Abstract
Pullulanases are endo-acting enzymes capable of hydrolyzing α-1, 6-glycosidic linkages in starch, pullulan, amylopectin, and related oligosaccharides, while amylopullulanases are bifunctional enzymes with an active site capable of cleaving both α-1, 4 and α-1, 6 linkages in starch, amylose and other oligosaccharides, and α-1, 6 linkages in pullulan. The amylopullulanases are classified in GH13 and GH57 family enzymes based on the architecture of catalytic domain and number of conserved sequences. The enzymes with two active sites, one for the hydrolysis of α-1, 4- glycosidic bond and the other for α-1, 6-glycosidic bond, are called α-amylase-pullulanases, while amylopullulanases have only one active site for cleaving both α-1, 4- and α-1, 6-glycosidic bonds. The amylopullulanases produced by bacteria find applications in the starch and baking industries as a catalyst for one step starch liquefaction-saccharification for making various sugar syrups, as antistaling agent in bread and as a detergent additive.
Keywords: amylopullulanase, amylase-pullulanase, pullulan, starch, thermostability, sugar syrups, site directed mutagenesis
Introduction
Amylopullulanases (E.C. 3.2.1.1/41) form a class of debranching enzymes belonging to the family of glycoside hydrolases (GHs) organized in the sequence-based classification of carbohydrate-active enzymes.1 Among glycoside hydrolases, GH13 represents the major α-amylase family comprising more than 30 different enzyme specificities and forms a clan GH-H along with GH-57, GH70 and GH77; these account for a small portion of α-amylase families.2 Both the families of enzymes utilize the retaining mechanism for the hydrolysis of α-glycosidic bond. The GH-13 family enzymes fold into a (β/α)8 TIM barrel with the strand β4-aspartate (catalytic nucleophile), β5-glutamate (proton donor) and β7-aspartate (transition-state stabilizer) and possess four to seven conserved sequences (CSs).3-5 While the GH-57 family enzymes adopt a (β/α)7 TIM barrel (i.e., an incomplete TIM barrel) with the catalytic machinery comprising the strand β4-glutamate (catalytic nucleophile) and β7-aspartate (proton donor) and contains five CSs. All the amylopullulanases from mesophiles come under GH13 family, while the thermostable amylopullulanases belong to either GH57 or GH13 family.6
Amylopullulanase is a bifunctional, endo-acting enzyme capable of hydrolyzing both α-1,4 and α-1,6-glycosoidic bonds in starch, pullulan, amylopectin and glycogen. Amylopullulanases are classified into two subgroups based on the number of active sites within the protein.9 The thermophilic anaerobes produce amylopullulanases possessing a single active site,10-12 while amylopullulanases from aerobic microbes contain either one13,14 or two active sites for hydrolyzing α-1,4 and α-1,6 glycosidic linkages.8,16 Due to its bifunctionality, the enzyme is also referred to as α-amylase-pullulanase based on the presence of two active sites, each being specific for one bond type.7,8
In recent years, amylopullulanases have emerged as useful starch processing enzymes17 for the production of maltose and maltotriose syrups, and thus, replacing α-amylases in the conventional starch liquefaction process.18,19 Besides their application in the industrial starch conversion, the enzyme finds application as a detergent additive too.20
Microorganisms Producing Amylopullulanases
Amylopullulanase is produced by both aerobic and anaerobic bacteria, the latter being the highest producers.10 Among aerobes, certain species of Bacillus and Geobacillus are known to produce amylopullulanase, most of which are thermophilic. Bacillus sp 3183,22 Bacillus sp TS-23,23 Bacillus subtilis,24 Bacillus sp XAL 601,13 Bacillus circulans F-2,26-28 Bacillus sp KSM-1378,9 Bacillus sp DSM 405,14 Geobacillus thermoleovorans NP33,30 Bacillus sp US149,31 and Geobacillus stearothermophilus L1432 have been reported to produce amylopullulanases. Among thermoanaerobes, Clostridium thermohydrosulfuricum,33-37 C. thermohydrosulfuricum Z 21–109,36 Clostridium sp strain EM1 (now Thermoanaerobacterium thermosulfurogenes),37,38 Thermoanaerobium brockii,10 Thermoanaerobium Tok6-B1,40,41 Thermoanaerobacterium thermosaccharolyticum,42 Thermoanaerobacter ethanolicus 39E,43 Thermoanaerobacter finni,44,45 Thermobacteroides acetoethylicus,44,45 T. ethanolicus,44,45 Thermotoga maritime,46 and Thermococcus profundus47 are identified as producers of amylopullulanase. The GH57 family amylopullulanases2 have been produced from archaea Pyrococcus furiosus,48 P. woesei,49 Thermococcus litoralis,50 T. celer,51 T. hydrothermalis52,53 and T. siculi.19
Cloning and Expression of Amylopullulanases
As the production levels and often the specific activity of the starch hydrolyzing enzymes achieved with the native hosts is inadequate, the molecular cloning of the corresponding genes and their expression in a homologous and heterologous hosts, which are genetically modified, have opened up gates for improving the protein yield. Thus the higher yields would permit economic utilization of enzymes for biotechnological applications.51
A large number of amylopullulanase encoding genes from the bacterial genomes have been cloned and their expression aspects have been investigated (Table 1). Dating back to 1987, Coleman et al.10 cloned the amylopullulanase gene from a thermophilic anaerobe, T. brockii into Escherchia coli and Bacillus subtilis. Amylopullulanase genes have also been cloned from other thermophilic anaerobes including C. thermohydrosulfuricum,21 T. saccharolyticum,11 T. ethanolicus12,39 and expressed in E. coli. Among aerobes, the gene was cloned from Bacillus sp KSM-1378,16 Bacillus sp strain XAL601,13 Bacillus stearothermophilus TS-2325 and G. thermoleovorans NP3366 and the expression was checked in E. coli. The amylopullulanase gene from Bacillus sp KSM-1378 had also been expressed in B. subtilis.16 The amylopullulanase encoding gene from lactic acid bacterium Lactobacillus plantarum L137 had been cloned and expressed in L. plantarum NCL21.35,54
Table 1. Cloning of amylopullulanase genes from various bacteria.
|
Organism |
Molecular mass (kDa) |
Host | Remarks | Reference |
|---|---|---|---|---|
|
Bacillus sp KSM-1378 |
211 | E. coli and Bacillus subtilis | The N-terminal and carboxyl-terminal half of the enzyme constitutes the amylase and pullulanase domains, respectively and are separated by a 35 amino acid sequence tandem repeat. Asp550-Glu579-Asp645 and Asp1464-Glu1493-Asp1581 were reported as catalytic triads for the amylase and pullulanase activity, respectively. Transmission electron microscopy of the purified enzyme revealed a “castanetlike” or “bent dumbbell-like” structure. | Hatada et al. 16 |
| Bacillus sp strain XAL601 | 220 | E.coli | The α-amylase-pullulanase has been overexpressed in E.coli and was found to be alkaline in nature. The enzyme has been found to adsorb strongly to crystalline cellulose (Avicel) and raw corn starch. was analyzed for its binding efficiency to various carbohydrates and was found to have strong adsorbtion to crystalline cellulose (Avicel) and raw corn starch. | Lee et al.13 |
| Bacillus stearothermophilus TS-23 | 223 | E.coli | The expressed gene products obtained were found degenerate with the largest active polypeptide of 220kDa and the smallest one of about 105kDa. | Chen et al.25 |
| Clostridium thermohydrosulfur-icum DSM 3783 | 165, 130, 100 |
E.coli | Immunoblotting has revealed more than ten α-amylase-pullulanase specific polypeptides. The largest polypeptide was found to have a molecular weight of about 165kDa, while the smallest enzymatically active polypeptide was about 100kDa. The enzyme was found to be optimally active at 80–85°C, 5°C lower compared with that of the native strain. | Melasniemi and Paloheimo 21 |
| Geobacillus ther-moleovorans NP33 | 182 | E.coli | The 300 amino-acid truncation from the C-terminus enhanced the production, specific enzyme activity, thermostability and starch saccharification efficiency. | Nisha and Satyanarayana66 |
| Lactobacillus plantarum L137 | 200 |
L. plantarum NCL21 |
The N-terminal and C-terminal region of the enzyme was found to possess amino acid sequence repeats. The truncation of the 100 amino acid repeat region of the C-terminus enhanced the production and specific activity of the enzyme. | Kim et al.35; Kim et al.54 |
| Thermoanaerobac-ter ethanolicus 39E | 162 | E. coli | Asp597, Glu626 and Asp703 have been identified as the catalytic triad by hydrophobic cluster analysis and site-directed mutagenesis studies. The enzyme was found to hydrolyze pullulan at an efficient rate compared with other substrates. | Mathupala et al.;12 Lin and Leu39 |
| Thermoanaerobac-ter saccharolyti-cum B6A-R1 | 140 | E. coli | Highly conserved amino acid residues of the protein have been identified by hydrophobic cluster analysis and multiple sequence alignment. | Ramesh et al.11 |
| Thermoanaerobiu-m brockii | 70–100 | E. coli and Bacillus subtilis | Secretion of enzyme increased from 0.23U/ml (in T. brockii) to 0.80 to 1.0 U/ml, when B. subtilis was used as an expression host. | Coleman et al.10 |
Increased levels of amylopullulanase production have been attained with the recombinant strains as compared with the wild type bacterial strains (Table 2). Four-fold higher enzyme production was achieved with the recombinant L. plantarum NCL21 harboring the L. plantarum L137 amylopullulanase gene than that of the native strain.35 In the recombinant B. subtilis containing T. brockii amylopullulanase gene, the enzyme production was 3.4−4.3-fold higher than that in the native host.10
Table 2. Characteristics of the native GH13 amylopullulanases.
