Methods are given for a screen using whole-genome multiplex sequencing in the filamentous fungus Aspergillus nidulans. Novel alleles of the molecular motor cytoplasmic dynein identified in this screen reveal that different dynein cargoes have distinct requirements for motor performance.
Abstract
The timely delivery of membranous organelles and macromolecules to specific locations within the majority of eukaryotic cells depends on microtubule-based transport. Here we describe a screening method to identify mutations that have a critical effect on intracellular transport and its regulation using mutagenesis, multicolor-fluorescence microscopy, and multiplex genome sequencing. This screen exploits the filamentous fungus Aspergillus nidulans, which has many of the advantages of yeast molecular genetics but uses long-range microtubule-based transport in a manner more similar to metazoan cells. Using this method, we identified seven mutants that represent novel alleles of components of the intracellular transport machinery: specifically, kinesin-1, cytoplasmic dynein, and the dynein regulators Lis1 and dynactin. The two dynein mutations identified in our screen map to dynein's AAA+ catalytic core. Single-molecule studies reveal that both mutations reduce dynein's velocity in vitro. In vivo these mutants severely impair the distribution and velocity of endosomes, a known dynein cargo. In contrast, another dynein cargo, the nucleus, is positioned normally in these mutants. These results reveal that different dynein functions have distinct stringencies for motor performance.
INTRODUCTION
In most eukaryotic cells, microtubules and microtubule-based motors are required for the transport and delivery of proteins, RNAs, and organelles to their sites of action. Although many cargoes for microtubule-based motors have been identified, there are numerous unanswered questions regarding the mechanism of microtubule-based transport. For example, how are motors loaded onto designated cargo at specific locations within the cell? How is transport temporally coordinated with cell growth, division, and development? The budding yeast Saccharomyces cerevisiae is and has been an important model system for understanding the mechanism of many fundamental cell biological processes such as the secretory pathway, the cell cycle, and protein degradation. However, S. cerevisiae relies almost exclusively on the actin cytoskeleton for directed cytoplasmic transport of proteins, RNAs, and organelles (Hammer and Sellers, 2012). In contrast, filamentous fungi, such as Aspergillus nidulans, use the microtubule cytoskeleton for long-distance transport (Egan et al., 2012a), akin to metazoan cells.
As a model system for dissecting microtubule-based transport, A. nidulans has several advantages. First, A. nidulans hyphae are highly polarized and rely on microtubule-based transport for rapid growth (Horio and Oakley, 2005; Taheri-Talesh et al., 2008). Second, homologous recombination occurs at high frequencies in strains lacking the Ku70 homologue, nkuA (Nayak et al., 2006), making manipulation of the endogenous genome fast and easy. Third, A. nidulans has a well-characterized life cycle that includes both haploid and diploid stages, facilitating genetic analysis (Todd et al., 2007a, b). Finally, the A. nidulans genome is sequenced and has been well annotated (Galagan et al., 2005; Wortman et al., 2009).
Vegetatively growing A. nidulans forms long, branched, multinucleate hyphae. Studies of microtubule-based transport have largely focused on the hyphal tip, as microtubules are uniformly polarized in this region, with their minus ends embedded in the spindle pole body of the most distal nucleus and their dynamic plus ends extending toward the hyphal tip. The A. nidulans genome contains a single gene for the minus end–directed motor cytoplasmic dynein (nudA; referred to as “dynein” here) and 11 kinesin-encoding genes, three of which are expected to drive long-distance plus end–directed transport based on sequence homology (kinA, belonging to the kinesin-1 family, and uncA and uncB, belonging to the kinesin-3 family). Pioneering screens in A. nidulans based on mutagenesis and genetic complementation demonstrated that dynein and its regulatory factors, which include the dynactin complex and Lis1/nudF, are required for the proper positioning of nuclei within hyphae (Morris, 1975; Xiang et al., 1994, 1995, 1999). Kinesin-1 is also required for nuclear positioning, likely due to its role in recycling dynein back to microtubule plus ends (Requena et al., 2001; Zhang et al., 2003, 2010; Abenza et al., 2009; Egan et al., 2012b). In addition, recent studies show that dynein and kinesin-3/uncA are the opposite-polarity motors responsible for endosome and peroxisome transport in A. nidulans and other filamentous fungi (Lenz et al., 2006; Abenza et al., 2009; Zhang et al., 2010; Egan et al., 2012b; Seidel et al., 2013).
Here we present a screening method to identify mutations that have a critical impact on intracellular transport. This screen exploits the aforementioned advantages of A. nidulans as a model organism, as well as the power and falling cost of multiplex genome sequencing. We identified seven mutants in our screen that map to players in intracellular transport: kinesin-1, dynein, and the dynein regulators Lis1 and dynactin. The two mutations we identify in dynein severely impair endosome distribution and velocity but support dynein's function in nuclear positioning, revealing that different cargoes have distinct stringencies for motor performance.
RESULTS
Screening strategy and rationale
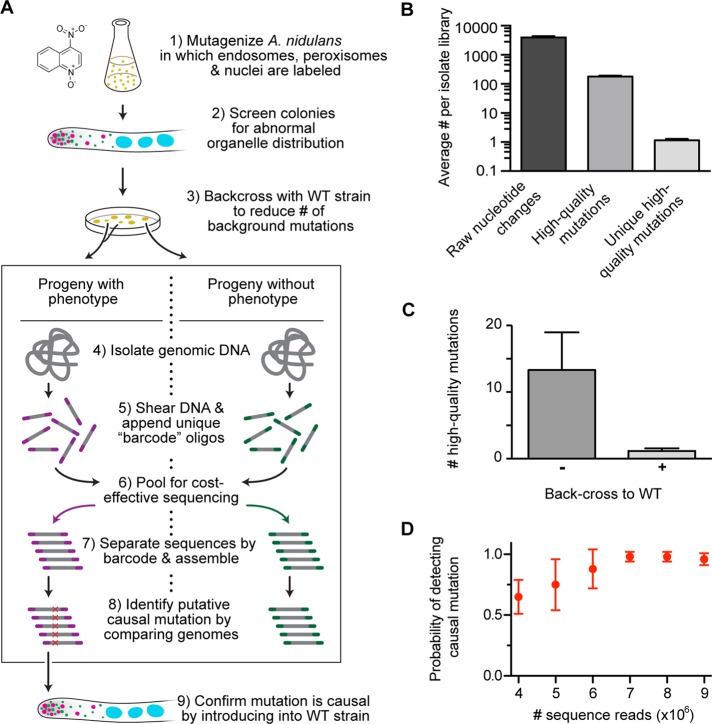
An outline of our screening method to identify mutations that perturb intracellular transport is shown in Figure 1A. A new reagent that we developed for this screen is an A. nidulans strain in which three types of organelle (endosomes, peroxisomes, and nuclei) are each labeled with a different-color fluorescent protein. The proper spatial distribution of endosomes, peroxisomes, and nuclei has been shown to depend on microtubule-based transport (Xiang et al., 1994; Abenza et al., 2009; Egan et al., 2012b). Thus, by allowing simultaneous visualization of these organelles’ distribution by live-cell microscopy, this A. nidulans strain provides a readout of the functionality in the underlying transport pathways.
