Abstract
Chronic obstructive pulmonary disease (COPD) is a respiratory disorder caused by extended exposure of the airways to noxious stimuli, principally cigarette smoke (CS). The mechanisms through which COPD develops are not fully understood, though it is believed that the disease process includes a genetic component, as not all smokers develop COPD. To investigate the mechanisms that lead to the development of COPD/emphysema, we measured whole genome gene expression and several COPD-relevant biological endpoints in mouse lung tissue after exposure to two CS doses for various lengths of time. A novel and powerful method, Reverse Engineering and Forward Simulation (REFS™), was employed to identify key molecular drivers by integrating the gene expression data and four measured COPD-relevant endpoints (matrix metalloproteinase (MMP) activity, MMP-9 levels, tissue inhibitor of metalloproteinase-1 levels and lung weight). An ensemble of molecular networks was generated using REFS™, and simulations showed that it could successfully recover the measured experimental data for gene expression and COPD-relevant endpoints. The ensemble of networks was then employed to simulate thousands of in silico gene knockdown experiments. Thirty-three molecular key drivers for the above four COPD-relevant endpoints were therefore identified, with the majority shown to be enriched in inflammation and COPD.
Keywords: Bayesian network, chronic obstructive pulmonary disease (COPD), reverse engineering and forward simulation (REFS™)
Introduction
COPD and smoking
Chronic obstructive pulmonary disease (COPD) refers to a group of pathologies that includes progressively worsening respiratory symptoms, non-reversible airway obstruction, chronic bronchitis, and emphysema.1–3 In the developed world, the most important risk factor for COPD is cigarette smoke (CS) and it has been estimated that tobacco use accounts for up to 70–95% of COPD in Western populations, 4,5 although pollution and occupational exposure to dust and chemicals are also important risk factors for this disease.6 Incidence estimates of COPD vary in smokers,7 and not all develop COPD, indicating that there may be genetic or host factors that predispose individuals to its development. The collection and analysis of clinical data has provided insights into the diagnosis and treatment of COPD, and it has been found advantageous to analyze molecular and genetic data collected from in vivo models to elucidate the underlying molecular mechanisms of COPD. Over the past few years, animal models of CS-induced lung damage that mimic human disease have been developed, including mouse models in which emphysematous changes in the lung can be observed after 5 months of exposure to CS.8 Whilst these models have provided information about the physiological outcomes of smoke inhalation, the molecular mechanisms underlying disease development remain unclear. To address this, we applied state of the art Bayesian network inference technology, Reverse Engineering and Forward Simulation (REFS™), to data generated from mouse models of CS-induced disease in an attempt to uncover gene regulatory networks involved in the development of COPD/emphysema.
Classical emphysema hypothesis
For the past few decades, the prevailing hypothesis regarding emphysema onset and progression has revolved around lung inflammation caused by CS and environmental pollutants. This is postulated to cause a protease/antiprotease imbalance, which ultimately results in the alveolar destruction seen in emphysema.9
One of the enzyme groups thought to play an important role in the development of emphysema is the matrix metalloproteinases (MMP). These enzymes are proteases that can be activated by reactive oxygen species via metalloproteinase precursors.10–16 In addition, they can be inhibited by so-called antiproteases.17 These proteases and antiproteases are thought to play a key role in CS-induced emphysema, and, at the most fundamental level, emphysema is caused by an imbalance of protease and antiprotease activities that result in lung parenchymal tissue destruction.18 This imbalance might result from an abnormal increase in pulmonary proteases or a decrease in antiproteases. For example, oxidation of methionine residues at the active sites of the antiprotease alpha-1 antitrypsin results in a dramatic reduction of its in vitro inhibitory ability, leading to an increase in proteases over antiproteases,17,19 which overwhelms the local antiproteolytic defense mechanism. The outcome is a breakdown of extracellular matrix components of the lung, which is postulated to result in pulmonary emphysema. Indeed, patients deficient in the serine elastase inhibitor a1-antitrypsin were shown to develop early-onset emphysema, particularly if they smoked.20
Matrix Metalloproteinases (MMPs)
MMPs are capable of degrading non-matrix proteins such as cytokines, chemokines, growth factors, and proteinase inhibitors, suggesting an indirect role in the development and progression of CS-induced pulmonary emphysema as well as chronic obstructive bronchiolitis and chronic bronchitis.21–23 Increased evidence for the involvement of MMPs in CS-related emphysema comes from several human as well as animal studies. Patients with emphysema showed increased concentrations of MMP-1, MMP-8, and MMP-9 in bronchoalveolar lavage fluid (BALF),11,24 while the expression and activity of MMP-1, MMP-2, and MMP-9 was found to be increased in the lung parenchyma of emphysematous patients compared with healthy controls.10,12,13 Although MMP-9 −/− mice were not protected against CS-induced pulmonary emphysema, they were protected from small airway fibrosis, which is another feature of COPD.25 Churg et al recently found that an MMP-9/12 inhibitor prevented the development of pulmonary emphysema in CS-exposed guinea pigs.26 Moreover, MMP-2, MMP-9, and MMP-12 were increased in BALF from mice with CS-induced pulmonary emphysema.27–29
Although, many human and animal studies suggest that MMP-9 may prove to be a useful biomarker for CS-induced pulmonary emphysema,24,27,29–31 current knowledge is still limited. Therefore, to investigate the protease/antiprotease balance, we measured the overall enzyme activity of MMP and the amount of MMP-9 protein together with levels of the anti-protease tissue inhibitor of metalloproteinase (TIMP-1).