| Organism | Mol mass (kDa) |
Purification strategy | Opt. Tem (°C) |
Opt. pH |
Specific activity (U/mg) | Fold purific- ation/ yield (%) |
Inhibitors | Stabil-isers | Additional properties | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus sp KSM-1378 | 210 | Ultrafiltration, DEAE-cellulose (pH 8.0), α-cyclodextrin coupled with Sepharose 6B (pH 8.0), Sephacryl S-200 | 50 | 8.5 (α-amylase) 9.5 (pullula-nase) |
42.6 (α-amylase) 83.1 (pullulanase) |
203/11 (α-amylase) 346/18 (pullulan-ase) |
Hg 2+, Cd 2+, Pb 2+ and Mn 2+ (pullulanase) Cd 2+, Pb 2+ and Mn 2+ (α-amylase) Diethyl pyro-carbonate(DEP), phenylmethane-sulphonyl fluoride and N-bromosucci-nimide |
Co2+ (pullulanase) |
Km: pullulan 0.72 (mg/ml) amylose 0.27 Vmax: pullulan 169 [µg(as amylose 51 glucose)/ ml/ min] |
Ara et al. 20 |
| Bacillus circulans F-2 | 220 | (NH4)2SO4 (80% satura-tion), starch adsorption, DEAE-Toyopearl650M ion exchange chromate-graphy (pH 8.0), hydrophobic interaction chromatographies using TSK Gel Phenyl-51AV (pH 7.5) | 50 | 7.0–8.5 (α-amylase) 7.0 (pullula-nase) |
81.7 (α-amylase) 84.2 (pullulanase) |
167/4 (α-amylase) 1400/31.8 (pullulan- ase) |
Nd | Nd | Km: pullulan 0.77 (mg/ml) soluble starch 0.53 amylose 0.56 Vmax: pullulan 5.3 (µg/ml/ soluble starch 2.6 min) amylose 3.1 |
Sata et al. 15 |
| Bacillus sp DSM 405 | 126 | Corn starch adsorption (pH 6.0), hydrophobic interaction chromate-graphy using phenyl-Sepharose CL-4B (pH 7.0), ultrafiltration and gel filtration | 70 | 6.5 (α-amylase) 6.0 (pullula-nase) |
11000 | 1400 | EDAC | Nd | Km: pullulan 0.41 (mg/ml) amylopectin 0.64 amylose 1.39 Vmax: pullulan 788 (U/ml) soluble starch 544 |
Brunswick et al.89 |
| Lactobacillus amylophilus GV6 | 90 | (NH4)2SO4 (pH 6.5) and sephacryl S-200 column chromatography | 37 | 6.5 | 15.238 (α-amylase) 7.142 (pullulanase) |
11.4 (α-amylase) 10 (pullulan-ase) |
Cu2+, Ni2+, Cd2+, Fe2+, Ba2+, Zn2+, EDTA, SDS and Tween 80 | Ca2+ | Nd | Vishnu et al. 28 |
| Geobacillus stearothermophilus L14 | 100 | 75% (NH4)2SO4, Q-sephar-ose column (pH 9.0), DEAE-sepharose column (pH 8.5), ultrafiltration and sephadex G-100 gel filtration column | 65 | 5.5 | 967 | 10/30 | Nd | Nd | Km: pullulan 0.48 (g/l) soluble starch 5 Vmax: pullulan 44 (µmol/ml/ soluble starch 80 min) |
Zareian et al. 32 |
| Thermoanaerobacter- ethanolicus 39E (Clostridium thermohydrosulfuric-um 39E) | 133 | Q-sepharose, β-cyclo-dextrin sepharose(pH 6.0) and ultrafiltration | 60 | 5.5 | 175 (α-amylase) 480 (pullulanase) |
2400/29 | DEP, DEAC, β-cyclodextrin | Nd | Km: pullulan 0.35 (mg/ml) amylose 1 Vmax: Nd |
Mathupala and Zeikus 43 |
| Thermoanaerobacter-ium thermosaccharo-lyticum DSM 571 | 150 | Butyl-sepharose column, POROS ether and superdex | 65 | 5.0–5.5 | 4.3 (α-amylase) 14 (pullulanase) |
140/1 | Nd | Nd | Nd | Ganghofner et al. 42 |
| Thermoanaerobacter strain B6A | 450 | DEAE-sepharose CL-6B column chromatography, gel filtration using high-pressure liquid chroma-tography and pullulan-sepharose affinity chromatography. | 65 | 5 | 215 | Nd | Fe3+, Zn2+, N-bromosuccinimide, β- and γ-cyclodextrin | - | Km: pullulan 0.43 (mg/ml) soluble starch 0.37 Vmax: Nd |
Saha et al. 36 |
Molecular Mass of the Recombinant Amylopullulanases
The amylopullulanases of both GH-13 and GH-57 family vary in gene sequence and length. The protein is usually of a high molecular weight among other glycosyl hydrolases. The molecular mass of the protein ranges between 80 and 250 kDa, and furthermore, some are glycoproteins.
Domain Architecture
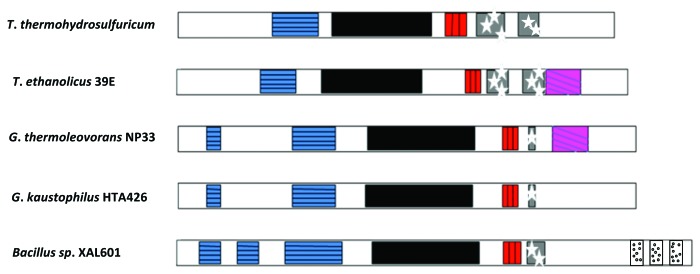
The amylopullulanases are multidomain proteins. GH13 amylopullulanases possess the cyclodextrin and pullulan degrading enzyme N-terminus domain, the (α/β)8 barrel core and the C-terminal region containing one α-amylase C-terminal all-β domain (AamyC), one or two fibronectin type III (FnIII) domains and one putative carbohydrate-binding module 20 domain (CBM20) (Fig. 1).
Figure 1. Schematic representation of conserved domain structure in the amylopullulanases from T. hydrosulfuricum (gi: 114076), T. ethanolicus 39E (gi: 728871), G. thermoleovorans NP33 (gi: JQ437895), G. kaustophilus HTA426 (gi: 56421715) and Bacillus sp XAL601 (gi: 460687). Symbols are: horizontal lines, cyclodextrin and pullulan-degrading enzymes’ N-terminus domain; black bar, α amylase catalytic domain; vertical lines, amyC domain; starred, fibronectin type III domain; diagonal lines, CBM20 domain; dotted, S-layer homology domain.
An α-amylase catalytic domain present in the form of a barrel of eight parallel β-strands surrounded by eight α-helices is common to all members of the α-amylase family. These β-strands are linked to the adjacent α-helices by irregular structures in the form of loops. These loops carry the catalytic amino acid residues of the active site (for details see ref. 55). A distinct N-terminal domain is present preceding the (α/β)8 catalytic domain and the amino acid residues of the N-terminal domain may not form part of the active sites unlike that reported for isoamylase.56
The AamyC is an all β-domain of α-amylase present in the C-terminus. The possible functions of AamyC is in disrupting the starch granule and separating the α-glucan chains together with the other substrate binding site in the α-amylase catalytic domain. The domain is considered to secure proper orientation of the active site of the enzyme on the substrate chains.57 It has also been suggested that the AamyC domain stabilizes the (α/β)8 catalytic domain by shielding the hydrophobic residues of the domain from the solvent.
