Abstract
Background
Hormone receptors, including the estrogen receptor and progesterone receptor, human epidermal growth factor receptor 2 (HER2), and other biomarkers like Ki67, epidermal growth factor receptor (EGFR, also known as HER1), the androgen receptor, and p53, are key molecules in breast cancer. This study evaluated the relationship between HER2 and hormone receptors and explored the additional prognostic value of Ki67, EGFR, the androgen receptor, and p53.
Methods
Quantitative determination of HER2 and EGFR was performed in 240 invasive breast cancer tissue microarray specimens using quantum dot (QD)-based nanotechnology. We identified two subtypes of HER2, ie, high total HER2 load (HTH2) and low total HER2 load (LTH2), and three subtypes of hormone receptor, ie, high hormone receptor (HHR), low hormone receptor (LHR), and no hormone receptor (NHR). Therefore, breast cancer patients could be divided into five subtypes according to HER2 and hormone receptor status. Ki67, p53, and the androgen receptor were determined by traditional immunohistochemistry techniques. The relationship between hormone receptors and HER2 was investigated and the additional value of Ki67, EGFR, the androgen receptor, and p53 for prediction of 5-year disease-free survival was assessed.
Results
In all patients, quantitative determination showed a statistically significant (P<0.001) negative correlation between HER2 and the hormone receptors and a significant positive correlation (P<0.001) between the estrogen receptor and the progesterone receptor (r=0.588), but a significant negative correlation (P<0.001, r=−0.618) with the HHR subtype. There were significant differences between the estrogen receptor, progesterone receptor, and HER2 subtypes with regard to total HER2 load and hormone receptor subtypes. The rates of androgen receptor and p53 positivity were 46.3% and 57.0%, respectively. Other than the androgen receptor, differences in expression of Ki67, EGFR, and p53 did not achieve statistical significance (P>0.05) between the five subtypes. EGFR and Ki67 had prognostic significance for 5-year disease-free survival in univariate analysis, but the androgen receptor and p53 did not. Multivariate analysis identified that EGFR expression had predictive significance for 5-year disease-free survival in hormone-receptor positive patients and in those with the lymph node-positive breast cancer subtype.
Conclusion
Hormone receptor expression was indeed one of the molecular profiles in the subtypes identified by quantitative HER2 and vice versa. EGFR status may provide discriminative prognostic information in addition to HER2 and hormone receptor status, and should be integrated into routine practice to help formulate more specific prediction of the prognosis and appropriate individualized treatment.
Keywords: quantum dots, breast cancer, molecular classification, prognosis, prediction
Introduction
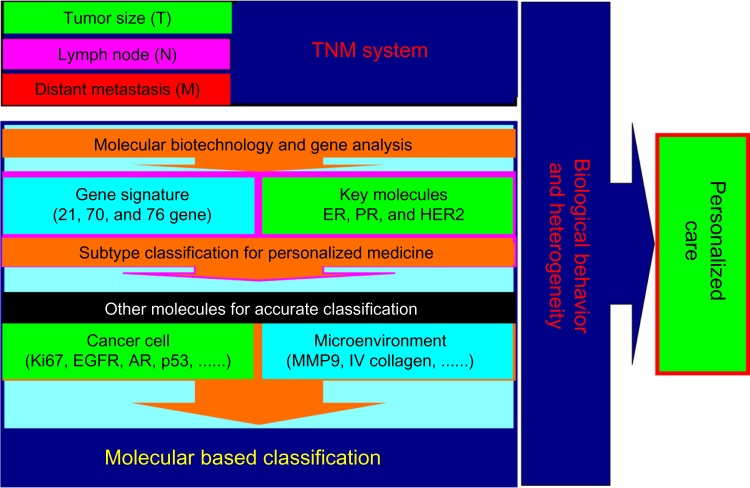
Breast cancer is a heterogeneous malignancy, and better understanding of its heterogeneity is essential for personalized care.1–3 Over the past two decades, classification of breast cancer based on key molecules and/or gene profiles has improved our understanding of the heterogeneity of this disease and revolutionized treatment concepts, resulting in improved survival and quality of life1,3–5 (Figure 1). A typical example is the ever increasing understanding of the molecular-based classification of breast cancer according to three common key molecules, including human epidermal growth factor receptor 2 (EGFR2, also known as HER2), the estrogen receptor, and the progesterone receptor.1
Figure 1.
Advancement of breast cancer classification. Accurate classification is essential for personalized care of breast cancer. The traditional classification method is the TNM staging system, which is important but not sufficient in the era of personalized care for breast cancer. With the development of biotechnology and gene analysis, the profiles of some key molecules have been identified and taken into the breast cancer classification system, resulting in revolutionized treatment of the disease. Other molecules in cancer cells as well as in the tumor microenvironment should be evaluated and integrated to understand further the biological behavior of breast cancer and improve the molecular-based classification by new approaches like quantum dot-based nanotechnology. In the future care of breast cancer, each patient with the disease would have personalized care based on combination of a molecular-based classification and the TNM classification.
