Abstract
Rapid and reversible methods for altering the level of endogenous proteins are critically important for studying biological systems and developing therapeutics. Here, we describe a membrane permeable targeting peptide-based method that rapidly and reversibly knocks down endogenous proteins through chaperone-mediated autophagy in vitro and in vivo. We demonstrated the specificity, efficacy and generalizability of the method by showing efficient knockdown of various proteins including death associated protein kinase 1 (160kDa), scaffolding protein PSD-95 (95kDa) and α-synuclein (18kDa) with their respective targeting peptides in a dose-, time- and lysosomal activity-dependent manner in neuronal cultures. More significantly, we showed that when given systemically, the peptide system efficiently knocked down the targeted protein in the brain of intact rats. Our study provides a robust and convenient research tool to manipulate endogenous protein levels, and may also lead to the development of protein knockdown-based novel therapeutics for treating various human diseases.
Within the biomedical research community, techniques that regulate the level of protein expression by targeting genes at the DNA or RNA level have proven to be powerful strategies in the drive to understand protein expression and function1–3. However, due to their very nature, these techniques are restricted in terms of speed, specificity and reversibility4. For instance, these genetic methods of disrupting protein expression can take days to weeks; consequently cellular and molecular compensation may take place, thereby obscuring expected phenotypes. In addition, as these genetic manipulations result in the eradication of all mRNA splice isoforms as well as post-translationally modified versions of targeted proteins5, these methods lack specificity and are largely limited to studying context-dependent protein function. Finally, the reversibility of these genetic manipulations, including many recently developed on-and-off inducible methods, is relatively slow (being achieved on a timescale of days to weeks), and incomplete. These limitations in turn severely constrain the potential of these methods as both research tools and clinical therapeutics.
To overcome shortcomings of DNA- and mRNA-based protein manipulations, several laboratories have recently attempted to harness cellular protein degradation systems to reduce levels of proteins-of-interest (POI)6–11. However, most of these proposed systems require genetic manipulations of the proteins to facilitate their targeting and degradation via specific cellular protein degradation systems. Hence, their usefulness for knocking down unmodified native proteins in primary cells is limited. Moreover, the majority of these methods require viral delivery in vivo, thus limiting their potential for clinical use. Here we propose a simple non-viral mediated, cell membrane permeable, targeting peptide-based system to rapidly and reversibly knockdown an endogenous POI by targeting it for lysosomal degradation. We demonstrate its efficacy, specificity and broad utility by targeting three native proteins in primary neuronal cultures. In each case, degradation was significant within hours of applying the targeting peptides. Depending on the nature of the interaction between the targeting peptide and the protein target, the knockdown could be either constitutive or conditional. More significantly, we found that our peptide system can efficiently knockdown a native neuronal protein in the brain of intact animals when the targeting peptide was given systemically. Our method can be easily generalized to degrade any native cytosolic POI. It offers a simpler, faster and more easily reversible alternative to current chemical-genetic protein degradation methods, and is suitable for both in vitro and in vivo use and for translation into clinically relevant therapies.
Results
Knockdown of recombinant proteins in HEK cells
CMA is a type of autophagy specific for proteins containing a pentapeptide motif biochemically related to KFERQ12. This CMA-targeting motif (CTM) is found in all substrate proteins of CMA to date13. Previous studies with KFERQ-containing fusion proteins demonstrated that the attachment of CTM is necessary and sufficient to make non-CMA fluorescent substrates amenable to CMA, so far as it is exposed14. Thus, to utilize CMA as our degradation pathway to knockdown endogenous proteins, we designed a targeting peptide consisting of three domains (Supplementary Fig. 1a; top): a cell membrane penetrating domain (CMPD) that allows the peptide to bypass the blood-brain barrier (BBB) and plasma membrane following peripheral delivery, a protein binding domain (PBD) that specifically binds to the endogenous POI through peptide-protein interaction, and the CTM that targets the peptide-protein pair for degradation through the lysosomal proteolytic machinery. We hypothesize that the targeting peptide, when applied to a cellular environment in vitro or in vivo, will enter the cell via its CMPD, where it specifically recognizes and forms a stable complex with its target protein via the PBD, and through interactions between the CTM and CMA machinery, deliver the peptide-protein complex into the lysosomal compartments for rapid knockdown (Supplementary Fig. 1a; bottom).
Using an overexpression system, we first tested if our proposed CTM, when fused to a non-CMA substrate protein, efficiently direct the protein into the lysosome for degradation. To increase targeting efficiency, we simultaneously tagged Green Fluorescent Protein (GFP) with three CTMs identified from CMA substrate proteins: RNaseA (KFERQ)15, hsc70 (QKILD)16 and hemoglobin (QRFFE)17 (Fig. 1a). We then transiently expressed the CTM-GFP construct, along with wild type GFP construct (WT-GFP) as a control, in Human Embryonic Kidney (HEK) or COS-7 cells. Consistent with the ability of the CTM to direct CTM-GFP for lysosomal degradation, immunoblot analysis showed that CTM-GFP protein levels were decreased (Fig. 1b) and this decrease was in a time-dependent manner (Supplementary Fig. 1b). As shown in Fig. 1c, co-immunofluorescent staining revealed diffuse expression of WT-GFP (n=17) in most compartments of the cell including the nucleus. Conversely, CTM-GFP (n=16) was predominantly directed into the lysosome, as evidenced by its high degree of co-localization with lysosome marker protein LAMP-1. Quantification of co-localization demonstrated a significant increase in co-localization of GFP and LAMP-1 signals from 33.6±3.6% to 79.1±1.4% by inclusion of the CTM (mean value±s.e.m.; p<0.001; Mann-Whitney Rank Sum Test, T=408.000; cells from 3 separate transfections).
Figure 1. Chaperone-mediated autophagy (CMA) targeting motif (CTM)-directed protein degradation.
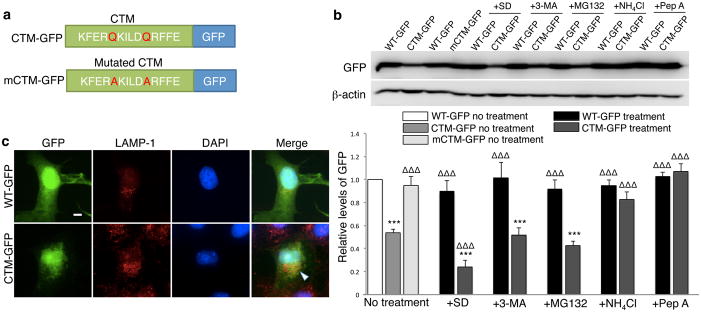
(a) Linear representation of constructs CTM-GFP and mCTM-GFP. (b) Quantification of CTM-GFP and mCTM-GFP levels relative to WT-GFP in HEK cells 24hrs after individual transfections with WT-GFP, mCTM-GFP (n=6) or CTM-GFP. Cells transfected with WT-GFP or CTM-GFP were treated without (No treatment; n=13) or with serum deprivation (+SD; n=5; to enhance CMA activity) or macroautophagy inhibitor (+3-MA; 10mM; n=5), proteasome inhibitor (+MG132; 5μM; n=6), or lysosome inhibitor ammonium chloride (+NH4Cl; 20mM; n=6) or pepstatin A (+PepA; 10μM; n=5). Top panels are representative immunoblotting of cell lysates for GFP. Membranes re-probed for β-actin were used as a loading control. Bars in bottom panel represent relative protein levels, normalized to WT-GFP (arbitrarily set as 1; white bar). Sample size (n) represents the number of independent experiments from at least 4 separate cultures. One-way ANOVA with Fischer LSD was used for comparison, F(12,73)=20.939. ***p<0.001 versus WT-GFP (white bar); ΔΔΔ p<0.001 relative to non-treated CTM-GFP levels (grey bar). Bars represent mean values ± s.e.m. Full-length blots are presented in Supplementary Figure 9. (c) Representative confocal images of co-localization of GFP with the lysosome marker LAMP-1 in COS-7 cells transfected with either wild type GFP (WT-GFP; top; n=17) or CTM-tagged GFP (CTM-GFP; bottom; n=16). Quantification of co-localization demonstrated a significant increase in co-localization of GFP and LAMP-1 signals from 33.6±3.6% (WT-GFP) to 79.1±1.4% (CTM-GFP) by inclusion of the CTM. Mann-Whitney Rank Sum Test, T=408.000 p<0.001. ***p<0.001, scale bar: 20μm.
Several lines of evidence further support that the reduction of CTM-GFP levels is a result of increased lysosomal targeting and degradation. First, CTM-GFP degradation was fully prevented by treatment with ammonium chloride, which inhibits lysosome degradation18 (NH4Cl; 20mM; 82.65%±6.28%; n=6; p<0.001) and pepstatin A (Pep A; 10μM; 106.98%±6.68%; n=5; p<0.001), which inhibits the two primary lysosomal proteases, cathepsins D and E19,20 (Fig. 1b). In contrast, inhibition of macroautophagy with 3-methyladenine (3-MA; 10mM; p=0.951) or proteasomal degradation with MG132 (MG132; 5μM; p=0.194) did not prevent reduction of CTM-GFP (Fig. 1b). Second, CMA can be activated or enhanced under conditions of stress, such as starvation induced by removal of serum in culture media21. Indeed, serum deprivation (SD) enhanced CTM-GFP degradation (SD; 23.95%±5.94% of WT-GFP level; n=5; p<0.001) (Fig. 1b). Third, mutation of the glutamine residue (Q) in the CTM of CMA substrate proteins impairs their targeting into the lysosomal lumen and degradation through CMA22. Consistently, mutating 2Qs into alanines (2A) within the CTM of CTM-GFP (mCTM-GFP) prevented its degradation (Fig. 1b; mCTM-GFP; 86.85%±6.18% of WT-GFP; n=6; p=0.075) Finally, overexpressing CTM-GFP did not significantly alter the stability of the endogenous CMA substrate glyceraldehyde 3-phosphate dehydrogenase (GAPDH)23 (Supplementary Fig. 1b), suggesting that CTM-GFP overexpression did not induce pan-activation of CMA in transfected cells. Together, this data is in line with recent evidence14 showing that the addition of CTM to a non-CMA substrate protein is sufficient to specifically channel the protein for degradation through the CMA pathway.