The fibronectin type III (FnIII) domain is a small folding unit of about 100 amino acid residues and possesses a seven-stranded β sandwich structure. The β-sandwich structure of FN3 is similar to that of immunoglobulin domains. No specific role has been assigned to the FnIII domain. It is possibly involved in the binding of the enzyme and the polysaccharide substrates.58-60
The carbohydrate-binding domain is composed of 40 to 200 amino acids and is characterized by a discrete fold that possesses carbohydrate binding property. The CBM facilitates the interaction between the insoluble substrate and the enzyme by bringing the substrate to the catalytic domain, and thereby improving the substrate hydrolysis. Currently CBMs are divided into 39 families based on the amino acid sequence similarity. The family 20 carbohydrate-binding module (CBM20) is also known as the starch-binding domain and is found in a large number of starch hydrolyzing enzymes including α-amylase, β-amylase, glucoamylase, amylopullulanases and CGTase (cyclodextrin glucanotransferase). CBM20 adopts an antiparallel β-barrel structure with two starch binding sites (SBS1 and SBS2) (for a review, see ref. 63). The two tryptophans and a lysine forms the evolutionary conserved SBS1, while SBS2 possesses a tryptophan residue (for a review, see refs. 61 and 62). These two sites are thought to differ functionally with SBS1 involved in the initial starch recognition site and SBS2 participate in the specific recognition of appropriate regions of starch.
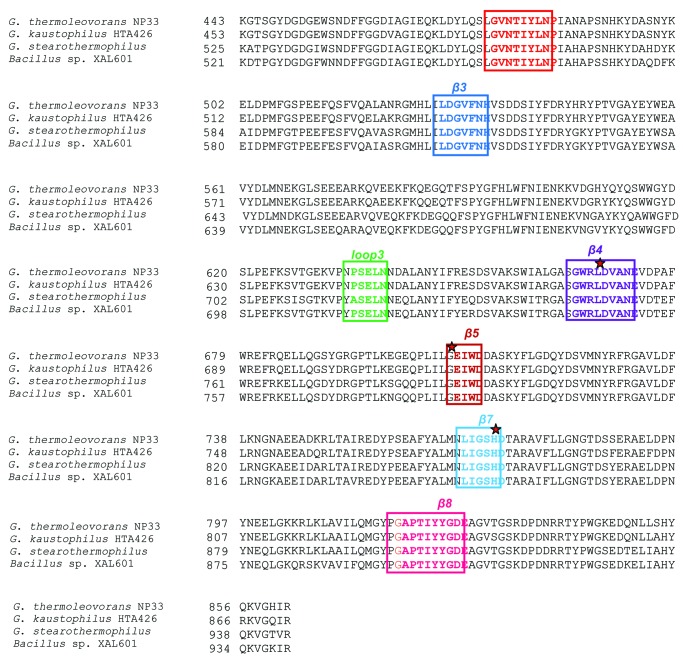
Conserved Sequences
The short stretches of sequences are well conserved within the amylopullulanases of GH13 family. According to the definition proposed by Takata et al.,64 the members of the α-amylase family contain four highly conserved sequences (I to IV). The conserved sequences I, II, III, and IV are located on strands β-3, β-4, β-5 and β-7, respectively.64,65 The catalytically important amino acid residues corresponding to two aspartic acids and one glutamate are present within the conserved sequences on the strands β-4, β-5 and β-7 (Fig. 2).
Figure 2. The sequence alignment of the α-amylase catalytic domain, (α/β)8 barrel of amylopullulanases of selected GH13 family members. The multiple sequence alignment of the amino acid sequence of protein was generated using the software, ClustalW2 of the European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The seven conserved regions around the strands β-2, β-3, β-4, β-5, β-7, β-8 and on loop 3 are highlighted in boxes with different colors. Numbers on the left side of the amino acid sequence indicate amino acid positions of the sequences. The catalytic triad residues, 2Asp and 1Glu are marked with a red star.
Three additional conserved sequences have been identified in the α-amylase family enzymes (for a review, see ref. 5). The fifth conserved sequence region of the amylopullulanases is characterized by the presence of calcium binding aspartate and are located on loop 3 that protrudes from the (α/β)8 catalytic domain and connects the β-3 strand and the third α-helix. The cyclomaltodextrinases, maltogenic amylases and neopullulanases, are unable to bind to the calcium ion as Asp is substituted by Lys in this region. The sixth conserved sequence region present on β-2 strand has been identified by the evolutionary conserved glycine and proline residues linked by seven or eight amino acid residues. Amylopullulanase and other α-amylase family members have seven amino acid residues, while CGTases have eight residues separating glycine and proline residues. The seventh conserved sequence region is differentiated by a well conserved glycine residue at the start of the region followed by a proline residue.
Active Site and Catalytic Mechanism
The bacterial amylopullulanases from both GH13 and GH57 familis possess one or two active sites for hydrolyzing α-1,4 and α-1,6 glycosidic linkages. A single active site is responsible for the bifunctionality of the enzyme in case of T. ethanolicus 39E,12,39 C. thermohydrosulfuricum,34 Bacillus sp strain XAL 60113 and G. thermoleovorans NP33.66 Kinetic experiments on competitive inhibition with mixed substrates and chemical modification with different inhibitors were used by Brunswick et al.14 to determine the number of active sites in the amylopullulanase of Bacillus sp DSM 405. Both these approaches suggested the presence of a single active site for the dual hydrolytic activities.
Dual hydrolytic activities associated with different active sites have been reported in α amylase-pullulanase from Bacillus sp KSM 1378.16 The partial hydrolysis of the enzyme with papain has revealed the presence of two functional domains for α-1, 4- and α-1, 6- hydrolytic activity.9 The amylose and pullulan-hydrolyzing polypeptides were visualized as a mixture of differently sized globular molecules joined by a thin short linker region under transmission electron microscope.
The role of acidic amino acids at the active site has been shown by Mathupala and Zeikus43 for T. ethanolicus 39E amylopullulanase. The two conserved aspartate residues on strands β4 and β7 act as catalytic nucleophile and proton donor, respectively, and one β5-glutamate residue involved in the transition state stabilizer have been found to play a catalytic role in GH13 amylopullulanases. The catalytic residues were identified at the C-terminal end of the barrel β-strands by using hydrophobic cluster analysis (HCA) in case of amylopullulanases from Bacillus sp KSM-1378,16 T. ethanolicus 39E,12 and T. saccharolyticum B6A-RI.11 HCA is a 2-D illustration of the amino acid sequences of the protein and is used for comparing the shapes and relative location of hydrophobic clusters in proteins. The shape of hydrophobic clusters predicts the secondary structure of the protein and the similarity of the clusters suggests the similarity in the polypeptide folding of the proteins.29,67
In Bacillus sp KSM-1378 amylopullulanase, the putative catalytic triads were identified as Asp550-Glu579-Asp645 for the amylase activity and Asp1464-Glu1493-Asp1581 for the pullulanase activity.16 The amylopullulanases of T. ethanolicus 39E12 and T. saccharolyticum B6A-RI11 have Asp597-Glu626-Asp703 and Asp594-Asp700-Glu623 as catalytic triads, respectively for amylase and pullulanase activities.
Site directed mutagenesis of the three catalytic amino acid residues to amides has led to complete loss of both α-amylase and pullulanase activities, suggesting a single active site for the dual catalytic activity in T. ethanolicus 39E.12 The three residues are located in close proximity with each other, forming a single active site for the dual activities in both T. ethanolicus 39E and T. saccharolyticum B6A-RI, in contrast to the dual active sites for the α-amylase-pullulanase of alkaline amylopullulanase from Bacillus sp KSM-1378.11,12,16 The GH13 amylopullulanases have also been found to contain two histidine residues that are critical for the transition state stabilization.
MacGregor et al.55 pointed out that the active site of an enzyme is made up of subsites, each capable of binding to a monosaccharide residue. The subsites are the amino acid side chains present in the loops of the enzyme structure that links the C-terminal ends of β-strands to the N-terminal ends of the adjacent α-helices of the catalytic domain. The catalytic activity of an enzyme requires the interaction of a glucose residue of the substrate to -1 subsite. The different enzyme specificities vary with the nature of the substrate portion binding onto the subsites +1 and +2. The three catalytic triads, 2 Asp and 1Glu as well as the 2His residues involved in the transition state stabilization has been reported to occupy the subsite -1, while the amino acid residues of the conserved sequence regions III and IV on strands β-4 and β-5, respectively constitute subsites +1 and +2. The flexible nature of the subsite +1 at the active site of the amylopullulanases or an α-amylase-pullulanase might be responsible for the action of the enzyme on more than one type of linkage. It has also been shown that the enzymes having amino acid sequence VANE at the conserved region III on β-4 strand can act on both α-1,4 and α-1,6 glycosidic linkages. The mutation of the residues led to a change in the bond specificity.