Causal modeling
Typically, high-throughput genome-wide molecular data are analyzed using multi-dimensional statistical approaches and machine-learning techniques to identify correlations between variables in a high dimensional space. Such analyses rank variable links according to statistical confidence values derived from the experimental observations. However, the identification of statistical links between variables alone cannot determine causal directions. Different mathematical modeling techniques use information “learned” from previous experimentation, which is often deemed to be the “true” knowledge of an existing physical property. These models are employed to establish a predictable dependency over time and to model the flux of variables in a physical/biological system. To extrapolate new knowledge from such models, comprehensive experimentation and sound statistical, correlation-based analyses of data are required.
The identification of interactions between molecular entities within cells is key to understanding the biological processes involved.32–34 Bayesian network theory provides a convenient framework for systematizing data to deduce probabilistic cause-and-effect relationships and for modeling systems of increasingly large numbers of interacting variables35–37; thus, Bayesian network simulations can be used to predict the effects of specific interventions. The REFS™ technology is an efficiently parallelized implementation of Bayesian network inference and simulation. REFS™ allows users to rapidly infer interaction networks from high dimensional data through the parallelization of high-intensity computations across an immensely large number of processing units. REFS™ combines two mathematical techniques: Bayesian network inference and simulations of ensembles of Bayesian networks. Previous approaches made predictions based on a single network topology,38,39 whereas REFS™ algorithms differ in two distinctive ways: the first is that REFS™ generates a statistical sample, or ensemble, of network structures consistent with collected data; the second is that REFS™ enables quantitative predictions to be made of perturbation effects and additionally accounts for uncertainties in network topology and in the parameter estimates of local statistical models. With the ability to predict the impact of specific interventions that may not be part of the collected data, REFS™ organizes the data into a rational model and also predicts unseen events. In this regard, the model captures the essence of the physical process as it is observed in the data.
The REFS™ platform has been utilized and validated in collaborations between GNS healthcare, the pharmaceutical industry, and academia to learn novel biology ab initio from experimental and clinical data sets.40 For example, Xing et al. used REFS™ to identify new therapeutic targets for rheumatoid arthritis.41 In a project studying a new cancer drug in cancer cell lines, six of seven key genes identified by REFS™ were validated by siRNA knockdown and other findings were validated in the literature.40 Moreover, in a collaboration between GNS Healthcare and an academic group, REFS metabolic disease mouse model data composed of genetic variation (single-nucleotide polymorphisms), gene expression, and endpoint data made accurate out-of-sample predictions about endpoints for mice not trained upon and yielded both existing and novel targets for metabolic disease.40
Results
CS-induced mouse model of COPD progression
To investigate the mechanism leading to the development of COPD/emphysema, the transcriptome of parenchymal tissue obtained from the lungs of CS-exposed A/J mice was measured as a function of CS exposure duration and smoking cessation as well as a function of CS dose. CS dose was defined as the number of hours per day that the animal was exposed to CS, and exposure duration was defined as the total number of days of exposure. The mice were exposed to two CS doses: 2 h per day for the low-dose group and 4 h per day for the high-dose group. Gene expression profiles were obtained from CS-exposed and control animals after various exposure times: 1 day, 7 days, 1 month, and 5 months (Table 1). Smoking cessation was modeled as 5 months of exposure followed by 2 months without CS exposure. The CS-exposed groups were exposed to mainstream CS generated from the standard 3R4F reference cigarette (University of Kentucky, Lexington, KY, USA) and control animals were exposed to filtered ambient air (sham exposure).