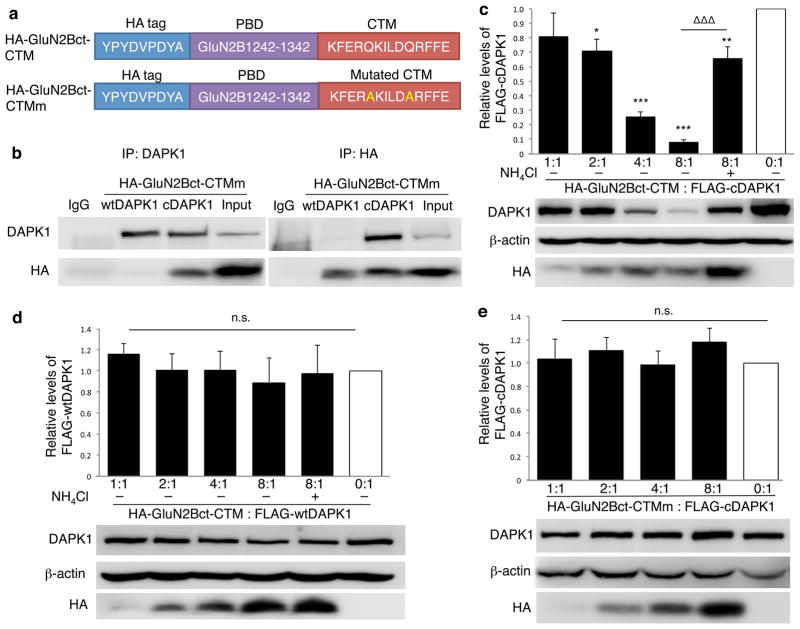
We next examined if a non-CMA substrate protein can be indirectly tagged for CMA lysosomal degradation by a targeting peptide containing the CTM and the PBD through the interaction between the protein and the targeting peptide. Death-associated protein kinase 1 (DAPK1) is a calcium-calmodulin regulated protein kinase normally inactive in the brain24. Under certain pathological conditions such as excitotoxic stimulation with N-methyl-D-aspartate (NMDA) or cerebral ischemia25, DAPK1 can be activated and recruited into NMDA receptor complex by its interaction with the C-terminal residues 1292-1304 of the GluN2B subunit (GluN2Bct1292-1304)26. Because GluN2B can only bind to the active, but not inactive form of DAPK1, we hypothesize that a targeting peptide composing of DAPK1 binding domain of the GluN2B and CTM would specifically knockdown the active DAPK1 by targeting it for CMA lysosomal degradation. Therefore, we designed two GluN2B carboxyl tail constructs bearing an HA tag, the C-terminal fragment of GluN2B (GluN2Bct1242-1342) containing the required sequence for binding to DAPK1 and either a functional CTM (HA-GluN2Bct-CTM) or the non-functional, mutated form of CTM (HA-GluN2Bct-CTMm) (Fig. 2a).
Figure 2. DAPK1 targeting peptide knocks down active DAPK1 in HEK cells.
(a) Linear representation of DAPK1-targeting peptide HA-GluN2Bct-CTM and its non-functional control peptide HA-GluN2Bct-CTMm. (b) Reciprocal co-immunoprecipitation followed by immunoblotting revealed that GluN2Bct specifically interacted with cDAPK1, but not wtDAPK1. Flag-tagged wild type (inactive) DAPK1 (wtDAPK1) or constitutively active mutant of DAPK1 (cDAPK1) was co-expressed with either HA-GluN2Bct-CTM or HA-GluN2Bct-CTMm at various ratios in HEK cells, and co-immunoprecipitation and/or immunoblotting was performed 24hrs thereafter. Anti-HA was used to detect HA-GluN2Bct-CTM and HA-GluN2Bct-CTMm, while anti-FLAG was used to detect wtDAPK and cDAPK. (c) HA-GluN2Bct-CTM specifically and dose-dependently decreased the level of cDAPK1 (n=4 independent experiments from 4 separate cell cultures and transfections; p<0.001; F(5,18)=18.27), but not wtDAPK1 (d; n=3 independent experiments from 3 separate cell cultures and transfections; p=0.933; F(5,12)=0.249). HA-GluN2Bct-CTM mediated cDAPK1 knockdown was significantly reduced by NH4Cl (c; 20mM; n=4;ΔΔΔ p<0.001, compared to HA-GluN2Bct-CTM:cDAPK1=8:1 group) and by mutational inactivation of CTM (e; HA-GluN2Bct-CTMm; p=0.785; F(4,35)=0.432, 8 independent experiments from 8 separate cell cultures and transfections). Levels of cDAPK1 or wtDAPK1 co-transfected with pcDNA3.0 vector (0:1, white bar) represent the control values arbitrarily set as 1. Membranes re-probed for β-actin were used as a loading control. One-way ANOVA was used with Fischer LSD. *p<0.05 ** p<0.01 ***p<0.001, compared with the control. Bars represent relative mean values±s.e.m. Full-length blots are presented in Supplementary Figure 9.
We first co-expressed HA-GluN2Bct-CTMm (lysosomal degradation resistant, and hence more stable than HA-GluN2Bct-CTM) with either a Flag-tagged constitutively active form of DAPK1 (cDAPK1)27 or wild-type DAPK1 (wtDAPK1) in HEK cells to assess DAPK1 activity-dependent binding between DAPK1 and GluN2B C-terminal. Reciprocal co-immunoprecipitation (Fig. 2b) revealed interaction between HA-GluN2Bct-CTMm and cDAPK1, but not wtDAPK1, confirming previous results that GluN2B and DAPK1 binding is conditionally dependent on the activation of the latter26. Co-expression of HA-GluN2Bct-CTM at various ratios efficiently decreased the levels of cDAPK1 in a dose-dependent manner assessed 24hrs after transfection (Fig. 2c). At a 1:2 transfection ratio, cDAPK1 levels were decreased by 29.18±1.91% of cDAPK1 transfected alone (n=4; p=0.021), whereas at 8:1, cDAPK1 levels were reduced by 92.85%±2.30% (n=4; p=0.001). In contrast, co-expression of the HA-GluN2Bct-CTM with wtDAPK1 even at the highest ratio (8:1) did not significantly affect wtDAPK1 protein levels (Fig. 2d; n=3; p=0.933), strongly suggesting that the ability of the targeting peptide to interact with DAPK1 is required for the peptide-induced cDAPK1 degradation. cDAPK1 degradation appears to be mediated by the lysosome, as it was significantly prevented by lysosomal inhibitor NH4Cl (Fig. 2c; n=4) or by mutating CTM within the targeting peptide (Fig. 2e; HA-GluN2Bct-CTMm; from at least 7 individual experiments; p=0.785). Taken together, these data suggest that the specific reduction of cDAPK1, but not wtDAPK1, is dependent on the targeting peptide-protein interaction and an intact CTM within the targeting peptide, and is mediated by the lysosome.
Knockdown of native proteins in primary neuronal cultures
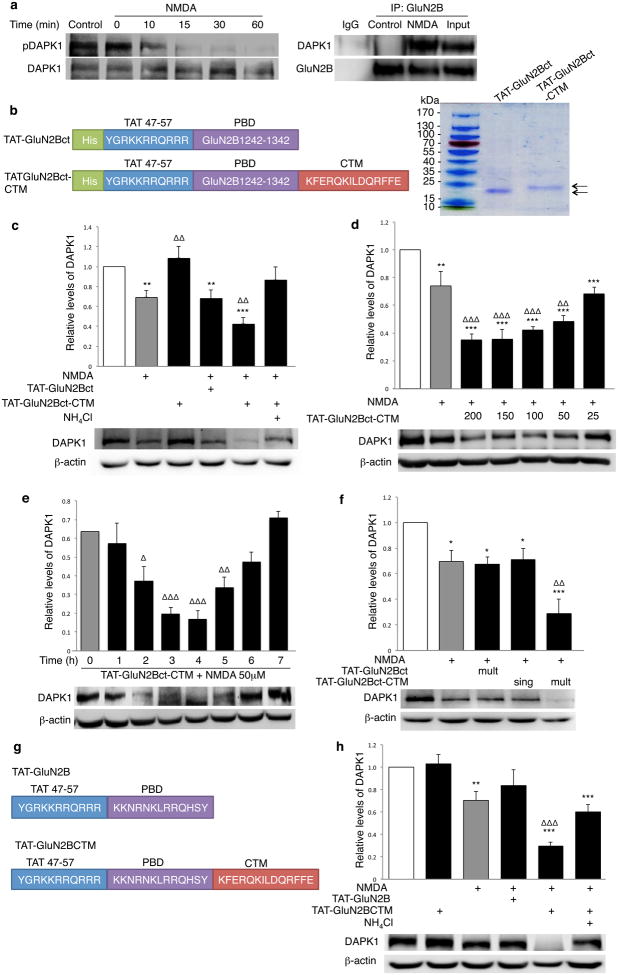
The above results demonstrate that a targeting peptide comprising a specific PBD and CTM can target its binding partner for lysosomal degradation in a dose-, time- and condition-dependent manner in recombinant expression systems. We next tested if targeting peptides can efficiently knock down non-genetically modified, native proteins in cultured primary neurons in situ. As previously reported26, treatment with 50μM NMDA for 30min resulted in the significant activation of DAPK1 (Fig. 3a; left), as evidenced by a decrease in the phosphorylated form of DAPK126, as well as a notable reduction in the total amount of DAPK1 (consistent with a previous report)25. No obvious association between GluN2B and DAPK1 was found under basal conditions using co-immunoprecipitation. However, NMDA treatment resulted in complex formation between the two proteins (Fig. 3a; right), consistent with previously reported activation and recruitment of the DAPK1 to NMDARs by NMDA stimulation26.
