higgs, 2006), released from beads by incubation with 20 nM PreScission Protease (GE Healthcare) for 30 min, stored at 4°C, and used in pyrene-actin assembly assays within 5 d.
Bulk actin assembly kinetic assays
Monomeric actin (2 μM; 5% pyrene labeled) in G-buffer (10 mM Tris-Cl−, pH 8.0, 0.2 mM ATP, 0.2 mM CaCl2, and 0.2 mM dithiothreitol) was converted to Mg-ATP-actin immediately before each reaction. Actin was mixed with equivalent volumes of proteins and/or control buffers, and then 3 μl of 20× initiation mix (40 mM MgCl2, 10 mM ATP, 1 M KCl) was added. Pyrene fluorescence was monitored over time at excitation 365 nm and emission 407 nm at 25°C in a fluorimeter (Photon Technology, Lawrenceville, NJ).
Protein pull down
SKBr3 cells were transfected with EGFP-C2, EGFP-FH2, EGFP-WIK2 EGFP-mDia1-FH2, EGFP-mDia2-FH2, or EGFP-mDia3-FH2 constructs using FuGENE (Roche), selected with Geneticin (Life Technologies, Saint Aubin, France), and sorted by flow cytometry to obtain cell populations with similar fluorescence levels. Stable cell lines were transfected with mDia1, mDia2, or mDia3 siRNA using Lipofectamine RNAiMAX (Life Technologies) for 72 h, before stimulation with 5 nM HRG for 20 min. Cells were collected in lysis buffer, and 1.5- to 7-mg protein extracts were incubated with 20 μl of GFP-Trap agarose beads (ChromoTek, Planegg-Martinsried, Germany). Beads were washed and suspended in sample buffer for mass spectrometry analysis.
Mass spectrometry analysis
Protein extracts were loaded on NuPAGE 4–12% bis–Tris acrylamide gels (Life Technologies) to stack proteins in a single band that was stained with Imperial Blue (Pierce, Rockford. IL), cut from the gel, and digested with high-sequencing-grade trypsin (Promega, Madison, WI). Samples (injected in quadruplicate) were analyzed by liquid chromatography (LC)–tandem mass spectrometry (MS/MS) in an LTQ-Orbitrap-Velos (Thermo Electron, Bremen, Germany) online with a nanoLC Ultimate 3000 chromatography system (Dionex, Sunnyvale, CA). Peptides were separated on a Dionex Acclaim PepMap RSLC C18 column. Samples were measured in a data-dependent acquisition mode. The peptide masses were measured in a survey full scan. In parallel to the high-resolution full scan, the data-dependent collision-induced dissociation scans of the 10 most intense precursor ions were fragmented and measured in the linear ion trap to have maximum sensitivity and maximum amount of MS/MS data.
Protein identification and quantification
Relative intensity–based label-free quantification was processed using Progenesis LC-MS software, version 4.1 (Nonlinear Dynamics, Newcastle, United Kingdom). First, raw LC Orbitrap MS data were imported, and LC-MS heatmaps of retention time and m/z were generated. MS-MS spectra were exported into peak list as Mascot generic files that were used to search data via in-house Mascot server, version 2.3.0 (Matrix Science, London, United Kingdom) against the human database subset of the SwissProt database, version 2012.02. Only peptides adjusted to 5% false discovery rate (identity) and with ion score cut-off of 20 were exported from Mascot results and imported back to Progenesis LC-MS for protein grouping and quantification. The raw abundances of GFP peptides were used to normalize LC-MS-MS intensities. Total ion intensity signal from each of the individual peptides generated protein quantification. Univariate one-way analysis of variance (ANOVA) was performed within Progenesis LC-MS to calculate the protein p value according to the sum of the normalized abundances across all runs.
Supplementary Material
Acknowledgments
This work was supported by Fondation ARC and INCa-DGOS-INSERM 6038 (A.B.). P.D. was supported by fellowships from the Ministère de l'Enseignement Supérieur et de la Recherche and Fondation ARC. We are grateful to D. Isnardon and M. L. Thibult for assistance with the use of the Cell Imaging and the Cell Sorting facility, respectively. We thank S. Narumiya, T. Svitkina, S. Gasman, B. Goud, and H. N. Higgs for providing cDNA or antibodies.
Abbreviations used:
- APC
adenomatous polyposis coli
- FH1/2
formin-homology domain 1/2
- Rab6IP2
Rab6-interacting protein 2
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0482) on January 8, 2014.