Abbreviations: AR, androgen receptor; EGFR, epidermal growth factor receptor; ER, estrogen receptor; MMP-9, matrix metallopeptidase 9; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Identification of distinct biological subtypes of breast cancer according to three key molecules, ie, the HER2 and hormone (estrogen and progesterone) receptors, has become increasingly important for selection of specific treatment strategies, prediction of prognosis, and monitoring of the disease course in clinical practice.1 Nevertheless, molecular-based subtype classification of breast cancer using these three molecules alone is insufficient to reflect the complex biological characteristics of the disease or meet the need for personalized care.1,6–9 For example, about one third of hormone receptor-positive patients benefit little from endocrine therapy according to current criteria.1,5,10 Further, some hormone receptor-positive patients with a potentially better prognosis are subjected to intensive chemotherapy but suffer from toxicity (overtreatment), while other patients at high risk of recurrence receive inadequate treatment, leading to compromised efficacy (undertreatment). Another convincing example is the limited utility of the current criteria in guiding molecular-targeted HER2 therapy. Nearly half of patients identified as HER2-positive do not respond to targeted trastuzumab injection (Herceptin®) therapy alone,1,8 while some patients in the nearly 80% of patients with breast cancer defined as HER2-negative might benefit from targeted therapy but cannot be identified by current methods. Moreover, the appropriate dosing and optimal schedule of conventional combination HER2 and hormone receptor therapy and other targeted agents need to be comprehensively investigated.
These observations indicate that more molecular information is needed to maximize the benefit from treatment.3,8 This requires establishing a new approach with a more rational, comprehensive breast cancer classification system based on the three key molecules to understand further the heterogeneity of the disease in individual patients.1 Two aspects should be considered in this new prediction system. One is to develop a accurate classification by improving the methodology and/or algorithm for the three key molecules,10 as demonstrated in our recent series of studies11–14 using quantum dot (QD)-based nanotechnology with highly sensitive, quantitative, and real-time molecular imaging in situ for quantitative analysis of HER2 and the hormone receptors. Our results have shown that incorporation of quantitative determination of HER2 and hormone receptors, along with a holistic approach for assessing HER2 status by integrating quantitative HER2 determination and tumor size (total HER2 load), could reveal the heterogeneity of breast cancer and identify five subtypes with different 5-year prognoses. Moreover, five molecular subtypes based on quantitative information from simple HER2 and hormone (estrogen and progesterone) receptor status could be as informative as multigene analysis, which in turn might be helpful for formulating more personalized comprehensive therapeutic strategies and prediction of breast cancer prognosis, but the intrinsic mechanisms involved should be evaluated further. The other aspect is to explore complementary molecules and/or genes and incorporate them into the current classification system. Gene signature assays are usually considered to be the gold standard, but the expense and technical complexities involved limit their routine clinical use. Further, classification of breast cancer according to certain key molecules could be as informative as multigene analysis according to a recent report15 and our own previous work.14,16,17 Therefore, the most convenient and cost-effective method is to incorporate other key complementary molecules into the current classification.1,18,19 In our previous studies, we had identified two additional molecules, ie, Ki67 and the epidermal growth factor receptor 1 (HER1) that can help to predict the prognosis.20,21 However, whether these molecules have a predictive value additional to that of HER2 and the hormone receptors needs further evaluation. This study assessed the quantitative relationship between HER2 and the hormone receptors and explored the added prognostic value of Ki67, EGFR, the androgen receptor, and p53.
Materials and methods
Patients and materials
Formalin-fixed, paraffin-embedded specimens from 240 patients with invasive breast cancer were collected from Hubei Cancer Hospital, People’s Republic of China, between January 2002 and December 2006. Information on type of surgery and adjuvant treatment (chemotherapy, radiotherapy, endocrine therapy) was obtained from the medical records of each patient. HER2-positive patients did not receive molecular targeted therapy. The major pathological parameters for these patients have been reported elsewhere.11–14,20
Detection of HER2, EGFR, and hormone receptor status by QD nanotechnology
Tissue microarrays from the breast cancer specimens were constructed using standard procedures as previously described.13,14,20 The antibodies, QD-conjugated streptavidin probes with an emission wavelength of 605 nm, and related reagents for detection of HER2, EGFR, and estrogen and progesterone receptor status were the same as before, with use of the following major procedures: deparaffinizing → antigen retrieval → blocking → primary antibody → washing and blocking → biotinylated IgG → washing and blocking → 605-QD-conjugated streptavidin probes → washing → mounting and detection. QD fluorescence signal information on these key molecules was acquired and analyzed using a fluorescence microscope (Olympus BX51 fluorescence microscope, Olympus Optical Co, Ltd, Tokyo, Japan) and multispectral imaging systems (Cambridge Research and Instrumentation, Inc., Woburn, MA, USA) as previously described.13,14 Quantitative determination of hormone receptor, HER2, and EGFR status in the 240 invasive breast cancer tissue microarray specimens was performed previously using QD nanotechnology. Two breast cancer subtypes, ie, high total HER2 load (HTH2) and low total HER2 load (LTH2), were identified by quantitative analysis of HER2, and three breast cancer subtypes, ie, high hormone receptor (HHR), low hormone receptor (LHR), and no hormone receptor (NHR), were identified by quantitative analysis of hormone receptors using QD nanotechnology. Therefore, the patients could be divided into five breast cancer subtypes by HER2 and hormone receptor status (HHR, LHR-LTH2, LHR-HTH2, NHR-LTH2, and NHR-HTH2).13,14
Detection of Ki67, androgen receptor, and p53 status by immunohistochemistry
Ki67 (purified anti-MKI67 mouse monoclonal antibody; clone 3D11, 1:100 dilution; Beijing ZSGB-BIO, Beijing, People’s Republic of China), the androgen receptor (mouse anti-human monoclonal antibodies against androgen receptor; clone AR27, 1:150 dilution; Beijing ZSGB-BIO), and p53 (purified TP53 mouse monoclonal antibody; clone 3G9; 1:100 dilution; Beijing ZSGB-BIO) were determined by conventional immunohistochemistry (IHC) methods, with the following major procedures: deparaffinizing → antigen retrieval → blocking → primary antibody (phosphate-buffered saline for control group) → washing → blocking → biotinylated secondary antibody → washing → blocking → DAB → washing → mounting and observation. Images of expression of these molecules on IHC were captured and evaluated using an Olympus BX51 fluorescence microscope equipped with an Olympus DP72 camera (Olympus Optical Co, Ltd, Tokyo, Japan, Figure 3).