Figure 3. DAPK1-targeting peptide specifically degrades activated endogenous DAPK1 in neuronal culture.
(a) Left: NMDA (50μM; 30min) activated DAPK1, resulting in a time- dependent decrease in its phosphorylation levels (pDAPK1, n=4). Right: Co-immunoprecipitation with anti-GluN2B and sequential immunoblotting for DAPK1 and GluN2B showed an NMDA-induced association between DAPK1 and GluN2B (n=3). (b) Design and production of TAT-GluN2Bct-CTM and TAT-GluN2Bct peptides (Left) using E. coli expression system. Coomassie blue staining of SDS-PAGE assessed their purity (Right). (c) Bath application of TAT-GluN2Bct-CTM (200μM; n=9), but not TAT-GluN2Bct (200μM; n=6), knocked down activated DAPK1, which was prevented by NH4Cl (20mM; n=5; One-way ANOVA; P<0.001, F(5,36)=10.891), and dose- (d; n=4; p<0.001; F(6,21)=18.14) and time-dependent (e; p<0.001; F(8,44)=12.074). (f) A single pretreatment of TAT-GluN2Bct-CTM (sing; 200μM, 60 min prior to and during the 30min NMDA stimulation) produced a transient reduction of DAPK1, returning to baseline within 7hrs (n=4; p=0.888) and an additional dose of the peptide after NMDA washout resulted in a persistent decrease in DAPK1 up to 7hrs (mult; n=4; ΔΔp=0.002). One way ANOVA; p<0.001, F(4,15)=10.389. (g) Schematic illustration of synthetic peptides TAT-GluN2B and TAT-GluN2BCTM. (h) TAT-GluN2BCTM (25μM; n=5; p=0.001), but not control TAT-GluN2B (25μM; n=4; p=0.223) decreased native DAPK1, which was prevented by NH4Cl (20mM; n=5; p=0.302). One-way ANOVA, p<0.001, F(5,24)=13.591. Relative levels of DAPK1 were normalized to those in non-treated naïve and compared to naïve (white bar, *) or NMDA-treated group (grey bar, Δ). Membranes re-probed for β-actin were used as a loading control. Sample size represents number of individual experiments. Full-length blots are presented in Supplementary Figure 9.
This activity-dependent association between GluN2B and DAPK1 suggests a potential use for our GluN2Bct-CTM construct (described above) to knockdown DAPK1 in an NMDA-dependent manner. Thus, we subcloned the GluN2Bct-CTM (GluN2Bct as control) along with the cell membrane-penetrating sequence TAT28 (as the CMPD) into bacterial expression vectors, and then expressed and purified them as His-tagged recombinant peptides (TAT-GluN2Bct-CTM and TAT-GluN2Bct; Fig. 3b). When bath applied on its own, TAT-GluN2Bct-CTM (200μM; n=4; p=0.47) produced no observable effect on the basal level of DAPK1 (Fig. 3c). However, when co-applied with NMDA, TAT-GluN2Bct-CTM (200μM; 60min prior to and during 30min NMDA treatment) decreased the level of DAPK1 by 57.50%±6.70%, 2hrs after NMDA washout (n=9; p=0.007; Fig. 3c); a process blocked by concomitant treatment with the lysosome inhibitor NH4Cl. In contrast, co-application of the same amount of control peptide TAT-GluN2Bct with NMDA did not produce any notable reduction of DAPK1 relative to NMDA treatment alone (Fig. 3c). These results strongly suggest that the observed decrease in DAPK1 is mediated by lysosomal degradation, and that it requires the specific interaction of GluN2Bct with the activated DAPK1 and the presence of the CTM in the targeting peptide.
More detailed analyses revealed that following NMDA stimulation (Fig. 3d), increasing TAT-GluN2Bct-CTM from 25μM to 200μM produced dose-dependent DAPK1 degradation (n=4). Furthermore, a single dose of TAT-GluN2Bct-CTM (200μM; 60min prior to and 30min during NMDA stimulation) resulted in a time-dependent reduction in DAPK1 levels, which became significant by 2hrs, peaked at 4hrs, and then gradually returned to baseline levels within 7hrs post-treatment (Fig. 3e). However, administrating a second dose of TAT-GluN2Bct-CTM immediately after NMDA washout allowed DAPK1 degradation to persist up to 7hrs (Fig. 3f; n=4; p=0.002). Importantly, the levels of DAPK1 mRNA were not affected (Supplementary Fig. 2). The reversibility and time-dependency of DAPK1 degradation strongly support the supposition that the degradation is not due to non-specific cellular processes, such as toxicity-induced cell death, but is a result of TAT-GluN2Bct-CTM mediated knockdown. Importantly, our results indicate that varying the concentrations and/or times of targeting peptide applications can easily control the degree and the duration of POI degradation, allowing fine-tuned control of expression levels of the endogenous POI.
Although TAT-GluN2Bct-CTM can be easily produced in recombinant expression systems, the 100 amino acids within the GluN2B C-terminal contain multiple proteolytic sites29 making it vulnerable to fast clearance from the cell, lowering its bioavailability, and increasing the chance of off-target effects. Since the binding of GluN2B to activated DAPK only requires 18 amino acid residues of GluN2B1292-134026, we next hypothesized that a short synthetic form of the targeting peptide containing this binding sequence, the membrane-permeable TAT and the CTM should be sufficient to target active DAPK1 to lysosomes for degradation. Hence we synthesized the targeting peptide TAT-GluN2BCTM and its control TAT-GluN2B (Fig. 3g). Bath application of TAT-GluN2BCTM (25μM) resulted in significant reduction of DAPK1 following NMDA-treatment (n=5; p=0.001 compared to NMDA-treated group) and was rescued by NH4Cl, while TAT-GluN2B did not affect DAPK1 levels (Fig. 3h). Furthermore, subcellular fractionation analysis showed that despite a rapid increase of DAPK1 in the mitochondria following NMDA stimulation, TAT-GluN2BCTM was able to reduce the levels of DAPK1 in nuclear, cytosolic and mitochondrial fractions (Supplementary Fig. 3). siRNA-mediated knockdown of LAMP-2A, a crucial component of the CMA machinery, significantly rescued DAPK1 knockdown (Supplementary Fig. 4), further supporting the role of lysosome-dependent degradation. To determine if other cell-penetrating peptides could replace TAT to make our method more generalizable, we further synthesized the small peptide GluN2B-CTM (Supplementary Fig. 5a) without a physically linked CMPD. To deliver GluN2B-CTM into the cell interior, we mixed GluN2B-CTM with the intracellular delivering carrier peptide Pep-130 at a 1:4 ratio for 30min to form a plasma membrane permeable peptide complex30,31. We then bath applied the complex into neurons for 60min prior to and during NMDA treatments (GluN2B-CTM 50μM; 30min). Bath application of the peptide, along with its carrier Pep-1, degraded activated DAPK1 in a dose- and time-dependent manner through the lysosome (Supplementary Figs. 5b–d). Taken together, the above data illustrate the reliability and specificity of DAPK1-targeting peptides (His-tagged recombinant peptide, TAT- or Pep-1 mediated synthetic peptides) to rapidly and reversibly degrade its endogenous binding partner DAPK1 in primary neuronal cultures in a dose, time and activity-dependent manner.
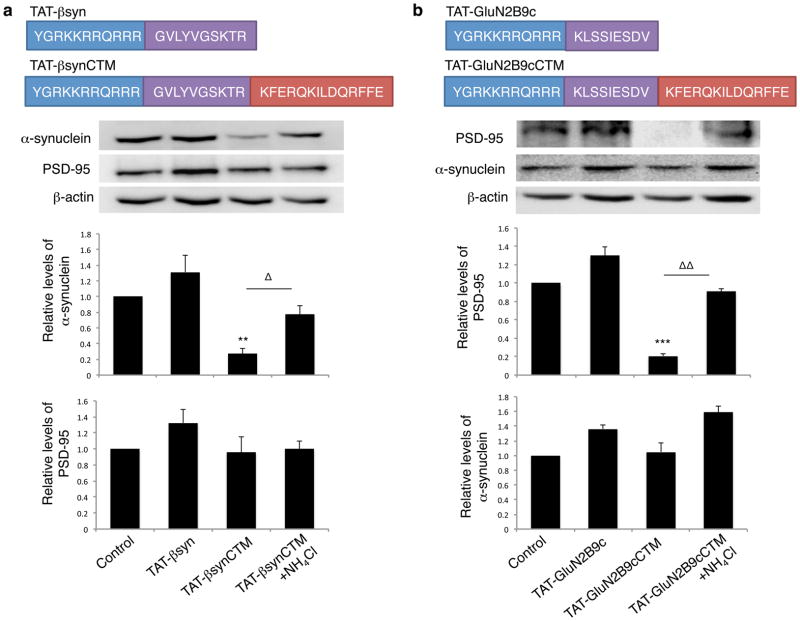
Having confirmed the success of our targeted peptide-mediated DAPK1 degradation approach, we next assessed the generalizability of our method by targeting two other native neuronal proteins, α-synuclein and Post Synaptic Density Protein 95 (PSD-95). α-synuclein, a protein implicated in neurodegenerative synucleinopathies such as Parkinson’s disease32, was recently found to strongly interact with a short amino acid stretch (between amino acids 36–45) of β-synuclein (βsyn36)33. Therefore, we designed a targeting peptide containing βsyn36 and the CTM (TAT-βsynCTM; Fig. 4a top) to target α-synuclein. PSD-95 is a membrane-associated guanylate kinase (MAGUK) concentrated at glutamatergic synapses and is involved in synapse stabilization and plasticity. PSD-95 acts as a scaffold to assemble a specific set of signaling proteins around the NMDAR, and binds to 9 amino acids at the GluN2B subunit C-terminal tail34 (GluN2B9c). We therefore synthesized the PSD-95 targeting peptide TAT-GluN2B9cCTM (Fig. 4b, top). We hypothesized that TAT-βsynCTM and TAT-GluN2B9cCTM, but not their respective controls (TAT-βsyn and TAT-GluN2B9c) would be sufficient to target α-synuclein and PSD-95 respectively for degradation. Indeed, TAT-βsynCTM (25μM; n=5; p<0.001), but not its control TAT-βsyn (25μM; n=5), when bath applied, significantly degraded native α-synuclein in primary cultured neurons, without affecting the non-intended target PSD-95 (Fig. 4a, middle and bottom panels). The knockdown was rescued by concomitant treatment with the lysosome inhibitor NH4Cl (Fig. 4a, middle), but not by inhibition of macroautophagy with 3-methyladenine (Supplementary Fig. 6). Conversely, TAT-GluN2B9cCTM (25μM; n=4; p<0.01), but not its control TAT-GluN2B9c (25μM; n=4) significantly reduced PSD-95 but not α-synuclein levels in a lysosome-dependent fashion (Fig. 4b). As α-synuclein harbors an endogenous CMA targeting motif (95VKKDQ9922), TAT-βsynCTM could be enhancing its accessibility or functioning in an additive manner. To determine if the targeting peptide sufficiently directs α-synuclein for lysosomal degradation independent of its endogenous CMA targeting motif, we mutated the endogenous motif to produce α-synuclein(ΔDQ). Following transfection into HEK293 cells, treatment with TAT-βsynCTM significantly reduced the level of α-synuclein(ΔDQ), which was rescued by NH4Cl (Supplementary Fig. 7). Finally, TAT-βsynCTM also decreased the levels of the A53T variant of α-synuclein (Supplementary Fig. 7), which is present in some forms of familial Parkinson’s Disease. Importantly, none of the peptides showed apparent toxicity at the concentrations used, even after 24hrs of application (Supplementary Fig. 8). Together, these results indicate that the targeting peptide–mediated degradation is specific for its targeted protein and that targeting peptide can be generalized to degrade in situ most, if not all, native cytosolic proteins, as well as some of their pathological variants.