Table 1.
Number of animals used to capture each measured endpoint (gene expression, MMP activity, MMP-9 and TIMP-1 abundance, lung weight) per condition (dose and time point).
| EXPOSURE TIME | 1 DAY | 7 DAYS | 1 MONTH | 5 MONTHS | 5 MONTHS + 2 MONTHS PE | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|||||||||
| SMOKE EXPOSURE GROUPS | SHAM | LOW | HIGH | SHAM | LOW | HIGH | SHAM | LOW | HIGH | SHAM | LOW | HIGH | SHAM | HIGH |
| Gene expression | 8 | 7 | 8 | 8 | 8 | 8 | 7 | 7 | 8 | 8 | 8 | 8 | 8 | 8 |
|
| ||||||||||||||
| MMP activity | ND | 9 | 9 | 10 | 10 | 8 | 10 | 9 | 10 | 10 | 10 | 10 | 5 | 9 |
|
| ||||||||||||||
| MMP-9 | ND | 10 | 10 | 10 | 10 | 10 | 10 | 9 | 9 | 10 | 10 | 10 | 8 | 10 |
|
| ||||||||||||||
| TIMP-1 | ND | 10 | 10 | 10 | 10 | 10 | 10 | 9 | 9 | 9 | 9 | 8 | 8 | 10 |
|
| ||||||||||||||
| Lung weight | 10 | 10 | 10 | 10 | 9 | 10 | 10 | 10 | 10 | 18 | ND | 24 | 20 | 20 |
Note: ND, not determined.
To identify key molecular regulators of COPD/emphysema progression in A/J mice, BALF from lung tissue was examined for the presence of the putative emphysema-related biological endpoints MMP-9, TIMP-1 (the levels of both), and MMP activity (measured by gelatinase activity). In addition to these molecular endpoints, animal lung weight was included as a crude indicator of the accumulated mass of connective tissue matrix material, structural cells, inflammatory cells, and edema fluid in the lung. Acute (1-day and 7-day) and chronic (1-month and 5-month) responses to CS exposure were investigated in the lung tissue at both high and low exposure duration levels.
We found that MMP activity was higher in BALF from CS-exposed A/J mice compared with sham-exposed at the 7-day, 1-month, and 5-month exposure time points. The high-dose (4-h exposure) group showed a greater increase than the low-dose (2-h exposure) group. In the smoking cessation group, MMP activity was almost completely reversed (Fig. S1). A similar trend was seen for MMP-9, with levels dose-dependently increased at each of the time points. MMP-9 levels increased with continuous exposure to CS at the high-exposure dose, and decreased after cessation of exposure back to a level lower than that observed in the 7-day exposure group. TIMP-1 levels dose-dependently increased at each time point, and decreased to levels seen at 1-day exposure after 2 months of cessation. The mean absolute lung weight, including the larynx and trachea, was dose- and time-dependently increased at the 7-day, 1-month, and 5-month exposure time points in both the low- and high-exposure groups compared with sham animals at the same time points and with exposed animals at the previous timepoint (Fig. S1). Following the 2-month cessation period, the lung weight of smoke-exposed animals decreased but remained heavier than in sham animals (Fig. S1).
Building the REFS™ BioModel™ of molecular interactions and COPD/emphysema endpoints
Bayesian causal networks were inferred from experimental observations to gain a more comprehensive understanding of the molecular mechanisms underpinning COPD/emphysema progression in the A/J mouse. REFS™ technology was used to generate an ensemble of Bayesian networks from gene expression, MMP activity, MMP-9 expression, TIMP-1 expression, and lung weight data. The REFS™ BioModel™ (hereafter called BioModel™) was built as a collection of 10 ensembles comprising 100 Bayesian networks from 10 data frames constructed using bootstrap sampling of endpoint values and endpoint measurements matched with microarray data within experimental groups. The lung weight values were imputed for one of the experimental groups (5-month, low dose) because of missing measurements (Fig. S2).