FIGURE 1:
Multiplexed whole-genome sequencing to identify mutations in the A. nidulans transport machinery. (A) A flowchart of our experimental approach. 1) Spores from a strain harboring TagGFP-Rab5/RabA–labeled endosomes, mCherry-PTS1–labeled peroxisomes, and TagBFP-HH1–labeled nuclei were mutagenized with 4NQO at doses that generated either 10 or 50% survival rate. 2) These mutants were screened for abnormal distribution of endosomes, peroxisomes, or nuclei using fluorescence microscopy. 3) Each mutant was crossed to a wild-type nonmutagenized strain. The organelle distribution phenotype of progeny from each cross was examined, and progeny were sorted and pooled into two categories: progeny with phenotype and progeny without phenotype. 4) All progenies from each category were grown as a pool, and genomic DNA was then isolated from each pool. 5) This DNA was sheared, and 8–base pair unique barcodes were appended to the sheared ends. 6) Libraries containing distinct barcodes were pooled and sequenced using Illumina sequencing. 7) The sequencing data were separated by barcode and assembled. 8) Candidate causal mutations were identified. 9) Causal mutations were confirmed by introducing the mutation into a wild-type nonmutagenized strain. (B) Bar graph showing average number of nucleotide changes per isolate library after data processing. Average (± SEM) number of nucleotide changes was 3948.29 ± 411.66 before data processing (raw nucleotide change);177.71 ± 12.91 for nucleotide changes that occurred at least five times in our mutant data sets, did not appear in the wild-type A. nidulans reference sequences (Galagan et al., 2005; Wortman et al., 2009), and were not synonymous mutations (high-quality mutations); and 1.14 ± 0.14 after subtracting all nucleotide changes also found in progenies lacking the phenotype (unique high-quality mutations). All mean values were statistically different from each other (p < 0.0001, unpaired t test; n = 7 mutants). (C) Bar graph showing average (± SEM) unique high-quality mutations with (+) or without (–) a backcross to a nonmutagenized strain. A backcross significantly reduced the number of unique high-quality mutations from 13.33 ± 5.65 to 1.17 ± 0.17 (mean ± SEM; p = 0.015, Kolmogorov–Smirnov test). (D) Graph showing the probability of detecting a causal mutation as a function of the number of sequence reads. Using a bootstrap methodology, we randomly sampled mutant data sets (n = 6 mutants) for 4, 5, 6, 7, 8, and 9 million reads (with each read containing ∼43 base pairs of useful sequence data). This resampling process was repeated 10 times for each mutant. Error bars show the SD of the probability of detection.
To generate mutations within this strain, we treated haploid spores with various doses of 4-nitroquinoline 1-oxide (4NQO), which generates nucleotide substitutions by inducing stable, bulky DNA adducts on guanine and adenine residues (Ikenaga et al., 1975; Bailleul et al., 1989). The survival rate of the cells was 50% at a 4NQO concentration of 0.24 μg/ml and 10% at a concentration of 0.45 μg/ml (Supplemental Figure S1A). We determined the average number of mutations generated at each mutagen dose for comparison (Supplemental Figure S1, B and C).
We analyzed the organelle distribution in the mutagenized strains manually using fluorescence microscopy. A “hit” was judged to be a strain in which the distribution of one or more organelle was perturbed when compared with a nonmutagenized strain. For example, peroxisomes and endosomes accumulate at the hyphal tip when minus end–directed transport is defective (Lenz et al., 2006; Abenza et al., 2009; Zhang et al., 2010; Egan et al., 2012b). The frequency of finding an isolate with an organelle mislocalization phenotype was higher (1.1%) at the elevated mutagen dose of 0.45 μg/ml 4NQO than with the lower 4NQO dose of 0.24 μg/ml (0.5% of isolates with a phenotype; Supplemental Figure S1D). From a total of 12,456 mutated hyphae, we identified 76 (0.6%) with an organelle distribution phenotype. We identified defects in the distribution of only one organelle (46%), two organelles (38%), or all three organelles (16%; Supplemental Figure S2A).
These mutagenized strains harbor one or more nucleotide change that causes the organelle misdistribution phenotype and many more noncausal “background” mutations. Classically, causal mutations in A. nidulans have been identified using linkage mapping and genetic complementation. However, the rapidly falling cost and increased throughput of whole-genome sequencing using multiplexing now make this a viable method for mutation mapping. With proper quality controls, we find that multiplex whole-genome sequencing can quickly and reliably identify putative causal mutations.
To reduce the number of background mutations before sequencing, we genetically crossed each isolate to a nonmutagenized strain and used fluorescence microscopy to sort the progenies based on organelle distribution. The progenies from the cross either carry the phenotype-causing mutation(s) or do not, and we find that sequencing both types of progeny provides useful information. In particular, sequence data from progenies lacking the phenotype reveal background mutations that can subsequently be eliminated from the list of candidate causal mutations.
For sequencing, we generated pools of either progenies with the phenotype or progenies without the phenotype for each isolate. Genomic DNA libraries were then prepared in each case by appending a unique eight–base pair “barcode” to the ends of sheared genomic DNA. These barcodes enable Illumina sequencing to be performed in a multiplex manner (i.e., sequencing multiple genomic libraries in the same reaction).
To identify putative phenotype-causing mutations from the sequence data, we first defined a “high-quality” mutation as a nucleotide change relative to the wild-type A. nidulans reference sequences (Galagan et al., 2005; Wortman et al., 2009) that occurred at least five times in our mutant data sets and was not a synonymous mutation. The average number of high-quality mutations in each isolate library is large (177.71 ± 12.91 [± SEM]). Figure 1B shows the utility of sequencing progenies without the phenotype: upon subtraction of nucleotide changes that appear in these progenies, the number of putative causal mutations is dramatically reduced. Indeed, in the majority of cases there is only one unique mutation remaining (1.14 ± 0.14). However, when the backcross step is omitted, the average number of unique mutations rises to 13.33 ± 5.65, demonstrating the value of reducing the number of background mutations before sequencing (Figure 1C).
Finally, to establish the depth of sequencing necessary to determine the putative causal mutation, we resampled our data sets (Figure 1D) and found that 7 million reads (representing ∼10-fold genome coverage for the 30-Mb A. nidulans genome) was sufficient to recover the causal mutation with high (∼98%) probability.
Hits from the screen represent novel alleles of microtubule-based motors and their regulators
The putative causal mutations from 33 initially sequenced hits mapped to genes encoding motors and motor regulators (24%), chaperones (9%), proteins involved in the endocytic pathway (6%), cell division and cytokinesis (6%), the peroxisome pathway (3%), and transcription factors (3%). The remaining 49% of hits corresponded to proteins with unknown function and cases in which there was more than one plausible candidate (Supplemental Figure S2). We focused on seven initial hits showing abnormal organelle distributions whose putative causal mutations mapped to well-characterized motors or motor regulators. To validate these mutations, we introduced them into a nonmutagenized strain with labeled endosomes, peroxisomes, and nuclei and found that the organelle distribution phenotypes closely matched the original hits from the screen, confirming that the identified mutations were causal.