Figure 4. Target peptide-mediated respective degradation of α-synuclein and PSD-95 in cultured neurons.
(a) Top: Schematics of the synthetic cell-penetrating α-synuclein targeting peptide TAT-βsynCTM and its control TAT-βsyn. Middle: Immunoblots demonstrate that TAT-βsynCTM (25μM; n=5), but not the CTM-lacking control peptide TAT-βsyn (25μM; n=5), specifically decreased the targeted endogenous α-synuclein (One way-ANOVA Tukey post-hoc, p<0.001, F(3,16)=12.435), without affecting the level of unrelated control proteins PSD-95 at 4 hours (bottom), and this reduction was prevented in the presence of lysosomal inhibitor NH4Cl (20mM; n=5). Sample size represents individual experiments from at least 3 separate primary cultures. (b) Top: Schematics of PSD-95 targeting peptide TAT-GluN2B9cCTM and control TAT-GluN2B9c. Middle: TAT-GluN2B9cCTM (25μM; n=4), but not Tat-GluN2B9c (25μM; n=4), effectively degraded endogenous PSD-95 (One-way ANOVA Tukey post-hoc, p<0.001, F(3,12)=18.154) without perturbing untargeted protein α-synuclein (Bottom). NH4Cl rescued PSD-95 degradation. Sample size represents individual experiments from at least 2 separate primary cultures. Membrane re-probing for β-actin was used as additional specificity and loading controls. * p<0.05, **,ΔΔ p<0.01 and ***p<0.001; bars represent relative mean values±s.e.m. normalized to the naïve non-treated control (arbitrarily set as 1). Full length blots were presented in Supplementary Figure 9.
Neuroprotection of DAPK1 knockdown in vitro and in vivo
Having shown the reliability of a targeting peptide to degrade of a variety of native proteins, we next asked if targeting peptide-mediated knockdown can have physiologically and/or pathologically relevant phenotypes. As mentioned above, DAPK1 is a cell death promoting protein kinase in many cell types, and is known to be required for cell death under pathological conditions such as stroke. It has previously been shown that following ischemic insult, overactivation of NMDARs activates and recruits DAPK1 to the GluN2B C-terminal of the NMDAR. Dissociation of DAPK1 from the NMDAR by administration of the membrane permeable GluN2B peptide without CTM (TAT-GluN2B) not only prevents NMDA-induced neuronal damage, but also partially reduce ischemic brain injuries in a stroke animal model26. Stroke-induced ischemic neuronal damage can also occur through mechanisms other than NMDAR-mediated excitotoxicity, including oxidative stress35. Previous results have revealed that DAPK1 is a central mediator of oxidative-stress induced cell death36. Therefore, we reasoned that, in comparison with the TAT-GluN2B (without CTM) that reduces NMDAR-mediated excitotoxicity by disrupting the DAPK1-NMDAR interaction26, our targeting peptide TAT-GluN2BCTM, by knocking down DAPK1, may protect neurons against not only NMDAR-mediated excitotoxicity, but also other neuronal damage signaling pathways including oxidative stress.
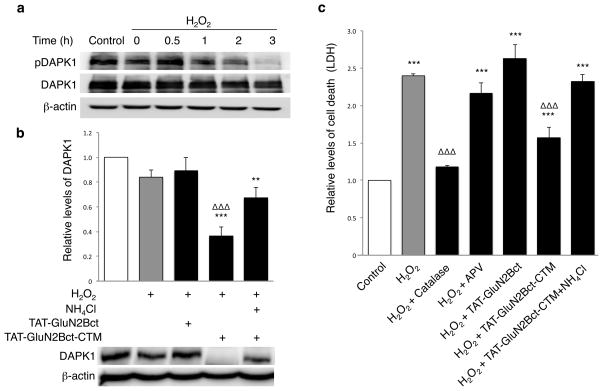
We first tested the idea that knocking down DAPK1 can protect neurons against NMDAR-independent, oxidative stress in primary neuronal cultures. As shown in Fig. 5a, exposure of cultured cortical neurons to H2O2 (300μM; 30min), a potent generator of oxidative stress-inducing reactive oxygen species37, significantly activated DAPK1, as demonstrated by time-dependent decrease in levels of DAPK1 phosphorylation. Pretreatment of these neurons with TAT-GluN2Bct-CTM (100μM), but not TAT-GluN2Bct, resulted in a more than 60% reduction in DAPK1 level in H2O2 -treated (but not control) neurons (reduced to 36.54±7.1%; n=8; p=0.001 compared with control). This reduction was prevented by co-application of NH4Cl (Fig. 5b). As depicted in Fig. 5c, a Lactate Dehydrogenase (LDH)-based cell death assay revealed that H2O2 exposure significantly increased neuronal death (n=4; 2.38±2.70; p=0.001) which was rescued by catalase, an enzyme that breaks down H2O237. On the other hand, treatment with the selective NMDAR antagonist (2R)-amino-5-phosphonovaleric acid (APV; 1mM) did not protect against H2O2-induced neurotoxicity, consistent that this H2O2-mediated neuronal damage is not mediated by NMDARs. In support of our hypothesis, TAT-GluN2Bct-CTM (50μM; 60min prior to and continuously present during and following H2O2 washout; n=9), but not TAT-GluN2Bct, significantly reduced H2O2-induced neurotoxicity. Furthermore, concurrent treatment with the lysosome inhibitor NH4Cl inhibited the neuroprotective effect of TAT-GluN2Bct-CTM (n=4), supporting neuroprotection mediated by TAT-GluN2Bct-CTM induced lysosomal degradation of DAPK1. However, as NH4Cl may inhibit the degradation of DAPK1 in the lysosomes without affecting its translocation into the lysosomal lumen, an un-related toxic effect from NH4Cl remains to be ruled out. Together, the results indicate that TAT-GluN2Bct-CTM is neuroprotective against non-NMDAR dependent oxidative stress and this neuroprotective effect is a result of the peptide-mediated lysosomal degradation of DAPK1.
Figure 5. TAT-GluN2Bct-CTM knocks down H2O2-activated DAPK1, protecting neurons against H2O2-induced neurotoxicity in neuronal cultures.
(a) Immunoblotting for phosphorylated DAPK1 (pDAPK1) revealed a time-dependent activation (dephosphorylation) of DAPK1 by H2O2 treatment (300μM; 30min; n=4). (b) Bath application of 100μM TAT-GluN2Bct-CTM (36.54±7.1% of control; n=8; p=0.001), but not TAT-GluN2Bct (89.13±10.78%; n=7; p=0.311), 60min prior to and during H2O2 treatment (300μM; 30min) knocked down DAPK1 at 2hrs post-washout, which was rescued by NH4Cl (20mM; n=8; p=0.003 to control*; p=0.106 to H2O2-treatedΔ). One-way ANOVA, F(4,34)=11.628, p<0.001. Bars represent DAPK1 levels relative to naïve group. β-actin was used as a loading control. (c) LDH assay revealed that H2O2 treatment (300μM; 30min) resulted in a significant increase in neuronal death 12hrs after treatment (n=8; 2.50±0.12; p<0.001 to control), which was rescued by breaking down H2O2 with catalase (100U; n=4; 1.17±0.02; p=0.001 to H2O2 group). H2O2-induced neurotoxicity was significantly reduced by TAT-GluN2Bct-CTM (50μM; applied 60min prior to and maintained throughout the experiments; n=9; 1.56±0.08; p=0.001 to H2O2 group), but not by TAT-GluN2Bct (50μM; n=9; 2.63±0.10; p=0.105 to H2O2 group) or the NMDAR antagonist APV (1μM; n=4; 2.16±0.14; p=0.169 to H2O2 group). NH4Cl abolished the neuroprotective effect of TAT-GluN2Bct-CTM (n=4; 2.39±0.27; p=0.538 compared to H2O2 group). One-way ANOVA, p<0.001, F(6,41)=26.842. *,Δ p<0.05, **,ΔΔ p<0.01 and ***, ΔΔΔp<0.001; bars represent relative mean values±s.e.m. normalized to the naïve control (white bar, arbitrarily set as 1). n represents individual experiments from at least 3 separate primary cultures. Full-length blots are presented in Supplementary Figure 9.