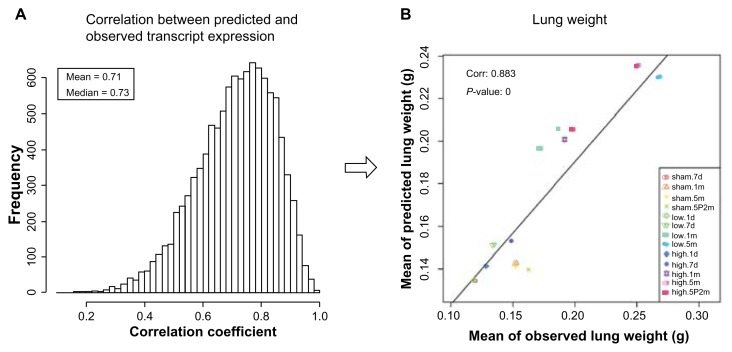
The BioModel™ of COPD/emphysema progression was assessed for accuracy by simulating variables for all experimental conditions. The simulation results for each of the 10 ensembles (data frames) of Bayesian networks were compared with observed variables. After the BioModel™ was built and successfully tested, the only inputs used were CS exposure time, total exposure, and CS cessation. Given these, the expression values of 10,643 probe sets were predicted, from which the four endpoints were predicted. The prediction/simulation was run 30 times for each of the 100 networks belonging to a specific data frame; therefore, a total of 3,000 predicted values were made for every probe set and every endpoint. Figure 1 shows representative comparison charts for one of the 10 data frames. Figure 1A shows the empirical distribution of the correlation coefficient between predicted and observed transcript expression for 10,643 probe sets; the mean of this is 0.71, which is significantly greater than zero. Figure 1B displays the scatter plot of the mean of observed and predicted lung weights. The correlation coefficient between the mean of the observed and predicted lung weights is 0.883, which is significant based on the P-value. We tested the null hypothesis that the predicted value is equal to the observed value. A paired t-test gave a P-value of 0.79, so we cannot reject the null hypothesis that the predicted value is equal to the observed value. The scatter plots of the other three endpoints, MMP-9, TIMP-1, and MMP activity, are similar to the scatter plot of lung weight with varying correlation coefficients. These results show that the predictions given by BioModel™ correlate well with the observations.
Figure 1.
Correlation between BioModel™-predicted values and observations. A. Empirical distribution of the correlation coefficients between predicted and observed transcript expression for 10,643 probe sets, the mean of which is 0.71. B. Scatter plot between means of observed and predicted lung weights, with a correlation of 0.883. The BioModel™ predictions correlate well with the observations.
BioModel™ identified molecular drivers of COPD/emphysema-related endpoints
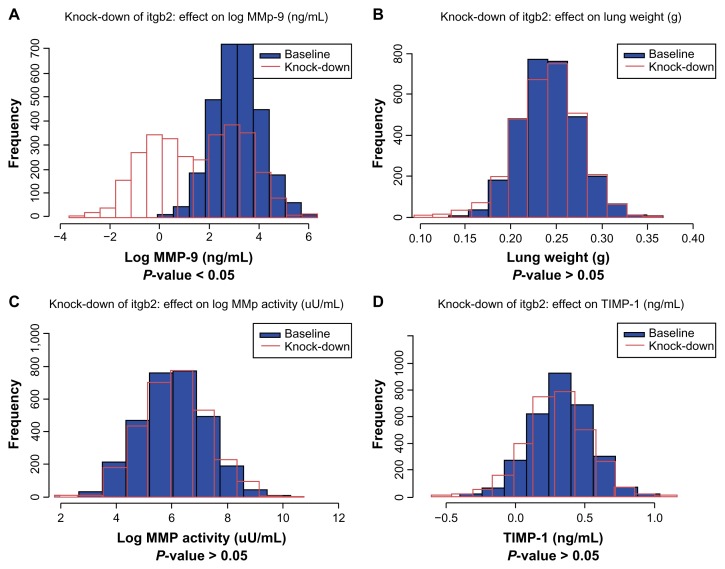
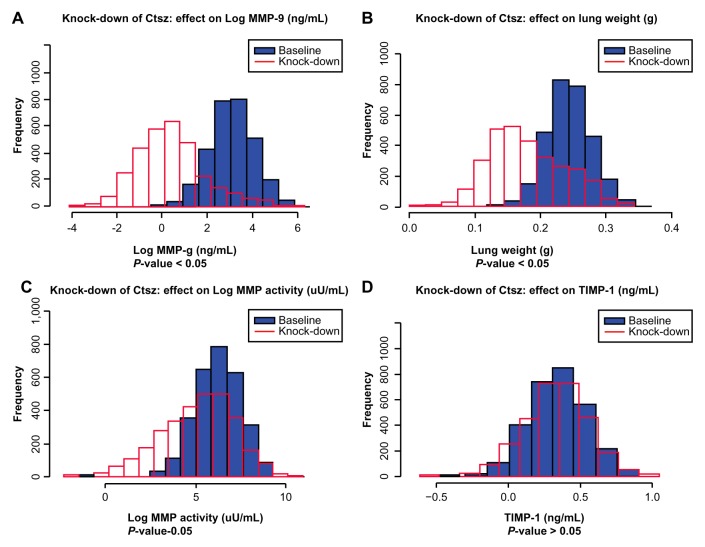
A REFS™ model is an ensemble of Bayesian networks that captures not only the most likely topology of variable interactions but also derives mathematical relationships of such interactions and their corresponding uncertainties. Extensive model simulations were therefore performed to identify gene expression variables that have a measurable effect on MMP activity, MMP-9 and TIMP-1 expression, and lung weight changes. By analyzing network topology and selecting variables upstream of the endpoints identified, causal transcripts could be obtained. The gene expression of every causal transcript was knocked-down in silico by 10-fold. Differences in the endpoint posterior distributions across the 10 data frames were processed to assess the effects of these perturbations. Molecular drivers for the endpoints were identified under high-dose CS exposure for 1 and 5 months, as well as CS cessation. Key molecular drivers for a