Biophysical and Biochemical Characteristics
Temperature
The amylopullulanases from L. plantarum L13735 and Bacillus sp KSM-137816 were reported to be optimally active at 45–50°C, while that of T. saccharolyticum B6A-RI11 and Bacillus sp strain XAL60113 were active at an optimum temperature of 65°C to 70°C. The amylopullulanases from T. ethanolicus 39E had temperature optimum at 90°C and 80°C for α-amylase and pullulanase activities, respectively, representing the highest temperature range reported for any GH13 amylopullulanase (Table 3). C. thermohydrosulfuricum DSM 3783 α-amylase-pullulanase was found to have the temperature optimum for α-amylase activity at 85°C and 80°C for the pullulanase activity, in the presence of 10mM Ca2+.39,41 The current industrial starch conversion process requires the highly thermostable amylopullulanases for the starch liquefaction-saccharification process as the starch bioprocessing at elevated temperatures would improve the starch solubility, minimize the microbial contamination and reduces its viscosity and reaction time, making the process economical.50
Table 3. Characteristics of the recombinant GH13 amylopullulanases.
| Organism | Molecular mass (kDa) |
Purification strategy | Opt. Tem (°C) |
Opt. pH |
Specific activity (U/mg) | Fold purific- ation/ yield (%) | Inhibitors | Stabilizers | Additional properties | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Lactobacillus plantarum L137 | 215.6 | Ultrafiltration, DEAE-sepharose CL-4B and superose 6 | 40–45 | 4.0–4.5 | 431 | 1.5/ 25 | Hg2+, Cu2+, N-bromosuccinimide, guanidine-HCl, urea, moderately by α-cyclodextrin, γ-cyclodextrin |
Co2+ | Km: pullulan 6.9 (g/l) soluble starch 7.7 amylose 2.5 Vmax: pullulan 37.9 (U/mg) soluble starch 53.4 amylose 32.3 |
Kim et al.35,54 |
|
Lactobacillus plantarum L137 (C-terminal truncation) |
150 -250 |
Ultrafiltration, DEAE-sepharose CL-4B and superose 6 | 40–45 | 4.0–4.5 | 596 | 1.5/29 | Hg 2+, Cu 2+, N-bromosuccinimide, guanidine-HCl and urea, moderately by α-cyclodextrin and γ-cyclodextrin | Co2+ | Km: pullulan 2.6 (g/l) soluble starch 5.0 amylose 3.3 |
Kim et al.54 |
|
Thermoanaero-bacterium saccharolyticum B6A-RI, |
142 ± 2 | Heat treatment of the recombinant E. coli cells, Q-sepharose and β-cyclodextrin-coupled sepharose affinity chromatography | 65 | 6.0 | 498 | 17/15.2 | Nd | Nd | Km: pullulan 0.49 (mg/ml) soluble starch 0.43 |
Ramesh et al.11 |
| Thermoanaero-bacter ethanolicus 39E | 109 (thermostable region) | Ni-nitriloacetic acid affinity purification using His-bind resin | 90 | 6.0 | Nd | Nd | EDTA, N-bromosuccinimide, and α-cyclodextrin |
Ca2+, Mn2+, Ba2+ | Km: pullulan 3.79 (mg/ml) soluble starch 1.38 Vmax: pullulan 98 μmol/(min.mg) starch 39 |
Mathupala et al.;12 Lin and Leu39 |
| Bacillus sp KSM-1378 | 210 | DEAE-cellulose, affinity chromatography on sepharose 6B-a-cyclodextrin and gel filtration on sephacryl S-200 | 50 | 9.5 | 47 (for soluble starch) 84 (for pullulan) | Nd | Nd | Nd | Nd | Hatada et al. 16 |
| Bacillus sp strain XAL601 | 225 | Ammonium sulfate precipitation, mono S HR5/5, superdex 200HR 10/30 | 70 | 9.0 | 56.7 U/ml (for soluble starch) and 57.3 U/ml (for pullulan) |
6.6 | Nd | Nd | Nd | Lee et al.13 |
| Geobacillus thermoleovorans NP33 | 182 | Nickel NTA affinity chromatography | 60 | 7.0 | 851 (for soluble starch) and 795 (for pullulan) | Nd | Hg2+, Cu2+, EDTA, EGTA, N-bromosuccinimide, guanidine HCl and EDAC |
Zn2+ | Km: pullulan 3.3 (mg/ml) soluble starch 0.833 Vmax: pullulan 640 soluble starch 666.4 |
Nisha and Satyanarayana66 |
| Geobacillus thermoleovorans NP33(C-terminal truncated) | 150 | Nickel NTA affinity chromatography | 60 | 7.0 | 1260 (for soluble starch) and 1169 (for pullulan) | Nd | Hg2+, Cu2+, EDTA, EGTA, N-bromosuccinimide, guanidine HCl and EDAC |
Zn2+ | Km: pullulan 2.8 (mg/ml) soluble starch 0.588 Vmax: pullulan 1192 solublestarch 1333.2 |
Nisha and Satyanarayana66 |
pH
Amylopullulanases act over a wide range of pH from acidic to alkaline. The optimum pH range for amylopullulanases from L. plantarum L137, T. saccharolyticum B6A-RI and T. ethanolicus 39E were found to be 4.0–6.0.11,35,39 The amylopullulanase from G. thermoleovorans NP33 was reported to have neutral pH optimum, while that from Bacillus sp strain XAL601 and Bacillus sp KSM-1378 was optimally active at pH 9.0–9.5 for both α-amylase and pullulanase activities. 13,16,66
Thermostability and pH stability
A relatively less thermostability was observed for the amylopullulanase from L. plantarum L137, retaining 50% activity after 30 min of incubation at 45°C than its C-terminal truncated variant (100 amino acids) losing 50% activity at 41°C.35,54 Similar observations were recorded for the T. ethanolicus 39E amylopullulanase (100 amino acids from C-terminus), with higher activation energy for the truncated enzyme compared with its full length form.6,39 The 300 amino acid deletion from the C-terminus of the amylopullulanase of G. thermoleovorans NP33 has also been found to enhance thermostability.66 The half-lives of the full-length and truncated G. thermoleovorans NP33 amylopullulanase were 30 min and 75 min, respectively for the α-amylase activity and 20 min and 30 min for the pullulanase activity at 90°C. A 20% loss in the enzyme activity was attained in the amylopullulanase from T. saccharolyticum B6A-RA1 on incubation at 65°C for 1 h.11 The α-amylase-pullulanase from C. thermohydrosulfuricum DSM 3783 retained 60% of the enzyme activity, even after 2 h at 85°C.21
The L. plantarum L137 amylopullulanase was found stable at pH 2.5−6.5 and was reported to be less stable above neutral pH in comparison with its C-terminal truncated variant.35,54 The truncated amylopullulanase of G. thermoleovorans NP33 had been shown to have high pH stability than the full-length variant.66
Effect of metal ions
The amylopullulanases from different bacterial sources have shown diverse behavior toward metal ions for their activity. Some enzymes require metal ions, some are inhibited by metal ions and some are unaffected by their presence. The α-amylase and pullulanase activities of the T. ethanolicus 39E amylopullulanase were found to be stimulated by Ca2+, Mn2+, Ba2+, while Hg2+, Ni2+, Zn2+ and Fe2+ were strong enzyme inhibitors.39 Hg2+ and Cu2+ ions inhibited both α-amylase and pullulanase activities of the L. plantarum L137 amylopullulanase, while Co2+ ions stimulated the activity.35 Zn2+ stimulated the enzyme activities of G. thermoleovorans NP33 amylopullulanase, while the enzyme activity was almost completely lost in the presence of Hg2+ and Cu2+ ions. A moderate loss of the enzyme activity was observed by Fe2+ and Mg2+, while Co2+, Mn2+ and Ca2+ ions did not affect the enzyme activity.66
Effect of inhibitors
EDTA, N-bromosuccinimide and α-cyclodextrin strongly inhibited the amylopullulanase activity in T. ethanolicus 39E.39 The inhibition by EDTA suggested that either the enzyme is metal dependent or the EDTA is changing the conformation of the protein instead of acting as a chelating agent. Enzyme inhibition by N-bromosuccinimide suggests the involvement of tryptophan residues in the catalytic activity. Cyclodextrins act by forming inclusion complexes between aromatic amino acid residues of amylopullulanase and cyclodextrin. The L. plantarum L137 amylopullulanase was strongly inhibited by N-bromosuccinimide, guanidine-HCl and urea and moderately by α-cyclodextrin and γ-cyclodextrin; the enzyme activity was unaffected by EDTA and β-cyclodextrin.35 Both α-amylase and pullulanase activities of G. thermoleovorans NP33 have been significantly affected by EDTA, EGTA, 1-ethyl–3-(3-dimethylaminopropyl)- carbodiimide (EDAC), N-bromosuccinimide, Gdn-HCl, β-Cyclodextrin and γ-cyclodextrin.66 The enzyme inhibition by EDAC revealed the irreversible modification of carboxyl groups or the presence of tyrosine groups in the enzyme.89
End Product Analysis
The action of amylopullulanases of T. ethanolicus 39E, Bacillus sp strain XAL601 and G. thermoleovorans NP33 on starch, amylose, amylopectin and glycogen liberated maltose, maltotriose and maltotetraose as the end products.39,66 The amylopullulanase of L. plantarum L137 hydrolyzed amylose to maltotriose, maltotetraose and maltopentaose. Maltose or glucose was not detected in amylose hydrolysate.35 T. saccharolyticum B6A-R1 amylopullulanase efficiently degraded starch, amylose, amylopectin, glycogen and pullulan.11 The action of most of the bacterial amylopullulanases on pullulan forms maltotriose as the only hydrolysis product unlike that reported for the native amylopullulanase from G. stearothermophilus L14 that produced glucose from pullulan hydrolysis.32
Circular Dichroism and Fluorescence Spectrometry
The effect of truncation and the comparative analysis of the secondary structures of some amylopullulanases have been studied by using circular dichroism spectroscopy and fluorescence spectrometry studies. The circular dichroism spectroscopy is based on the proteins’ unequal absorption of right- and left-handed circularly polarized light, while fluorescence spectroscopy measures the intrinsic fluorescence generated from aromatic amino acids such as tryptophan, phenylalanine and tyrosine.