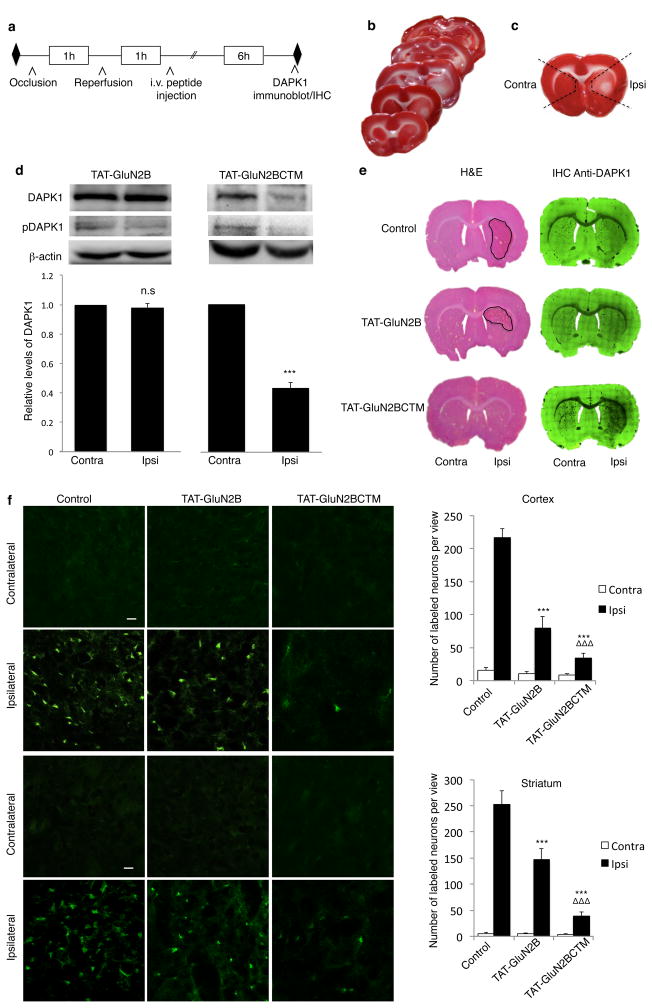
We next examined if systemic application of TAT-GluN2BCTM can knockdown DAPK1, thereby producing neuroprotection against ischemic insult in vivo using a well-characterized rat model of focal ischemia (Middle Carotid Artery Occlusion; MCAo), that was previously shown to reliably activate DAPK1 in vivo26. In order to measure the efficacy of DAPK1 knockdown using both immunoblot and immunocytochemistry, we used a relatively a minor (60min unilateral occlusion) ischemic insult. As shown in Fig. 6a, rats subjected to 60min of MCAo were injected intravenously (i.v.) with either saline, TAT-GluN2BCTM or TAT-GluN2B 1hr following reperfusion. 2,3,5-triphenyltetrazolium chloride (TTC) staining of transverse brain sections from saline-treated rats revealed that unilateral MCAo reliably induced ischemic brain damage mostly in the ipsilateral striatum (Fig. 6b). As DAPK1 is activated by MCAo-induced ischemic stimulation26 and the TAT-GluN2BCTM knockdown of DAPK1 is DAPK1 activation dependent, we reasoned that DAPK1 would be maximally activated and degraded in regions most affected by ischemic insult. To determine ischemia-induced knockdown of DAPK1 by TAT-GluN2BCTM, we excised tissues from both MCAo-challenged and contralateral sides of the striatum and nearby cortex as indicated in Fig. 6c and immunoblotted for DAPK1 levels. While TAT-GluN2B (10mg/kg, i.v.) produced no obvious change in the levels of DAPK1 in the brain tissues of either ischemic or contralateral side, TAT-GluN2BCTM (10mg/kg; i.v.) resulted in a significant reduction in DAPK1 levels only in the ischemic side of the brain; the level of DAPK1 was reduced to 43.3% of that on the contralateral side (Fig. 6d; p<0.001, n=3; two-tailed student’s t-test). Given that the collected tissues included some non-infarct areas (Fig. 6c), the actual efficiency of targeting peptide-mediated knockdown would be expected to be even greater. To further assess region-specific DAPK1 degradation in a more straightforward manner, we used immunohistochemistry to probe for DAPK1 in brain transverse brain sections (Fig. 6e, right). As expected, DAPK1 knockdown was specific to stroke-damaged areas, as visualized with Hematoxylin & Eosin (H&E) staining (Fig. 6e, left) of the adjacent sections. This further confirms our in vitro results demonstrating that TAT-GluN2BCTM is capable of specific knockdown of the active (but not inactive) form of DAPK1. To our knowledge, this is the first evidence for such a disease-specific protein knockdown in intact animals. The specific knockdown of DAPK1 by TAT-GluN2BCTM was associated with a much more significant reduction of the infarct area, in comparison with TAT-GluN2B, which partially decreased ischemic damage by uncoupling DAPK1 from the GluN2B receptor signaling complex as previously reported26. We further confirmed the more prominent neuroprotective effects of TAT-GluN2BCTM-mediated DAPK1 knockdown by quantifying numbers of Fluorojade B stained degenerating neurons in both striatum and cortex. Fluorojade B is a common stain that labels degenerating cells; it is suitable for assessment of cellular injury following MCAo38,39. Consistent with the results of H&E staining (Fig. 6e, left), Fluorojade B staining showed that while both TAT-GluN2B and TAT-GluN2BCTM significantly reduced the numbers of degenerating neurons in both striatum and cortical areas, the neuroprotective effects of the later was significantly more prominent (Fig. 6f). Thus, it appears that knocking down active DAPK1 (by TAT-GluN2BCTM) is much more neuroprotective than just simply uncoupling the GluN2B-DAPK1 association (by TAT-GluN2B).
Figure 6. TAT-GluN2BCTM specifically knocks down DAPK1 in ischemic brain areas and reduces neuronal damage in the MCAo model of focal ischemia in rats.
(a) Timeline of tissue collection for analysis of DAPK1 degradation in rats. (b) 2,3,5-triphenyltetrazolium chloride (TTC) staining of a series of transverse brain sections showed reliable damage in the ipsilateral side following unilateral MCAo. (c) Black-dashed lines represent brain areas removed for immunoblotting. (d) Immunoblots demonstrate specific DAPK1 knockdown in the infract (Ipsi), but not contralateral (Contra) side following application of TAT-GluN2BCTM (10mg/kg, i.v.; n=3; t(4)=14.459, p<0.001), but not TAT-GluN2B (10mg/kg, i.v.; n=3; t(4)=0.739, p=0.501). β-actin was used as a loading control, two-tailed student’s t-test ***p<0.001 (e) Images of Hematoxylin & Eosin (left) and immunohistochemical DAPK1 staining (right) of adjacent brain sections show that compared with saline (top) and TAT-GluN2B treated (middle) controls, TAT-GluN2BCTM treatment (bottom) selectively reduced infarct area (left) and DAPK1 levels (right) ipsilaterally. (f) Left, images of brain sections stained with Fluorojade B in rats injected with saline (n=6), TAT-GluN2B (n=5) or TAT-GluN2BCTM (n=5) after treatment as shown in a, scale bar 20μm. Right, quantification of cellular damage by counting the number of Fluorojade B-positive cells in each image at 10X magnification. TAT-GluN2BCTM (10mg/kg) displayed more prominent neuroprotection in the cortex (top p<0.001) and striatum (bottom p<0.001) as compared to TAT-GluN2B (10mg/kg). Cortex: H(2)=41.235; p<0.001; striatum: H(2)=38.808; p<0.001. Kruskal-Wallis ANOVA on ranks with Dunn’s post-hoc, bars represent relative mean values±s.e.m, ***,ΔΔΔp<0.001. n represents tissue from 3 animals collected from at least 2 litters. Full-length blots are presented in Supplementary Figure 9.
Together, these data provide proof-of-concept evidence for the feasibility of our targeting peptide-based protein knockdown strategy in vivo. Furthermore, these data illustrate the possibility of achieving region- and/or disease-specific knockdown of endogenous proteins, depending on the nature of the interaction between the targeting peptide and its protein substrate. Finally, given that efficient protein knockdown was obtained following systemic application of the targeting peptide, our evidence suggests that targeting peptide may be suitable for use in designing clinically relevant therapeutics.
Discussion
In this study, we demonstrated a novel targeting peptide-based method to knockdown native proteins in vitro and in vivo. Such a method offers a robust, reversible, dose- and time-dependent and possibly conditional way to degrade native proteins. The efficient knockdown of both small (19kDa, α-synuclein) and large (160kDa, DAPK1) cytoplasmic proteins, as well as the synaptic scaffolding protein PSD-95 and the A53T pathological variant of α-synuclein, further demonstrate the feasibility and versatility of this targeting peptide-based method, providing proof-of-concept evidence for using it to efficiently knockdown most, if not all, endogenous cytosolic proteins.
This method has several advantages over other currently used methods such as genetic knockout and siRNA-mediated protein knockdown. First, our method is much faster; knockdown can be realized as fast as 2hrs after treatment. Such speed cannot be achieved with any previously described protein manipulations at either DNA or mRNA levels. Therefore, our method is expected to have fewer issues with the compensation often associated with these slower DNA or mRNA based protein knockdown methods. Second, the reversibility and dose-dependency of our targeting peptide-based method means that the level and duration of the protein knockdown can be easily controlled by varying the dose and/or times of the peptide applications, thereby making it a useful and effective research tool in biomedical studies aimed at elucidating the functions of a protein in physiological processes and in the pathogenesis of diseases. Third, the peptide can be generated through multiple means. It can be overexpressed following cDNA plasmid transfection, expressed and purified as a recombinant protein using common bacterial expression systems, or commercially synthesized as short synthetic peptides. Therefore, it is a versatile system that can be widely utilized by almost any biomedical laboratory, even those without sophisticated molecular biological facilities.