One of the mutations mapped to the first intron of the dynein regulator Lis1/nudF and caused a defect in the distribution of endosomes, peroxisomes, and nuclei similar to that produced by Lis1 deletion (Supplemental Figure S3). Similarly, two mutations within kinesin-1’s motor domain and one near the autoinhibitory element in kinesin-1’s tail each produced a defect in organelle positioning that resembled the kinesin-1–null strain (Supplemental Figure S4). Finally, one of the mutations mapped to the Arp1/nudK subunit of the dynein regulator, dynactin, and the other two lay within the motor domain of dynein itself. All of these nucleotide changes represent new alleles of these genes.
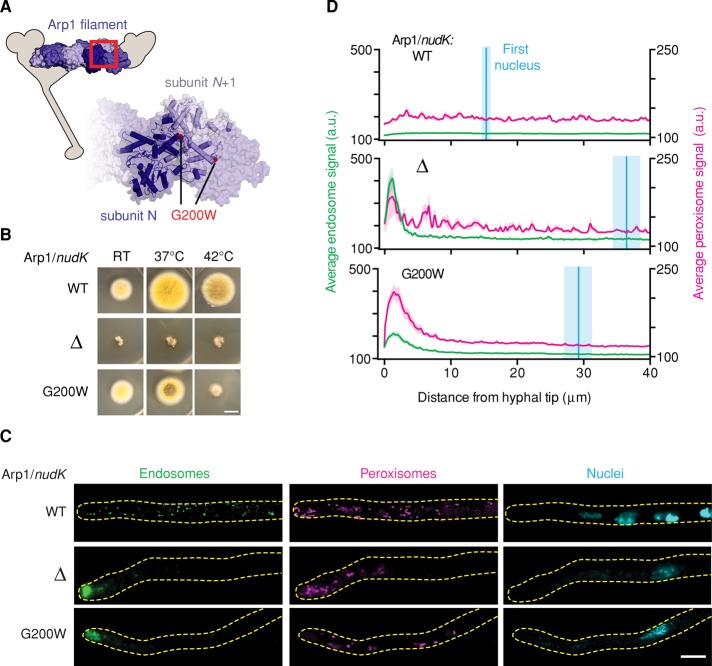
The Arp1 allele is temperature sensitive
Arp1 is an actin-related protein that forms short filaments and is an integral part of the dynactin complex, which regulates dynein's intracellular localization, processivity, and cargo binding (Schafer et al., 1994; Eckley et al., 1999; Muller et al., 2005). The Arp1 mutation from our screen results in the substitution of a glycine residue (G200)—whose position is highly conserved within the actin family—for a bulky tryptophan. Based on an actin filament model (Murakami et al., 2010), this mutation would lie at the end of an α-helix near the interface between Arp1 subunits (Figure 2A).
FIGURE 2:
The Arp1 allele is temperature sensitive. (A) Structural model of the Arp1 minifilament based on the structure of Oryctolagus cuniculus skeletal muscle F-actin (PDB 3G37; Murakami et al., 2010), illustrating the predicted position of the G200W mutation in Arp1. Inset, enlargement of the region indicated by the red box. (B) Images of colony morphology show that the Arp1G200W mutation is temperature sensitive. Wild-type, Arp1Δ, and Arp1G200W strains were grown at room temperature, 37°C, or 42°C. The wild-type strain forms large colonies that conidiate at all temperatures (top). The Arp1Δ strain forms small colonies that do not conidiate at any temperature tested (middle). The Arp1G200W mutant conidiates at room temperature and shows a minor conidiation defect at 37°C, but the colonies are small and nonconidiating at 42°C (bottom). Scale bar, 1 cm. (C) Representative micrographs of strains grown at 42°C. In wild-type hyphae, endosomes, peroxisomes, and nuclei were distributed (top). In both Arp1Δ and Arp1G200W hyphae, endosomes and peroxisomes accumulated at the hyphal tip, and the distance between the distal nucleus and hyphal tip was increased (middle and bottom, respectively). Scale bar, 5 μm. (D) Endosome (green) and peroxisome (magenta) distribution was quantified using line scans of fluorescence micrographs. Mean (green or magenta lines) ± SEM (green or magenta shading) fluorescence intensity as a function of distance from the hyphal tip. Arp1Δ and Arp1G200W hyphae showed a pronounced peak of fluorescence intensity for endosomes and peroxisomes in the hyphal tip, whereas wild-type cells did not. Nuclear distribution was quantified by measuring the distance from the distal nucleus to the hyphal tip. The mean (cyan line) ± SEM (cyan shading) distance was 15.30 ± 0.79 μm (n = 19) in wild-type, 36.48 ± 5.74 μm (n = 24) in Arp1Δ, and 29.23 ± 2.19 μm (n = 32) in Arp1G200W hyphae. The nuclear distributions in both Arp1Δ and Arp1G200W hyphae were significantly different from that in wild-type hyphae (p < 0.001, unpaired t test).
We found that this Arp1 mutant is temperature sensitive: at room temperature the Arp1G200W strain had a colony morphology similar to wild type; however, when grown at 42°C the colony phenotype resembled an Arp1-null strain (Figure 2B). We next examined the distribution of endosomes, peroxisomes, and nuclei in wild-type, Arp1∆, and Arp1G200W strains at the restrictive temperature (42°C; Figure 2C). To quantify the endosome and peroxisome distribution phenotype, we performed line scans of fluorescence micrographs and plotted the average fluorescence intensity as a function of distance from the hyphal tip (Figure 2D). In wild-type cells the fluorescence intensity of endosomes and peroxisomes was similar throughout the length of the hyphal tip region, whereas in Arp1∆ and Arp1G200W hyphae the fluorescence intensity of endosomes and peroxisomes showed a pronounced peak at the hyphal tip, indicative of defective transport toward the microtubule minus end. To quantify the nuclear distribution phenotype, we measured the distance from the most distal nucleus to the hyphal tip and plotted this position on the line scan graph (Figure 2D). In wild-type hyphae, the distal nucleus was significantly closer to the hyphal tip than with Arp1∆ and Arp1G200W hyphae. Such line scan analysis and measurement of nuclear position may be useful in automating the identification of hits in future screening efforts (see Discussion).
