Multicellular development of Dictyostelium is induced by starvation and is crucial for its long-term survival. Coronin A mediates the transition from growth to development of the cells and initiates the cAMP-dependent relay by regulating the response to secreted cell density and nutrient deprivation factors.
Abstract
Many biological systems respond to environmental changes by activating intracellular signaling cascades, resulting in an appropriate response. One such system is represented by the social amoeba Dictyostelium discoideum. When food sources become scarce, these unicellular cells can initiate a cAMP-driven multicellular aggregation program to ensure long-term survival. On starvation, the cells secrete conditioned medium factors that initiate cAMP signal transduction by inducing expression of genes such as cAMP receptors and adenylate cyclase. The mechanisms involved in the activation of the first pulses of cAMP release have been unclear. We here show a crucial role for the evolutionarily conserved protein coronin A in the initiation of the cAMP response. On starvation, coronin A–deficient cells failed to up-regulate the expression of cAMP-regulated genes, thereby failing to initiate development, despite a normal prestarvation response. Of importance, external addition of cAMP to coronin A–deficient cells resulted in normal chemotaxis and aggregate formation, thereby restoring the developmental program and suggesting a functional cAMP relay in the absence of coronin A. These results suggest that coronin A is dispensable for cAMP sensing, chemotaxis, and development per se but is part of a signal transduction cascade essential for system initiation leading to multicellular development in Dictyostelium.
INTRODUCTION
The survival and development of a complex organism requires intracellular and intercellular communication via a plethora of signaling events. In particular, cells need to be able to sense environmental signals in order to adapt their intracellular physiology to a changing environment. An exquisite example of a eukaryotic organism relying for its survival on environmental cues is the social amoeba Dictyostelium discoideum. This unicellular eukaryote initiates a multicellular developmental program as a survival mechanism when nutrients are depleted. Nutrient starvation induces the aggregation of individual amoebae into a multicellular structure, which ultimately forms a fruiting body containing two major differentiated cell types—stalk cells and spores (Devreotes, 1989; Kimmel and Firtel, 1991; Weijer, 1999).
Before aggregation, Dictyostelium cells secrete autocrine factors for sensing cell density and nutrient deprivation (Mann and Firtel, 1991), leading to the expression of early developmental genes such as discoidin A (dscA) and the initiation of the cAMP pathway via activation of G protein–dependent processes (Yuen et al., 1995). Among the secreted signaling molecules active at the transition from growth to development, the glycosylated protein prestarvation factor (PSF; Clarke et al., 1988) and conditioned medium factor (CMF; Gomer et al., 1991) are essential. During growth, cells continuously secrete PSF, which allows them to estimate the density of the population (Clarke et al., 1988). On starvation, the level of PSF decreases and a second density-sensing pathway is activated via CMF secretion. CMF regulates the induction of cAMP signaling during early development by regulating the lifetime of the cAMP-stimulated Gα2-GTP configuration and controlling cAMP-dependent gene expression through plasma membrane receptors that stimulate both a G protein–independent signaling pathway (CMF receptor 1 [CMFR1]) and a Gα1-dependent pathway (Jain et al., 1992; Jain and Gomer, 1994; Brazill et al., 1998; Deery and Gomer, 1999; Deery et al., 2002; Bakthavatsalam et al., 2007; Ray et al., 2011).
After a few hours of starvation, some cells start secreting pulses of cAMP, which are then relayed by neighboring cells, thus initiating a positive feedback loop that further increases cell differentiation. Chemotactic motility of cells is guided by these cAMP waves and drives the formation of multicellular aggregates (Chisholm and Firtel, 2004). At the molecular level, interaction of the secreted cAMP with the G protein–coupled cAMP receptor 1 (cAR1) on the plasma membrane induces a series of molecular and morphological events (Swaney et al., 2010), including enhanced expression of early developmental genes such as carA and adenylyl cyclase A (aca), leading to polarization and chemotaxis of the cells (Johnson et al., 1992; Pitt et al., 1992; Insall et al., 1994).
The mechanisms involved in the transfer of environmental cues to the intracellular response and the generation of cAMP pulses are incompletely understood. We here report that coronin A, a member of the highly conserved coronin protein family, plays an essential role in the earliest stages of development by regulating the expression of genes required for cAMP synthesis and sensing.
Coronin protein family members are characterized by tryptophan–aspartic acid (WD) repeats that form a β-propeller, followed by a region unique to each member and a coiled-coil domain that mediates oligomerization (Pieters et al., 2013). Coronins have sequence homology to the β subunit of trimeric G proteins, and bioinformatic and structural analysis showed that mammalian coronin 1 adopts a seven-bladed β-propeller structure similar to Gβ (de Hostos et al., 1991; Gatfield et al., 2005; Appleton et al., 2006). D. discoideum coronin A was originally isolated as a copurifying protein in a preparation of actomyosin and was localized in crown-shaped cortical structures (de Hostos et al., 1991). Coronin A–deficient cells show pleiotropic phenotypes, including defects in cytokinesis, yeast particle uptake, and chemotaxis (de Hostos et al., 1991; Maniak et al., 1995; Shina et al., 2011). Accordingly, coronin A is considered to be primarily an F-actin–binding and modulatory protein (de Hostos, 1999). The mammalian homologue for Dictyostelium coronin A, coronin 1, however, appears to be dispensable for F-actin–dependent processes, instead being involved in the activation of intracellular signal transduction cascades after cell surface stimulation. In both T-cells and macrophages, coronin 1 is essential for the activation of signal transduction after T-cell receptor stimulation and mycobacterial uptake, respectively (Jayachandran et al., 2007, 2008; Mueller et al., 2008, 2011; Combaluzier et al., 2009; Combaluzier and Pieters, 2009).
The findings described in this article show that Dictyostelium coronin A is required for initiation of cAMP pulsing upon starvation, leading to multicellular development. Of importance, supplying external cAMP to coronin A–deficient cells restored chemotaxis and development. However, conditioned medium, which is known to contain factors required for the initiation of the developmental program upstream of the cAMP pathway, failed to trigger development in cells lacking coronin A. This suggests that Dictyostelium coronin A is dispensable for these latter processes but instead is involved in activation of intracellular signal transduction after cell surface receptor triggering during the early phase of starvation-induced development.
RESULTS
Essential role for coronin A in multicellular development
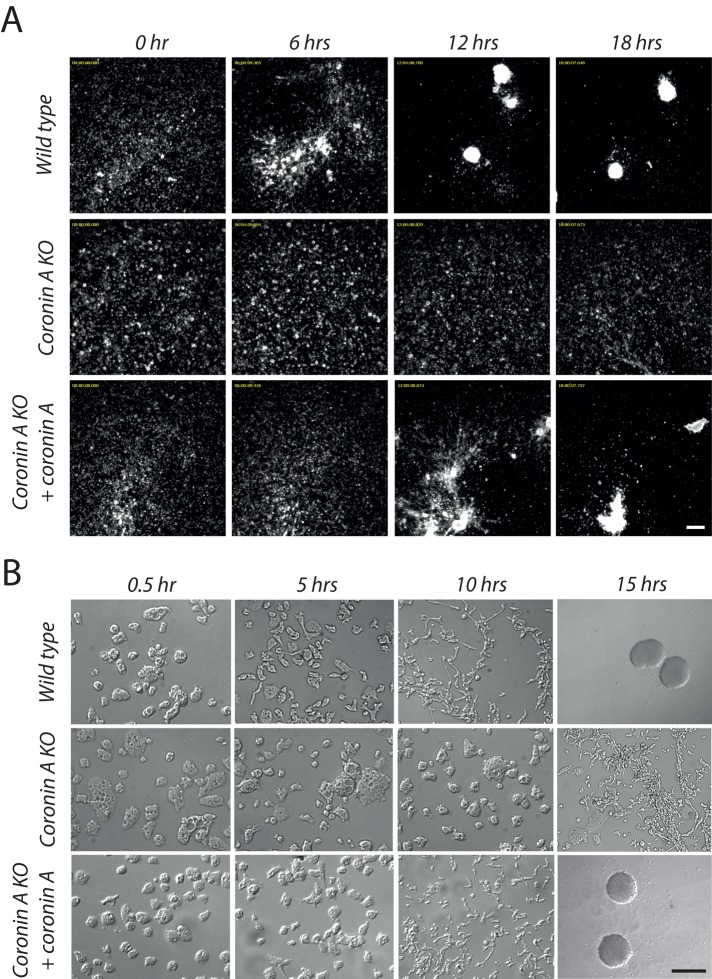
In the course of analyzing the phenotype of D. discoideum cells lacking coronin A, we noticed that upon starvation, strains lacking coronin A failed to form aggregates. To investigate the role of coronin A during development more systematically, we starved wild-type and coronin A–deficient cells (generated in the DH1-10 strain; see Materials and Methods and Supplemental Figure S1) in Bonner's salt solution (BSS) and cells were analyzed over a time period of 20 h. As shown in Figure 1A and Supplemental Movies S1 and S2, whereas wild-type cells initiated aggregate formation ∼4–6 h after starvation, a process that was completed around 10 h, coronin A–deficient cells failed to aggregate; however, aggregation was restored by coronin A expression (Figure 1 and Supplemental Movie S3). Defective aggregation in the absence of coronin A was similar for newly generated coronin A–knockout cells (in the DH1-10 background) as well as for the previously described coronin A–deficient cells HG1569 and HG1570 (de Hostos et al., 1993; Supplemental Figure S2 and data not shown). Although at high densities coronin A–deficient cells were able to aggregate, this occurred at dramatically delayed time points (Supplemental Table S1).
FIGURE 1:
Early development of wild-type and coronin A–deficient cells. (A) Dictyostelium cells were seeded into multiwell plates at a density of 2 × 105 cells/cm2, starved in BSS, and imaged over a period of 20 h. The images shown are taken from a series of frames during the aggregation stage of the wild-type, coronin A–deficient, and complemented coronin A–deficient cells. Bar, 200 μm. See also Supplemental Movies S1–S3. (B) Growth phase cells were washed, resuspended in BSS, and inoculated in a plastic dish. Images are taken at the indicated time points. Bar, 50 μm, except for wild-type and coronin A–complemented, coronin A–deficient cells at 15 h, bar, 0.5 mm.
Given that one of the earliest signs of cellular differentiation, preceding aggregation, is a change in cellular morphology resulting in cellular polarization (Kimmel and Firtel, 2004), we analyzed the capacity of coronin A–deficient Dictyostelium cells to polarize. As shown in Figure 1B, whereas the majority of wild-type cells were elongated ∼5 h after starvation initiation, elongation was markedly delayed in coronin A–deficient cells. Thus deletion of coronin A results in an inability of cells to polarize, resulting in a deficiency in initiating cellular differentiation and aggregation.
Absence of cAMP-dependent oscillation and cAMP production upon coronin A deletion
Aggregation is caused by periodic cAMP secretion, amplified by surrounding cells, resulting in cell polarization. These propagating waves of cAMP guide the chemotactically moving cells toward the aggregation center, where they accumulate into a multicellular structure (Weijer, 1999; Kimmel and Firtel, 2004). Thus, transient activation of the cAMP-producing enzyme adenylate cyclase and subsequent production and secretion of cAMP in response to the extracellular cAMP signal are necessary for polarization and chemotactic movement of starving Dictyostelium cells.