specific endpoint were defined as those genes that act as molecular drivers of the particular endpoint at all three time points. Table 2 lists these for the four endpoints: MMP activity, MMP-9 and TIMP-1 expression, and lung weight. As an example, the distribution of baseline expression and knockdown expression in silico for integrin (Itgb2; Fig. 2) and cathepsin Z (Ctsz; Fig. 3) genes under high-dose CS exposure for 5 months was plotted. Knocked-down simulations were performed for the high-dose CS group as more genes are known to be perturbed. The in silico 10-fold knockdown of Itgb2 gene expression is predicted to significantly down-regulate endpoint MMP-9 but not to affect the other three endpoints (threshold for adjusted P-value, 0.05). In silico 10-fold knockdown in gene expression of the protease Ctsz is predicted to down-regulate the MMP-9 and lung weight endpoints, but not MMP activity or TIMP-1.
Table 2.
Key molecular drivers identified by REFS™ for the four measured endpoints.
| GENE SYMBOL | NAME | MMP-9 | MMP ACTIVITY | TIMP-1 | LUNG WEIGHT |
|---|---|---|---|---|---|
| Csf2rb | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | × | |||
| Csf2rb2 | colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) | × | |||
| Cyba | cytochrome b-245, alpha polypeptide | × | × | ||
| Rnh1 | ribonuclease/angiogenin inhibitor 1 | × | × | ||
| Ctsz | cathepsin Z | × | × | ||
| Hal | histidine ammonia lyase | × | |||
| Gusb | glucuronidase, beta | × | × | ||
| Itgb2 | integrin beta 2 | × | |||
| Fuca1 | fucosidase, alpha-L-1, tissue | × | × | × | |
| Psmd8 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | × | |||
| Clec7a | C-type lectin domain family 7, member a | × | |||
| Itgax | integrin alpha X | × | |||
| Nceh1 | arylacetamide deacetylase-like 1 | × | |||
| Macf1 | microtubule-actin crosslinking factor 1 | × | |||
| Ceacam1; Ceacam2 | carcinoembryonic antigen-related cell adhesion molecule 1; carcinoembryonic antigen-related cell adhesion molecule 2 | × | |||
| Slc9a3r2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | × | |||
| Ubxn2a | UBX domain protein 2A | × | |||
| Pltp | phospholipid transfer protein | × | |||
| Kif5b | kinesin family member 5B | × | |||
| Gstp1 | glutathione S-transferase, pi 1 | × | |||
| Zranb1 | zinc finger, RAN-binding domain containing 1 | × | |||
| Pcf11 | cleavage and polyadenylation factor subunit homolog (S. cerevisiae) | × | |||
| Slc11a1 | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | × | |||
| Npy | neuropeptide Y | × | |||
| Trem2 | triggering receptor expressed on myeloid cells 2 | × | |||
| Hvcn1 | hydrogen voltage-gated channel 1 | × | |||
| Orm1 | orosomucoid 1 | × | |||
| Ctsb | cathepsin B | × | |||
| Msr1 | macrophage scavenger receptor 1 | × | |||
| Pla2g2d | phospholipase A2, group IID | × | |||
| Atp6v0c | ATPase, H+ transporting, lysosomal V0 subunit C | × | |||
| Hpse | heparanase | × |
Figure 2.
In silico 10-fold knockdown of Itgb2 modulates MMP-9. Plots of simulated concentrations of A. MMP-9, B. lung weight, C. MMP activity, and D. TIMP-1 in response to a 10-fold knockdown of Itgb2 expression. Baseline (without knockdown), blue; knockdown, red. The time point shown here is 5 months and the conditions are high-dose CS.