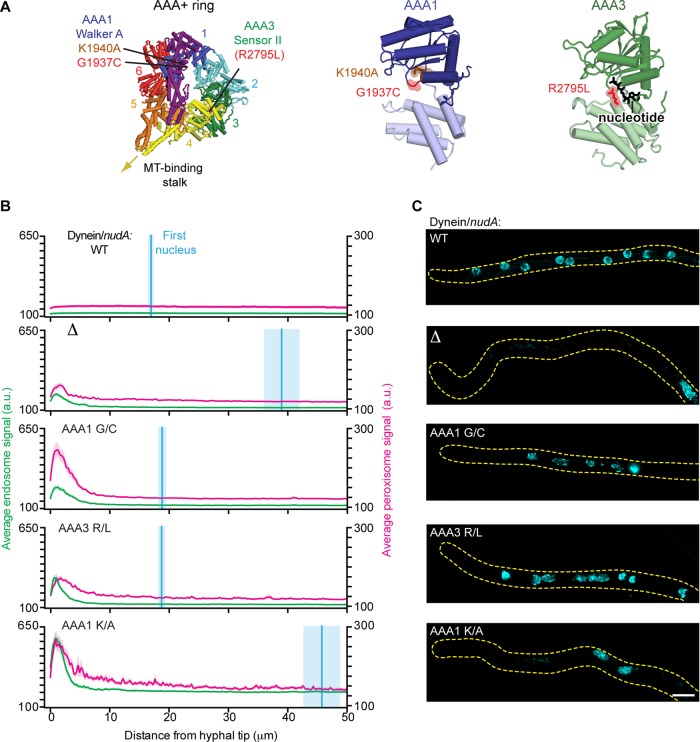
Mutations in dynein that severely impair the positioning of endosomes and peroxisomes but not nuclei
The two dynein mutations from the screen lie within the ring of six AAA+ modules that forms dynein's catalytic core (Figure 3A). Specifically, the first mutation (G1937C) is located within the highly conserved Walker A motif of AAA1, the main site of ATP hydrolysis in dynein's motor domain. Within AAA1’s Walker A consensus sequence (GPAGGKT), mutation of the lysine is believed to severely impair ATP binding and has been shown to abolish dynein motility in vitro (Kon et al., 2004) and impair endosome distribution in vivo (Zhang et al., 2010). However, mutations of the second glycine (corresponding to G1937 in A. nidulans dynein) have not been previously reported. Here we refer to the glycine mutation identified in our screen as AAA1 G/C and the lysine mutation known to abolish dynein motility as AAA1 K/A (corresponding to K1940 in A. nidulans dynein). The other dynein mutation from our screen (R2795L) maps to the Sensor II motif of AAA3 in dynein's ring. ATP binding and hydrolysis at AAA3 are believed to serve important regulatory functions in cytoplasmic dynein's mechanochemical cycle. By analogy to related AAA+ proteins, the Sensor II motif is expected to be important for ATP hydrolysis in AAA3 (Erzberger and Berger, 2006). However, Sensor II mutations in dynein were not previously reported. We refer to the Sensor II mutation as AAA3 R/L.
FIGURE 3:
Mutations in dynein that impair the positioning of endosomes and peroxisomes but not nuclei. (A) Structure of the S. cerevisiae dynein motor domain (left; PDB 4AKH; Schmidt et al., 2012). Expected positions of the A. nidulans dynein point mutations (G1937C and R2795L) identified in our screen, as well as the previously characterized K1940A mutation (Zhang et al., 2010) are highlighted. Closeups of AAA1 (middle) and AAA3 (right) are also shown. (B) Plots of endosome, peroxisome, and nuclear distribution in various dynein backgrounds, quantified as described in Figure 2D. DyneinΔ, dyneinAAA1 G/C, dyneinAAA3 R/L, and dyneinAAA1 K/A hyphae all showed defects in endosome and peroxisome distribution, as there was a pronounced peak of fluorescence intensity for endosomes and peroxisomes in the hyphal tip compared with wild-type hyphae. Nuclear distribution was perturbed in dyneinΔ and dyneinAAA1 K/A hyphae but not in dyneinAAA1 G/C, dyneinAAA3 R/L, or wild-type hyphae. The mean ± SEM distances from the distal nucleus to the hyphal tip were 16.95 ± 0.57 μm (n = 52) in wild-type, 38.97 ± 3.12 μm (n = 31) in dyneinΔ, 18.81 ± 0.78 μm (n = 46) in dyneinAAA1 G/C, 18.71 ± 0.71 μm (n = 36) in dyneinAAA3 R/L, and 45.75 ± 3.22 μm in dyneinAAA1 K/A (n = 45) hyphae. The distal nucleus position in dyneinAAA1 G/C and dyneinAAA3 R/L hyphae was not statistically different from that in wild-type hyphae (p > 0.05, unpaired t test), whereas the nuclear distribution in dyneinAAA1 K/A and dyneinΔ hyphae was statistically different from that in wild-type hyphae (p < 0.0001, unpaired t test). (C) Representative micrographs showing nuclear distribution in A. nidulans hyphae in the various dynein backgrounds. Scale bar, 5 μm.
We first characterized the effect of these mutations on organelle distribution in vivo. Line scans along the length of the hyphae revealed that endosomes and peroxisomes accumulated in hyphal tips of the dyneinAAA1 G/C and dyneinAAA3 R/L strains, similar to the dynein∆ strain (Figure 3B; Xiang et al., 1994; Abenza et al., 2009; Egan et al., 2012b). However, the position of the most distal nucleus in the dyneinAAA1 G/C and dyneinAAA3 R/L strains was similar to the wild-type strain. This contrasts with the dynein∆ strain, which showed a severe nuclear positioning defect, as expected (Figure 3, B and C). Thus these dynein mutations do not phenocopy the dynein-null strain: although the positioning of endosomes and peroxisomes is severely perturbed, the dynein-dependent nuclear positioning pathway is functional.
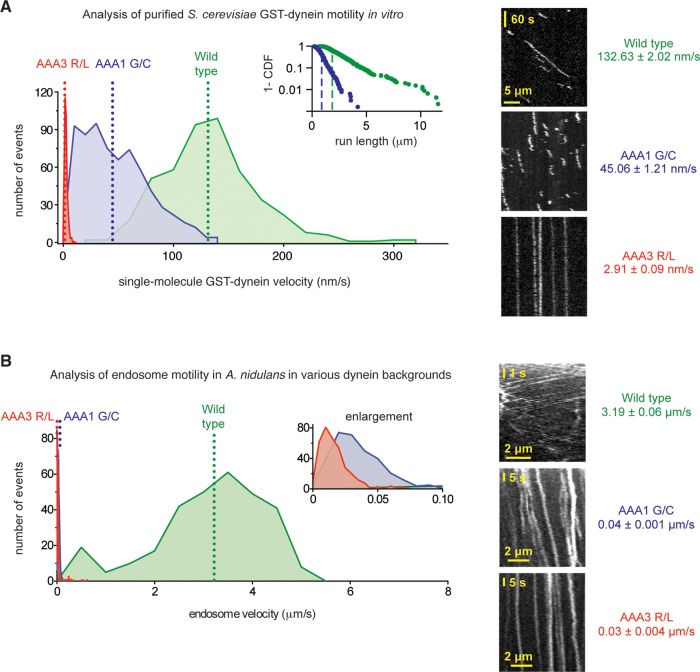
The dynein mutations support motility in vitro and in vivo but at severely reduced velocities
The dynein mutations could have a different effect on nuclear positioning than on endosome and peroxisome transport because these functions have different stringencies for motor function. For example, it is possible that dynein with limited motility is sufficient to correctly position the nucleus but not endosomes and peroxisomes. Alternatively, the distinct organelle phenotypes could arise from indirect effects, such as changes in how dynein interacts with specific regulators or is loaded onto cargo. To determine whether the mutations have a direct effect on dynein motor function, we used an in vitro assay that visualizes the motility of single purified dynein molecules.