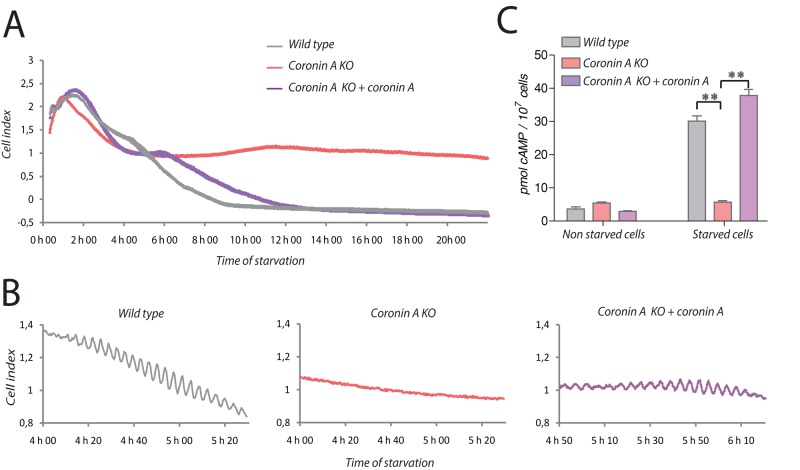
To analyze whether the inability of coronin A–deficient cells to undergo polarization and initiate chemotaxis was associated with a defective cAMP relay, we analyzed starving populations of wild-type or coronin A–deficient Dictyostelium by real-time cell analysis, using electric cell–substrate impedance sensing. This sensitive method allows us to follow the dynamics of cell–substrate and cell–cell contacts in real time (Schafer et al., 2011, 2013). As shown in Figure 2A and Supplemental Figure S3A, the seeding of Dictyostelium cells produced a transient increase in impedance within the first 2 h for both wild-type and coronin A–deficient strains. After 4–5 h of starvation, wild-type Dictyostelium showed oscillatory behavior as recorded by impedance, eventually decaying after 7 h (Figure 2, A and B, and Supplemental Figure S3, A and B), which is indicative of the generation of cAMP waves in the population (Gerisch, 1982; Weijer, 1999). At the end of this oscillation phase, the cells aggregated into small groups, which accumulated and caused an impedance drop. Strikingly, when we zoomed in on the signal produced at 4–6 h, we observed an oscillatory signal for wild-type and coronin A–complemented Dictyostelium cells, which was absent in cells lacking coronin A (Figure 2B and Supplemental Figure S3B).
FIGURE 2:
The cAMP signaling pathway in wild-type and coronin A–deficient cells. (A, B) Vegetative wild-type, coronin A–deficient, and coronin A–deficient cells complemented with coronin A were washed, resuspended in BSS, and seeded at 75,000 cells/well in a 96-well plate format adapted for the xCELLigence measurement. Impedance was measured every 30 s during 22 h and is represented as cell index. (C) Production of cAMP in response to starvation was determined in wild-type, coronin A–deficient, and coronin A–deficient cells complemented with coronin A after 6 h in a BSS suspension. The produced cAMP was determined using the HTRF cAMP assay (Cisbio) as described in Materials and Methods. Data in A and B are presented as a representative experiment out of three independent experiments. Results in C are presented as means ± SD of triplicate determinations of a representative experiment of three independent experiments. **p < 0.001 (t test).
To directly determine whether the inability to enter the oscillatory phase upon starvation was related to a defect in cAMP production in the absence of coronin A, we analyzed cAMP production using a competitive immunoassay. To that end, cells were starved for 6 h and lysed, and cAMP levels were analyzed. As expected, wild-type Dictyostelium responded to starvation with robust cAMP production. However, in cells lacking coronin A, starvation did not result in cAMP production but was fully restored by expression of coronin A in the coronin A–deficient cells (Figure 2C and Supplemental Figure S3C).
Chemotaxis in the presence and absence of coronin A
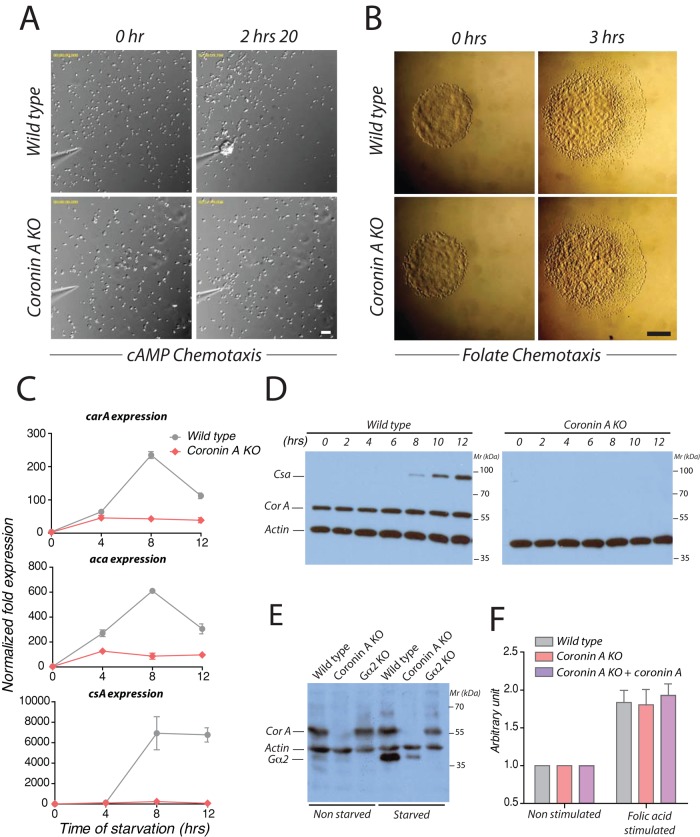
The foregoing results suggest a defective ability of cells lacking coronin A to produce cAMP upon starvation. As mentioned, Dictyostelium cells initiate the cAMP relay necessary for differentiation, aggregation, and development, including a positive feedback loop that further increases the expression of cAMP receptors (Bonner, 1947; Gross et al., 1976; Wang and Kuspa, 1997; Gregor et al., 2010). To investigate whether defective cAMP production in the absence of coronin A was due to defective sensing of cAMP, we analyzed chemotaxis. To that end, wild-type or coronin A–deficient cells were starved for 5–6 h, and their ability to chemotax toward a cAMP gradient generated by a micropipette was observed by time-lapse microscopy (Firtel and Chung, 2000). In addition to reduced cell motility in the absence of coronin A, as described before (de Hostos et al., 1993; Shina et al., 2011), coronin A–deficient cells failed to polarize and migrated with poor directionality. In contrast, wild-type cells chemotaxed with a coordinated motion toward the micropipette tip in a polarized manner (Figures 3A and 4B and Supplemental Movies S4 and S5).
FIGURE 3:
Chemotaxis and developmentally regulated gene expression profiles of wild-type and coronin A–deficient cells. (A) Vegetative wild-type or coronin A–deficient cells were starved for 5 h, resuspended in BSS, and seeded at a density of 1 × 105 cells/cm2. A cAMP gradient was established by injection of 1 μM cAMP through a micropipette at a pressure of 20 hPa. Cells were imaged every 20 s for 2.5 h. Bar, 50 μm. See also Supplemental Movies S4 and S5. (B) A 1-μl droplet of vegetative wild-type and coronin A–deficient cells was spotted on a 1.5% BSS agar Petri dish at a distance of 4 mm from a folate source (right part of the gel). Chemotaxis of the strains at 0 (left) and 3 h (right) after the drop deposition on agar. Bar, 1 mm. (C) The mRNA levels of acaA and carA, relative to the control Ig7 and gpdA at 0, 4, 8, and 12 h after the induction of starvation in BSS were determined by real-time PCR for wild-type and coronin A–deficient cells. (D) Starving cells in BSS in shaking cultures were collected at the indicated times after the initiation of starvation and boiled in SDS sample buffer. Lysates from an equal number of cells were separated on 10% SDS–PAGE, followed by immunoblotting for csA, coronin A, and actin expression. (E) Cells were grown in HL-5 or starved in BSS in shaking cultures and collected 5 h after the initiation of the starvation. Membranes were extracted as described in Materials and Methods and boiled in SDS sample buffer. Extracts from an equal number of cells were separated by 10% SDS–PAGE, followed by immunoblotting for Gα2, coronin A, and actin. (F) Vegetative wild-type, coronin A–deficient, and coronin A–deficient cells complemented with coronin A were washed and resuspended in BSS for 30 min. The cAMP production was determined as described in Figure 2C after a 6-min stimulation period with 50 μM folate. Results in C and F are presented as mean ± SD of triplicate determinations of a representative experiment of three independent experiments.
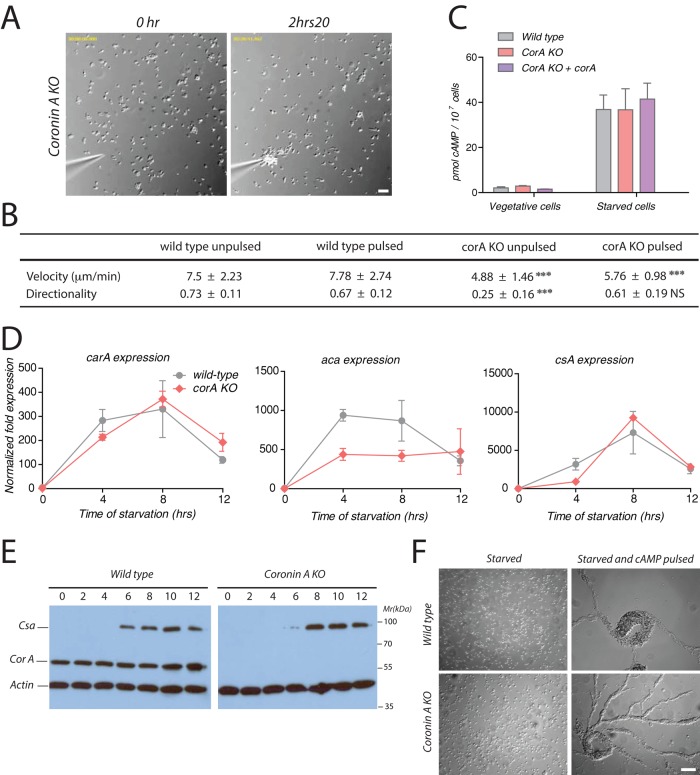
FIGURE 4:
cAMP sensing, production, and early developmental gene expression upon exogenous cAMP pulses. (A) Coronin A–deficient cells were washed, starved, and pulsed during 5 h with 50 nM cAMP every 6.5 min. Cells were then washed, resuspended in BSS, and seeded at a density of 1 × 105 cells/ml. A cAMP gradient was established as described in the legend to Figure 3A. Bar, 50 μm. See also Supplemental Movie S8. (B) Cells were prepared and processed as described in Figures 3A and 5A. Cell paths (from at least 30 cells per experimental condition from three independent experiments) were generated, and the velocity and directionality parameters were analyzed using the Chemotaxis and Migration Tool of ImageJ. Means ± SD; statistical significance between coronin A–deficient cells and corresponding wild-type cells are presented using a t test. ***p < 0 0.001; NS, not significant. (C) Cells were pulsed with cAMP as described in Materials and Methods and then assayed for cAMP production using the HTRF cAMP assay (Cisbio). (D) Vegetative wild-type and coronin A–deficient cells were washed, starved, and pulsed during 5 h with 50 nM cAMP every 6.5 min. Cells were then lysed at the indicated time points, and the mRNA levels of acaA, carA, and csA relative to the housekeeping control genes Ig7 and gpdA were determined at 0, 4, 8, and 12 h by real-time PCR. (E) Vegetative wild-type and coronin A–deficient cells were washed, starved, and pulsed with 50 nM cAMP every 6.5 min. Cells were collected at the indicated times after the initiation of the starvation and boiled in SDS sample buffer. Equal amounts of cells were separated by 10% SDS–PAGE and analyzed by Western blot for csA, coronin A, and actin expression. (F) Vegetative wild-type and coronin A–deficient cells were washed and starved with or without 50 nM cAMP pulsing during 5 h every 6.5 min in suspension. Cells were then washed, resuspended in BSS, and placed on a Petri dish at a density of 4 × 105 cells/cm2. Images were taken 4 h after seeding of the cells. Bar, 50 μm. Results in C are presented as means ± SD of triplicate determinations of a representative experiment of three independent experiments.