The secondary structural analysis of the amylopullulanase from T. ethanolicus 39E has been made and compared with that of its C-terminal truncated mutant (with deletion of 100 amino acids from C-terminus) using flourescence emission and CD spectrometry.6 The enzymes have been found to exhibit similar fluorescence spectra upon denaturation with urea and renaturation. The comparative analysis of the secondary structures of full length amylopullulanase and its C-terminal truncated mutant using far-UV CD spectroscopy has also revealed the CD spectra of equal intensity. Both the enzymes exhibited identical secondary structure.
The fluorescence and circular dichroism spectrometric methods have also revealed highly indistinguishable structure for the full-length amylopullulanase from T. saccharolyticum and its C-terminal truncated mutant.68 An identical active conformation was attained for both enzymes on fluorescence spectra. A similar thermal unfolding and a one-step melting curve were observed upon far UV-CD measurements. The truncation experiments on amylopullulanase from T. ethanolicus 39E suggested that a large part of the C-terminal carbohydrate-binding module family 20, a portion of the first fibronectin III motifs and the second fibronectin type III could be deleted without causing a significant change in the structure and action of the enzyme on soluble starch and pullulan,6 and a similar experiment on T. saccharolyticum NTOU1 amylopullulanase revealed non-essentiality of the C-terminal fibronectin typeIII (FnIII) motif.68
There is no information on the tertiary structure of the GH13 amylopullulanases, since crystal structures have not yet been resolved, and thus, further investigations are called for understanding the structure-function relationship of amylopullulanases.
Applications of Amylopullulanases
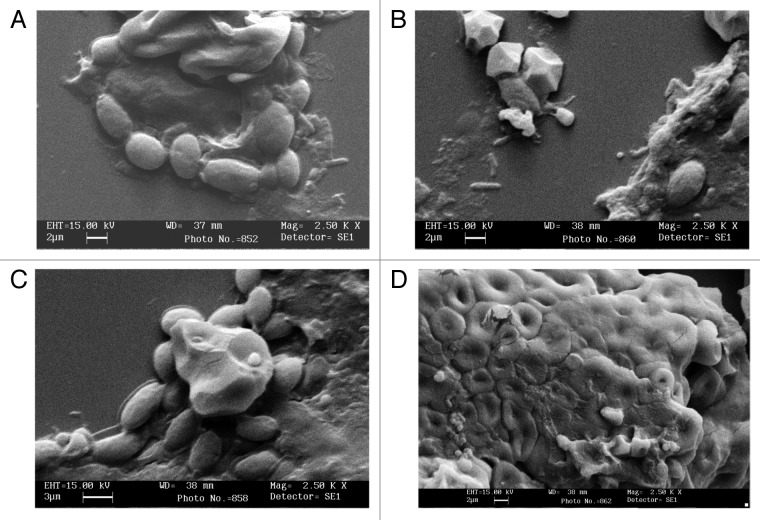
Amylopullulanases are one of the emerging enzymes for use in the industrial starch processing industry. Because of the enzymes’ debranching ability, bifunctionality as well as the calcium independence in some will make it extremely useful for the current starch conversion process (Fig. 3). The enzyme can be used as a catalyst for one step starch liquefaction-saccharification process, and therefore, can replace other amylolytic enzymes like α-amylases and β-amylases. Besides its application in the starch industry for the production of various sugar syrups including maltose, maltotriose and maltotetraose and as anti-stale in baking, the amylopullulanase and α-amylase-pullulanase find application in the detergent industry. The addition of the enzyme in starch liquefaction process has been found to increase the yield of maltose, thereby reducing the amount of branched oligosaccharides.
Figure 3. Scanning electron micrographs of the untreated and treated raw rice starch granules with the amylopullulanase of G. thermoleovorans NP33. (A) Untreated raw rice starch granule. (B) Hydrolyzed granules in 30 min of reaction with the enzymes; (C) hydrolyzed portion of the granule in 1 h; (D) almost completely hydrolyzed starch granules in 2 h.
The α-amylases employed in the current starch conversion process are calcium dependent and does not act at a pH below 5.9. Therefore, the process requires the addition of calcium and the pH adjustment to that of the starch slurry (pH 4.5). The reversion products such as maltose and isomaltose formed by glucoamylase at the expense of glucose need to be minimized. The amylopullulanases would, therefore, prove advantageous in the starch conversion process as it increases the production of maltose, reduces the reaction time, allows an increase in substrate concentration and limits the use of glucoamylase, thereby making the process economical.
Maltose and maltooligosaccharide syrups are employed in food, beverage, pharmaceutical and chemical industries.69 Maltose-containing syrups are used in the baking, soft drink, canning, confectionery and other food industries. Maltotriose syrup has been reported to have low freezing point depression and solution viscosity, good heat stability, mild sweetness, keeps in moisture, prevents the starch retrogradation in foodstuffs and forms less color as compared with maltose, glucose and sucrose syrups. These properties are very important in food and pharmaceutical industries.70 High maltotriose syrup also finds application in the food industry in making desserts and in baking and brewing. The maltose and maltotriose syrups are being used in pharmaceutical industry as a substitute for glucose in intravenous feeding.
Transgenic rice seeds containing a thermostable and bifunctional amylopullulanase from T. etha-nolicus 39E enzyme have also been generated, which would facilitate the industrial production of sweeteners and fermentation products. The granule-bound amylopullulanase activity has also been found to be associated with the reduction of amylose in developing transgenic rice seeds. High level of the amylose content results in dry, fluffy and non-sticky rice grains.71 The reduced amylose contents would be advantageous for improving starch quality, starch bioprocessing and production of protein-enriched flour from rice seeds, thereby significantly improving the nutritional value and reducing the production cost.72,73
The amylopullulanases have a potential role as antistaling agents in the baking industry. Staling refers to all the undesirable changes that occur upon storage of baked products, like the loss of crispness of the crust, increase of crumb firmness and decrease in the moisture content of the crumb with loss of bread flavor. Staling happens as a result of the retrogradation of the amylopectin fraction of starch.74,75 The thermostable amylopullulanases are considered to decrease the increased stickiness of the α-amylase treated bread associated with the production of branched maltodextrins, and therefore, can be used in the place of α-amylase as an antistaling agent.76
The use of amylopullulanase in the alcohol and brewing industries along with glucoamylase can increase the amount of fermentable sugars and may facilitate filtration steps. Amylopullulanases also find use in the production of low carbohydrate (low calorie) “Lite beer.” The enzyme can be added with fungal α-amylase or glucoamylase to the wort during fermentation instead of pullulanase.
The enzyme has also been used in the production of slowly digestible starch.77,78 Slowly digestible starch has an impact on human health, as it is associated with a low glycemic index for the treatment and prevention of various diseases, such as cardiovascular diseases,79 non-insulin diabetes,80 obesity81 and provides sustained and stable energy for athletes.82 Amylopullulanase has also been found to increase the resistant starch content.83 During starch hydrolysis process, the gelatinized starch may revert to a form that is highly resistant to α-amylase hydrolysis and is called resistant starch.84 The resistant starch has potential application in the food industry and has attracted much attention of the nutritionists as it causes reduced levels of plasma glucose and insulin, increased fecal bulk and short-chain fatty acid (SCFA) production through fermentation in the large intestine.85 RS has been produced by a heating-cooling process and chemical modification.85 However, the chemical modification may not be safe and the heating-cooling process alone may lower the RS content due to the structure of starch.