Because both the AAA1 G1937 and AAA3 R2795 amino acids are highly conserved among cytoplasmic dynein sequences, we introduced each point mutation into a S. cerevisiae dynein motor domain construct (glutathione S-transferase [GST]–Dyn1331 kDa). S. cerevisiae dynein is the evolutionarily closest motor to A. nidulans dynein for which the tools for single-molecule imaging exist (Reck-Peterson et al., 2006). Although dynein from S. cerevisiae is expected to be a slower motor than the A. nidulans enzyme, and hence the velocities determined from this assay are not directly applicable to A. nidulans dynein, qualitative trends in the effect of the mutations are expected to be conserved (Kon et al., 2004; Cho et al., 2008). Strikingly, both the AAA1 G/C and AAA3 R/L yeast dynein motors retained the ability to bind to and move along microtubules, as observed by total internal reflection fluorescence microscopy (TIRF). However, their motile characteristics were different from both each other and the wild-type dynein (Figure 4A). DyneinAAA1 G/C moved at a reduced velocity of ∼45 nm/s, a 66% reduction in velocity compared with the wild-type motor, and its average run length was also reduced (Figure 4A, inset). This behavior is distinct from that caused by mutation of the lysine (GPAGGKT) in the AAA1 Walker A sequence in Dictyostelium dynein, as this renders the motor immotile (Kon et al., 2004) with constitutively high microtubule affinity (Imamula et al., 2007). DyneinAAA3 R/L moved at an even slower velocity of ∼3 nm/s, a 98% reduction in velocity, and its microtubule encounters were extended in time compared with the wild-type and dyneinAAA1 G/C motors. The effect of this mutation in AAA3’s Sensor II motif is thus highly similar to other mutations that impair ATP binding or hydrolysis in AAA3 (Kon et al., 2004; Cho et al., 2008; Huang et al., 2012). These data indicate that the AAA1 G/C and AAA3 R/L mutations have a direct effect on dynein motor function, impairing but not abolishing dynein motility.
FIGURE 4:
The dynein mutations severely reduce velocities in vitro and in vivo. (A) Left, velocity histograms for tetramethylrhodamine-labeled wild-type S. cerevisiae GST-Dyn1331 kDa (green) and the dyneinAAA1 G/C (blue) and dyneinAAA3 R/L (red) mutants. In S. cerevisiae dynein, the AAA1 G/C mutation corresponds to G1799C and the AAA3 R/L mutation corresponds to R2620L. The dotted lines indicate average velocities (132.63 ± 2.02 nm/s for wild-type dynein [mean ± SEM; n = 457], 47.96 ± 1.16 nm/s for dyneinAAA1 G/C [n = 679], and 2.91 ± 0.09 nm/s for dyneinAAA3 R/L [n = 374]). Both mutant dyneins are statistically slower than wild-type dynein (p < 0.0001; unpaired t test). Inset, log plot of the 1 – cumulative probability distribution of run lengths for the wild-type (green dots) and dyneinAAA1 G/C (blue dots) constructs. Average run lengths are 1.70 μm for wild type (green dashed line; error of the fit = 0.012 μm; n = 457) and 0.69 μm for dyneinAAA1 G/C (blue dashed line; error of the fit = 0.006 μm; n = 679). Run lengths for the dyneinAAA3 R/L construct could not be determined, as the majority of molecules remained attached to the microtubule for the entire duration of image acquisition. Right, representative kymographs. (B) Left, velocity histograms of retrograde endosome movement in A. nidulans hyphae in various dynein backgrounds: wild type (green), dyneinAAA1 G/C (blue), and dyneinAAA3 R/L (red). Inset, enlarged view of the x-axis. The dotted lines indicate average velocities (3.19 ± 0.06 μm/s in wild type [mean ± SEM; n = 296], 0.04 ± 0.001 μm/s in dyneinAAA1 G/C [n = 327], and 0.03 ± 0.004 μm/s dyneinAAA3 R/L [n = 368] hyphae). Endosomes in both mutant dyneins are statistically slower than in wild type (p < 0.0001; unpaired t test). Right, representative kymographs of endosomes in wild-type, dyneinAAA1 G/C, and dyneinAAA3 R/L hyphae.
Because both mutants showed limited but measurable motility in vitro, we carefully examined the behavior of endosomes for evidence of directed movement in A. nidulans hyphae using fluorescence microscopy. In both mutants, the majority of endosomes appeared in an immotile accumulation at the distal tip of the hypha (Figure 3B), consistent with the endosome distribution phenotype observed in line scans. Proximal to the accumulation, however, endosomes could be observed moving slowly in the dynein direction away from the hyphal tip (Figure 4B). Quantification of endosome motility showed that the mutations had a stronger relative effect on endosome velocity in A. nidulans hyphae than with S. cerevisiae dynein in vitro, but the trend was indeed the same: the AAA3 R/L mutation had a more severe effect than the AAA1 G/C substitution. In contrast to the slow endosome movement in these mutants, endosomes in a strain bearing the previously described AAA1 K/A mutant were completely immobile (Supplemental Figure S5; Zhang et al., 2010). Moreover, the dyneinAAA1 K/A strain had a nuclear distribution phenotype that resembled dynein∆ hyphae (Figure 3B), again differing from the mutations identified in our screen. These results suggest that 1) the AAA1 G/C and AAA3 R/L dynein mutations impair but do not abolish dynein motility, and 2) this limited motility is necessary and sufficient for dynein to participate in nuclear positioning.
DISCUSSION
Here we describe a screening method to identify mutations that impair the intracellular transport machinery in the highly tractable model system A. nidulans. Our approach is based on multiplexed whole-genome sequencing and multicolor fluorescence microscopy, lending it several notable features. First, the screen gives a direct output of the putative nucleotide change associated with a given transport phenotype. Multiplex genome sequencing, which enables this direct output, has become dramatically more affordable in recent years, and its cost is continuing to decrease (Meyer et al., 2007; Lefrancois et al., 2009; Wong et al., 2013). Second, by using a strain with three types of organelle labeled with different-colored fluorescent proteins, this method is capable of differentiating between mutations that broadly impair intracellular transport and those that are organelle specific. In principle, this analysis can be extended to study the intracellular transport of any organelle or macromolecule that can be genetically tagged, and it will be possible to create strains with different permutations of labeled molecules through genetic crosses. Finally, the identification of hits in this method (through fluorescence microscopy of A. nidulans hyphae) is well suited for automation. In the course of this study, we developed methods to array A. nidulans colonies on 96-well plates, an important first step for automated microcopy. Moreover, because the hyphal tip is a highly polarized, rod-like structure, the long axis of the hyphae provides a useful and readily identifiable coordinate system onto which organelle distributions can be plotted (e.g., Figure 2D). In addition to plotting the average organelle distribution, it may also be possible to record the motile characteristics of individual cargo (such as velocity, run length, and reversal frequency) through automated tracking.
Here we characterized seven mutants that map to genes required for intracellular transport: kinesin-1, dynein, and the dynein regulators Lis1 and dynactin. Of interest, the two mutations we identified in dynein severely reduce velocity and cause strong defects in endosome and peroxisome transport but still support dynein's role in nuclear positioning. This indicates that different dynein functions have distinct stringencies for motor function. This also highlights the ability of our screen to uncover mutations that differentially affect specific organelles, something that was observed for dynein mutations in Neurospora crassa (Sivagurunathan et al., 2012) and deletion of different dynactin subunits in Neurospora, Aspergillus, and mammalian cell lines (Lee et al., 2001; Zhang et al., 2011; Yeh et al., 2012).