The authors declare no conflict of interest.
REFERENCES
- Alberts AS. Identification of a carboxyl-terminal Diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Aspenstrom P. Formin-binding proteins: modulators of formin-dependent actin polymerization. Biochim Biophys Acta. 2010;1803:174–182. doi: 10.1016/j.bbamcr.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Moseley JB, Schmoranzer J, Cassimeris L, Goode BL, Gundersen GG. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol. 2008;181:523–536. doi: 10.1083/jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Ramalingam N, Gundersen GG. Actin-capping protein promotes microtubule stability by antagonizing the actin activity of mDia1. Mol Biol Cell. 2012;23:4032–4040. doi: 10.1091/mbc.E12-05-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benseddik K, Sen Nkwe N, Daou P, Verdier-Pinard P, Badache A. ErbB2-dependent chemotaxis requires microtubule capture and stabilization coordinated by distinct signaling pathways. PLoS One. 2013;8:e55211. doi: 10.1371/journal.pone.0055211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336:1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JM, Ginsberg MH. CD98 at the crossroads of adaptive immunity and cancer. J Cell Sci. 2012;125:1373–1382. doi: 10.1242/jcs.096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello S, Monzo P, Vallee RB. NudC is required for interkinetic nuclear migration and neuronal migration during neocortical development. Dev Biol. 2011;357:326–335. doi: 10.1016/j.ydbio.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavnar PJ, Berthier E, Beebe DJ, Huttenlocher A. Hax1 regulates neutrophil adhesion and motility through RhoA. J Cell Biol. 2011;193:465–473. doi: 10.1083/jcb.201010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Colucci-Guyon E, Niedergang F, Wallar BJ, Peng J, Alberts AS, Chavrier P. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr Biol. 2005;15:2007–2012. doi: 10.1016/j.cub.2005.09.051. [DOI] [PubMed] [Google Scholar]
- Deguchi-Tawarada M, Inoue E, Takao-Rikitsu E, Inoue M, Ohtsuka T, Takai Y. CAST2: identification and characterization of a protein structurally related to the presynaptic cytomatrix protein CAST. Genes Cells. 2004;9:15–23. doi: 10.1111/j.1356-9597.2004.00697.x. [DOI] [PubMed] [Google Scholar]
- Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17:579–591. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Microtubules in cell migration. Annu Rev Cell Dev Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F, Kaibuchi K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- Gaillard J, Ramabhadran V, Neumanne E, Gurel P, Blanchoin L, Vantard M, Higgs HN. Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell. 2011;22:4575–4587. doi: 10.1091/mbc.E11-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasman S, Kalaidzidis Y, Zerial M. RhoD regulates endosome dynamics through Diaphanous-related formin and Src tyrosine kinase. Nat Cell Biol. 2003;5:195–204. doi: 10.1038/ncb935. [DOI] [PubMed] [Google Scholar]
- Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21:384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulimari P, Knieling H, Engel U, Grosse R. LARG and mDia1 link Galpha12/13 to cell polarity and microtubule dynamics. Mol Biol Cell. 2008;19:30–40. doi: 10.1091/mbc.E06-11-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I, et al. Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell. 2007;13:305–314. doi: 10.1016/j.devcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Harris ES, Higgs HN. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006;406:190–214. doi: 10.1016/S0076-6879(06)06015-0. [DOI] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T, Morishima Y, Okamoto M, Furuyashiki T, Kato T, Narumiya S. Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nat Cell Biol. 2001;3:8–14. doi: 10.1038/35050598. [DOI] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I, Straube A. Regulation of cell migration by dynamic microtubules. Semin Cell Dev Biol. 2011;22:968–974. doi: 10.1016/j.semcdb.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. ACF7: an essential integrator of microtubule dynamics. Cell. 2003;115:343–354. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Progressing actin: formin as a processive elongation machine. Nat Cell Biol. 2004;6:1158–1159. doi: 10.1038/ncb1204-1158. [DOI] [PubMed] [Google Scholar]