Figure 3.
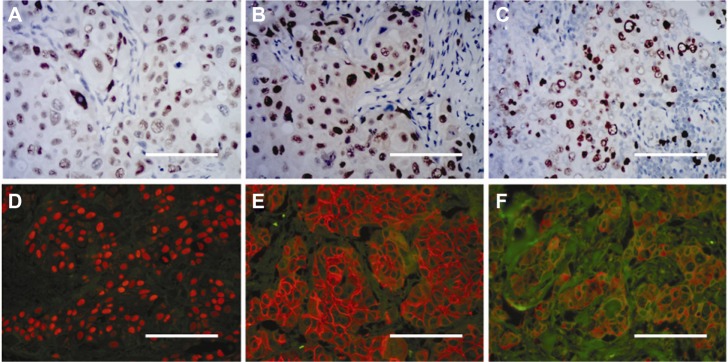
Molecular determination by conventional immunohistochemistry and quantum dot immunohistochemistry. Androgen receptor (A), p53 (B), and Ki67 (C) determination by conventional immunohistochemistry. Estrogen receptor (D), HER2 (E), and epidermal growth factor receptor (F) imaged by quantum dot immunohistochemistry.
Note: Scale bar, 50 μm.
Statistical analysis
Because all the quantitative variables for HER2 and the hormone receptors were non-normally distributed, comparisons of values in the different subtype groups were done using the Mann–Whitney U test or Kruskal–Wallis H test, and correlations were calculated using the Spearman’s rho test. The chi-square test was used to compare the positivity rates for molecules between the subtype groups. Five-year disease-free survival was the primary endpoint, and was analyzed using the Kaplan–Meier and log-rank tests. A multivariate Cox proportional hazards regression model was used to analyze the independent prognostic factors. Statistical analyses were performed using SPSS version 13 software (SPSS Inc., Chicago, IL, USA), and a two-tailed P<0.05 was considered to be statistically significant.
Results
Correlation between HER2, estrogen, and progesterone receptors by QD nanotechnology
Our previous work demonstrated a negative correlation between HER2 and the estrogen receptor.12 The present study shows a significant negative correlation between HER2 and the estrogen and progesterone receptors (Spearman’s rho, P<0.001; HER2 versus estrogen receptor, r=−0.324; HER2 versus progesterone receptor, r=−0.268) and a significant positive correlation between the estrogen and progesterone receptors (P<0.001, r=0.588). Further analysis showed similar correlations between the three molecules in hormone receptor-positive patients, but correlations between HER2 and the estrogen receptor and between the estrogen receptor and the progesterone receptor became less significant (HER2 versus the estrogen receptor, P=0.014, r=−0.184; the estrogen receptor versus the progesterone receptor, P<0.001, r=0.324), and the correlation between HER2 and the progesterone receptor was not statistically significant (P=0.151). In contrast, correlations between the three molecules were more significant (HER2 versus estrogen receptor, r=−0.433; HER2 versus progesterone receptor, r=−0.276; estrogen receptor versus the progesterone receptor, r=0.621) in the HTH2 group identified in a previous study.13 It should be noted that the estrogen receptor and the progesterone receptor showed a significant negative correlation (P<0.001, r=−0.618) in the HHR group identified earlier.14
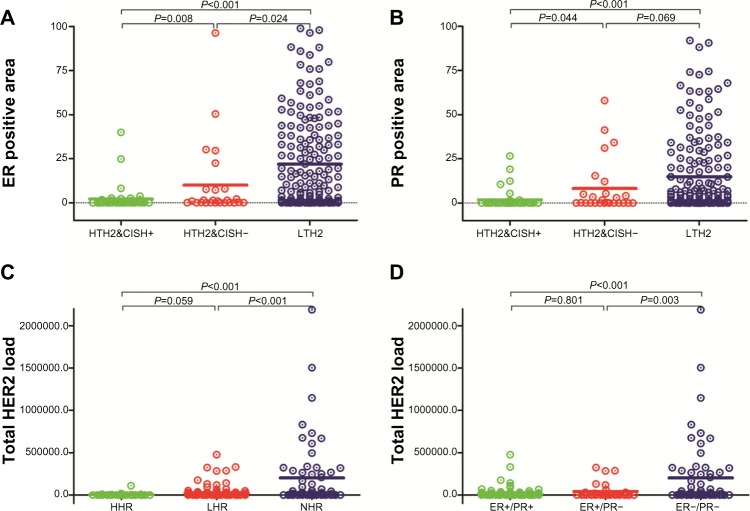
To study the correlations between the three molecules further, we compared differences in hormone receptor expression between the three subgroups based on total HER2 load (TH2) in our previous study13 (Figures 2 and 3). The results showed significant differences in hormone receptor expression between the three groups (P<0.0001). Further analysis showed that the difference in estrogen receptor expression between the groups was statistically significant (Figure 2A). In contrast, with the exception of a difference in progesterone receptor expression between HTH2–/chromogenic in situ hybridization (CISH)– and LHT2 groups (P=0.069), progesterone receptor expression between the other subgroups was also statistically significant (Figure 2B). Likewise, we also compared differences in TH2 expression between the three subgroups based on hormone receptor status14 (Figure 2C and D). The results showed significant differences in TH2 between the three groups (P<0.0001), and further analysis showed that other than the groups between HHR and LHR (P=0.059), the differences between the other two groups (HHR vs NHR; LHR vs NHR) achieved statistical significance (P<0.001).