The inability of coronin A–deficient cells to chemotax in a cAMP gradient could be a consequence of a failure to initiate a development program or may reflect a more general chemotactic defect. To address this question, we assayed the chemotaxis of vegetative wild-type and coronin A–deficient cells to folic acid. During vegetative growth, D. discoideum chemotaxes toward folate and other nutrients released by bacteria (Aubry and Firtel, 1999; Manahan et al., 2004; Lim et al., 2005). As shown in Figure 3B, wild-type cells, as well as coronin A–deficient cells, effectively moved toward a folic acid gradient using an agar-based assay. Similarly, under submerged conditions, wild-type cells and coronin A–deficient cells were able to chemotax through a gradient created by a micropipette filled with folic acid and clustered at the tip of the micropipette during the first hour (Supplemental Figure S4, A and B and Supplemental Movies S6 and S7). Together these results suggest that coronin A is dispensable for folate-mediated chemotaxis but is specifically required for sensing and producing cAMP after starvation.
Defective early developmental gene expression in the absence of coronin A
The cAMP relay and subsequent development depend on the expression of proteins required for cAMP-induced cAMP production, such as cAR1 and ACA. The pronounced defect in polarization, cAMP-dependent oscillation, and aggregation in the absence of coronin A could therefore be due to a defect in early developmental gene expression, similar to other mutants of key components of the cAMP signaling pathway (Parent and Devreotes, 1996; Manahan et al., 2004). To analyze the expression of the early developmental genes carA and aca, as well as the development marker contact site A (csA; Faix et al., 1992; Iranfar et al., 2003), we starved cells and performed quantitative PCR on mRNA isolated from lysates prepared from either wild-type or coronin A–deficient cells at the time points indicated in Figure 3C. Whereas in wild-type cells the expression of these genes increased dramatically, reaching a peak at 8 h after the initiation of starvation, in coronin A–deficient cells no or little induction of mRNA was detected for these early developmental genes. Analysis by immunoblotting confirmed the absence of induction of the contact site A protein (Figure 3D). Furthermore, Gα2 proteins, whose upregulated expression is necessary for the cAMP relay early upon starvation (Kumagai et al., 1989; Kimmel and Firtel, 2004), was significantly upregulated in wild-type cells upon starvation but barely detectable in coronin A–deficient cells (Figure 3E). These data indicate that genes involved in regulatory networks required to establish and maintain the cAMP relay fail to be upregulated in the absence of coronin A.
Given the suggested role for coronin A in the modulation of the F-actin cytoskeleton, we set out to determine whether a perturbation of the actin cytoskeleton could alter the expression of early developmental genes. To that end, we treated wild-type cells with the F-actin depolymerizing drug latrunculin A (Lat A) at a concentration of 1 μM (Parent et al., 1998) and analyzed gene expression during starvation. As shown in Supplemental Figure S5, A and B, the addition of Lat A resulted in cell rounding and a strong decrease in cell motility, as well as in a failure to aggregate (Supplemental Figure S5, A and B). However, Lat A–treated cells retained the ability to up-regulate aca and carA during the first 6 h of starvation (Supplemental Figure S5C), which is in accordance with previous results that Lat A does not have a detrimental effects on cAR1-mediated signal transduction (Parent et al., 1998; Kriebel et al., 2008).
Together these results show a role for coronin A in the induction of early developmental genes upon starvation, which likely explains the cAMP relay defect in cells lacking coronin A. To test the specificity of the cAMP production defect during the early developmental stage in coronin A–deficient cells, we measured cAMP production after folic acid stimulation in vegetative cells, which is known to be mediated via an extracellular regulated kinase-2 (ERK2)–dependent pathway (Devreotes, 1983; De Wit et al., 1986; Maeda and Firtel, 1997). Wild-type, coronin A–deficient, and coronin A–complemented cells were starved for 30 min and stimulated with folic acid, and the level of cAMP was quantified. Coronin A–deficient cells showed a similar increase in cAMP production in response to folate stimulation as wild-type and complemented cells (Figure 3F). These results indicate that coronin A is specifically required for the induction of early development gene expression, which is a crucial step for the establishment of any subsequent cAMP-dependent development program.
Rescue of cAMP relay of coronin A–deficient cells by exogenously supplied cAMP
Together the foregoing results suggest that coronin A is important for initiation of the cAMP-dependent starvation response that is required for polarization, chemotaxis, and aggregation. To analyze whether, in principle, coronin A–deficient cells can respond to extracellular cAMP, we provided cAMP as pulses to mimic the conditions arising during starvation (Darmon et al., 1975; Mann and Firtel, 1987). When coronin A–deficient cells were starved in a shaking culture with 50 nM cAMP pulses at a frequency of 6.5 min for 5 h before being seeded, most cells were able to chemotax in a polarized manner to a cAMP gradient generated by a micropipette (Figure 4A and Supplemental Movie S8). Although the directionality of cells lacking coronin A was restored, the motility of these cells was still largely defective as compared with wild-type cells (Figure 4B), indicating a cAMP-independent origin of the motility defect.
Furthermore, wild-type and coronin A–deficient cells produced comparable levels of cAMP after 5 h of starvation and pulsing (Figure 4C; compare with Figure 2C). In addition, quantitative PCR revealed that in pulsed coronin A–deficient cells, the early developmental gene expression necessary for cAMP relay was restored, although at various degrees for the different genes (Figure 4D). Similarly, as analyzed by immunoblotting, CsA protein expression was restored in coronin A–deficient cells upon cAMP pulsing (Figure 4E). Finally, cAMP pulsing rescued aggregation of coronin A–deficient cells (Figure 4F). These results show that the cAMP relay defect of coronin A–deficient cells can be overcome by exogenous signals released by cAMP pulses provided in trans.
Restoration of aggregation of coronin A–deficient cells by 8-Br-cAMP
The fact that cAMP-dependent polarization and aggregation in coronin A–deficient cells was restored by external cAMP pulses suggests that coronin A is dispensable for cellular processes downstream of the cAMP receptor and instead functions in the transduction of the initial signal required to activate the cAMP relay pathway. To assess independently whether or not coronin A is required in the cellular processes leading to chemotaxis and aggregation, we exposed cells to 8-Br-cAMP. This membrane-permeable cAMP analogue possesses a poor affinity for the extracellular cAMP receptors but enters the cells and directly activates the cAMP-dependent protein kinase A (Van Haastert et al., 1981; Van Haastert and Kien, 1983; Kay, 1982; Riley and Barclay, 1990). Incubation of coronin A–deficient cells with 8-Br-cAMP for 5 h and subsequent cell transfer in compound-free BSS was sufficient to restore the ability to form aggregates (Figure 5). Indeed, coronin A–deficient cells were able to polarize and readily aggregate after incubation with the cAMP analogue, whereas cells incubated in BSS were unresponsive (Supplemental Figure S6 and Supplemental Movies S9 and S10). Observation at a later time point showed similar aggregate formation for wild-type cells and coronin A–deficient cells after incubation with the 8-Br-cAMP analogue. These results, first, suggest that protein kinase A activation and downstream pathways are still functional in the absence of coronin A, and, second, confirm that coronin A is dispensable for chemotaxis and aggregation per se but is required for initiation of the starvation response.
FIGURE 5:
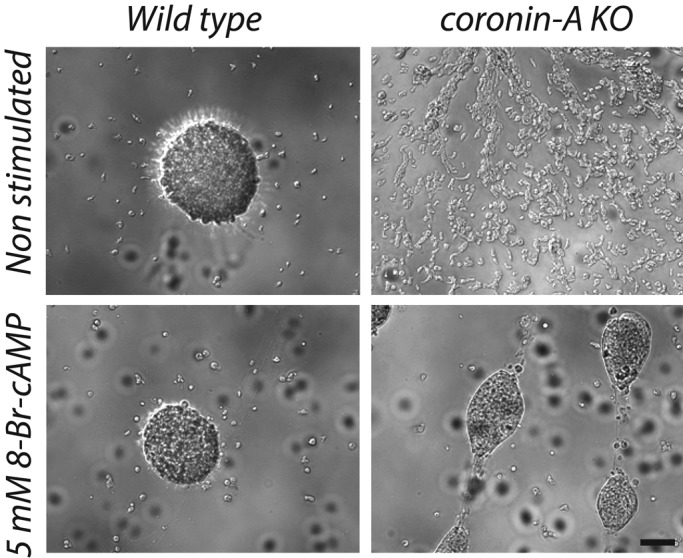
Aggregation of wild-type and coronin A–deficient cells in the presence of 8-Br-cAMP. Wild-type and coronin A–deficient cells were washed in BSS, seeded into multiwell plates at a density of 2 × 105 cells/cm2, and incubated in BSS or BSS + 5 mM 8-Br-cAMP during 5 h. Cells were washed and then imaged after 16 h. Bar, 200 μm.