Figure 3.
In silico 10-fold knockdown of Ctsz modulates MMP-9 and lung weight. Plots of simulated concentrations of A. MMP-9, B. lung weight, C. MMP activity, and D. TIMP-1 in response to a 10-fold knockdown of Ctsz expression. Baseline (without knockdown), blue; knockdown, red. The time point shown here is 5 months and the conditions are high-dose CS.
Literature support for the REFS™ findings
At least 15 of the identified key drivers were described in the context of inflammatory responses (Ingenuity Systems, www.ingenuity.com); eight of these (Ceacam1, Clec7a, Csf2rb, Itgb2, mMsr1, Npy, Sc11a1, and Trem2) were reported to be involved in the activation of leucocytes, and six (Csf2rb, Ctsz, Itgax, Itgb2, Msr1, and Pltp) were associated with immune cell adhesion. A major contributory factor to the development of COPD is the inflammatory response to CS.42 Neutrophils and macrophages have been implicated in this process through their ability to release proteolytic enzymes and generate oxidants, which cause tissue damage, as well as the release of cytokines and chemokines, which can potentiate inflammation and trigger an immune response. Therefore, it is comprehensible that nearly half of our identified key drivers are linked to this process.
Functional annotations were performed using the DAVID Bioinformatics Resources 6.7 database.43 Key molecular drivers Atp6v0c, Ctsb, Ctsz, Fuca1, Gusb, and Slc11a1 were assigned to lysosomal pathways that transport damaged proteins and organelles to lysosomes for degradation. This process of autophagy involves a highly conserved homeostatic pathway that was recently linked to the pathogenesis of COPD.44–46
The REFS™ approach identified several endpoint drivers for the protease Mmp-9, including Itgb2 (which encodes the integrin β2 chain, CD18) and Itgax (which encodes the integrin alpha X chain, CD11c) (Table 2). Integrins are integral cell-surface receptors that are involved in cell functions such as signaling, adherence, aggregation, and the regulation of shape, motility and the cell cycle. Thus, integrin α and β chains were identified as drivers for MMP-9. Interestingly, both integrin chains can form a complex (integrin αXβ2 or p150,95) known as complement receptor 4 (CR4).47 The complement receptors and phagocytic cells that express those receptors are likely to be of great importance in maintaining a degree of order in the smoker’s lung by triggering clearance through phagocytosis48 Indeed, several studies suggest that complement is activated in COPD.49,50 Moreover, a literature search revealed that integrin β2 chain functions as a substrate for MMP-9, thus linking them directly. These studies show that Mmp-9 induces the shedding of the integrin β2 chain subunit from macrophages, causing an efflux of macrophages from the inflammatory site.51 It was proposed that this mechanism contributes to the resolution of inflammation. The in silico 10-fold knockdown in gene expression of Itgb2 was predicted to down-regulate the endpoint MMP-9 (Fig. 2), which might indicate that Itgb2 regulates Mmp-9 feedback. When less Itgb2 is expressed, fewer macrophages or neutrophils would be attached to the sites of inflammation, which may lead to a reduced contribution of MMP9-mediated cleavage of the integrin β2 chain. Further, in silico 10-fold knockdown of Ctsz expression was predicted to down-regulate the endpoint MMP-9 (Fig. 3). It has been reported that the overexpression of Ctsz up-regulates Mmp-9 (both the precursor and active form) in hepatocellular carcinoma, 52 although to our knowledge no relationship with COPD has been identified. It is tempting to speculate that CtsZ has been identified as a new protease involved in COPD.
BioModel™ also identified β-glucuronidases as key drivers of Mmp-9 (and lung weight). β-glucuronidases catalyze the breakdown of complex carbohydrates,53 and the gelatinolytic activity in tracheal aspirates of horses suffering from COPD has been correlated with β-glucuronidase activity.54 These findings suggest that MMP-9-related gelatinases, possibly originating from neutrophils or macrophages, may play a role in the pathogenesis of equine respiratory diseases. In addition to the horse data, alveolar macrophages from COPD patients have significantly more β-glucuronidase than macrophages from patients without COPD.55
Colony stimulating factor 2 receptor β and the β-2 subunit were also identified as key drivers of MMP-9. The protein encoded by this gene is the common β chain of the high affinity receptor for interleukin (IL)-3, IL-5, and colony stimulating factor (CSF). Although no direct link with the COPD receptor has been reported, the CSF2 ligand granulocyte colony-stimulating factor 2 (granulocyte-macrophage), also known as GM-CSF, has been shown to play an important role in the pathogenesis of acute and chronic lung disease,56 and it has been suggested that neutralization of GM-CSF is a novel treatment modality for lung inflammation, particularly for COPD.56 However, only a few studies have suggested a connection between GM-CSF and MMP-9; for example, small murine cholangiocyte (bile duct epithelia) cultures treated with a combination of stem cell factor and GM-CSF showed significantly elevated Mmp2 and Mmp-9 levels,57 suggesting that GM-CSF may be involved in the expression of MMP-9, although the effects could have been caused by stem cell factor alone.