Compared to recently developed protein knockdown strategies, which typically need to genetically modify target proteins to render them susceptible to regulation by cellular degradation systems6–8, our method enables the study of native proteins in situ without prior modification, precluding potential artifacts arising from genetically manipulating the POI. Our proof-of-concept experiments with the TAT transducing domain as the CMPD clearly demonstrate that targeting peptides can readily cross the plasma membrane when bath applied in primary cell cultures in vitro or given peripherally in intact animals in vivo, eliminating the need for viral infection. Indeed, we and others have previously shown that TAT can deliver biologically active cargo across the blood brain barrier and plasma membrane into the cell interior in a highly efficient manner both in vitro and in vivo30,40,41. However, the CMPD is not limited to TAT. As Pep-1 can also efficiently deliver the targeting peptide, it appears that TAT can be replaced by alternative cell penetrating peptides (CPP). This is especially useful in cases where covalent linkage between CPP and cargo is not desirable. Compared to previous methods that use small chemicals to control protein levels7,11,42, peptides can be more easily designed to target a protein for which binding pockets amenable to small molecule inhibition cannot be found43. Here, we have successfully designed three targeting peptide PBDs by literature search and proven their efficacy. Methods like phage display and peptide arrays43 can further aid the discovery of appropriate PBDs, and rational design through computational modeling may increase the specificity and affinity of a candidate PBD to its target protein.
Specificity is of upmost importance in any system designed to perturb specific protein function. In our case, the specificity and effectiveness of targeting peptide to knockdown POIs would largely depend on the specificity and affinity of the interaction between the PBD and the POI. Although peptide-protein binding is mediated by a small number of contacts formed by the residues in linear motifs, these interactions can be extremely specific44. In isolation these motifs bind their target proteins with sufficient strength to establish a functional interaction44. Furthermore, these interactions are often limited by the biological context, as seen with DAPK1 and GluN2B interaction, which adds an additional layer of specificity and control. In our tests, each targeting peptide was specific for its own intended target proteins with no off-target effects. Importantly, TAT-GluN2BCTM was specific for the active, but not inactive form of DAPK1, illustrating a degree of specificity that cannot be achieved by either DNA or mRNA targeting.
Unlike other methods of protein degradation, our system utilizes the endosome-lysosome system in place of the proteasome. Like the ubiquitin-proteasome pathway, CMA activity is detected under basal conditions in the brain, and is further activated under conditions of stress45 and proteasome inhibition14. Hence a CMA-based degradation system would not only complement current protein knockdown methods, but also be especially powerful under pathological conditions where the cell is under stress and/or the proteasome is inhibited. The effectiveness of targeting peptides supports the conclusion that attachment of CTM facilitates the degradation of a non-CMA protein via CMA, as previously described14. We further show that a targeting peptide with CTM can also lead to degradation of its binding partner through the lysosome, regardless of the latter’s endogenous route of degradation. In line with a previous study describing degradation of mutant Huntington by viral infection of a CTM-containing adaptor protein through CMA19, our experiments with lysosomal and macroautophagy inhibitors and siRNA also argue for CMA as the main route of degradation. Hence, an inherent limitation to our strategy is that it cannot manipulate proteins involved in the CMA machinery or lysosomal integrity. A specific case to note is that certain pathological variants of disease-causing proteins, such as A53T α-synuclein, inhibit the CMA machinery by binding tightly to the LAMP-2A receptor and precluding degradation of other CMA substrates22. However, in the present work, we found that the α-synuclein targeting peptide was able to degrade A53T α-synuclein in a lysosomal activity-dependent manner (Supplementary Fig. 7). Nevertheless, in cases where lysosomal activity is compromised, these problems may be overcome by harnessing the proteasomal degradation system using a similar targeting peptide strategy. To this end, a short amino-acid based strong proteasome targeting signal has recently been described3. Furthermore, we can also predict that a combination of a peptide containing CTM and one with proteasome-targeting signal peptides may dramatically enhance knockdown of certain native proteins.
Perhaps the greatest potential of our system, compared to previously existing methods of regulating protein levels either at the DNA, mRNA or protein level, is its ready potential for clinical translation. In most cases, many previously described protein knockdown methods require expression of a pre-genetically modified cDNA into targeted cells with viral infection42, and are therefore not immediately practical for therapeutic use in human patients. However, the use of the TAT cell membrane-penetrating domain in our method easily delivers the targeting peptide into the interior of cells in various organs, after administration through a number of drug delivery routes including the commonly used i.v. application. The efficacy and safety of TAT-mediated transduction of therapeutic peptides to cross the BBB and cell membrane of neurons in the brain has recently been demonstrated in a successful phase 2B clinical trial46. Our proof-of-concept experiments with MCAo, a common model for focal ischemia, show that the targeting peptide is capable of knocking down death-inducing DAPK1 in the brain, but specifically in the damaged areas, leaving DAPK1 in non-ischemic regions intact. The ability to achieve such a disease-related, region-specific knockdown has the obvious advantage of reducing unwanted effects over a general genetic deletion of DAPK1. While the delivery and stability of peptides as therapeutic agents has historically been problematic, recent developments in a range of modification techniques, such as retro-inverso peptides47, can be applied to improve the delivery and pharmacological profile of a peptide43. Thus, the method described here may open the possibility for developing novel therapeutics for treating human diseases, particularly for those diseases in which the pathogenesis is at least in part caused by gain of function due to an overexpression and/or mutation of a particular protein.
Together, we present a novel peptide-based method that can rapidly and reversibly knockdown endogenous proteins in situ. It is suitable for both in vitro and in vivo use, can be easily generalized to potentially degrade any cytosolic POI, and offers a complementary approach to current genetic and chemical means of regulating the expression levels of native proteins. It should soon be proved as not only a powerful tool for scientific research, but also a means of facilitating the development of effective therapeutics for treating human diseases.
Online Methods
General antibodies and reagents
Anti-GFP (Clonetech, 632381), anti-LAMP-1(Abcam, ab13523), anti-GAPDH (Abcam, ab9485), anti-actin(Abcam, ab8227), anti-α-synuclein (BD Transduction Laboratories, 610786), anti-HA (Roche applied science, 11867431001), monoclonal anti-FLAG M2 antibody (Sigma-Aldrich, F1804-200UG), anti-DAPK1 (Sigma, D1319-200UL), monoclonal anti-phospho-DAPK1 (pSer308, Sigma, D4941), anti-GluN2B (lab generated), anti-LAMP-2A (Abcam, ab18528), anti-Labmin B1(Abcam, ab16048), anti-HSP90 (BD Transduction Laboratories, 610418), anti-VDAC1 (Porin) (MitoSciences,MSA03). Antibodies were validated for their intended purpose (immunoblotting, immunocytochemistry, immunohistochemistry and co-immunoprecipitation) as outlined in the product sheet or in lab. Ammonium chloride (Sigma, A0171), 3-methyladenine (Sigma, M9281), MG132 (Sigma, C2211), Pepstatin A (Sigma), N-Methyl-D-aspartic acid (NMDA, Tocris Asc-052), H2O2 (Sigma, 7722-84-1) Catalase (Sigma, C1345), (2R)-amino-5-phosphon-opentanoic acid (APV, Ascent Scientific, Asc-003). GluN2B-CTM and TAT-GluN2B were synthesized by Brain Research Centre peptide synthesis facility at UBC. All other synthetic peptides used were synthesized by GL Biochem.
Plasmid construction
CTM-GFP was constructed by introducing a BamHI fragment containing the CTM coding sequence into the pEGFP-N2 vector (Clontech #6081-1). The CTM coding sequence was prepared by annealing custom design oligonucleotides (Integrated DNA Technologies). mCTM-GFP was constructed by performing single point mutations to CTM-GFP plasmid. FLAG-cDAPK1 was constructed by deleting the autoinhibitory domain from WT-DAPK1 (789-936bp). GluN2Bct (1242-1342aa) was prepared by PCR using GluN2B expression vector. GluN2Bct-CTM fragment was obtained by inserting GluN2Bct fragment into CTM-GFP using EcoRI and BglII restriction sites (EcoRI, Fermentas, FD0274; BglII, Fermentas, FD0084), then PCR, introducing NcoI and EcoRI to the fragment (NcoI, Fermentas, FD0574). His-TAT-GluN2Bct-CTM was constructed by cloning GluN2Bct-CTM into the pTAT/pTAT-HA plasmids (generous gift of S. Dowdy, Washington University, St. Louis, MO34) using NcoI and EcoRI restriction sites. His-TAT-GluN2Bct was constructed by mutating the first amino acid in CTM sequence into stop codon. HA-GluN2Bct-CTM was constructed by PCR using TAT-GluN2Bct-CTM as template, with BamHI site in both forward and reverse primers, and then inserted into pcDNA3.0 with BamHI (Fermentas, FERFD0054). HA-GluN2Bct-CTMm and α-synuclein (ΔDQ) were constructed by performing point mutation PCR.
His peptide purification
TAT-GluN2Bct and TAT-GluN2Bct-CTM plasmids were transformed into BL21, plated onto Amp resilient plates and incubated overnight a 37°C. Single colony from each plasmid was resuspended in LB(Amp+) and incubated at 37°C until OD600 reached 0.5. Expression was induced by adding IPTG (1mM) and incubating for 5 hours. Pellets were then collected by centrifugation and discarding the medium. Pellets were sonicated and centrifuged before purification. His peptide purification was done according to the manufacturer’s protocols (Thermo Scientific, 88223). Briefly, Ni-NTA resin columns for equilibrated before prepared peptide extracts were added to the resin. The columns were then washed before eluted using elution buffer. The purified peptides were then monitored for purity using Coomassie Blue staining, and peptide concentration measured by absorbance at 280nm.