Coronin A–deficient cells fail to sense factors secreted in the conditioned medium
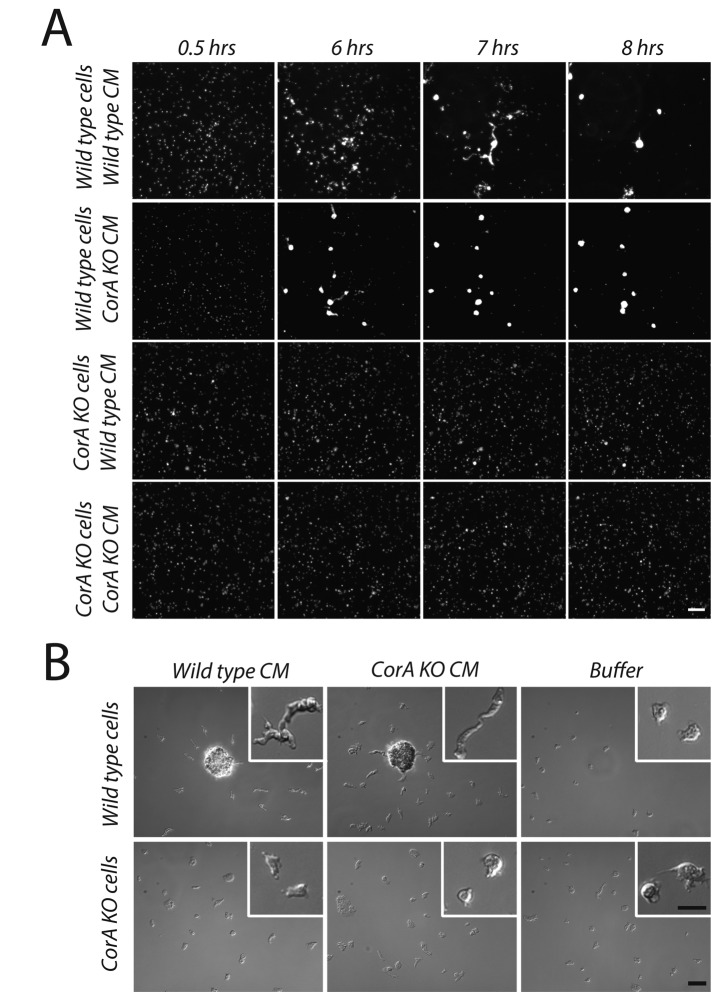
The rescue of polarization, chemotaxis, and development in cells lacking coronin A by either cAMP pulsing or inclusion of 8-Br-cAMP suggests that coronin A acts upstream of the cAMP relay. We therefore analyzed whether cells lacking coronin A could produce or sense the cell density and nutrient deprivation factors necessary for initiation of aggregation and development. First, to determine whether cells lacking coronin A can produce conditioned medium factors, we incubated cells at a low density with conditioned medium (CM) prepared from either wild-type or coronin A–deficient cells. At this density, cells failed to initiate development when cultured in buffer alone—that is, in the absence of secreted factors (Figure 6B and Supplemental Figure S7). Of importance, whereas wild-type cells initiated polarization and aggregation upon addition of conditioned medium from either wild-type or coronin A–deficient cells, coronin A–deficient cells failed to respond to either of these conditioned media (Figure 6, A and B, Supplemental Figure S7, and Supplemental Movies S11 and S12). This suggests that coronin A–deficient cells are able to secrete cell density–sensing factors but cannot respond to signals induced by conditioned medium factors, which are required for polarization and aggregation.
FIGURE 6:
Transition from growth to development induced by conditioned medium in wild-type but not coronin A–deficient cells. (A) Exponentially growing wild-type and coronin A–deficient cells were washed in PBM and seeded into multiwell plates at a density of 5 × 104 cells/cm2, incubated in conditioned medium obtained from wild-type or coronin A–deficient starving cells, and imaged over a period of 20 h. The images are taken from a series of frames during the conditioned medium incubation. Bar, 200 μm. See also Supplemental Movies S11 and S12. (B) Cells were prepared as described in A and inoculated in a plastic dish. Images are taken after 6 h of incubation of the cells in conditioned medium. Bar, 100 μm (large pictures), 50 μm (insets).
To test whether the defect of the coronin A–deficient cells to sense condition medium could be a result of impaired CMF receptor 1 (CMFR1; Deery and Gomer, 1999) expression, we analyzed membrane fractions prepared from nonstarved or 6-h-starved cells by immunoblotting for CMFR1 levels. In all conditions, the CMFR1 was similarly expressed in coronin A–deficient and wild-type cells, whereas cells deficient for CMFR1 lacked expression, as judged by immunoblot analysis (Figure 7A). To analyze plasma membrane expression of the CMFR1, we biotinylated and purified cell surface proteins and assessed the presence of CMFR1 by immunoblotting (Froquet et al., 2012). As shown in Figure 7B, CMFR1 is expressed at the surface of both wild-type and coronin A–deficient cells.
FIGURE 7:
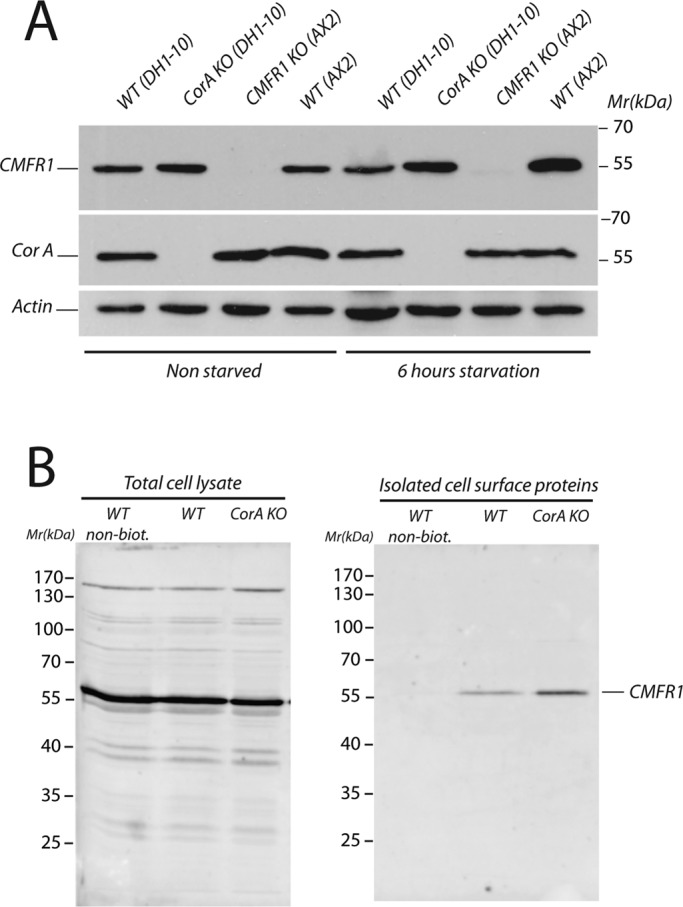
CMF receptor 1 expression in wild-type and coronin A–deficient cells. (A) Exponentially growing wild-type (DH1-10 and AX2), coronin A–deficient, and CMFR1-deficient cells were washed in BSS, and membranes were extracted immediately or after 6 h of starvation in BSS at a density of 10 × 106 cells/ml. Equal amounts of cells were separated by 10% SDS–PAGE and analyzed by Western blot for CMFR1, coronin A, and actin expression. (B) Wild-type and coronin A–deficient cells were cooled, pelleted, and washed twice. Cell surface proteins were then biotinylated and purified. A Western blot against CMFR1 was performed on total cell lysates (left) or isolated cell surface proteins (right).
The prestarvation response of coronin A–deficient cells is normal
Thus far, the data suggest that coronin A is required for the initiation of development upon starvation but is dispensable for cAMP-mediated signaling. However, whether coronin A is important for the prestarvation response is unclear. The prestarvation response is regulated by a factor released by vegetative cells called the PSF, which is responsible for the induction of several genes needed for cell aggregation. At high bacterium concentrations, the PSF response is inhibited, which allows the cells to monitor their density relative to the food supply (Clarke and Gomer, 1995). As the availability of bacteria decreases, PSF induces several genes, including the discoidin gene, which is required for initiation of the starvation response (Clarke et al., 1988; Rathi et al., 1991). We mimicked the prestarvation response by growing Dictyostelium cells in a suspension of reduced bacterial loads. Analysis of the discoidin expression showed a similar increase in wild-type and coronin A–deficient cells when the bacterial load was reduced (Figure 8A). Furthermore, for the same bacterial load, a cell density increase resulted in similar discoidin up-regulation in wild-type and coronin A–deficient cells (Figure 8B). We also observed a comparable induced discoidin expression when wild-type or coronin A–deficient cells were incubated in conditioned medium from either wild-type cells or coronin A–deficient cells (Figure 8C). Similarly, we found that discoidin expression was reduced in both wild-type and coronin A–deficient cells upon incubation with folate (data not shown). These data indicate that cells lacking coronin A are able to produce and respond to PSF to modulate discoidin expression. Together these results strongly suggest that coronin A is specifically required to respond to secreted factors needed for the initiation of development once starvation has been initiated, but is dispensable for the prestarvation response.
FIGURE 8:
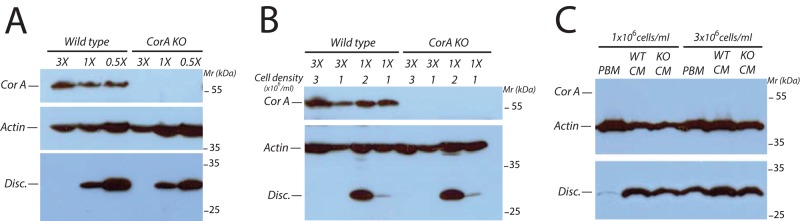
Response to PSF is not affected in coronin A–deficient cells. (A) Vegetative wild-type and coronin A–deficient cells were washed and resuspended at 1 × 104 cells/ml in PBM containing 3×, 1×, or 0.5× bacteria (see Materials and Methods). When cells reached a density of 1 × 106 cells/ml, they were then washed free of bacteria and boiled in SDS sample buffer. Equal amounts of cells were separated on a 10% SDS gel and analyzed by Western blot for discoidin, coronin A, and actin expression. (B) Cells were prepared as described in A but were washed and boiled in SDS sample buffer when they reached a density of 3 × 106, 2 × 106, or 1 × 106 cells/ml. Equal amounts of cells were separated on a 10% SDS gel and analyzed by Western blot for discoidin, coronin A, and actin expression. (C) Coronin A–deficient cells were prepared as described in A in 1× density bacteria in PBM or CM from wild-type or coronin A–deficient cells. Cells were then washed free of bacteria and boiled in SDS sample buffer when they reached a density of 1 × 106 or 3 × 106 cells/ml. Equal amounts of cells were separated on a 10% SDS gel and analyzed by Western blot for discoidin, coronin A, and actin expression.
FIGURE 9:

Proposed model for initiation of the cAMP pathway by coronin A. In response to nutrient deprivation, cells secrete factors that bind to cell surface receptors and subsequently activate the Gα2βγ-dependent pathway upon cAMP binding (cAMP relay). The data presented in this article suggest a role for coronin A immediately downstream of the receptor(s) activated by secreted factors and upstream of the activation of the cAMP relay. Coronin A could also mediate the expression of the developmental genes via a cAMP-independent pathway, which would then be necessary for the amplification of the cAMP relay and thus aggregation.
DISCUSSION
Survival of complex organisms relies on the ability to adapt to a changing environment. For the soil-dwelling social amoeba D. discoideum, when a threat is posed by nutrient depletion, the cells enter a developmental program in order to form aggregates that develop into fruiting body–carrying spores. This developmental program involves, first, the sensing of the cell density in order to ensure that cell differentiation can be initiated when nutrient availability decreases, and, second, the induction of genes that allow an amplification of cAMP signaling via a so-called cAMP relay (Weijer, 1999; Kimmel and Firtel, 2004), followed by cAMP-mediated chemotaxis. The results presented here indicate that coronin A plays an essential role in the initiation of this developmental program by being involved in the response to factors secreted during the transition from growth to development of the cells (Figure 9). Of importance, although Dictyostelium cells lacking coronin A were unable to initiate development, once stimulated through external sources of cAMP, coronin A–deficient cells were fully competent to undergo chemotaxis and aggregation, suggesting that coronin A is dispensable for the cAMP relay, as well as for the processes that occur downstream of the cAMP relay. We therefore conclude that coronin A is important for initiation of the cAMP relay by mediating signal transduction in response to cell density and nutrient deprivation (Figure 9).