Figure 2.
Expression of HER2 and hormone receptors in breast cancer subtypes by total HER2 load or hormone receptors. Expression of estrogen receptor (A) and progesterone receptor (B) in three subtypes identified by total HER2 load previously. Expression of TH2 in new subtype classification by quantitative analysis of hormone receptors based on quantum dot nanotechnology (C) and by conventional classification using traditional methods (D).
Abbreviations: CISH, chromogenic in situ hybridization; HTH2&CISH+, high total HER2 load (HTH2) and CISH-positive; HTH2&CISH−, HTH2 and CISH negative; LTH2, low TH2; HHR, high hormone receptor expression; LHR, low hormone receptor expression; NHR, negative hormone receptor expression; TH2, total HER2 load; HER2, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor.
Ki67, p53, EGFR, and androgen receptor expression by quantitative HER2 and hormone receptor analysis
We then investigated androgen receptor and p53 expression by IHC in our 240 breast cancer patients (Figure 3). Androgen receptor expression was successfully determined in 229 cases, and the positivity rate was 46.3% (106/229) in all patients, 53.2% (91/171) in hormone receptor-positive patients, and 25.9% (15/58) in hormone receptor-negative patients (P<0.001). The positivity rate was 43.8% (28/64) in HTH2 patients and 47.3% (78/165) in LTH2 patients (P=0.631). Similarly, p53 expression was successfully determined in 228 cases, and the positivity rate was 57.0% (130/228) in all patients, 55.6% (94/169) in hormone receptor-positive patients, and 61.0% (36/59) in hormone receptor-negative patients (P=0.471). The positivity rate was 67.7% (44/65) in HTH2 patients and 52.8% (86/163) in LTH2 patients (P=0.04). In our previous studies,20,21 we also detected EGFR expression by QD nanotechnology and Ki67 by IHC (Figure 3), as well as expression of the androgen receptor, p53, EGFR, and Ki67, among the five breast cancer subtypes by TH2 and hormone receptors, as listed in Table 1. The results show that other than the androgen receptor, differences in expression of p53, EGFR, and Ki67 between the five subtypes did not achieve statistical significance (P>0.05). Notably, the highest positivity rates for EGFR and Ki67 were in the NHR-LTH2 group, the highest positivity rate for the androgen receptor was in the HHR group (P<0.05 with other groups), and the lowest rate was in the NHR-LTH2 group (P<0.05 with other groups).
Table 1.
Expression of androgen receptor, p53, EGFR, and Ki67 molecules in five subtypes of breast cancer
| HHR n (%) | LHR-HTH2 n (%) | LHR-LTH2 n (%) | NHR-HTH2 n (%) | NHR-LTH2 n (%) | P-value | |
|---|---|---|---|---|---|---|
| EGFR | 0.381 | |||||
| Positive | 21 (52.5) | 20 (58.8) | 48 (46.2) | 21 (60.0) | 17 (63.0) | |
| Negative | 19 (47.5) | 14 (41.2) | 56 (53.8) | 14 (40.0) | 10 (37.0) | |
| Ki67 | 0.077 | |||||
| Positive | 17 (42.5) | 22 (64.7) | 49 (47.1) | 17 (48.6) | 19 (70.4) | |
| Negative | 23 (57.5) | 12 (35.3) | 55 (52.9) | 18 (51.4) | 8 (29.6) | |
| AR* | <0.001 | |||||
| Positive | 29 (76.3) | 17 (51.5) | 45 (45.0) | 11 (35.5) | 4 (14.8) | |
| Negative | 9 (23.7) | 16 (48.5) | 55 (55.0) | 20 (64.5) | 23 (85.2) | |
| p53** | 0.164 | |||||
| Positive | 24 (63.2) | 22 (66.7) | 48 (49.0) | 22 (68.8) | 14 (51.9) | |
| Negative | 14 (36.8) | 11 (33.3) | 50 (51.0) | 10 (31.2) | 13 (48.1) |
Notes: For Ki67, >13% tumor cell nuclei staining was defined as positive; for AR and p53, >10% tumor cell nuclei staining was defined as positive
AR status could not be obtained for 11 patients
p53 status could not be obtained for 12 patients.
Abbreviations: AR, androgen receptor; EGFR, epidermal growth factor receptor; HHR, high hormone receptor expression; LHR, low hormone receptor expression; NHR, negative hormone receptor expression; HTH2, high total HER2 load; LTH2, low total HER2 load.