Cell culture, transfection and treatments. HEK293 and COS7 cells were cultured in DMEM (Sigma, D6429-24X500ML) supplemented with 10% Fetal Bovine Serum (Invitrogen,12483020) Cells were grown to 80% confluence in 6-well plates before transiently transfected with Lipofectamine 2000 (Invitrogen, 11668019), as according to the manufacturer’s protocols. Cells were transfected for either 24hr or 48hrs at 37°C before harvesting for biochemical analyses.
Primary culture of cortical neurons
Dissociated cultures of rat cortical neurons were prepared from Sprague Dawley rat embryos collected from killed mothers 18d after fertilization as previously described39. Briefly, hippocampi and cerebral cortices were extracted from embryos and incubated for 30min in 0.25% trypsin-EDTA. Digested tissues were dissociated by trituration and plated on poly-D-lysine-coated (Sigma, P7280) plates. Plating medium consisted of Neurobasal Media (Invitrogen, 21103-049) supplemented with B27 (Invitrogen, 17504044), glutamic acid (Sigma, G8415) and GlutaMax (Invitrogen, 35050-061). After 2 days, 2/3 of the media was replaced with fresh Neurobasal feeding media consisting Neurobasal Media, B27 and GlutaMax. Cultures were maintained at 37°C in a humidified 5% CO2 atmosphere. Mature neurons (14–18 d in vitro (DIV)) were used for experiments.
Immunoblotting
Immunoblotting assays were carried out as previously described48. Briefly, proteins were extracted from neurons using a lysis buffer composed of 150 mM NaCl, 50 mM Tris, pH 7.4, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, 1 mM Na3VO4, and proteinase inhibitor mixture (Thermo Fisher, PI78442). Samples were separated on 10% SDS-PAGE gels, transferred to polyvinylidene difluoride (PVDF) membrane, and immunoblotted with respective antibodies. Blots were enhanced with chemiluminescence detection reagent kit (Fisher, 32106) and visualized with Bio-Rad imager and Quantity One software. Signal intensities from each band was quantified with Bio-Rad Image Lab software, and the bands were analyzed relative to their controls from the same membrane and experiment.
Co-immunoprecipitation
Co-immunoprecipitation (Co-IP) assays were performed as previously described with minor modifications49. Cortical neuronal cultures lysed in ice-cold lysis buffer sans SDS. The extract (0.5mg) were precleared for 1 hr with 10μl Protein A-sepharose beads (GE Life Sciences, 17-0780-01), then incubated with nonspecific IgG (4μg), poly-clonal anti-GluN2B (lab, 4μg) overnight at 4°C, followed by addition of 60ul Protein A-sepharose beads (Sigma) for 3 hr at 4°C. Samples were washed two times with lysis buffer, two times with sterile PBS and denatured with SDS sample buffer. SDS-PAGE and immunoblotting were subsequently performed as described above.
Immunocytochemistry
Immunocytomistry was carried out as previously described49. COS cells were washed with ice-cold PBS, then fixed in pre-warmed 4% PFA/PBS solution at 37°C for 60 min, permeabilized in 0.1% Triton X-100 for 5 min, and blocked with 5% fetal bovine serum (FBS) in PBS for 30 min at 37°C with extensive PBS washings between each step. Primary antibodies were diluted in 3% FBS. Cells were incubated with anti-LAMP-1 (1:50) for 24-48hr at 4°C, then washed 6X2min with PBS. Secondary antibody Alexa 555 was diluted in 3% FBS/PBS at 1:1000 and incubated for 30min at 37°C and then washed extensively. Nuclei were stained with DAPI (1:5000, 10min RT) prior to mounting on slides in ProLong Gold medium (Invitrogen, P36930). Captured images were obtained from a confocal microscope (Leica DMIRE2 & CTRMIC). Representative images have been adjusted to maximize the signal:noise ratio. Quantification of co-localization was performed with the Image J (National Institutes of Health) JACoP plugin.
Cellular fractionation
Cytoplasm/nuclei fractionation was performed on cultured cortical neurons (6.0 * 106cells/100 mm dish). Briefly, cells were washed with ice cold PBS and rocked in lysis buffer for 30min. Cells were then collected and centrifuged to obtain a rough cytoplasmic and nuclear fraction. Supernatant was collected and further centrifuged to obtain purified cytosolic fraction. The original pellet was washed and vortexed to obtain a nuclear lysis. Mitochondrial fractionation was performed as described in the Pierce Mitochondria Isolation Kit for Cultured Cells (Thermo Scientific, 89874) user guide. Purity was assessed by immunoblotting for the presence of LB1 (nucleus-only), HSP90 (cytosol-only) and VDAC1 (mitochondria-only).
Assessment of neuronal death
Cytotoxic damage of primary neuron cultures was assessed by measuring LDH released into culture media as previously described50. Cortical neurons were exposed to H2O2-induced neurotoxicity (300 μM for 30 min) in the presence or absence of TAT-GluN2Bct-CTM or control peptide TAT-GluN2Bct (50μM, 1hr pretreatment and throughout the experiment), APV (1mM, 30min pretreatment and throughout experiment) or catalase (100U, 15 min pretreatment and throughout experiment), their culture media were collected 12hr post insult for LDH enzymatic activity. Peptide toxicity was assessed by treating neurons with 25μM synthetic peptide or 200μM recombinant peptide for 24hrs. Media was collected for LDH assay. Positive controls were obtained by lysing the cells with 100% Trixton X-100 prior to media collection. The amount of LDH in the medium was determined using a LDH cytotoxicity detection kit (Sigma, TOX7) according to the manufacturer’s instructions. The absorbance at 490 nm was determined using a microplate reader (μQuant, Bio-TEK instruments), which was adjusted by background reading reduction.
Middle cerebral arterial occlusion (MCAo)
All animal experiments were performed according to protocols approved by the University of British Columbia Committee on Animal Care. Adult naïve male Sprague-Dawley rats (300–350g, Charles River) were group housed (3–4 animals/cage) in 12hr:12hr light-dark cycles and had free access to rat pellet chow and water prior to surgery. Reversible MCAo with suture-insertion method was described previously39. Briefly, a nylon suture with a blunted tip was entered through the right external carotid artery of anesthetized rats and advanced to the right internal carotid artery until the right MCA was occluded. After 60min of occlusion, the rat was re-anesthetized to facilitate the removal of the occlusion. Body temperature was maintained between 36.5 and 37.5 °C throughout the surgical procedure with a heating pad. Peptides (10 mg/kg) or vehicle control (saline; 1 ml/kg) were injected via the jugular vein. Rats were then sutured and allowed to recover until tissue collection.
Immunohistochemistry
Rats were anesthetized and perfused with double-filtered saline and 4% paraformaldehyde (PFA) in PBS. Brains were collected, immersed in 4% PFA before subjected to cryoprotection by 30% sucrose/PBS. After the brains had sunk, they were flash frozen with dry ice before overnight freezing in -80°C. They were then sliced at 30μm with a cryostat, and stored in 0.1M PB (sodium phosphate dibasic and sodium phosphate monobasic). Prior to staining, slices were washed 3X10min with 0.1M PB, permeabilized and blocked in 0.1M PB with 1%BSA and 0.2% Triton X-100 for 30min, and stained with anti-DAPK1 (1:100) at 4°C for 3days. They were then washed and stained with Alexa 488 (1:1000) at 4°C overnight before washing and mounting.
Hematoxylin & Eosin (H&E) staining
Slices were mounted and dried on glass slides prior staining. Slides were immersed in Hematoxylin solution (Sigma, MHS1-100ML) for 15min away from light, followed by 5min blueing under tap water. They were then counterstained with 0.5% Eosin Y (Sigma, E4009-5G) and dipped in ddH2O until the eosin stopped streaking. They were then dehydrated with EtOH (50%, 70%, 95% and 100%) and cleared 2 times with Xylene. Permount (Fischer Scientific, SP15-500) was used for coverslips.
Fluorojade B staining
PFA-perfused slices were treated with 100% ethanol for 3min, followed by 70% ethanol and dH2O for 1min each. Slices were then transferred to 0.06% potassium permanganate (P279-500, Fischer) solution for 15min on a shaker, followed by staining with 0.0001% Fluorojade B (Millipore, AG310) in a 0.1% acetic acid aqueous solution for 1hr. Slices were washed with dH2O 3 times, 1min each before mounting. Glass slides were air-dried in a drawer overnight, and then cleared by dipping into xylene 3X 1min each. Permount (Fisher Scientific SP12-500) was used for coverslips.
RNA extraction and cDNA preparation
Total RNA was extracted using TRIzol (Invitrogen 15596-026) and isopropyl alchohol, followed by washing with 75% ethanol. RNA was re-dissolved in RNase-free water and DNA was digested with Invitrogen DNAse I Amplification Grade (Invitrogen 18068-015) per manufacture’s instructions. Following inactivation with 25mM EDTA, RNA was reverse-transcribed into cDNA using SuperScript II RT kit (Invitrogen, 18064-022) as described by the manufacturer.
Conventional PCR and q-PCR
Conventional PCR was performed with Taq (Invitrogen, 10342-053) with the following parameters: 1X 95°C 2min, 20X 95°C 30s, 52°C 30s, 72°C 1min, 1X 72°C 10min. Primers specific for LAMP-2A used were: 5′-GGTCTCAAGCGCCATCATAC-3′ and its reverse compliment. PCR products were run on a 2% agarose gel and visualized with Biorad Gel Dox XR system. For q-PCR, cDNA was mixed with SYBR Green (Applied Biosystems, 4309155) per manufacturer’s instructions. q-PCR was performed with ABI 7300 Real Time PCR system with the following parameters: 1X 50°C 2min, 1X 95°C 10min, 40X 95°C 15s, 40X 60°C 1min, 1X 95°C 15s, 60°C 1min, 95°C 15s, 60°C 15s. Primers specific for DAPK1 used were: 5′ –CTCAGTGGTGTCCCGGTG-3′ and its reverse compliment. Primers for ACTB were a generous gift from Dr. Max Cynader, University of British Columbia.