Coronins are highly conserved molecules expressed in all eukaryotes, with a recent analysis listing 723 coronin molecules from 358 species (Eckert et al., 2011). A core structure of all coronin molecules is the N-terminal WD repeat region, which folds into a seven-bladed β-propeller domain (Gatfield et al., 2005; Appleton et al., 2006; Pieters et al., 2013) and is fused to a coiled-coil region via a unique domain. D. discoideum expresses only two coronins: coronin A, consisting of a WD repeat domain, and coronin B, which essentially represents a “tandem” WD repeat region separated by a unique domain but lacks a coiled coil (Clemen et al., 2008). In contrast to coronin A–deficient cells, disruption of the coronin B gene displayed no significant alterations in motility and polarization and caused an accelerated development at early time points (Shina et al., 2011), suggesting that coronin B is dispensable for chemotaxis, which rules out a redundant role of both coronins during development.
Our results are in accordance with previous studies showing defective fruiting body formation and a defect in the directional migration of cells lacking coronin A toward a cAMP gradient. Indeed, coronin A–deficient cells were shown to be defective in motility during the vegetative state as well as for cAMP chemotaxis (de Hostos et al., 1993; Shina et al., 2011). Of importance, as we show here, when cells lacking coronin A were pulsed with cAMP, this rescued chemotaxis. Of interest, velocity was only partially rescued by cAMP pulsing of the coronin A mutants, suggesting that the velocity defect of coronin A–deficient cells may be independent of the cAMP pathway. These findings likely indicate impaired ability of the mutant cells to reach a chemotaxis-competent state.
Together the results presented here suggest a signaling role for coronin A in early development, independent of a role for coronin A in the direct modulation of F-actin. First, the defect in the cAMP-elicited response is specific for the starvation-induced development, as shown by the normal level of cAMP production in vegetative coronin A–deficient cells after folic acid stimulation. Second, folic acid chemotaxis is normal in vegetative coronin A–deficient cells, arguing against a general chemotactic defect. Third, coronin A–deficient cells can up-regulate early gene expression and undergo chemotaxis and development when exogenous cAMP pulses are applied. Fourth, incubation of the mutant cells with the membrane-permeable cAMP analogue 8-Br-cAMP restored aggregation.
Related to this, recent work showed that cells lacking cortexilin I and II, which are involved in modulation of F-actin and also showing impaired cAMP-mediated signaling, including defective development gene expression, defective chemotaxis, and aggregation, cannot be rescued by cAMP pulsing (Shu et al., 2012). This is in marked contrast to the ability of cells lacking coronin A to fully restore cAMP signaling through cAMP pulses. Furthermore, the fact that cells treated with latrunculin A at a concentration that impairs motility and aggregation still are able to up-regulate early developmental gene expression supports the notion of a role for coronin A in signaling rather than in the modulation of F-actin to explain the defect of coronin A–lacking cells to enter into development.
The fact that, in vegetative cells, coronin A is not required for folate-mediated chemotaxis and cAMP production, both of which depend on an intact adenylate cyclase, suggests a proper functioning of the molecules involved in cAMP production, including a proper expression, localization, and transport of adenylate cyclase. Furthermore, the restoration of chemotaxis and aggregation by exogenous pulses of cAMP in the absence of coronin A also suggests that coronin A is dispensable for these processes once they have been initiated and suggests that both cAMP receptor functionality and all processes downstream of cAMP production, including the activity of Ras, target of rapamycin complex, and phosphoinositide 3-kinase, are functional in the absence of coronin A. These results are therefore consistent with a function for coronin A at an early stage in the initiation of the developmental response.
Of interest, the phenotype of cells lacking coronin A is reminiscent of the phenotypes observed for other mutants lacking molecules involved in the regulation of the growth-to-development transition. Indeed, a similar aggregation deficiency was described for cells lacking adenylate cyclase activity (Wang and Kuspa, 1997) or a component of the YakA/PKA pathway keaA (Mantzouranis et al., 2010).
Exactly how coronin A is involved in the initiation of Dictyostelium development remains to be established, but our finding that cells lacking coronin A fail to respond to conditioned medium from wild-type cells strongly suggests a role for coronin A in the signaling pathway involved in cell density and/or nutrient sensing. Among the secreted signaling molecules involved in the growth-to-development transition, PSF accumulates in the medium in proportion to cell density and regulates discoidin expression in a dose-dependent manner (Rathi and Clarke, 1992). The fact that induction of discoidin expression in growing cells lacking coronin A was similar to that in wild-type cells indicates that coronin A is required at the transition phase of growth to starvation and not during the prestarvation phase. It also indicates that receptor(s) to PSF are properly exported at the plasma membrane and activated by PSF, showing that general receptor trafficking is unaltered in the absence of coronin A, which is also suggested by a similar pattern of plasma membrane molecules as analyzed by cell surface biotinylation (Supplemental Figure S8). Another early secreted factor active at the transition from growth to development is the glycosylated protein called CMF, which is rapidly released when the cells are starving (Jain et al., 1992; Jain and Gomer, 1994; Yuen et al., 1995; Brazill et al., 1998; Bakthavatsalam et al., 2007; Ray et al., 2011). This factor is essential to induce early differentiation of the cells into development-competent cells (Gomer et al., 1991). Whether coronin A functions downstream of CMF or any other factor present in the conditioned medium remains to be elucidated. Of note, the phenotype of coronin A–deficient cells phenocopies that of null mutants of other CMF signaling components such as CMF or one of its receptors, CMFR1 (Jain and Gomer, 1994). Of interest, CMFR1 is expressed at the cell surface of cells lacking coronin A, indicating normal CMFR1 trafficking and in accordance with a potential role of coronin A downstream of the CMF receptor.
In addition, cells deficient in the G protein–coupled receptor RpkA, which is involved in CMF signal transduction, show similar phenotypes to coronin A–deficient cells, with a density-dependent defect in early and late development, a cAMP chemotaxis defect, an altered expression of developmentally regulated gene expression, as well as a normal prestarvation response (Bakthavatsalam et al., 2007). Whether coronin A functions downstream of the RpkA receptor or in a parallel pathway that senses conditioned medium factors remains to be established.
Coronin molecules are expressed in all eukaryotic organisms and have been implicated in regulation of a wide variety of cellular activities. However, the precise function for most coronins is largely unknown. The work presented here suggests that in the lower eukaryote D. discoideum, coronin A is an essential component in the signal transduction cascade initiating multicellular development that is crucial for long-term survival. It is interesting to note that mammalian coronin 1 has also been implicated in signal transduction (Jayachandran et al., 2007; Mueller et al., 2008; Pieters et al., 2013), and perhaps the signaling role of the Dictyostelium homologue reflects a conserved role shared by a common ancestor.
MATERIALS AND METHODS
Growth and development conditions
DH1-10 (Cornillon et al., 2000) and AX2 wild-type, HG1569, HG1570 (coronin A knockout [KO]), gpbA− (GβKO), cmfB− (CMFR1KO), and gpaD– (Gα2KO) D. discoideum cells were acquired from http://dictybase.org/. Vegetative cells of D. discoideum were grown in shaking culture at 160 RPM, 22°C, in HL5 medium (Watts and Ashworth, 1970) and cultured from (0.2–2) × 106 cells/ml. Development was initiated by harvesting cells and washing twice in BSS (10 mM NaCl, 10 mM KCl, 2.5 mM CaCl2) unless another buffer is specified. Cells were then resuspended to 1 × 107 cells/ml in BSS. The cultures were shaken (160 RPM) at 22°C for the indicated time with or without externally applied pulses of a final concentration of 50 nM cAMP delivered every 6.5 min by an IPC Microprocessor–controlled dispensing pump (ISMATEC, Wertheim, Germany).
Reagents and antibodies
Anti–coronin A antibodies were raised in rabbits (Eurogentec, SA, Seraing, Belgium) against a synthesized peptide corresponding to a unique C-terminal portion of the coronin A amino acid sequence (GGFVKKASAVEFKPV, residues 388–402) using a 28-d immunization protocol. The antibody was purified from the serum of immunized rabbits by affinity chromatography using the same peptide as affinity matrix. Anti-Gα2 was raised in rabbits (Thermo Scientific, Waltham, MA) against a synthesized peptide (CASSMEGEKTNTDINLSIEK, residues 4–24; Kumagai et al., 1989) using a 70-d immunization protocol. The mouse monoclonal antibody against CsA (123-353-1) and anti–discoidin 1 (80-52-13) developed by G. Gerisch (Max Planck Institute of Biochemistry, Martinsried, Germany) were obtained through the Developmental Studies Hybridoma Bank at the University of Iowa (Iowa City, IA) and the National Institute of Child Health and Human Development. Mouse anti-actin clone c4 was purchased from Millipore (Temecula, CA). The cyclic AMP and the 8-bromo-cAMP analogue were obtained from Sigma-Aldrich (St. Louis, MO). Latrunculin A was from Calbiochem (Billerica, MA). Antibodies against CMFR1 were a kind gift from Richard Gomer, Texas A&M University (College Station, TX) and were purified using a protein A column (HiTrap protein A HP; GE Healthcare, Uppsala, Sweden).
Generation of coronin A–deficient and complemented strains
All restriction enzymes and the T4 DNA ligase used in the cloning procedures were purchased from New England Biolabs (NEB, Ipswich, MA). Primer production and sequencing reactions were done by Microsynth (Windisch, Switzerland).
The coronin A deletion mutant was generated from the DH1-10 parental strain (Cornillon et al., 2000) via homologous recombination, using intragenic regions of coronin A flanking a blasticidin resistance cassette to generate the coronin A deletion mutant. For generation of the coronin A homologous regions, primers were used that added SacI and BamHI sites to the 5′ fragment (forward primer: 5′-ATGAGCTCCAGGTAAAACCACATCAG-3′; reverse primer: 5′-ATGGATCCTGACAAACGACTTCGTTGAC-3′) and a BamHI site to the 3′ fragment (forward primer: 5′-ATGGATCCTGCCATTCTATGATGCTGAC-3′; reverse primer: 5′-ACTAACAGTCTTTGGTTCAGCATTGGTACC-3′) of coronin A. Fragments were amplified from DH1-10 genomic DNA via PCR using the polymerase Easy A (Promega, Madison, WI) that combines proofreading activity with adenosine overhang addition, purified and ligated separately via T/A-cloning into the cloning vector T-easy (Promega). The 5′ CorA fragment was digested with SacI and BamHI and the 3′ CorA fragment with BamHI and SalI, and both fragments were sequentially ligated into the multiple cloning site of pUC19. The blasticidin resistance cassette (Bsr) was obtained by digesting pBsr50 (Puta and Zeng, 1998) with BamHI and ligated in between the two coronin A fragments. Electroporation was performed as described (Alibaud et al., 2003).