The two dynein mutations identified in our screen slowed the velocity of S. cerevisiae dynein in vitro. In A. nidulans, both dynein mutations caused the accumulation of endosomes and peroxisomes at hyphal tips, indicative of a strong transport defect. Indeed, the velocity of endosome movement was decreased >80-fold. By contrast, nuclear positioning, which is well known to require dynein in A. nidulans (Xiang et al., 1994), was still functional in dynein-mutant backgrounds that lacked full motor activity. However, a mutation that completely blocks dynein motility (Kon et al., 2004; Zhang et al., 2010) did lead to strong defects in nuclear positioning, demonstrating that some degree of motor activity is required for this function.
The mechanism of nuclear positioning in filamentous fungi is not well understood. It is possible that cortically anchored dynein pulls on the distal nucleus by walking along microtubules that are attached to the spindle pole body, as is the case in S. cerevisiae (Moore et al., 2009). Because nuclear positioning occurs over relatively long time scales (15–22 min at 19°C; Fernandez-Abalos et al., 1998), very slow movement may be sufficient for this activity. Alternatively, it is possible that the main driving force for nuclear positioning comes from microtubule depolymerization, and cortically anchored dynein acts as a dynamic tether that couples depolymerizing microtubule ends to the cell cortex (Ligon et al., 2001; Malone et al., 2003; Vogel et al., 2009; Hendricks et al., 2012; Laan et al., 2012; Perlson et al., 2013). In this scenario, it could be that the AAA1 G/C and AAA3 R/L mutations support dynein's ability to maintain a dynamic connection with the microtubule end, whereas the AAA1 K/A mutation does not because it converts dynein to a “rigor” state with constitutively high microtubule affinity (Imamula et al., 2007; Laan et al., 2012). Further studies are required to investigate these possibilities.
We anticipate that screening methods such as those described here will lead to other new avenues of investigation into the many functions of cytoplasmic dynein, as well as into the mechanism and regulation of microtubule-based intracellular transport in general.
MATERIALS AND METHODS
Fungal growth conditions
A. nidulans strains were grown in liquid or on plates in yeast and glucose (YG) medium (Szewczyk et al., 2006) or minimal medium (MM) (Nayak et al., 2006), supplemented with 1 mg/ml uracil, 2.4 mg/ml uridine, 2.5 μg/ml riboflavin, 1 μg/ml para-aminobenzoic acid, and 0.5 μg/ml pyridoxine as required. Glufosinate was used as described (Nayak et al., 2006). The strains were grown at 37°C unless indicated otherwise.
A. nidulans strain construction
Strains of A. nidulans used in this study are listed in Supplemental Table S1. DNA constructs to delete genes, make point mutations, or tag endogenous genes with A. nidulans codon–optimized fluorescent proteins were generated using yeast homologous recombination (Orr-Weaver et al., 1983) or isothermal assembly (Gibson et al., 2009). To create the triple-labeled strain for mutagenesis and screening, early endosomes were labeled by fusing Rab5/RabA with TagGFP (Subach et al., 2008). Nuclei were labeled by fusing histone H1 with TagBFP (Subach et al., 2008). Peroxisomes were labeled by fusing a peroxisome targeting sequence (PTS1) to mCherry (Shaner et al., 2004). Constructs were integrated using homologous recombination in strains lacking the ku70 homologue (Nayak et al., 2006) at the endogenous loci, with the exception of PTS1-mCherry, which was integrated at the yA locus. All new strains were verified by PCR analyses using genomic DNA isolated as described (Lee and Taylor, 1990). Some strains were created through genetic crosses as described (Todd et al., 2007b).
4NQO mutagenesis and fluorescence microscopy–based screening
A. nidulans spores were diluted to 1 × 107 spores/ml in 0.1 M KPO4 (pH 7) and incubated for 30 min at 37°C with 4NQO. To determine appropriate 4NQO doses, various concentrations of mutagen were used, and the spores were plated on YG plates to achieve a density of 30–150 colonies/plate (Barbour et al., 2006). The survival rate was determined by calculating the percentage of spores that survived mutagenesis. For our large-scale mutagenesis we chose doses of 4NQO resulting in 10 and 50% survival rates (Supplemental Figure S1A).
Mutagenized spores were grown on YG plates for 48 h and then transferred onto a fresh YG plate for indexing. For microscopy, spores (or hyphae, in the case of nonconidiating mutants) were inoculated on MM plates containing the appropriate auxotrophic supplements and grown at 37°C for 12–24 h. Colony edges were excised from plates as agar squares, inverted onto 10 μl of liquid MM in 96-well MatriPlates (Matrical Bioscience, Spokane, WA), and observed at 22°C using a Nikon Ti epifluorescence microscope with Perfect Focus, a Nikon PlanApo 100×/1.40 oil objective, and a Hamamatsu ORCA-ER charge-coupled device camera, controlled by Elements (Nikon, Melville, NY) software. TagBFP, TagGFP, and mCherry were excited with a Prior Lumen 200 mercury lamp and a filter wheel system housing 405-, 495-, and 585-nm emission filters (Chroma Technology, Bellows Falls, VT).
Backcrosses, genomic DNA library preparation, and barcoding
Isolates with organelle mislocalization phenotypes were crossed to a nonmutagenized strain, and the progeny were sorted based on whether they exhibited the mutant phenotype by fluorescence microscopy. For genomic DNA preparation >12 progeny with the phenotype or without the phenotype were grown separately on YG plates at 37°C as individual colonies. Colonies with or without the phenotype were then pooled and used to coinoculate 100 ml of liquid YG at 37°C, with shaking at 100 rpm for 16 h. This was the only pooling step during the procedure. The cultures were filtered and dried with paper towels, and genomic DNA was isolated as described (Lee and Taylor, 1990). The genomic libraries were prepared from each category for whole-genome sequencing following the protocol of the genomic DNA sample preparation handbook (www.illumina.com) with the following changes. Genomic DNA (5 μg) was suspended in 500 μl of Tris-EDTA buffer and sonicated (Misonix Sonicator 3000, Cole-Parmer Instrument Company, Vernon Hills, IL) for 10–15 min with 10-s cooling intervals, producing DNA fragments that were predominantly 500 base pairs or smaller. To phosphorylate 5′ ends, 2 μg of sonicated DNA was incubated with 5 μl of T4 DNA polymerase (3000 U/ml; NEB, Ipswich, MA), 5 μl of T4 polynucleotide kinase (10,000 U/ml; NEB), 1 μl of DNA polymerase I, and large (Klenow) fragment (5000 U/ml, NEB) for 30 min at room temperature. The resulting DNA fragments were purified using QIAquick PCR Purification kits (Qiagen, Valencia, CA) and eluted with EB buffer to a final volume of ∼40 μl, thus removing enzymes and buffer components from the 5′-end repair step. An adenine deoxyribonucleotide was added to the 3′ ends of the DNA fragments by adding 3 μl of Klenow fragment (3′ → 5′ exo-, 5000 U/ml, NEB), 5 μl of 10× NEB Buffer 2, and 1 μl of 10 mM dATP (NEB) to 41 μl of end-repaired DNA fragments, followed by incubation at 37°C. The DNA fragments were then purified using MinElute PCR Purification kits (Qiagen).