The enzyme has also been used for the production of branched cyclodextrins.86 Branched cyclodextrins possess one or more saccharide chains such as glucose, maltose and other saccharides linked to cyclodextrins by an α-1, 6 bond.87 The branched cyclodextrins are more soluble in water and organic solvents than cyclodextrins which have no branches and thus are likely to form more soluble inclusion complexes with various chemicals. Cyclodextrins are used in the food, cosmetic, pharmaceutical and plastic industries as emulsifiers, antioxidants and stabilizing agents. Ara et al.88 reported the applicability of an alkaline pullulanase from alkaliphilic Bacillus sp KSM-1876 in dishwashing and laundry detergents.
Conclusions
Amylopullulanases are receiving considerable attention as bifunctional and debranching enzymes for the industrial starch saccharification process. Attempts have been made in cloning amylopullulanase encoding genes and overexpressing them for attaining high enzyme yields and to characterize the proteins. The truncation experiments on many amylopullulanases enabled to enhance specific enzyme activity and thermostability. The primary structures of bacterial amylopullulanases revealed the domain architecture distinct from that of other amylolytic enzymes. The tertiary structure of the enzyme needs to be studied for understanding the catalytic mechanism and possible role of different domains and demarcation from other amylolytic enzymes.
Acknowledgments
We gratefully acknowledge financial assistance from the Council of Scientific and Industrial Research (CSIR) and Department of Science and Technology, Government of India and University of Delhi while writing this review.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/24629
References
- 1.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janecek S. Amylolytic families of glycoside hydrolases: focus on the family GH-57. Biologia (Bratisl) 2005;60(suppl 16):177–84. [Google Scholar]
- 3.Matsuura Y, Kusunoki M, Harada W, Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984;95:697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- 4.Kuriki T, Imanaka T. The concept of the α-amylase family: structural similarity and common catalytic mechanism. J Biosci Bioeng. 1999;87:557–65. doi: 10.1016/S1389-1723(99)80114-5. [DOI] [PubMed] [Google Scholar]
- 5.Janecek S. How many conserved sequence regions are there in the α-amylase family? Biologia (Bratisl) 2002;57(suppl 11):29–41. [Google Scholar]
- 6.Lin HY, Chuang HH, Lin FP. Biochemical characterization of engineered amylopullulanase from Thermoanaerobacter ethanolicus 39E-implicating the non-necessity of its 100 C-terminal amino acid residues. Extremophiles. 2008;12:641–50. doi: 10.1007/s00792-008-0168-4. [DOI] [PubMed] [Google Scholar]
- 7.Melasniemi H. Purification and some properties of the extracellular α-amylase-pullulanase produced by Clostridium thermohydrosulfuricum. Biochem J. 1988;250:813–8. doi: 10.1042/bj2500813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim C-H, Kim YS. Substrate specificity and detailed characterization of a bifunctional amylase-pullulanase enzyme from Bacillus circulans F-2 having two different active sites on one polypeptide. Eur J Biochem. 1995;227:687–93. doi: 10.1111/j.1432-1033.1995.tb20189.x. [DOI] [PubMed] [Google Scholar]
- 9.Ara K, Igarashi K, Saeki K, Ito S. An alkaline amylopullulanase from alkaliphilic Bacillus sp. KSM-1378; kinetic evidence for two independent active sites for the α- 1,4 and α- 1,6 hydrolytic reactions. Biosci Biotechnol Biochem. 1995;59:662–5. doi: 10.1271/bbb.59.662. [DOI] [Google Scholar]
- 10.Coleman RD, Yang SS, McAlister MP. Cloning of the debranching-enzyme gene from Thermoanaerobium brockii into Escherichia coli and Bacillus subtilis. J Bacteriol. 1987;169:4302–7. doi: 10.1128/jb.169.9.4302-4307.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramesh MV, Podkovyrov SM, Lowe SE, Zeikus JG. Cloning and sequencing of the Thermoanaerobacterium saccharolyticum B6A-RI apu gene and purification and characterization of the amylopullulanase from Escherichia coli. Appl Environ Microbiol. 1994;60:94–101. doi: 10.1128/aem.60.1.94-101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathupala SP, Lowe SE, Podkovyrov SM, Zeikus JG. Sequencing of the amylopullulanase (apu) gene of Thermoanaerobacter ethanolicus 39E, and identification of the active site by site-directed mutagenesis. J Biol Chem. 1993;268:16332–44. [PubMed] [Google Scholar]
- 13.Lee SP, Morikawa M, Takagi M, Imanaka T. Cloning of the aapT gene and characterization of its product, alpha-amylase-pullulanase (AapT), from thermophilic and alkaliphilic Bacillus sp. strain XAL601. Appl Environ Microbiol. 1994;60:3764–73. doi: 10.1128/aem.60.10.3764-3773.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunswick JM, Kelly CT, Fogarty WM. The amylopullulanase of Bacillus sp. DSM 405. Appl Microbiol Biotechnol. 1999;51:170–5. doi: 10.1007/s002530051378. [DOI] [PubMed] [Google Scholar]
- 15.Sata H, Umeda M, Kim C, Taniguchi H, Maruyama Y. Amylase-pullulanase enzyme by B. circulans F-2. Biochim Biophys Acta. 1989;991:388–94. doi: 10.1016/0304-4165(89)90062-7. [DOI] [Google Scholar]
- 16.Hatada Y, Igarashi K, Ozaki K, Ara K, Hitomi J, Kobayashi T, et al. Amino acid sequence and molecular structure of an alkaline amylopullulanase from Bacillus that hydrolyzes α-1,4 and α-1,6 linkages in polysaccharides at different active sites. J Biol Chem. 1996;271:24075–83. doi: 10.1074/jbc.271.39.24075. [DOI] [PubMed] [Google Scholar]
- 17.Kim TJ, Kim MJ, Kim BC, Kim JC, Cheong TK, Kim JW, et al. Modes of action of acarbose hydrolysis and transglycosylation catalyzed by a thermostable maltogenic amylase, the gene for which was cloned from a Thermus strain. Appl Environ Microbiol. 1999;65:1644–51. doi: 10.1128/aem.65.4.1644-1651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy I, Gupta MN. Hydrolysis of starch by a mixture of glucoamylase and pullulanase entrapped individually in calcium alginate beads. Enzyme Microb Technol. 2004;34:26–32. doi: 10.1016/j.enzmictec.2003.07.001. [DOI] [Google Scholar]
- 19.Jiao Y, Wang S, Lv M. [Structural and functional analysis of GH57 family thermostable amylopullulanase--a review] Wei Sheng Wu Xue Bao. 2011;51:21–8. [PubMed] [Google Scholar]
- 20.Ara K, Saeki K, Igarashi K, Takaiwa M, Uemura T, Hagihara H, et al. Purification and characterization of an alkaline amylopullulanase with both α-1,4 and α-1,6 hydrolytic activity from alkalophilic Bacillus sp. KSM-1378. Biochim Biophys Acta. 1995;1243:315–24. doi: 10.1016/0304-4165(94)00148-Q. [DOI] [PubMed] [Google Scholar]
- 21.Melasniemi H, Paloheimo M. Cloning and expression of the Clostridium thermohydrosulfuricum α-amylase-pullulanase gene in Escherichia coli. J Gen Microbiol. 1989;135:1755–62. doi: 10.1099/00221287-135-6-1755. [DOI] [PubMed] [Google Scholar]
- 22.Shen GJ, Srivastava KC, Saha BC, Zeikus JG. Physiological and enzymatic characterization of a novel pullulan degrading thermophilic Bacillus strain 3183. Appl Microbiol Biotechnol. 1990;33:340–4. doi: 10.1007/BF00164533. [DOI] [Google Scholar]
- 23.Lin LL, Tsau MR, Chu WS. Purification and properties of a 140 kDa amylopullulanase from thermophilic and alkaliphilic from Bacillus sp strain TS-23. Appl Biochem Biotechnol. 1996;24:101–7. [Google Scholar]
- 24.Takasaki Y. Pullulanase-amylase complex enzyme from Bacillus subtilis. Agric Biol Chem. 1987;51:9–16. doi: 10.1271/bbb1961.51.9. [DOI] [Google Scholar]
- 25.Chen JT, Chen MC, Chen LL, Chu WS. Structure and expression of an amylopullulanase gene from Bacillus stearothermophilus TS-23. Biotechnol Appl Biochem. 2001;33:189–99. doi: 10.1042/BA20010003. [DOI] [PubMed] [Google Scholar]