For generation of the complemented strain, coronin A was amplified by PCR using genomic DNA isolated from DH1-10 D. discoideum using polymerase Easy A (Promega). Primers were designed to amplify the full gene and to add BamHI restriction sites to either end of the coronin A ORF. Forward primer: 5′-AGAGCGGATCCATGTCTAAAGTAGTCCG-3′; reverse primer: 5′-AGAGCGGATCCTTAGTTGGTGAGTTCTTTG-3′. The resulting product was ligated into T-easy cloning vector (Promega) via T-A cloning. Ampicillin-resistant clones were isolated. We used pBIG (Egelhoff et al., 1993; from dictybase.org) as expression vector. The coronin A insert and the pBIG vector were both digested with BamHI and ligated. A Dictyostelium actin15 promoter (taken from the pTX-GFP vector sequence available on dictybase.org) synthesized by GenScript with the addition of XbaI restriction sites to either end of the product was added just upstream of the coronin A sequence. The promoter sequence and the pBIGCorA vector were both digested using XbaI and then ligated. For transfections, cells were electroporated with three exponential pulses from a GenePulser Xcell (Biorad, Hercules, CA) at a voltage of 650 V and a capacitance of 10 μF. The pulses were applied in a 0.1 mm electrocuvette at 5-s intervals at a cell density of 1 × 108 cells/ml in H-50 buffer (20 mM HEPES, pH 7, 50 mM KCl, 10 mM NaCl, 1 mM MgSO4, 5 mM NaHCO3, 1 mM NaH2PO4). Cells were then transferred in 15 ml HL-5 to a T-flask and left overnight to recover. The next day G418 was added to a concentration of 10 μg/ml to select for transfected cells.
Analysis of cell impedance
The xCELLigence system was used according to the instructions of the supplier (Roche Applied Science, Mannheim, Germany), using E-plates 96 as disposable devices for performing cell-based assays. The E-plate incorporates gold cell sensor arrays in the bottom, and the electronic impedance of sensor electrodes is measured to allow detection of physiological changes of the cells on the electrodes. The data are expressed in cell index (CI) units.
Immunofluorescence microscopy
For immunofluorescence microscopy, cells were seeded onto eight-well Teflon slides at a density of 10,000 cells/well. The cells were washed in BSS and fixed in ice-cold methanol for 4 min at −20°C. Fixed cells were subsequently blocked (phosphate-buffered saline [PBS], pH 7.4, 1% bovine serum albumin, 2% fetal calf serum) for 20 min. Cells were stained at room temperature with anti–coronin A primary antibody at 1:2000 in blocking solution for 1 h and washed three times for 5 min with blocking solution, followed by incubation in anti-rabbit Alexa Fluor 488 (1:1000; Invitrogen, Carlsbad, CA) and phalloidin 568 for 1 h. Finally, the slides were washed again with blocking solution, incubated in Prolong Gold antifade with 4′,6-diamidino-2-phenylindole (Invitrogen) to prevent bleaching and stain the DNA, and sealed with a coverslip and nail polish. All images were acquired with a Zeiss Axioplan 2 fluorescence microscope at an exposure time of 600 ms for the red channel and 20 ms for the blue channel.
cAMP assay
Vegetative or 6-h-starving cells (5-h cAMP pulsed and unpulsed) were washed in BSS and resuspended in a buffer mix as described by the manufacturer (Cisbio Bioassays, Codolet, France). Then 50,000 cells were seeded in a 384-well plate (Greiner Bio-One, Essen, Germany), and 10 mM 1.4-dithiothreitol and 100 μM 3-isobutyl-1-methylxanthine (final concentration) were then added to the wells to inhibit the secreted and cytosolic phosphodiesterases. After 30-min incubation the cells were lysed in the wells according to the manufacturer's protocol (the one-step procedure) and the fluorescence resonance energy transfer fluorescence measured on a Tecan spectrophotometer (Tecan Group, Männedorf, Switzerland) according to the homogeneous time-resolved fluorescence (HTRF) protocol.
Cell aggregation, chemotaxis, and development
To examine cell aggregation, log-phase-growing cells were washed, resuspended in BSS, and plated at (5, 10, 20, or 40) × 104 cells/cm2 in a 24-well plate (Multiwell; Falcon) and allowed to adhere for 1 h. Aggregation was visualized by time-lapse microscopy, taking images every 135 s with a Zeiss Cellobserver microscope (Carl Zeiss, Jena, Germany) equipped with 5× objective and an electron-multiplying charge-coupled device camera (Evolve) automated by AxioVision software (Carl Zeiss). For aggregation of cells treated with Lat A, cells were plated at 20 × 104 cells/cm2 in an Ibidi eight-chamber slide and allowed to adhere for 30 min before starting movies. The drug was added 20 min after initiation of image acquisition. The microscope used was as described, except that a 10× objective was used. For cell polarization analysis, vegetative cells were washed, resuspended in BSS, and seeded in a 35-mm Petri dish at the indicated densities.
To evaluate chemotaxis of the cells in response to cAMP, a micropipette assay of cAMP-induced Dictyostelium chemotaxis was performed as described (Parent et al., 1998). Starved (cAMP pulsed or unpulsed) cells were resuspended in BSS on a 35-mm Petri dish, and a chemoattractant gradient was generated with a microinjector (Transjector 5246) attached to a micropipette (Femtotips II; Eppendorf, Hamburg, Germany) filled with 1 μM cAMP. Chemotactic migration was continuously recorded at intervals of 20 s with a 10× objective and AxioVision software. Tracking of cell migration was performed using the manual tracking tool in ImageJ (http://rsb.info.nih.gov/ij/plugins/index.html), and the velocity and directionality parameters were determined using the chemotaxis and migration tool (www.ibidi.com/applications/ap_chemo.html) plug-ins for ImageJ.
For the folic acid chemotaxis experiment, cells were cultured in the presence of Escherichia coli bacteria in DB buffer (5 mM Na2HPO4, 5 mM KH2PO4, 2 mM MgCl2, 0.2 mM CaCl2) for 40 h before washing and resuspension in DB. Cells were then seeded in a 35-mm Petri dish at a density of 1 × 104 cells/cm2, and a chemoattractant gradient was generated as described above with 250 μM folic acid solution (97% dehydrate; Sigma-Aldrich) from a 10 mM stock solution in H2O.
Conditioned medium assay
Conditioned medium was freshly prepared as described (Gomer et al., 1991). Log-phase wild-type or coronin A–deficient cells were collected from suspension cultures and washed threefold in PBM (0.02 M potassium phosphate, 10 μM CaCl2, and l mM MgCl2, pH 6.1). The cells were then resuspended in PBM at a density of 10 × l06 cells/ml and shaken for 20 h at 110 RPM/22°C. The conditioned medium was collected after centrifugation at 400 × g for 3 min and then clarified by centrifugation at 8000 × g for 15 min at 4°C. The conditioned medium was then filtered through a 0.45-μm pore and diluted threefold in PBM before incubation with the cells.
Real-time PCR
For total RNA isolation, developing cells were lysed using 1 ml of ice-cold Qiazol Lysis Reagent (79306; Qiagen) for 1 × 107 cells at 0, 4, 8, and 12 h after the initiation of the development in BSS and lysates were stored at −80°C. Upon thawing, 700 μl of lysate was mixed with 700 μl ethanol. Tubes were homogenized by shaking and spinning. The supernatant was loaded on a Zymo-Spin IIC column, and further steps were realized according to the protocol provided by the Direct-zol RNA MiniPrep Kit (R2050; Zymo Research, Irvine, CA) including DNase treatment. Elution was performed using 50 μl of RNase-free water (AM9937; Ambion, Life Technologies, Carlsbad, CA). We denaturated 700 ng of total RNA for 8 min at 70°C, followed by incubation on ice in the presence of 140 ng of Random Decamers (AM5722G; Ambion) and 47 ng of Anchored Oligo(dT)20 Primer (12577-011; Invitrogen). The following values are final concentrations or quantities in the reverse transcriptase reactions: cDNA was generated in a 20-μl reaction using 14 U of Super RT Enzyme (HT-RT01a; HT Biotechnology, Cambridge, UK) in the presence of buffer provided, 1 mM dNTPs (HT-SB23; HT Biotechnology), 0.6 U of human placental ribonuclease inhibitor (HT-RI01a; HT Biotechnology). A control lacking ribonuclease inhibitor and reverse transcriptase (–RT) was included for each sample to ensure the absence of genomic DNA amplification during PCR. On completion of the reaction, the volume was adjusted to 80 μl, which was considered as the starting point (not diluted samples) for the standard curve performed by real-time PCR to assess PCR efficiency.
The following oligonucleotides were obtained from Microsynth. Sequences are as follows. For carA (Mantzouranis et al., 2010), forward primer, 5′-ATGTTTCCACCAGCACTCAA-3′, and reverse primer, 5′-AAATGTGACAGATGCCCAAA-3′; aca (Teo et al., 2010), forward primer, 5′-CATTCTAGAGGCGGTATTGGC-3′, and reverse primer, 5′ -GGAGAAAATGTCTGATTTCGCTT-3′; csA (Lucas et al., 2009), forward primer, 5′-GAAAGCTGGTATCTCAAATGTTGTCAC-3′, and reverse primer, 5′-GGAATCTGGAGCACAAACTATATCAGTAG-3′; dscA (Mantzouranis et al., 2010), forward primer, 5′-GGTGCTGCTGTTACTGGTGT-3′, and reverse primer, 5′- GGTGGATAGCAATTGAACGA-3′; gpda (Lucas et al., 2009), forward primer, 5′-GGTTGTCCCAATTGGTATTAATGG-3′, and reverse primer, 5′-CCGTGGGTTGAATCATATTTGAAC-3′; and ig7 (Teo et al., 2010), forward primer, 5′-TCCAAGAGGAAGAGGAGAACTGC-3′, and reverse primer, 5′-TGGGGAGGTCGTTACACCATTC-3′.
PCR was performed using the SyBr Fast Kit (KK4602; Kapa Biosystems, Woburn, MA) according to standard recommendations in a 17.5-μl final volume of reaction, using 5 μl of cDNA template (diluted four times after reverse transcription). Final concentration of each primer was 400 nM. Real-time PCR was performed in a Corbett Research RG-6000A instrument in 100-μl reaction tubes running under Rotor-Gene software, version 1.7. Wavelengths of source and detection were set at 470 and 510 nm, respectively. Gain was set at 8.33. The PCR program was set as follows: 95°C, 60 s, 45× (95°C, 3 s; 60°C, 8 s; 72°C, 8 s), followed by a melting curve analysis (61 to 95°C, rising by 0.7°C each step of 3 s) to attest to amplification specificity. Expression levels were normalized to gpdA and ig7 using Qbase PLUS2 software, including PCR efficiency correction determined by cDNA dilution series.
Cell surface biotinylation
Cells (30 × 106) were cooled to 4°C, pelleted, and washed twice with 10 ml of ice-cold phosphate buffer, 120 mM sorbitol, pH 6.0. For all the washes, cells were centrifuged at 1600 rpm for 4 min at 4°C. The cells were then resuspended in 1.5 ml of phosphate-buffer, 120 mM sorbitol, pH 8.0 (SB sorbitol; see Cornillon et al., 1994), and 500 μl of biotinylation reagent was added (1 mg sulfo-NHS-SS-biotin dissolved in 500 μl SB sorbitol, pH 8.0; see also Froquet et al., 2012). After 10 min of incubation, cells were centrifuged and resuspended in 10 ml of PBS containing 100 mM glycine and incubated for 5 min on ice. Four washes with 10 ml of SB sorbitol were performed to eliminate excess of free biotin. Cells were then incubated for 15 min in 900 μl of lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) plus protease inhibitors (10 μg/ml aprotinin, 10 μg/ml leupeptin, 18 μg/ml phenylmethylsulfonyl fluoride, 1.8 mg/ml iodoacetamide). The lysate was centrifuged for 5 min at 9300 × g, the pellet was discarded, and the supernatant was added to 100 μl of NeutrAvidin Ultralink resin beads (Thermo Scientific, Waltham, MA), incubated overnight, and centrifuged for 1 min at 10,000 rpm. Beads were washed four times in 1 ml of lysis buffer, and 800 μl 6 M urea was added, followed by 15 min of incubation. Beads were washed three more times with 1 ml of the lysis buffer. Biotinylated proteins were eluted by adding 50 μl of 4× sample buffer plus 12% β-mercaptoethanol and incubated for 15 min at room temperature and 5 min at 60°C. Supernatant, 20 μl, was loaded on a 10% SDS–polyacrylamide gel and processed for silver staining (Switzer et al., 1979) or for Western blot analysis using anti-CMFR1 antibodies.