Additional prognostic value of p53 Ki67, EGFR, and the androgen receptor
The 5-year disease-free survival rates in the androgen receptor-positive and androgen receptor-negative groups were 63.2% (39/106) and 60.2% (49/123), respectively (Kaplan–Meier analysis, log-rank test, P=0.511). Likewise, 5-year disease-free survival rates in the p53-positive and p53-negative subgroups were 60.0% (52/130) and 65.3% (34/98), respectively (P=0.288). Further analysis showed that the 5-year disease-free survival rates between the androgen receptor-positive group and androgen receptor-negative group and between the p53-positive group and p53-negative group were not significantly different in the hormone receptor-positive or hormone receptor-negative subgroups (P>0.05). In our previous study,20,21 we confirmed the ability of EGFR and Ki67 to predict 5-year disease-free survival using univariate survival analysis, so we added these two molecules for further multivariate analysis. The results (Table 2) show that the significance of the subtypes by combination of TH2 and hormone receptor status are similar to previous findings,14 especially for the predictive subtypes of LHR-HTH2 and NHR-HTH2, but EGFR and Ki67 were not. Further analysis showed that EGFR expression was predictive of 5-year disease-free survival in both hormone receptor-positive and lymph node-positive patients (Table 3).
Table 2.
Multivariate analysis by 5-year disease-free survival
| Items | Hazard ratio (95% CI) | P-value |
|---|---|---|
| Overall subtypes | 0.028 | |
| HHR* | Reference | – |
| LHR-LTH2 | 1.984 (0.866–4.543) | 0.105 |
| LHR-HTH2 | 2.824 (1.148–6.947) | 0.024 |
| NHR-LTH2 | 2.190 (0.851–5.637) | 0.104 |
| NHR-HTH2 | 3.853 (1.605–9.252) | 0.003 |
| EGFR | 1.418 (0.901–2.232) | 0.131 |
| Ki67 | 1.149 (0.719–1.836) | 0.56 |
| Tumor size | 1.518 (1.042–2.212) | 0.03 |
| Tumor grade | 2.626 (1.719–4.011) | <0.001 |
| Lymph nodes | 3.605 (2.073–6.270) | <0.001 |
Note:
HHR subgroup used as reference state in multivariate analysis.
Abbreviations: EGFR, epidermal growth factor receptor; CI, confidence interval; HHR, high hormone receptor expression; LHR, low hormone receptor expression; NHR, negative hormone receptor expression; HTH2, high total HER2 load; LTH2, low total HER2 load.
Table 3.
Multivariate analysis by 5-year disease-free survival in patients with hormone receptor-positive or lymph node-positive breast cancer
| Items | HR-positive patients
|
Lymph node-positive patients
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Overall subtypes | 0.042 | 0.003 | ||
| HHR* | Reference | – | Reference | – |
| LHR-LTH2 | 2.071 (0.900–4.767) | 0.087 | 1.917 (0.775–4.744) | 0.159 |
| LHR-HTH2 | 3.214 (1.292–7.992) | 0.012 | 3.953 (1.514–10.318) | 0.005 |
| NHR-LTH2 | – | – | 1.806 (0.607–5.370) | 0.288 |
| NHR-HTH2 | – | – | 4.746 (1.842–12.227) | 0.001 |
| EGFR | 1.847 (1.064–3.206) | 0.029 | 1.720 (1.023–2.895) | 0.041 |
| Ki67 | 1.144 (0.642–2.038) | 0.647 | 1.151 (0.680–1.950) | 0.601 |
| Tumor size | 1.290 (0.770–2.159) | 0.334 | 1.518 (0.863–1.989) | 0.205 |
| Tumor grade | 2.358 (1.385–4.016) | 0.002 | 1.982 (1.256–3.130) | 0.003 |
| Lymph node | 4.506 (2.150–9.445) | <0.001 | – | – |
Note:
HHR subgroup used as reference state in multivariate analysis.
Abbreviations: HHR, high hormone receptor expression; LHR, low hormone receptor expression; NHR, negative hormone receptor expression; HTH2, high total HER2 load; LTH2, low total HER2 load; HR, hazards ratio; CI, confidence interval; EGFR, epidermal growth factor receptor.
Discussion
Molecular classification is playing an increasingly important role in the personalized care of breast cancer, and three key molecules, ie, HER2, the estrogen receptor, and the progesterone receptor, have been evaluated in routine clinical practice.1,5,18,22 However, the biological significance and internal interactions of these three molecules should be investigated further to determine their heterogeneity and guide treatment in the complexities of the clinical setting, which is one of the key issues for personalized care of breast cancer.1,3,14,23 In this study, we investigated these relationships based on quantitative determination of HER2 and hormone receptor status by QD nanotechnology, and found them to be consistent with our own previous results12 and other reports.1,9 In addition, expression of the three molecules according to the different subtypes identified by TH2 and hormone receptor status previously,13,14 indeed achieved statistical significance, suggesting intrinsic interactions between the HER2, estrogen, and progesterone receptors evaluated by our new approach. Moreover, the relationships between the three molecules displayed different patterns on further subtype analysis, being weak in hormone receptor-positive patients and strong in HTH2 patients. Of note, the estrogen receptor and progesterone receptor even showed an inverse correlation in HHR patients, suggesting potential biological behavior in this subtype with simple endocrine therapy or less intensive chemotherapy regimens as supported by other clinical studies.1,10
An increasing number of studies have demonstrated that the current breast cancer classification based merely on HER2 and hormone receptor status cannot provide enough information for clinical practice (Figure 1). In this molecular era of personalized medicine, other biomarkers, such as the androgen receptor, P53, Ki67 and EGFR, might provide additional information and/or deeper insights into clinical care for breast cancer.1,18,24–27 In our previous studies,20,21 we evaluated the value of EGFR and Ki67, which could have potential prognostic value in breast cancer, but the additional value of these two molecules in relation to HER2 and the hormone receptors should be further investigated. Therefore, in a subsequent study, EGFR and Ki67, as well as the androgen receptor and p53, were selected to assess their additional value for prediction of 5-year disease-free survival.