- 26.Saha BC, Zeikus JG. Characterization of pullulanase and α-amylase activities of a Thermus sp. AMD33. Trends Biotechnol. 1989;7:234–8. doi: 10.1016/0167-7799(89)90013-9. [DOI] [Google Scholar]
- 27.Kim CH, Kim DS, Taniguchi H, Maruyama Y. Purification of α amylase-pullulanase bifunctional enzyme by high-performance size exclusion and hydrophobic-interaction chromatography. J Chromatogr A. 1990;512:131–7. doi: 10.1016/S0021-9673(01)89479-6. [DOI] [Google Scholar]
- 28.Vishnu C, Naveena BJ, Altaf M, Venkateshwar M, Reddy G. Amylopullulanase -a novel enzyme of L. amylophilus GV6 in direct fermentation of starch to L (+) lactic acid. Enzyme Microb Technol. 2006;38:545–50. doi: 10.1016/j.enzmictec.2005.07.010. [DOI] [Google Scholar]
- 29.Lemesle-Varloot L, Henrissat B, Gaboriaud C, Bissery V, Morgat A, Mornon JP. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2-D-representation of protein sequences. Biochimie. 1990;72:555–74. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- 30.Noorwez SM, Ezhilvannan M, Satyanarayana T. Production of a high maltose forming, hyperthermostable and Ca2+ independent amylopullulanase by an extreme thermophile Geobacillus thermoleovorans in submerged fermentation. Indian J Biotechnol. 2006;5:337–45. [Google Scholar]
- 31.Roy A, Messaoud EV, Bejar S. Isolation and purification of an acidic pullulanase type II from newly isolated Bacillus sp. US149. Enzyme Microb Technol. 2003;33:720–4. doi: 10.1016/S0141-0229(03)00212-6. [DOI] [Google Scholar]
- 32.Zareian S, Khajeh K, Ranjbar B, Dabirmanesh B, Ghollasi M, Mollania N. Purification and characterization of a novel amylopullulanase that converts pullulan to glucose, maltose, and maltotriose and starch to glucose and maltose. Enzyme Microb Technol. 2010;46:57–63. doi: 10.1016/j.enzmictec.2009.09.012. [DOI] [Google Scholar]
- 33.Hyun HH, Zeikus JG. General biochemical characterization of thermostable pullulanase and glucoamylase from Clostridium thermohydrosulfuricum. Appl Environ Microbiol. 1985;49:1168–73. doi: 10.1128/aem.49.5.1168-1173.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melasniemi H. Characterization of alpha-amylase and pullulanase activities of Clostridium thermohydrosulfuricum. Evidence for a novel thermostable amylase. Biochem J. 1987;246:193–7. doi: 10.1042/bj2460193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JH, Sunako M, Ono H, Murooka Y, Fukusaki E, Yamashita M. Characterization of gene encoding amylopullulanase from plant-originated lactic acid bacterium, Lactobacillus plantarum L137. J Biosci Bioeng. 2008;106:449–59. doi: 10.1263/jbb.106.449. [DOI] [PubMed] [Google Scholar]
- 36.Saha BC, Lamed R, Lee C-Y, Mathupala SP, Zeikus JG. Characterization of an endo-acting amylopullulanase from Thermoanaerobacter strain B6A. Appl Environ Microbiol. 1990;56:881–6. doi: 10.1128/aem.56.4.881-886.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antranikian G, Herzberg C, Mayer F, Gottschalk G. Changes in the cell envelope structure of Clostridium sp. strain EM1 during massive production of α-amylase and pullulanase. FEMS Microbiol Lett. 1987;41:193–7. [Google Scholar]
- 38.Spreinat A, Antranikian G. Purification and properties of a thermostable pullulanase from Clostridium thermosulfurogenes EM1 which hydrolyzes both α-1, 6 and α-1, 4-glycosidic linkages. Appl Microbiol Biotechnol. 1990;33:511–8. doi: 10.1007/BF00172543. [DOI] [Google Scholar]
- 39.Lin FP, Leu KL. Cloning, expression, and characterization of thermostable region of amylopullulanase gene from Thermoanaerobacter ethanolicus 39E. Appl Biochem Biotechnol. 2002;97:33–44. doi: 10.1385/ABAB:97:1:33. [DOI] [PubMed] [Google Scholar]
- 40.Plant AR, Clemens RM, Daniel RM, Morgan HW. Purification and preliminary characterization of an extracellular pullulanase from Thermoanaerobium Tok6-B1. Appl Microbiol Biotechnol. 1987;26:427–33. doi: 10.1007/BF00253526. [DOI] [Google Scholar]
- 41.Plant AR, Clemens RM, Morgan HW, Daniel RM. Active-site- and substrate-specificity of Thermoanaerobium Tok6-B1 pullulanase. Biochem J. 1987;246:537–41. doi: 10.1042/bj2460537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganghofner D, Kellermann J, Staudenbauer WL, Bronnenmeier K. Purification and properties of an amylopullulanase, a glucoamylase, and an α-glucosidase in the amylolytic enzyme system of Thermoanaerobacterium thermosaccharolyticum. Biosci Biotechnol Biochem. 1998;62:302–8. doi: 10.1271/bbb.62.302. [DOI] [PubMed] [Google Scholar]
- 43.Mathupala SP, Zeikus JG. Improved purification and biochemical characterization of extracellular amylopullulanase from Thermoanaerobacter ethanolicus 39E. Appl Microbiol Biotechnol. 1993;39:487–93. doi: 10.1007/BF00205038. [DOI] [Google Scholar]
- 44.Koch R, Zablowski P, Antranikian G. Highly active and thermostable amylases and pullulanases from various anaerobes. Appl Microbiol Biotechnol. 1987;27:192–8. doi: 10.1007/BF00251944. [DOI] [Google Scholar]
- 45.Koch R, Antranikian G. The action of amylolytic and pullulytic enzymes from various anaerobic thermophiles on linear and branched glucose polymers. Starch. 1990;42:397–403. doi: 10.1002/star.19900421007. [DOI] [Google Scholar]
- 46.Bibel M, Brettl C, Gosslar U, Kriegshäuser G, Liebl W. Isolation and analysis of genes for amylolytic enzymes of the hyperthermophilic bacterium Thermotoga maritima. FEMS Microbiol Lett. 1998;158:9–15. doi: 10.1111/j.1574-6968.1998.tb12793.x. [DOI] [PubMed] [Google Scholar]
- 47.Kwak YS, Akiba T, Kudo T. Purification and characterization of α-amylase from hyperthermophilic archaeon Thermococcus profundus, which hydrolyzes both α-1, 4 and α-1, 6 glucosidic linkages. J Ferment Bioeng. 1998;86:363–7. doi: 10.1016/S0922-338X(99)89005-9. [DOI] [Google Scholar]
- 48.Dong G, Vieille C, Zeikus JG. Cloning, sequencing, and expression of the gene encoding amylopullulanase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol. 1997;63:3577–84. doi: 10.1128/aem.63.9.3577-3584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rüdiger A, Jorgensen PL, Antranikian G. Isolation and characterization of a heat-stable pullulanase from the hyperthermophilic archaeon Pyrococcus woesei after cloning and expression of its gene in Escherichia coli. Appl Environ Microbiol. 1995;61:567–75. doi: 10.1128/aem.61.2.567-575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown SH, Kelly RM. Characterization of amylolytic enzymes, having both a-1,4 and a-1,6 hydrolytic activity, from the thermophilic archaea Pyrococcus furiosus and Thermococcus litoralis. Appl Environ Microbiol. 1993;59:2614–21. doi: 10.1128/aem.59.8.2614-2621.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Canganella F, Andrade CM, Antranikian G. Characterization of amylolytic and pullulytic enzymes from thermophilic archaea and from a new Fervidobacterium species. Appl Microbiol Biotechnol. 1994;42:239–45. [Google Scholar]
- 52.Erra-Pujada M, Debeire P, Duchiron F, O’Donohue MJ. The type II pullulanase of Thermococcus hydrothermalis: molecular characterization of the gene and expression of the catalytic domain. J Bacteriol. 1999;181:3284–7. doi: 10.1128/jb.181.10.3284-3287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zona R, Chang-Pi-Hin F, O’Donohue MJ, Janecek S. Bioinformatics of the glycoside hydrolase family 57 and identification of catalytic residues in amylopullulanase from Thermococcus hydrothermalis. Eur J Biochem. 2004;271:2863–72. doi: 10.1111/j.1432-1033.2004.04144.x. [DOI] [PubMed] [Google Scholar]