Unique barcode oligonucleotides were attached to the 3′ ends of the DNA fragments (10 μl) using 5 μl of Quick T4 DNA ligase (400,000 U/ml; NEB), 10 μl of 10 μM barcode oligonucleotide, and 25 μl of Quick ligase buffer (NEB), followed by incubation for 15 min at room temperature (www.illumina.com; Wong et al., 2013). The DNA fragments were purified using QIAquick PCR purification kits. The resulting mixture was separated by 2% agarose gel electrophoresis, and 200– to 400–base pair DNA fragments were excised and purified using Gel Extraction kits (Qiagen). This gel-purification procedure was repeated. PCR amplification of the barcode-modified DNA was performed, and fragments between 200 and 400 base pairs were isolated using DNA SizeSelector-I Solid Phase Reversible Immobilization magnetic beads (Aline Biosciences, Woburn, MA; Wong et al., 2013).
Illumina sequencing and data analysis
The barcoded libraries were combined, and their concentrations were determined as described (Wong et al., 2013). The pooled libraries were again purified by 2% agarose gel electrophoresis, and DNA fragments between 200 and 400 base pairs were excised. A final round of size selection was performed with DNA SizeSelector-I beads (Wong et al., 2013). Single-end whole-genome sequencing was performed using the Illumina Genome Analyzer HiSeq 2000 platform, generating sequence reads ∼50 base pairs in length. In addition to isolates from the screen, we sequenced three wild-type libraries as references (RPA216, RPA478, and RPA496). After sequencing, the data sets were sorted by barcode using a Python script available on our website (https://reck-peterson.med.harvard.edu/protocols). The data were then aligned to the A. nidulans reference sequence (Wortman et al., 2009; Galagan et al., 2005; www.aspergillusgenome.org/download/sequence/A_nidulans_FGSC_A4/current/A_nidulans_FGSC_A4_current_chromosomes.fasta.gz) using the Burrows–Wheeler Aligner (BWA; Li and Durbin, 2009). After file format conversion with SAMtools (Li et al., 2009), the Genome Analysis Toolkit (GATK; McKenna et al., 2010) was used for realignment of gapped regions. We wrote scripts in Unix to navigate between BWA, SAMtools, and GATK (https://reck-peterson.med.harvard.edu/protocols). Nucleotide changes were determined by comparison to the Fungal Genetics Stock Center (FGSC) A. nidulans genomic sequence. A Python script (https://reck-peterson.med.harvard.edu/protocols) was used to identify and remove synonymous mutations, as well as nucleotide changes common to the nonmutagenized parent strains, progeny with the phenotype, and progeny lacking the phenotype. Nucleotide changes were labeled by genomic feature, and “high-quality” mutations—those appearing five or more times in the mutant data sets but not in the reference sequence—were written to a spreadsheet by the script.
Live-cell fluorescence microscopy
Multicolor wide-field images were generated at 22°C using a custom-built DeltaVision OMX microscope (Applied Precision, Issaquah, WA) equipped with a PlanApo N 60×/1.12 oil objective lens (Olympus, Center Valley, PA). TagBFP, TagGFP, and mCherry were excited using 405-, 488-, and 568-nm laser lines, respectively. A series of dichroic mirrors in the OMX light path (Applied Precision Standard Filter Set v4) were used to split the emission light from the three fluorophores to three different Photometrics CoolSnap HQ2 cooled charge-coupled device cameras. A narrowband emission filter in front of each camera was used to select appropriate wavelengths (436/31, 528/48, and 609/37 nm for TagBFP, TagGFP, and mCherry, respectively). Images were deconvolved using the enhanced ratio method in softWoRx 6.0 image processing software.
The line scan plots shown in Figures 2 and 3 and Supplemental Figures S3 and S4 were generated by projecting the average fluorescence intensity over a 10-s time-lapse sequence using ImageJ (National Institutes of Health, Bethesda, MD). Images were acquired every 100 ms for both endosomes and peroxisomes. The nuclear distribution measurement shown in the same figures was determined by measuring the distance from the most distal nucleus to the hyphal tip from a single frame with a 500-ms exposure time.
Endosome velocities were measured using ImageJ 1.46e. Maximum-intensity projections were created from time-lapse series to define the trajectory of endosomes. The segmented line tool was used to trace the trajectories perpendicular to and 10 μm away from the hyphal tip. The trajectories were then mapped onto the original movie sequence and “resliced” to generate a kymograph. The velocities of individual endosome particles were calculated from the inverse of the slopes of the kymograph traces. The trajectories were selected for analysis based on image quality and the clarity of discrete endosome movements.
Dynein mutant construction, purification, and single-molecule microscopy
The S. cerevisiae strains used in this study for dynein purification are listed in Supplemental Table S2. Mutations generating the AAA1 G/C and AAA3 R/L substitutions (corresponding to G1799C and R2620L in S. cerevisiae dynein, respectively) were introduced into the genomic copy of DYN1 (encoding the dynein heavy chain) using homologous recombination. Transformations with phenol-chloroform–purified PCR products were performed using the lithium acetate method, and transformants were screened by colony PCR. Point mutations were introduced using PCR primers and verified by DNA sequencing.
S. cerevisiae cultures for protein purification were grown, harvested, and frozen as previously described (Reck-Peterson et al., 2006). Dynein was purified via a ZZ tag, labeled with HaloTag-tetramethylrhodamine, and visualized on immobilized microtubules (containing ∼10% biotin tubulin) in flow chambers using an Olympus IX-81 microscope with a 100×/1.45 numerical aperture oil immersion TIRF objective (Olympus) as described (Reck-Peterson et al., 2006; Huang et al., 2012; Qiu et al., 2012). Images were collected every 1–2 s for 5–10 min. Velocities were calculated from kymographs generated in ImageJ 1.46e.
Graphs and statistical analyses
Statistical analyses and graphs were generated using Prism 6.0b (GraphPad, La Jolla, CA), Excel version 14.3 (Microsoft, Redmond, WA), and ImageJ 1.46e.
Supplementary Material
Acknowledgments
We thank Jennifer Wortman, Brian Haas, and Gustavo Cerqueira (Broad Institute, Cambridge, MA) for advice and discussions about data processing; Koon Ho Wong for advice on preparing genomic DNA libraries for Illumina sequencing; Pei-Hsin Hsu and François Aguet for advice on statistical analysis; Michael Wu, Natalie Heer, and Marisa Di-Gregorio for help generating strains and reagents; Bret Redwine and Mark McClintock for insightful comments on the manuscript; and Gaudenz Danuser and the Nikon Imaging Center at Harvard Medical School for microscopy support. This work was supported by a National Institutes of Health New Innovator award (OD004268) to S.R-P. A.J.R. is funded by a Sir Henry Wellcome Postdoctoral Fellowship (092436/Z/10/Z) sponsored by Peter J. Knight and Stan A. Burgess (University of Leeds, Leeds, United Kingdom).