Like the estrogen and progesterone receptors, the androgen receptor is a steroid hormone receptor and has been found to be expressed in approximately 58.1%–78.7% of breast cancers, suggesting potential prognostic value and the possibility of specific treatment protocols for patients with certain subtypes of breast cancer.24,28–34 Therefore, the androgen receptor is potentially an important molecule in the clinical care of breast cancer. In this study, androgen receptor positivity rates were significantly different in the subtype groups, ranging from 76.3% in the HHR group to 14.8% in the NHR-LTH2 group, indicating that the androgen receptor plays different roles in these different subtypes and necessitates diverse androgen receptor-targeted therapy.28–34 For example, androgen receptor signaling might be one of the major mechanisms involved in stimulating the action of hormone receptors, or vice versa. Androgen receptor-targeted therapy in addition to endocrine therapy might be a sensible approach for this group. Several studies have also suggested that the androgen receptor might be a good predictor of survival in patients with breast cancer.30,31,33 In this study, 5-year disease-free survival in androgen receptor-positive patients was better than that in androgen receptor-negative patients, but the difference did not achieve statistical significance in univariate survival analysis.
p53, a tumor suppressor protein, is similarly important for the personalized care of patients with breast cancer,25,35–37 including for prediction of prognosis, response to chemotherapy and endocrine therapy, and selection of appropriate treatment strategies. In this study, the p53 positivity rate was more than 50% in the NHR-LTH2 group, so p53 might be an effective target for this aggressive breast cancer subtype, as suggested elsewhere.25 However, p53 had no statistically significant ability to predict 5-year disease-free survival in all patients or in hormone receptor-positive patients. Therefore, the value of the androgen receptor and p53 for prediction of the prognosis in patients with breast cancer remains controversial in this study, possibly due to the small number of patients, short duration of follow-up, and composition of the different subtypes. Further evaluations are needed in this regard.
We evaluated the additional prognostic value of Ki67 and EGFR by multivariate analysis. Ki67, a key molecule associated with tumor cell proliferation, has recently emerged as a prognostic indicator and a guide to treatment selection, and has become an integral part of clinicopathological practice in the treatment of breast cancer.1,18,21,27,38 In this study, Ki67 was a prognostic indicator in univariate analysis, but was not predictive in multivariate analysis, either in patients with hormone receptor-positive breast cancer or in those with lymph node-positive breast cancer. This result suggests that Ki67 had no value additional to that of HER2 and the hormone receptors, but might have had some value for treatment selection; for example, patients in the NHR-LTH2 and LHR-HTH2 groups with the highest Ki67 positivity rate might obtain benefit from chemotherapy, as suggested in other reports.22,39,40
EGFR, a key molecule in the EGF receptor family, has an important role in the development and progression of breast cancer.41,42 Moreover, it may be an excellent therapeutic target, as has been increasingly reported in recent years, especially with regard to lapatinib, an agent targeted to HER2 and HER1.23,41–43 In this study, EGFR had prognostic value in univariate analysis and in the hormone receptor-positive and lymph node-positive subgroups in multivariate analysis, indicating additional value for prediction of 5-year disease free survival. These results might have some implications for clinical practice. In hormone receptor-positive patients, lower hormone receptor levels may contribute to the less favorable response to trastuzumab seen in HER2-positive patients, which has been one of the key issues to be resolved in targeted therapy. In our study, more than 45% patients in the LHR-HTH2 and LHR-LTH2 groups had overexpression of EGFR, and these patients might achieve additional benefit from therapy targeted to EGFR, as suggested by others.23,41–44 In lymph node-positive patients, those with HTH2 might derive the most benefit from EGFR-targeted therapy.
We acknowledge some limitations to our study. First, the results should be evaluated further in a larger multicenter breast cancer patient population with long-term follow-up. Second, the intrinsic molecular mechanisms involved merit further exploration. Further, the relationships and additional values of these molecules expressed on tumor cells with other molecules expressed in the tumor microenvironment (displays dynamic interactions with tumor for continued tumor growth and progression) should be further investigated. Recently, we have undertaken a comprehensive molecular characterization in the tumor microenvironment45 and studied the complex dynamic interactions between tumor tissue and the microenvironment using QD-based multicolor imaging,46,47 providing new perspectives in understanding the biological behavior of tumors and potentially enabling more accurate tumor assessment for personalized decision-making. Therefore, the relationships between molecules and intrinsic mechanisms in tumor cells (like HER2, the estrogen and progesterone receptors, and EGFR) and in the tumor microenvironment (eg, the proteolytic enzyme matrix metallopeptidase 9, type IV collagen, and chemokine ligand 5) will be evaluated further in future research (Figure 1).
In summary, we have evaluated further the correlations between quantitative HER2 and the hormone receptors using QD nanotechnology and the additional prognostic value of four further key molecules (the androgen receptor, p53, EGFR, and Ki67). Significant expression of the estrogen and progesterone receptors was indeed one of the molecular profiles in the subtypes identified by quantitative HER2, and vice versa, and further demonstrates the value of quantitative evaluation of HER2 and hormone receptor expression for subtype classification and potential selection of a treatment strategy. Further, quantitative analysis of EGFR by QD nanotechnology could provide discriminative information in addition to HER2 and the hormone receptors, and could be integrated into routine practice to enable more specific prediction of prognosis and appropriate personalized treatment.