Based on the REFS™ approach, none of the above-mentioned transcripts appear to cause changes in TIMP-1 expression. Pro-inflammatory factors such as MMP-9 are broadly over-expressed and secreted by macrophages and lymphocytes whereas their inhibitors (for example TIMP-1) are typically over-expressed by alveolar epithelial cells in response to acute and chronic inflammation.30,58,59 Thus, the REFS™ data-driven approach has confirmed that the pro-inflammatory molecular regulation pathways are distinct from the protease inhibition or anti-inflammatory regulation pathways. That the Mmp9 and Timp-1 genes themselves were not identified as key drivers for MMP activity may be explained as follows: the key drivers are not direct interactions, and/or the expression levels of MMP genes do not necessarily correlate with the amount of protein and even less with enzyme activity, especially because the MMPs are synthesized as inactive pro-enzymes or zymogens.
The literature search revealed that some of the key drivers identified by BioModel™ were not yet reported to be associated with COPD/emphysema and could therefore represent new findings that might be valuable for experimental confirmation. The reports that link the identified key drivers to COPD/emphysema (see above) provide confidence in the relevance of our REFS™ simulation approach. Furthermore, the agreement between the knockdown predictions and some of the published experiments suggests that BioModel™ has the ability to identify key molecular drivers from large-scale systems biology experiments relevant for disease investigations.
BioModel™ simulates intermediate time points when no experiments were performed
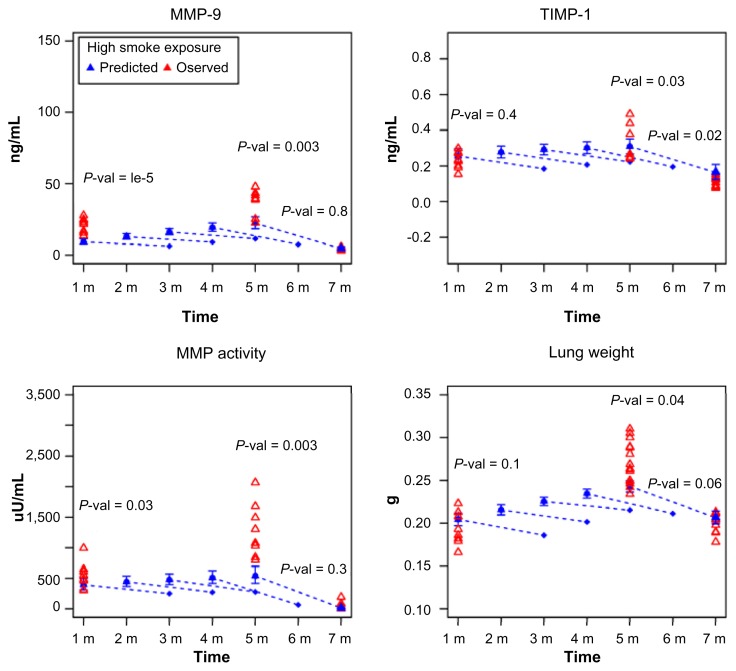
We examined whether BioModel™ could model intermediate time points when no experiments were performed. Although traditional statistical regression is a useful technique for creating predictive models of response variables and enumerating the response variable dependence on experimental factors, it is less useful for the reconstruction of molecular interaction networks and the inference of mathematical hierarchy between causal and end-point variables. Compared with statistical regression, Bayesian networks are causal, hierarchical models that can reflect biological mechanisms as well as equations from a set of independent variables. Figure 4 shows the BioModel™-predicted (all time points after 1 month) and experimentally observed (1, 5, and 7 months) values under high dose CS exposure for the four endpoints. The model links experimental factors to the endpoints by fitting mathematical models for gene expression variables and endpoint changes through modeled changes in gene expression.
Figure 4.