SDS–PAGE and immunoblotting
For Gα2 and CMFR1 expression analysis, membrane fractions were obtained from vegetative or starving cells resuspended in homogenizing buffer (20 mM Tris-HCl, pH 7.5, 200 mM sucrose, 5 mM EDTA) supplemented with protease inhibitors (Roche cocktail). Cells were then immediately pushed through a 5-μm-pore filter (Millex; Millipore) and centrifuged for 15 min at 12,000 × g. The pellet was washed once with the homogenizing buffer and resuspended in SDS sample buffer.
For discoidin expression analysis, cells were cultured in the presence of three different densities of E. coli bacteria in PBM buffer or conditioned medium for 40 h. Cells were washed to remove bacteria when they reached the density of 1 × 106 cells/ml or as indicated in the figures. Cells were then lysed in Laemmli (1970) sample buffer.
Proteins were separated on 10% SDS–polyacrylamide gels and transferred to nitrocellulose membranes using a semidry system (Bio-Rad). Antibodies were diluted in 5% milk, PBS-Tween-20 at 1:15,000 for anti–coronin A, 1:10,000 for anti-actin, 1:500 for anti-Gα2, 1:1000 for anti-CMFR1, and 1:200 for anti-CsA and anti-discoidin. The membrane was soaked in this primary antibody solution either at room temperature for 2 h or overnight at 4°C. Secondary antibodies were horseradish peroxidase (HRP)–coupled anti-rabbit or anti-mouse antibodies purchased from Southern Biotech (Birmingham, AL) and used at dilutions of 1:20,000 and 1:10,000, respectively. Membranes were soaked in secondary antibody solution for 1 h and then washed in 5% milk, PBS-Tween-20, and PBS-Tween-20 consecutively. SuperSignal PicoWest chemiluminescence substrate (Thermo) was used as a substrate for HRP.
Supplementary Material
Acknowledgments
We thank the Dictyostelium Stock Center for strains and reagents, Richard Gomer for antibodies, Richard Gomer and Derrik Brazill for discussions, Philippe Demougin for the quantitative PCR analysis, and members of the laboratory for discussions. This study was financed by grants from the Swiss National Science Foundation (J.P. and P.C.) and the Canton of Basel. A.F.V. is a recipient of a Long Term EMBO Fellowship.
Abbreviations used:
- ACA
adenylyl cyclase A
- BSS
Bonner's salt solution
- cAMP
cyclic adenosine monophosphate
- cAR1
cAMP receptor 1
- CM
conditioned medium
- CMF
conditioned medium factor
- CMFR1
conditioned medium factor receptor 1
- CorA
coronin A
- CsA
contact site A
- dscA
discoidin A
- HRP
horseradish peroxidase
- Lat A
latrunculin A
- PBM
phosphate buffer medium
- PBS
phosphate-buffered saline
- PSF
prestarvation factor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-04-0219) on January 8, 2014.
*These authors contributed equally.
REFERENCES
- Alibaud L, Cosson P, Benghezal M. Dictyostelium discoideum transformation by oscillating electric field electroporation. Biotechniques. 2003;35:78–80. doi: 10.2144/03351st03. [DOI] [PubMed] [Google Scholar]
- Appleton BA, Wu P, Wiesmann C. The crystal structure of murine coronin-1: a regulator of actin cytoskeletal dynamics in lymphocytes. Structure. 2006;14:87–96. doi: 10.1016/j.str.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Aubry L, Firtel R. Integration of signaling networks that regulate Dictyostelium differentiation. Annu Rev Cell Dev Biol. 1999;15:469–517. doi: 10.1146/annurev.cellbio.15.1.469. [DOI] [PubMed] [Google Scholar]
- Bakthavatsalam D, Brazill D, Gomer RH, Eichinger L, Rivero F, Noegel AA. A G protein-coupled receptor with a lipid kinase domain is involved in cell-density sensing. Curr Biol. 2007;17:892–897. doi: 10.1016/j.cub.2007.04.029. [DOI] [PubMed] [Google Scholar]
- Bonner JT. Evidence for the formation of cell aggregates by chemotaxis in the development of the slime mold Dictyostelium discoideum. J Exp Zool. 1947;106:1–26. doi: 10.1002/jez.1401060102. [DOI] [PubMed] [Google Scholar]
- Brazill DT, Lindsey DF, Bishop JD, Gomer RH. Cell density sensing mediated by a G protein-coupled receptor activating phospholipase C. J Biol Chem. 1998;273:8161–8168. doi: 10.1074/jbc.273.14.8161. [DOI] [PubMed] [Google Scholar]
- Chisholm RL, Firtel RA. Insights into morphogenesis from a simple developmental system. Nat Rev. Mol Cell Biol. 2004;5:531–541. doi: 10.1038/nrm1427. [DOI] [PubMed] [Google Scholar]
- Clarke M, Gomer RH. PSF and CMF, autocrine factors that regulate gene expression during growth and early development of Dictyostelium. Experientia. 1995;51:1124–1134. doi: 10.1007/BF01944730. [DOI] [PubMed] [Google Scholar]
- Clarke M, Yang J, Kayman SC. Analysis of the prestarvation response in growing cells of Dictyostelium discoideum. Dev Genet. 1988;9:315–326. doi: 10.1002/dvg.1020090413. [DOI] [PubMed] [Google Scholar]
- Clemen CS, Rybakin V, Eichinger L. The coronin family of proteins. Subcell Biochem. 2008;48:1–5. doi: 10.1007/978-0-387-09595-0_1. [DOI] [PubMed] [Google Scholar]
- Combaluzier B, Mueller P, Massner J, Finke D, Pieters J. Coronin 1 is essential for IgM-mediated Ca2+ mobilization in B cells but dispensable for the generation of immune responses in vivo. J Immunol. 2009;182:1954–1961. doi: 10.4049/jimmunol.0801811. [DOI] [PubMed] [Google Scholar]
- Combaluzier B, Pieters J. Chemotaxis and phagocytosis in neutrophils is independent of coronin 1. J Immunol. 2009;182:2745–2752. doi: 10.4049/jimmunol.0801812. [DOI] [PubMed] [Google Scholar]
- Cornillon S, Foa C, Davoust J, Buonavista N, Gross JD, Golstein P. Programmed cell death in Dictyostelium. J Cell Sci. 1994;107:2691–2704. doi: 10.1242/jcs.107.10.2691. [DOI] [PubMed] [Google Scholar]
- Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, Bruckert F, Cosson P. Phg1p is a nine-transmembrane protein superfamily member involved in Dictyostelium adhesion and phagocytosis. J Biol Chem. 2000;275:34287–34292. doi: 10.1074/jbc.M006725200. [DOI] [PubMed] [Google Scholar]
- Darmon M, Brachet P, Da Silva LH. Chemotactic signals induce cell differentiation in Dictyostelium discoideum. Proc Natl Acad Sci USA. 1975;72:3163–3166. doi: 10.1073/pnas.72.8.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deery WJ, Gao T, Ammann R, Gomer RH. A single cell density-sensing factor stimulates distinct signal transduction pathways through two different receptors. J Biol Chem. 2002;277:31972–31979. doi: 10.1074/jbc.M204539200. [DOI] [PubMed] [Google Scholar]
- Deery WJ, Gomer RH. A putative receptor mediating cell-density sensing in Dictyostelium. J Biol Chem. 1999;274:34476–34482. doi: 10.1074/jbc.274.48.34476. [DOI] [PubMed] [Google Scholar]
- de Hostos EL. The coronin family of actin-associated proteins. Trends Cell Biol. 1999;9:345–350. doi: 10.1016/s0962-8924(99)01620-7. [DOI] [PubMed] [Google Scholar]
- de Hostos EL, Bradtke B, Lottspeich F, Guggenheim R, Gerisch G. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein beta subunits. EMBO J. 1991;10:4097–4104. doi: 10.1002/j.1460-2075.1991.tb04986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, Rehfuess C, Bradtke B, Waddell DR, Albrecht R, Murphy J, Gerisch G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P. Dictyostelium discoideum: a model system for cell-cell interactions in development. Science. 1989;245:1054–1058. doi: 10.1126/science.2672337. [DOI] [PubMed] [Google Scholar]
- Devreotes PN. The effect of folic acid on cAMP-elicited cAMP production in Dictyostelium discoideum. Dev Biol. 1983;95:154–162. doi: 10.1016/0012-1606(83)90014-3. [DOI] [PubMed] [Google Scholar]
- De Wit RJ, Bulgakov R, Rinke de Wit TF, Konijn TM. Developmental regulation of the pathways of folate-receptor-mediated stimulation of cAMP and cGMP synthesis in Dictyostelium discoideum. Differentiation. 1986;32:192–199. doi: 10.1111/j.1432-0436.1986.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Eckert C, Hammesfahr B, Kollmar M. A holistic phylogeny of the coronin gene family reveals an ancient origin of the tandem-coronin, defines a new subfamily, and predicts protein function. BMC Evol Biol. 2011;11:268. doi: 10.1186/1471-2148-11-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff TT, Lee RJ, Spudich JA. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- Faix J, Gerisch G, Noegel AA. Overexpression of the csA cell adhesion molecule under its own cAMP-regulated promoter impairs morphogenesis in Dictyostelium. J Cell Sci. 1992;102(Pt 2):203–214. doi: 10.1242/jcs.102.2.203. [DOI] [PubMed] [Google Scholar]
- Firtel RA, Chung CY. The molecular genetics of chemotaxis: sensing and responding to chemoattractant gradients. BioEssays. 2000;22:603–615. doi: 10.1002/1521-1878(200007)22:7<603::AID-BIES3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Froquet R, le Coadic M, Perrin J, Cherix N, Cornillon S, Cosson P. TM9/Phg1 and SadA proteins control surface expression and stability of SibA adhesion molecules in Dictyostelium. Mol Biol Cell. 2012;23:679–686. doi: 10.1091/mbc.E11-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield J, Albrecht I, Zanolari B, Steinmetz MO, Pieters J. Association of the leukocyte plasma membrane with the actin cytoskeleton through coiled coil-mediated trimeric coronin 1 molecules. Mol Biol Cell. 2005;16:2786–2798. doi: 10.1091/mbc.E05-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch G. Chemotaxis in Dictyostelium. Annu Rev Physiol. 1982;44:535–552. doi: 10.1146/annurev.ph.44.030182.002535. [DOI] [PubMed] [Google Scholar]
- Gomer RH, Yuen IS, Firtel RA. A secreted 80 × 10(3) Mr protein mediates sensing of cell density and the onset of development in Dictyostelium. Development. 1991;112:269–278. doi: 10.1242/dev.112.1.269. [DOI] [PubMed] [Google Scholar]
- Gregor T, Fujimoto K, Masaki N, Sawai S. The onset of collective behavior in social amoebae. Science. 2010;328:1021–1025. doi: 10.1126/science.1183415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JD, Peacey MJ, Trevan DJ. Signal emission and signal propagation during early aggregation in Dictyostelium discoideum. J Cell Sci. 1976;22:645–656. doi: 10.1242/jcs.22.3.645. [DOI] [PubMed] [Google Scholar]