Abbreviations used:
- 4NQO
4-nitroquinoline 1-oxide
- MM
minimal medium
- YG
yeast and glucose medium
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-09-0557) on January 8, 2014.
REFERENCES
- Abenza JF, Pantazopoulou A, Rodriguez JM, Galindo A, Penalva MA. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic. 2009;10:57–75. doi: 10.1111/j.1600-0854.2008.00848.x. [DOI] [PubMed] [Google Scholar]
- Bailleul B, Daubersies P, Galiegue-Zouitina S, Loucheux-Lefebvre MH. Molecular basis of 4-nitroquinoline 1-oxide carcinogenesis. Jpn J Cancer Res. 1989;80:691–697. doi: 10.1111/j.1349-7006.1989.tb01698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour L, Hanna M, Xiao W. Mutagenesis. In: Xiao W, editor. Yeast Protocols. Totowa, NJ: Humana Press; 2006. pp. 121–127. [DOI] [PubMed] [Google Scholar]
- Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem. 2008;283:25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J Cell Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, McClintock MA, Reck-Peterson SL. Microtubule-based transport in filamentous fungi. Curr Opin Microbiol. 2012a;15:637–645. doi: 10.1016/j.mib.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012b;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Fernandez-Abalos JM, Fox H, Pitt C, Wells B, Doonan JH. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol Microbiol. 1998;27:121–130. doi: 10.1046/j.1365-2958.1998.00664.x. [DOI] [PubMed] [Google Scholar]
- Galagan J, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Hammer JA, 3rd, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nat Rev. Mol Cell Biol. 2012;13:13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- Hendricks AG, Lazarus JE, Perlson E, Gardner MK, Odde DJ, Goldman YE, Holzbaur EL. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr Biol. 2012;22:632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T, Oakley B. The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol Biol Cell. 2005;16:918–926. doi: 10.1091/mbc.E04-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenaga M, Ishii Y, Tada M, Kakunaga T, Takebe H. Excision-repair of 4-nitroquinolin-1-oxide damage responsible for killing, mutation, and cancer. Basic Life Sci. 1975;5B:763–771. doi: 10.1007/978-1-4684-2898-8_54. [DOI] [PubMed] [Google Scholar]
- Imamula K, Kon T, Ohkura R, Sutoh K. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc Natl Acad Sci USA. 2007;104:16134–16139. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Kumar S, Plamann M. Null mutants of the Neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol Biol Cell. 2001;12:2195–2206. doi: 10.1091/mbc.12.7.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Taylor JW. Isolation of DNA from fungal mycelia and single spores. In: Innis DGM, Sninsky J, White T, editors. PCR Protocols: A Guide to Methods and Applications. Orlando, FL: Academic Press; pp. 282–287. [Google Scholar]
- Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon LA, Karki S, Tokito M, Holzbaur EL. Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat Cell Biol. 2001;3:913–917. doi: 10.1038/ncb1001-913. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Myles S, Prufer K, Hofreiter M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 2007;35:e97. doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Stuchell-Brereton MD, Cooper JA. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil Cytoskeleton. 2009;66:546–555. doi: 10.1002/cm.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR. Mitotic mutants of Aspergillus nidulans. Genet Res. 1975;26:237–254. doi: 10.1017/s0016672300016049. [DOI] [PubMed] [Google Scholar]
- Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B. Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell. 2005;16:5736–5748. doi: 10.1091/mbc.E05-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Yasunaga T, Noguchi TQ, Gomibuchi Y, Ngo KX, Uyeda TQ, Wakabayashi T. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell. 2010;143:275–287. doi: 10.1016/j.cell.2010.09.034. [DOI] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani S, Oakley B. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Perlson E, Hendricks AG, Lazarus JE, Ben-Yaakov K, Gradus T, Tokito M, Holzbaur EL. Dynein interacts with the neural cell adhesion molecule (NCAM180) to tether dynamic microtubules and maintain synaptic density in cortical neurons. J Biol Chem. 2013;288:27812–27824. doi: 10.1074/jbc.M113.465088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Derr ND, Goodman BS, Villa E, Wu D, Shih W, Reck-Peterson SL. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat Struct Mol Biol. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena N, Alberti-Segui C, Winzenburg E, Horn C, Schliwa M, Philippsen P, Liese R, Fischer R. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol Microbiol. 2001;42:121–132. doi: 10.1046/j.1365-2958.2001.02609.x. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-A crystal structure. Nat Struct Mol Biol. 2012;19:492–497, S1. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel C, Moreno-Velasquez SD, Riquelme M, Fischer R. Neurospora crassa NKIN2, a kinesin-3 motor, transports early endosomes and is required for polarized growth. Eukaryot Cell. 2013;12:1020–1032. doi: 10.1128/EC.00081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sivagurunathan S, Schnittker RR, Razafsky DS, Nandini S, Plamann MD, King SJ. Analyses of dynein heavy chain mutations reveal complex interactions between dynein motor domains and cellular dynein functions. Genetics. 2012;191:1157–1179. doi: 10.1534/genetics.112.141580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Gruenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani S, Oakley B, Oakley B. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Taheri-Talesh N, Horio T, Araujo-Bazán L, Dou X, Espeso EA, Peñalva M, Osmani S, Oakley B. The tip growth apparatus of Aspergillus nidulans. Mol Biol Cell. 2008;19:1439–1449. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RB, Davis MA, Hynes MJ. Genetic manipulation of Aspergillus nidulans: heterokaryons and diploids for dominance, complementation and haploidization analyses. Nat Protoc. 2007a;2:822–830. doi: 10.1038/nprot.2007.113. [DOI] [PubMed] [Google Scholar]
- Todd RB, Davis MA, Hynes MJ. Genetic manipulation of Aspergillus nidulans: meiotic progeny for genetic analysis and strain construction. Nat Protoc. 2007b;2:811–821. doi: 10.1038/nprot.2007.112. [DOI] [PubMed] [Google Scholar]
- Vogel SK, Pavin N, Maghelli N, Julicher F, Tolic-Norrelykke IM. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009;7:e1000087. doi: 10.1371/journal.pbio.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Jin Y, Moqtaderi Z. Multiplex Illumina sequencing using DNA barcoding. 2013. Curr Protoc Mol Biol Chapter 7, Unit 7.11. [DOI] [PubMed]
- Wortman JR, et al. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genet Biol. 2009;46(Suppl 1):S2–S13. doi: 10.1016/j.fgb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Zuo W, Efimov VP, Morris NR. Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr Genet. 1999;35:626–630. doi: 10.1007/s002940050461. [DOI] [PubMed] [Google Scholar]
- Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. Dynactin's pointed-end complex is a cargo-targeting module. Mol Biol Cell. 2012;23:3827–3837. doi: 10.1091/mbc.E12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Fischer R, Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell. 2003;14:1479–1488. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yao X, Fischer L, Abenza JF, Penalva MA, Xiang X. The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J Cell Biol. 2011;193:1245–1255. doi: 10.1083/jcb.201011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhuang L, Lee Y, Abenza JF, Penalva MA, Xiang X. The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J Cell Sci. 2010;123:3596–3604. doi: 10.1242/jcs.075259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.