- 54.Kim JH, Sunako M, Ono H, Murooka Y, Fukusaki E, Yamashita M. Characterization of the C-terminal truncated form of amylopullulanase from Lactobacillus plantarum L137. J Biosci Bioeng. 2009;107:124–9. doi: 10.1016/j.jbiosc.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 55.MacGregor EA, Janecek S, Svensson B. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim Biophys Acta. 2001;1546:1–20. doi: 10.1016/S0167-4838(00)00302-2. [DOI] [PubMed] [Google Scholar]
- 56.Katsuya Y, Mezaki Y, Kubota M, Matsuura Y. Three-dimensional structure of Pseudomonas isoamylase at 2.2 A resolution. J Mol Biol. 1998;281:885–97. doi: 10.1006/jmbi.1998.1992. [DOI] [PubMed] [Google Scholar]
- 57.Robert X, Haser R, Gottschalk TE, Ratajczak F, Driguez H, Svensson B, et al. The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs. Structure. 2003;11:973–84. doi: 10.1016/S0969-2126(03)00151-5. [DOI] [PubMed] [Google Scholar]
- 58.Kataeva IA, Seidel RD, 3rd, Shah A, West LT, Li XL, Ljungdahl LG. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl Environ Microbiol. 2002;68:4292–300. doi: 10.1128/AEM.68.9.4292-4300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki K, Taiyoji M, Sugawara N, Nikaidou N, Henrissat B, Watanabe T. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem J. 1999;343:587–96. doi: 10.1042/0264-6021:3430587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe T, Ito Y, Yamada T, Hashimoto M, Sekine S, Tanaka H. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J Bacteriol. 1994;176:4465–72. doi: 10.1128/jb.176.15.4465-4472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christiansen C, Abou Hachem M, Janecek S, Viksø-Nielsen A, Blennow A, Svensson B. The carbohydrate-binding module family 20--diversity, structure, and function. FEBS J. 2009;276:5006–29. doi: 10.1111/j.1742-4658.2009.07221.x. [DOI] [PubMed] [Google Scholar]
- 62.Janeček S, Svensson B, MacGregor EA. Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enzyme Microb Technol. 2011;49:429–40. doi: 10.1016/j.enzmictec.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 63.Southall SM, Simpson PJ, Gilbert HJ, Williamson G, Williamson MP. The starch-binding domain from glucoamylase disrupts the structure of starch. FEBS Lett. 1999;447:58–60. doi: 10.1016/S0014-5793(99)00263-X. [DOI] [PubMed] [Google Scholar]
- 64.Takata H, Kuriki T, Okada S, Takesada Y, Iizuka M, Minamiura N, et al. Action of neopullulanase. Neopullulanase catalyzes both hydrolysis and transglycosylation at alpha-(1----4)- and alpha-(1----6)-glucosidic linkages. J Biol Chem. 1992;267:18447–52. [PubMed] [Google Scholar]
- 65.Nakajima R, Imanaka T, Aiba S. Comparison of amino acid sequences of eleven different α-amylases. Appl Microbiol Biotechnol. 1986;23:355–60. doi: 10.1007/BF00257032. [DOI] [Google Scholar]
- 66.Nisha M, Satyanarayana T. Characterization of recombinant amylopullulanase (gt-apu) and truncated amylopullulanase (gt-apuT) of the extreme thermophile Geobacillus thermoleovorans NP33 and their action in starch saccharification. Appl Microbiol Biotechnol. 2012 doi: 10.1007/s00253-012-4538-6. [DOI] [PubMed] [Google Scholar]
- 67.Woodcock S, Mornon JP, Henrissat B. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 1992;5:629–35. doi: 10.1093/protein/5.7.629. [DOI] [PubMed] [Google Scholar]
- 68.Lin FP, Ma HY, Lin HJ, Liu SM, Tzou WS. Biochemical characterization of two truncated forms of amylopullulanase from Thermoanaerobacterium saccharolyticum NTOU1 to identify its enzymatically active region. Appl Biochem Biotechnol. 2011;165:1047–56. doi: 10.1007/s12010-011-9319-7. [DOI] [PubMed] [Google Scholar]
- 69.Fogarty WM, Kelly CT. Microbial Enzymes and Biotechnology. London: Elsevier Applied Science Publishers; 1990. [Google Scholar]
- 70.Zoebelein H, Böllert V. Dictionary of renewable resources. Weinheim: Wiley-VCH, 2001:181. [Google Scholar]
- 71.Juliano B. Rice chemistry and technology American Association of Cereal Chemists. 3rded. St Paul (MN); AAC International Press; 1985. [Google Scholar]
- 72.Hansen LP, Hosek R, Callan M, Joens FT. The development of high-protein rice flour for early childhood feeding. J Food Technol. 1981;35:38–43. [Google Scholar]
- 73.Morita T, Kiriyama S. Mass production method for rice protein isolate and nutritional evaluation. J Food Sci. 1993;58:1393–7. doi: 10.1111/j.1365-2621.1993.tb06190.x. [DOI] [Google Scholar]
- 74.Kulp K, Ponte JG., Jr. Staling white pan bread: fundamental causes. Crit Rev Food Sci Nutr. 1981;15:1–48. doi: 10.1080/10408398109527311. [DOI] [PubMed] [Google Scholar]
- 75.Champenois Y, Della Valle G, Planchot V, Buleon A, Colonna P. Influence of alpha-amylases on the bread staling and on retrogradation of wheat starch models. Acta Sci Pol Technol Aliment. 1999;19:471–86. [Google Scholar]
- 76.De Stefanis VA, Turner EW. Modified enzyme system to inhibit bread firming method for preparing same and use of same inbread and other bakery products. US Patent 1981; 4:299-848.
- 77.Guraya HS, James C, Champagne ET. Effect of enzyme concentration and storage temperature on the formation of slowly digestible starch from cooked debranched rice starch. Starch. 2001;53:131–9. doi: 10.1002/1521-379X(200104)53:3/4<131::AID-STAR131>3.0.CO;2-M. [DOI] [Google Scholar]
- 78.Miao M, Jiang B, Zhang T. Effect of pullulanase debranching and recrystallization on structure and digestibility of waxy maize starch. Carbohydr Polym. 2009;76:214–21. doi: 10.1016/j.carbpol.2008.10.007. [DOI] [Google Scholar]
- 79.Ells LJ, Seal CJ, Kettlitz B, Bal W, Mathers JC. Postprandial glycaemic, lipaemic and haemostatic responses to ingestion of rapidly and slowly digested starches in healthy young women. Br J Nutr. 2005;94:948–55. doi: 10.1079/BJN20051554. [DOI] [PubMed] [Google Scholar]
- 80.Shin SI, Kim HJ, Ha HJ, Lee SH, Moon TW. Effect of hydrothermal treatment on formation and structural characteristics of slowly digestible nonpasted granular sweet potato starch. Starch. 2005;57:421–30. doi: 10.1002/star.200400377. [DOI] [Google Scholar]
- 81.Wolf BW, Bauer LL, Fahey GC., Jr. Effects of chemical modification on in vitro rate and extent of food starch digestion: an attempt to discover a slowly digested starch. J Agric Food Chem. 1999;47:4178–83. doi: 10.1021/jf9813900. [DOI] [PubMed] [Google Scholar]
- 82.Eliasson AC. Starch in food: Structure, function and applications. Woodhead Publishing Limited. Cambridge, 2004:477-505. [Google Scholar]
- 83.Zhang H, Jin Z. Preparation of resistant starch by hydrolysis of maize starch with pullulanase. Carbohydr Polym. 2011;83:865–7. doi: 10.1016/j.carbpol.2010.08.066. [DOI] [Google Scholar]
- 84.Annison G, Topping DL. Nutritional role of resistant starch: chemical structure vs physiological function. Annu Rev Nutr. 1994;14:297–320. doi: 10.1146/annurev.nu.14.070194.001501. [DOI] [PubMed] [Google Scholar]
- 85.Mun SH, Shin M. Mild hydrolysis of resistant starch from maize. Food Chem. 2006;96:115–21. doi: 10.1016/j.foodchem.2005.02.015. [DOI] [Google Scholar]
- 86.Watanabe N, Yamamoto K, Tsuzuki W, Ohya T, Kobayashi S. A Novel Method to Produce Branched α-cyclodextrins: pullulanase-glucoamylase-mixed method. J Ferment Bioeng. 1997;83:43–7. doi: 10.1016/S0922-338X(97)87325-4. [DOI] [Google Scholar]
- 87.French D, Pulley AO, Effenberger JA, Rougvie MA, Abdullah M. Studies on the Schardinger dextrins. XII. The molecular size and structure of the δ-, ε-, ζ-, and η-dextrins. Arch Biochem Biophys. 1965;111:153–60. doi: 10.1016/0003-9861(65)90334-6. [DOI] [PubMed] [Google Scholar]
- 88.Ara K, Igarashi K, Saeki K, Kawai S, Ito S. Purification and some properties of an alkaline pullulanase from alkalophilic Bacillus sp. KSM-1876. Biosci Biotechnol Biochem. 1992;56:62–5. doi: 10.1271/bbb.56.62. [DOI] [Google Scholar]
- 89.Brunswick JM, Kelly CT, Fogarty WM. The amylopullulanase of Bacillus sp. DSM 405. Appl Microbiol Biotechnol. 1999;51:170–5. doi: 10.1007/s002530051378. [DOI] [PubMed] [Google Scholar]