- Insall RH, Soede RD, Schaap P, Devreotes PN. Two cAMP receptors activate common signaling pathways in Dictyostelium. Mol Biol Cell. 1994;5:703–711. doi: 10.1091/mbc.5.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranfar N, Fuller D, Loomis WF. Genome-wide expression analyses of gene regulation during early development of Dictyostelium discoideum. Eukaryot Cell. 2003;2:664–670. doi: 10.1128/EC.2.4.664-670.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Gomer RH. A developmentally regulated cell surface receptor for a density-sensing factor in Dictyostelium. J Biol Chem. 1994;269:9128–9136. [PubMed] [Google Scholar]
- Jain R, Yuen IS, Taphouse CR, Gomer RH. A density-sensing factor controls development in Dictyostelium. Genes Dev. 1992;6:390–400. doi: 10.1101/gad.6.3.390. [DOI] [PubMed] [Google Scholar]
- Jayachandran R, Gatfield J, Massner J, Albrecht I, Zanolari B, Pieters J. RNA interference in J774 macrophages reveals a role for coronin 1 in mycobacterial trafficking but not in actin-dependent processes. Mol Biol Cell. 2008;19:1241–1251. doi: 10.1091/mbc.E07-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran R, Sundaramurthy V, Combaluzier B, Mueller P, Korf H, Huygen K, Miyazaki T, Albrecht I, Massner J, Pieters J. Survival of mycobacteria in macrophages is mediated by coronin 1-dependent activation of calcineurin. Cell. 2007;130:37–50. doi: 10.1016/j.cell.2007.04.043. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Gundersen R, Hereld D, Pitt GS, Tugendreich S, Saxe CL, Kimmel AR, Devreotes PN. G-protein-linked signaling pathways mediate development in Dictyostelium. Cold Spring Harbor Symp Quant Biol. 1992;57:169–176. doi: 10.1101/sqb.1992.057.01.022. [DOI] [PubMed] [Google Scholar]
- Kay RR. cAMP and spore differentiation in Dictyostelium discoideum. Proc Natl Acad Sci USA. 1982;79:3228–3231. doi: 10.1073/pnas.79.10.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel AR, Firtel RA. cAMP signal transduction pathways regulating development of Dictyostelium discoideum. Curr Opin Genet Dev. 1991;1:383–390. doi: 10.1016/s0959-437x(05)80304-1. [DOI] [PubMed] [Google Scholar]
- Kimmel AR, Firtel RA. Breaking symmetries: regulation of Dictyostelium development through chemoattractant and morphogen signal-response. Curr Opin Genet Dev. 2004;14:540–549. doi: 10.1016/j.gde.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kriebel PW, Barr VA, Rericha EC, Zhang G, Parent CA. Collective cell migration requires vesicular trafficking for chemoattractant delivery at the trailing edge. J Cell Biol. 2008;183:949–961. doi: 10.1083/jcb.200808105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Pupillo M, Gundersen R, Miake-Lye R, Devreotes PN, Firtel RA. Regulation and function of G alpha protein subunits in Dictyostelium. Cell. 1989;57:265–275. doi: 10.1016/0092-8674(89)90964-1. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lim CJ, Zawadzki KA, Khosla M, Secko DM, Spiegelman GB, Weeks G. Loss of the Dictyostelium RasC protein alters vegetative cell size, motility and endocytosis. Exp Cell Res. 2005;306:47–55. doi: 10.1016/j.yexcr.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Lucas J, Bilzer A, Moll L, Zundorf I, Dingermann T, Eichinger L, Siol O, Winckler T. The carboxy-terminal domain of Dictyostelium C-module-binding factor is an independent gene regulatory entity. PLoS One. 2009;4:e5012. doi: 10.1371/journal.pone.0005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Firtel RA. Activation of the mitogen-activated protein kinase ERK2 by the chemoattractant folic acid in Dictyostelium. J Biol Chem. 1997;272:23690–23695. doi: 10.1074/jbc.272.38.23690. [DOI] [PubMed] [Google Scholar]
- Manahan CL, Iglesias PA, Long Y, Devreotes PN. Chemoattractant signaling in Dictyostelium discoideum. Annu Rev Cell Dev Biol. 2004;20:223–253. doi: 10.1146/annurev.cellbio.20.011303.132633. [DOI] [PubMed] [Google Scholar]
- Maniak M, Rauchenberger R, Albrecht R, Murphy J, Gerisch G. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell. 1995;83:915–924. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- Mann SK, Firtel RA. Cyclic AMP regulation of early gene expression in Dictyostelium discoideum: mediation via the cell surface cyclic AMP receptor. Mol Cell Biol. 1987;7:458–469. doi: 10.1128/mcb.7.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SK, Firtel RA. A developmentally regulated, putative serine/threonine protein kinase is essential for development in Dictyostelium. Mech Dev. 1991;35:89–101. doi: 10.1016/0925-4773(91)90060-j. [DOI] [PubMed] [Google Scholar]
- Mantzouranis L, Bagattini R, Souza GM. KeaA, a Dictyostelium Kelch-domain protein that regulates the response to stress and development. BMC Dev Biol. 2010;10:79. doi: 10.1186/1471-213X-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P, Liu X, Pieters J. Migration and homeostasis of naive T cells depends on coronin 1-mediated prosurvival signals and not on coronin 1-dependent filamentous actin modulation. J Immunol. 2011;186:4039–4050. doi: 10.4049/jimmunol.1003352. [DOI] [PubMed] [Google Scholar]
- Mueller P, et al. Regulation of T cell survival through coronin-1-mediated generation of inositol-1,4,5-trisphosphate and calcium mobilization after T cell receptor triggering. Nat Immunol. 2008;9:424–431. doi: 10.1038/ni1570. [DOI] [PubMed] [Google Scholar]
- Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. Molecular genetics of signal transduction in Dictyostelium. Annu Rev Biochem. 1996;65:411–440. doi: 10.1146/annurev.bi.65.070196.002211. [DOI] [PubMed] [Google Scholar]
- Pieters J, Muller P, Jayachandran R. On guard: coronin proteins in innate and adaptive immunity. Nat Rev Immunol. 2013;13:510–518. doi: 10.1038/nri3465. [DOI] [PubMed] [Google Scholar]
- Pitt GS, Milona N, Borleis J, Lin KC, Reed RR, Devreotes PN. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell. 1992;69:305–315. doi: 10.1016/0092-8674(92)90411-5. [DOI] [PubMed] [Google Scholar]
- Puta F, Zeng C. Blasticidin resistance cassette in symmetrical polylinkers for insertional inactivation of genes in Dictyostelium. Folia Biol Prague. 1998;44:185–188. [PubMed] [Google Scholar]
- Rathi A, Clarke M. Expression of early developmental genes in Dictyostelium discoideum is initiated during exponential growth by an autocrine-dependent mechanism. Mech Dev. 1992;36:173–182. doi: 10.1016/0925-4773(92)90068-u. [DOI] [PubMed] [Google Scholar]
- Rathi A, Kayman SC, Clarke M. Induction of gene expression in Dictyostelium by prestarvation factor, a factor secreted by growing cells. Dev Genet. 1991;12:82–87. doi: 10.1002/dvg.1020120115. [DOI] [PubMed] [Google Scholar]
- Ray S, Chen Y, Ayoung J, Hanna R, Brazill D. Phospholipase D controls Dictyostelium development by regulating G protein signaling. Cell Signal. 2011;23:335–343. doi: 10.1016/j.cellsig.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BB, Barclay SL. Conditions that alter intracellular cAMP levels affect expression of the cAMP phosphodiesterase gene in Dictyostelium. Proc Natl Acad Sci USA. 1990;87:4746–4750. doi: 10.1073/pnas.87.12.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer E, Tarantola M, Polo E, Westendorf C, Oikawa N, Bodenschatz E, Geil B, Janshoff A. Chemotaxis of Dictyostelium discoideum: collective oscillation of cellular contacts. PLoS One. 2013;8:e54172. doi: 10.1371/journal.pone.0054172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer E, Westendorf C, Bodenschatz E, Beta C, Geil B, Janshoff A. Shape oscillations of Dictyostelium discoideum cells on ultramicroelectrodes monitored by impedance analysis. Small. 2011;7:723–726. doi: 10.1002/smll.201001955. [DOI] [PubMed] [Google Scholar]
- Shina MC, Muller-Taubenberger A, Unal C, Schleicher M, Steinert M, Eichinger L, Müller R, Blau-Wasser R, Glöckner G, Noegel AA. Redundant and unique roles of coronin proteins in Dictyostelium. Cell Mol Life Sci. 2011;68:303–313. doi: 10.1007/s00018-010-0455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S, Liu X, Kriebel PW, Daniels MP, Korn ED. Actin cross-linking proteins cortexillin I and II are required for cAMP signaling during Dictyostelium chemotaxis and development. Mol Biol Cell. 2012;23:390–400. doi: 10.1091/mbc.E11-09-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer RC, 3rd, Merril CR, Shifrin S. A highly sensitive silver stain for detecting proteins and peptides in polyacrylamide gels. Anal Biochem. 1979;98:231–237. doi: 10.1016/0003-2697(79)90732-2. [DOI] [PubMed] [Google Scholar]
- Teo R, Lewis KJ, Forde JE, Ryves WJ, Reddy JV, Rogers BJ, Harwood AJ. Glycogen synthase kinase-3 is required for efficient Dictyostelium chemotaxis. Mol Biol Cell. 2010;21:2788–2796. doi: 10.1091/mbc.E09-10-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJ, Kien E. Binding of cAMP derivatives to Dictyostelium discoideum cells. Activation mechanism of the cell surface cAMP receptor. J Biol Chem. 1983;258:9636–9642. [PubMed] [Google Scholar]
- Van Haastert PJ, Van Der Meer RC, Konijn TM. Evidence that the rate of association of adenosine 3’,5’-monophosphate to its chemotactic receptor induces phosphodiesterase activity in Dictyostelium discoideum. J Bacteriol. 1981;147:170–175. doi: 10.1128/jb.147.1.170-175.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Kuspa A. Dictyostelium development in the absence of cAMP. Science. 1997;277:251–254. doi: 10.1126/science.277.5323.251. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Ashworth JM. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem J. 1970;119:171–174. doi: 10.1042/bj1190171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijer CJ. Morphogenetic cell movement in Dictyostelium. Semin Cell Dev Biol. 1999;10:609–619. doi: 10.1006/scdb.1999.0344. [DOI] [PubMed] [Google Scholar]
- Yuen IS, Jain R, Bishop JD, Lindsey DF, Deery WJ, Van Haastert PJ, Gomer RH. A density-sensing factor regulates signal transduction in Dictyostelium. J Cell Biol. 1995;129:1251–1262. doi: 10.1083/jcb.129.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.