Acknowledgments
This research was supported by grants from the National Science Foundation of China (81201196, 81230031), the Fundamental Research Funds for the Central Universities (121004), the Key Scientific Research Project of Hubei Provincial Department of Education (D20126102), the Natural Science Foundation of Hubei Province, People’s Republic of China (301130851, 2011CBD489), and the Research Foundation of Public Health Bureau of Hubei Province (JS-2011018, JX4B19, JX3A14).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C, Peng J, Sun SR, Peng CW, Li Y, Pang DW. Tapping the potential of quantum dots for personalized oncology: current status and future perspectives. Nanomedicine. 2012;7(3):411–428. doi: 10.2217/nnm.12.9. [DOI] [PubMed] [Google Scholar]
- 3.Tamimi RM, Colditz GA, Hazra A, et al. Traditional breast cancer risk factors in relation to molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;131(1):159–167. doi: 10.1007/s10549-011-1702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufmann M, Pusztai L. Use of standard markers and incorporation of molecular markers into breast cancer therapy: Consensus recommendations from an International Expert Panel. Cancer. 2011;117(8):1575–1582. doi: 10.1002/cncr.25660. [DOI] [PubMed] [Google Scholar]
- 6.Nahta R, O’Regan RM. Therapeutic implications of estrogen receptor signaling in HER2-positive breast cancers. Breast Cancer Res Treat. 2012;135(1):39–48. doi: 10.1007/s10549-012-2067-8. [DOI] [PubMed] [Google Scholar]
- 7.Montemurro F, Rossi V, Cossu Rocca M, et al. Hormone-receptor expression and activity of trastuzumab with chemotherapy in HER2-positive advanced breast cancer patients. Cancer. 2012;118(1):17–26. doi: 10.1002/cncr.26162. [DOI] [PubMed] [Google Scholar]
- 8.Saxena R, Dwivedi A. ErbB family receptor inhibitors as therapeutic agents in breast cancer: current status and future clinical perspective. Med Res Rev. 2012;32(1):166–215. doi: 10.1002/med.20209. [DOI] [PubMed] [Google Scholar]
- 9.Pinhel I, Hills M, Drury S, et al. ER and HER2 expression are positively correlated in HER2 non-overexpressing breast cancer. Breast Cancer Res. 2012;14(2):R46. doi: 10.1186/bcr3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brouckaert O, Paridaens R, Floris G, Rakha E, Osborne K, Neven P. A critical review why assessment of steroid hormone receptors in breast cancer should be quantitative. Ann Oncol. 2013;24(1):47–53. doi: 10.1093/annonc/mds238. [DOI] [PubMed] [Google Scholar]
- 11.Chen C, Peng J, Xia HS, et al. Quantum dots-based immunofluorescence technology for the quantitative determination of HER2 expression in breast cancer. Biomaterials. 2009;30(15):2912–2918. doi: 10.1016/j.biomaterials.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Peng J, Xia HS, et al. Quantum dots-based immunofluorescent imaging of HER2 and ER provides new insights into breast cancer heterogeneity. Nanotechnology. 2010;21(9):95–101. doi: 10.1088/0957-4484/21/9/095101. [DOI] [PubMed] [Google Scholar]
- 13.Chen C, Xia HS, Gong YP, et al. The quantitative detection of total HER2 load by quantum dots and the identification of a new subtype of breast cancer with different 5-year prognosis. Biomaterials. 2010;31(33):8818–8825. doi: 10.1016/j.biomaterials.2010.07.091. [DOI] [PubMed] [Google Scholar]
- 14.Chen C, Sun SR, Gong YP, et al. Quantum dots-based molecular classification of breast cancer by quantitative spectroanalysis of hormone receptors and HER2. Biomaterials. 2011;32(30):7592–7599. doi: 10.1016/j.biomaterials.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Bartlett JM, Bloom KJ, Piper T, et al. Mammostrat as an immunohistochemical multigene assay for prediction of early relapse risk in the tamoxifen versus exemestane adjuvant multicenter trial pathology study. J Clin Oncol. 2012;30(36):4477–4484. doi: 10.1200/JCO.2012.42.8896. [DOI] [PubMed] [Google Scholar]
- 16.Brouckaert O, Laenen A, Vanderhaegen J, et al. Applying the 2011 St Gallen panel of prognostic markers on a large single hospital cohort of consecutively treated primary operable breast cancers. Ann Oncol. 2012;23(10):2578–2584. doi: 10.1093/annonc/mds062. [DOI] [PubMed] [Google Scholar]
- 17.Cuzick J, Dowsett M, Pineda S, et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol. 2011;29(32):4273–4278. doi: 10.1200/JCO.2010.31.2835. [DOI] [PubMed] [Google Scholar]
- 18.Sheri A, Dowsett M. Developments in Ki67 and other biomarkers for treatment decision making in breast cancer. Ann Oncol. 2012;23(Suppl 10):x219–x227. doi: 10.1093/annonc/mds307. [DOI] [PubMed] [Google Scholar]
- 19.Millar EK, Graham PH, McNeil CM, et al. Prediction of outcome of early ER+ breast cancer is improved using a biomarker panel, which includes Ki-67 and p53. Br J Cancer. 2011;105(2):272–280. doi: 10.1038/bjc.2011.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang XQ, Chen C, Peng CW, et al. Quantum dot-based quantitative immunofluorescence detection and spectrum analysis of epidermal factor receptor in breast cancer tissue arrays. Int J Nanomed. 2011;6:2265–2273. doi: 10.2147/IJN.S24161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang XQ, Wang FB, Chen C, Peng CW, Zhang JF, Li Y. High Ki-67 expression is a poor prognostic indicator of 5-year recurrence free survival in patients with invasive breast cancer. Asian Pac J Cancer Prev. 2011;12(11):3101–3105. [PubMed] [Google Scholar]