- Lansbergen G, et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- Lewkowicz E, Herit F, Le Clainche C, Bourdoncle P, Perez F, Niedergang F. The microtubule-binding protein CLIP-170 coordinates mDia1 and actin reorganization during CR3-mediated phagocytosis. J Cell Biol. 2008;183:1287–1298. doi: 10.1083/jcb.200807023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone R, Hess D, Dankort D, Muller WJ, Hynes NE, Badache A. Memo mediates ErbB2-driven cell motility. Nat Cell Biol. 2004;6:515–522. doi: 10.1038/ncb1134. [DOI] [PubMed] [Google Scholar]
- Monier S, Jollivet F, Janoueix-Lerosey I, Johannes L, Goud B. Characterization of novel Rab6-interacting proteins involved in endosome-to-TGN transport. Traffic. 2002;3:289–297. doi: 10.1034/j.1600-0854.2002.030406.x. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, et al. Cast: a novel protein of the cytomatrix at the active zone of synapses that forms a ternary complex with RIM1 and munc13–1. J Cell Biol. 2002;158:577–590. doi: 10.1083/jcb.200202083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol. 2001;3:723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Radhika V, Onesime D, Ha JH, Dhanasekaran N. Galpha13 stimulates cell migration through cortactin-interacting protein Hax-1. J Biol Chem. 2004;279:49406–49413. doi: 10.1074/jbc.M408836200. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Boeda B, Etienne-Manneville S. APC binds intermediate filaments and is required for their reorganization during cell migration. J Cell Biol. 2013;200:249–258. doi: 10.1083/jcb.201206010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- Shimada A, Nyitrai M, Vetter IR, Kuhlmann D, Bugyi B, Narumiya S, Geeves MA, Wittinghofer A. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol Cell. 2004;13:511–522. doi: 10.1016/s1097-2765(04)00059-0. [DOI] [PubMed] [Google Scholar]
- Shinohara R, et al. A role for mDia, a Rho-regulated actin nucleator, in tangential migration of interneuron precursors. Nat Neurosci. 2012;15:373–380, S371–372. doi: 10.1038/nn.3020. [DOI] [PubMed] [Google Scholar]
- Spencer KS, Graus-Porta D, Leng J, Hynes NE, Klemke RL. ErbB2 is necessary for induction of carcinoma cell invasion by ErbB family receptor tyrosine kinases. J Cell Biol. 2000;148:385–397. doi: 10.1083/jcb.148.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Thumkeo D, Shinohara R, Watanabe K, Takebayashi H, Toyoda Y, Tohyama K, Ishizaki T, Furuyashiki T, Narumiya S. Deficiency of mDia, an actin nucleator, disrupts integrity of neuroepithelium and causes periventricular dysplasia. PLoS One. 2011;6:e25465. doi: 10.1371/journal.pone.0025465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston SF, Kulacz WA, Shaikh S, Lee JM, Copeland JW. The ability to induce microtubule acetylation is a general feature of formin proteins. PLoS One. 2012;7:e48041. doi: 10.1371/journal.pone.0048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, Meng W, Togashi K, Urano H, Alberts AS, Tominaga M. The Rho GTPase effector protein, mDia, inhibits the DNA binding ability of the transcription factor Pax6 and changes the pattern of neurite extension in cerebellar granule cells through its binding to Pax6. J Biol Chem. 2002;277:47686–47691. doi: 10.1074/jbc.M207539200. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon E. Positive feedback interactions between microtubule and actin dynamics during cell motility. Curr Opin Cell Biol. 1999;11:61–67. doi: 10.1016/s0955-0674(99)80008-8. [DOI] [PubMed] [Google Scholar]
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Wickstrom SA, et al. Integrin-linked kinase controls microtubule dynamics required for plasma membrane targeting of caveolae. Dev Cell. 2010;19:574–588. doi: 10.1016/j.devcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Cell motility: can Rho GTPases and microtubules point the way. J Cell Sci. 2001;114:3795–3803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Yamada M, et al. mNUDC is required for plus-end-directed transport of cytoplasmic dynein and dynactins by kinesin-1. EMBO J. 2010;29:517–531. doi: 10.1038/emboj.2009.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamana N, et al. The Rho-mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing Apc and c-Src. Mol Cell Biol. 2006;26:6844–6858. doi: 10.1128/MCB.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaoui K, Benseddik K, Daou P, Salaun D, Badache A. ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc Natl Acad Sci USA. 2010;107:18517–18522. doi: 10.1073/pnas.1000975107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaoui K, Honore S, Isnardon D, Braguer D, Badache A. Memo-RhoA-mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. J Cell Biol. 2008;183:401–408. doi: 10.1083/jcb.200805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.