Statistical analyses
No statistical methods were used to pre-determine sample sizes but were similar to those generally employed in the field. We did not perform formal randomization, although cell cultures and animals were chosen randomly for each experimental group. All procedures were preformed under the same conditions with internal controls. Data are expressed as means ± s.e.m. Quantifications were conducted using at least three independent experiments. Statistical significance was defined as * or Δ P < 0.05, ** or ΔΔ P < 0.01, *** or ΔΔΔ p<0.001. One way ANOVA (Fischer LSD method) was used unless otherwise specified, data was tested for normality (Shapiro-Wilk test, power 0.05) and equal variance (power 0.05) prior commencing ANOVA analysis.
Supplementary Material
Acknowledgments
We thank Yuping Li for technical support and Dr. Loren Oschipok for his excellent editorial assistance. We also thank Lin Luo for her help in obtaining whole-brain images of DAPK1 immunostaining in Fig. 6f. This work was supported by the Canadian Institutes of Health Research (CIHR) and Taiwan Department of Health Clinical Trial and Research Center of Excellence (DOH102-TD-B-111-004). YTW is the holder of Heart and Stroke Foundation of British Columbia and Yukon Chair in Stroke Research.
Footnotes
Author Contributions
XF and WYJ designed and performed experiments, as well as analyzed the data. XF also wrote the manuscript. JL and JW assisted in performing some of the molecular biochemical experiments. YTW designed the study, supervised the overall project and wrote the manuscript.
References
- 1.Houdebine LM. Transgenic animal models in biomedical research. Methods Mol Biol. 2007;360:163–202. doi: 10.1385/1-59745-165-7:163. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto A, Hen R, Dauer WT. The Ons and Offs of Inducible Transgenic Technology: A Review. Neurobiology of Disease. 2001;8:923–932. doi: 10.1006/nbdi.2001.0452. [DOI] [PubMed] [Google Scholar]
- 3.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. 2012:1–16. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banaszynski LA, Wandless TJ. Conditional Control of Protein Function. Chemistry & Biology. 2006;13:11–21. doi: 10.1016/j.chembiol.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nature Publishing Group. 2011;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- 7.Bonger KM, Chen LC, Liu CW, Wandless TJ. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nature Chemical Biology. 2011;7:531–537. doi: 10.1038/nchembio.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neklesa TK, et al. Small-molecule hydrophobic tagging–induced degradation of HaloTag fusion proteins. Nature Chemical Biology. 2011;7:538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AGL, Wandless TJ. A Rapid, Reversible, and Tunable Method to Regulate Protein Function in Living Cells Using Synthetic Small Molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto KM, et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dice FJ. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends in biochemical sciences. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 13.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends in Cell Biology. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koga H, Martinez-Vicente M, Macian F, Verkhusha VV, Cuervo AM. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nature Communications. 2011;2:386–10. doi: 10.1038/ncomms1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Backer J, Bourret L, Dice JF. Regulation of catabolism of microinjected ribonuclease A requires the amino-terminal 20 amino acids. Proc Natl Acad Sci. 1983;80:2166–2170. doi: 10.1073/pnas.80.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113(Pt 24):4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 17.Slot LA, Lauridsen AM, Hendil K. Intracellular protein degradation in serum-deprived human fibroblasts. Journal of Biochemistry. 1986;237:491–498. doi: 10.1042/bj2370491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seglen PO, Reith A. Ammonia inhibition of protein degradation in isolated rat hepatocytes. Quantitative ultrastructural alterations in the lysosomal system. Exp Cell Res. 1976;100:276–280. doi: 10.1016/0014-4827(76)90148-8. [DOI] [PubMed] [Google Scholar]
- 19.Bauer PO, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 20.Cataldo AM, Nixon RA. Enzymatically active lysosomal proteases are associated with amyloid deposits in Alzheimer brain. Proc Natl Acad Sci USA. 1990;87:3861–3865. doi: 10.1073/pnas.87.10.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neff NT, Bourret L, Miao P, Dice JF. Degradation of proteins microinjected into IMR-90 human diploid fibroblasts. The Journal of Cell Biology. 1981;91:184–194. doi: 10.1083/jcb.91.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuervo AM. Impaired Degradation of Mutant -Synuclein by Chaperone-Mediated Autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 23.Cuervo AM, Terlecky SR, Dice JF, Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J Biol Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- 24.Henshall DC, et al. Expression of death-associated protein kinase and recruitment to the tumor necrosis factor signaling pathway following brief seizures. J Neurochem. 2003;86:1260–1270. doi: 10.1046/j.1471-4159.2003.01934.x. [DOI] [PubMed] [Google Scholar]
- 25.Shamloo M, et al. Death-associated protein kinase is activated by dephosphorylation in response to cerebral ischemia. J Biol Chem. 2005;280:42290–42299. doi: 10.1074/jbc.M505804200. [DOI] [PubMed] [Google Scholar]
- 26.Tu W, et al. DAPK1 Interaction with NMDA Receptor GluN2B Subunits Mediates Brain Damage in Stroke. Cell. 2010;140:222–234. doi: 10.1016/j.cell.2009.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen O, Feinstein E, Kimchi A. DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. EMBO J. 1997;16:998–1008. doi: 10.1093/emboj/16.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vivès E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 29.Traynelis SF, et al. Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacological Reviews. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris MC, Depollier J, Mery J, Heitz F, Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y. Amino-3-hydroxy-5-methylisoxazole-4-propionic Acid Subtype Glutamate Receptor (AMPAR) Endocytosis Is Essential for N-Methyl-D-aspartate-induced Neuronal Apoptosis. Journal of Biological Chemistry. 2004;279:41267–41270. doi: 10.1074/jbc.C400199200. [DOI] [PubMed] [Google Scholar]
- 32.Spillantini MG, et al. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 33.Shaltiel-Karyo R, et al. Inhibiting α-Synuclein Oligomerization by Stable Cell-Penetrating β-Synuclein Fragments Recovers Phenotype of Parkinson’s Disease Model Flies. PLoS ONE. 2010;5:e13863. doi: 10.1371/journal.pone.0013863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aarts M, et al. Treatment of ischemic brain damage by perturbing NMDA receptor-PSD-95 protein interactions. Science. 2002;298:846. doi: 10.1126/science.1072873. [DOI] [PubMed] [Google Scholar]
- 35.Iadecola C, Anrather J. Stroke research at a crossroad: asking the brain for directions. Nature Neuroscience. 2011;14:1363–1368. doi: 10.1038/nn.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisenberg-Lerner A, Kimchi A. DAP kinase regulates JNK signaling by binding and activating protein kinase D under oxidative stress. Cell Death Differ. 2007;14:1908–1915. doi: 10.1038/sj.cdd.4402212. [DOI] [PubMed] [Google Scholar]
- 37.Ricart KC, Fiszman ML. Hydrogen peroxide-induced neurotoxicity in cultured cortical cells grown in serum-free and serum-containing media. Neurochem Res. 2001;26:801–808. doi: 10.1023/a:1011660001941. [DOI] [PubMed] [Google Scholar]
- 38.Liu F, Schafer DP, McCullough LD. TTC, fluoro-Jade B and NeuN staining confirm evolving phases of infarction induced by middle cerebral artery occlusion. J Neurosci Methods. 2009;179:1–8. doi: 10.1016/j.jneumeth.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taghibiglou C, et al. Role of NMDA receptor–dependent activation of SREBP1 in excitotoxic and ischemic neuronal injuries. Nature Medicine. 2009:1–9. doi: 10.1038/nm.2064. [DOI] [PubMed] [Google Scholar]
- 40.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tymianski M. Can Molecular and Cellular Neuroprotection Be Translated Into Therapies for Patients?: Yes, but Not the Way We Tried It Before. Stroke. 2010;41:S87–S90. doi: 10.1161/STROKEAHA.110.595496. [DOI] [PubMed] [Google Scholar]
- 42.Banaszynski LA, Sellmyer MA, Contag CH, Wandless TJ, Thorne SH. Chemical control of protein stability and function in living mice. Nature Medicine. 2008;14:1123–1127. doi: 10.1038/nm.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mason JM. Design and development of peptides and peptide mimetics as antagonists for therapeutic intervention. Future Medicinal Chemistry. 2010;2:1813–1822. doi: 10.4155/fmc.10.259. [DOI] [PubMed] [Google Scholar]
- 44.Stein A, Aloy P. Contextual Specificity in Peptide-Mediated Protein Interactions. PLoS ONE. 2008;3:e2524. doi: 10.1371/journal.pone.0002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends in Endocrinology & Metabolism. 2010;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill M. Evaluating Neuroprotection in Aneurysm Coiling Therapy (ENACT) Trial Final Results. International Stroke Conference; 2012. [Google Scholar]
- 47.Foy KC, Liu Z, Phillips G, Miller M, Kaumaya PTP. Combination treatment with HER-2 and VEGF peptide mimics induces potent anti-tumor and anti-angiogenic responses in vitro and in vivo. Journal of Biological Chemistry. 2011;286:13626–13637. doi: 10.1074/jbc.M110.216820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, et al. NMDA Receptor Subunits Have Differential Roles in Mediating Excitotoxic Neuronal Death Both In Vitro and In Vivo. Journal of Neuroscience. 2007;27:2846–2857. doi: 10.1523/JNEUROSCI.0116-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peineau S, et al. LTP Inhibits LTD in the Hippocampus via Regulation of GSK3β. Neuron. 2007;53:703–717. doi: 10.1016/j.neuron.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 50.Taghibiglou C, Lu J, Mackenzie IR, Wang YT, Cashman NR. Sterol regulatory element binding protein-1 (SREBP1) activation in motor neurons in excitotoxicity and amyotrophic lateral sclerosis (ALS): Indip, a potential therapeutic peptide. Biochemical and Biophysical Research Communications. 2011;413:159–163. doi: 10.1016/j.bbrc.2011.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.