- 22.Wang Y, Yin Q, Yu Q, et al. A retrospective study of breast cancer subtypes: the risk of relapse and the relations with treatments. Breast Cancer Res Treat. 2011;130(2):489–498. doi: 10.1007/s10549-011-1709-6. [DOI] [PubMed] [Google Scholar]
- 23.Arpino G, Wiechmann L, Osborne CK, Schiff R. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29(2):217–233. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hickey TE, Robinson JL, Carroll JS, Tilley WD. Minireview: the androgen receptor in breast tissues: growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol. 2012;26(8):1252–1267. doi: 10.1210/me.2012-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner N, Moretti E, Siclari O, et al. Targeting triple negative breast cancer: is p53 the answer? Cancer Treat Rev. 2013;39(5):541–550. doi: 10.1016/j.ctrv.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136(2):331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11(2):174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 28.Qi JP, Yang YL, Zhu H, et al. Expression of the androgen receptor and its correlation with molecular subtypes in 980 chinese breast cancer patients. Breast Cancer. 2012;6:1–8. doi: 10.4137/BCBCR.S8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni M, Chen Y, Lim E, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20(1):119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Q, Niu Y, Liu N, et al. Expression of androgen receptor in breast cancer and its significance as a prognostic factor. Ann Oncol. 2011;22(6):1288–1294. doi: 10.1093/annonc/mdq586. [DOI] [PubMed] [Google Scholar]
- 31.Park S, Koo JS, Kim MS, et al. Androgen receptor expression is significantly associated with better outcomes in estrogen receptor-positive breast cancers. Ann Oncol. 2011;22(8):1755–1762. doi: 10.1093/annonc/mdq678. [DOI] [PubMed] [Google Scholar]
- 32.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses’ Health Study. Mod Pathol. 2011;24(7):924–931. doi: 10.1038/modpathol.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu R, Dawood S, Holmes MD, et al. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin Cancer Res. 2011;17(7):1867–1874. doi: 10.1158/1078-0432.CCR-10-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park S, Koo J, Park HS, et al. Expression of androgen receptors in primary breast cancer. Ann Oncol. 2010;21(3):488–492. doi: 10.1093/annonc/mdp510. [DOI] [PubMed] [Google Scholar]
- 35.Coates AS, Millar EK, O’Toole SA, et al. Prognostic interaction between expression of p53 and estrogen receptor in patients with node-negative breast cancer: results from IBCSG Trials VIII and IX. Breast Cancer Res. 2012;14(6):R143. doi: 10.1186/bcr3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lara JF, Thor AD, Dressler LG, et al. p53 Expression in node-positive breast cancer patients: results from the Cancer and Leukemia Group B 9344 Trial (159905) Clin Cancer Res. 2011;17(15):5170–5178. doi: 10.1158/1078-0432.CCR-11-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HS, Yom CK, Kim HJ, et al. Overexpression of p53 is correlated with poor outcome in premenopausal women with breast cancer treated with tamoxifen after chemotherapy. Breast Cancer Res Treat. 2010;121(3):777–788. doi: 10.1007/s10549-009-0560-5. [DOI] [PubMed] [Google Scholar]
- 38.Dowsett M, Nielsen TO, A’Hern R, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer Working Group. J Natl Cancer Inst. 2011;103(22):1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klintman M, Bendahl PO, Grabau D, Lovgren K, Malmstrom P, Ferno M. The prognostic value of Ki67 is dependent on estrogen receptor status and histological grade in premenopausal patients with node-negative breast cancer. Mod Pathol. 2010;23(2):251–259. doi: 10.1038/modpathol.2009.167. [DOI] [PubMed] [Google Scholar]
- 40.Blows FM, Driver KE, Schmidt MK, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7(5):e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rimawi MF, Shetty PB, Weiss HL, et al. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer. 2010;116(5):1234–1242. doi: 10.1002/cncr.24816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foley J, Nickerson NK, Nam S, et al. EGFR signaling in breast cancer: bad to the bone. Semin Cell Dev Biol. 2010;21(9):951–960. doi: 10.1016/j.semcdb.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 2010;28(10):1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 44.Ferraro DA, Gaborit N, Maron R, et al. Inhibition of triple-negative breast cancer models by combinations of antibodies to EGFR. Proc Natl Acad Sci U S A. 2013;110(5):1815–1820. doi: 10.1073/pnas.1220763110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng CW, Liu XL, Chen C, et al. Patterns of cancer invasion revealed by QDs-based quantitative multiplexed imaging of tumor microenvironment. Biomaterials. 2011;32(11):2907–2917. doi: 10.1016/j.biomaterials.2010.12.053. [DOI] [PubMed] [Google Scholar]
- 46.Liu XL, Peng CW, Chen C, et al. Quantum dots-based double-color imaging of HER2 positive breast cancer invasion. Biochem Biophys Res Commun. 2011;409(3):577–582. doi: 10.1016/j.bbrc.2011.05.052. [DOI] [PubMed] [Google Scholar]
- 47.Fang M, Yuan JP, Peng CW, Pang DW, Li Y. Quantum dots-based in situ molecular imaging of dynamic changes of collagen IV during cancer invasion. Biomaterials. 2013;34(34):8708–8717. doi: 10.1016/j.biomaterials.2013.07.069. [DOI] [PubMed] [Google Scholar]