BioModel™ models intermediate time points when no experiment was performed. Observed (red triangle) and the mean of BioModel™-predicted values (blue triangle and diamond) for A. MMP-9 abundance, B. TIMP-1 abundance, C. MMP activity, and D. lung weight. The means of the predicted values of the four endpoints are denoted by blue filled triangles together with 95% confidence intervals for the different CS exposure regimens from 1–7 months. The dashed extension lines with the blue diamond at the end are model predictions of endpoint values with the addition of the 2-month smoking cessation period.
The expression values of more than 10,000 genes were predicted, from which the four endpoint values were simulated. For every endpoint at one specific time point, 101 predicted values were simulated. The null hypotheses that the predicted value is equal to the observed value at experimental time points were tested for all endpoints. Welch’s t-test was performed and P-values are shown in Figure 4. A stringent cutoff for the P-value, 0.01, was set as the sample size is large. Under this cutoff, reasonable consistency between the predictions and observations for all endpoints across all experimental time points was obtained; three exceptions were the predictions for MMP-9 at 1 and 5 months, and the prediction for MMP activity at 5 months. The deviation in the predicted value of MMP activity at 5 months could be caused by the large variations in experimental values that were obtained for this group. The dashed extension line beginning at the 5-month time point indicates predicted endpoint values for the effect of 2 months of CS cessation after 5 months of CS exposure. BioModel™ reasonably captures the cessation effect. In addition to the measured time points, endpoints and gene expression changes can be predicted at time points outside of the original experimental design by simulating the BioModel™ model. For example, Figure 4 shows the prediction of a 2-month cessation effect for 1-, 2-, 3-, and 4-month time points at high doses of CS exposure (dashed lines). The interpolation of the results of CS cessation should be used with caution because there is only one time point for cessation in the observed data.
Interaction networks of key molecular drivers
In addition to the identification of key molecular regulators of the study endpoints, BioModel™ allows the interpretation of key molecular driver interaction networks (hereafter referred to as molecular interaction networks). Gene expression of every key driver, for example driver A, was knocked-down in silico by 10-fold. Differences in the posterior distributions of key drivers other than driver A, say driver B, across the 10 data frames were processed to assess the effects of driver A perturbation. P-values were computed by a t-test under the null hypothesis that there is no difference between the means of posterior distributions for driver B before and after perturbation of driver A. An interaction between drivers A and B was defined when the difference caused by perturbation of driver A was significant (P-value cutoff, 0.05). Though each network in the ensemble is a directed acyclic graph (DAG), this is not necessarily true of the interaction network, and bi-directional arrows could be observed.
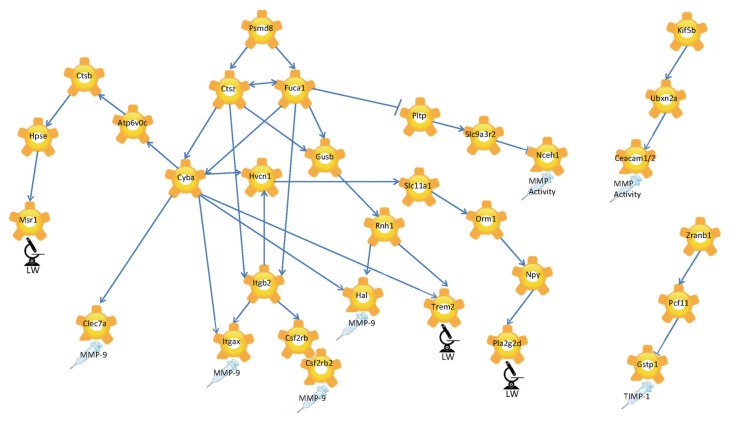
Figure 5 displays the molecular interaction network as a visualization of the interactions between key molecular drivers. The end of each network branch is marked with an endpoint icon that indicates the specific study observation predicted by BioModel™ to be regulated by the cascade of molecular mechanisms displayed within the sub-network. The model shows 26S proteasome non-ATPase regulatory subunit 8 (Psmd8) as the master regulator of the entire network because all the arrows associated with Psmd8 point outwards. Furthermore, Psmd8 is predicted to regulate fucosidase-1 (Fuca1) and Ctsz, which were predicted to interact with each other.
Figure 5.
Simulation-based causal network of molecular interactions for key molecular drivers of experimental endpoints. Microscope with LW label symbolizes the lung weights as experimentally determined, the syringe symbolizes endpoints determined from BALF. The direction of the arrows reflects causality.
Discussion and Conclusions
As emphysematous changes occur in the parenchyma tissue of the lung, we believe that this tissue is the most appropriate to analyze gene expression and end point evaluation. Progression of CS-induced emphysema was previously shown to associate with the expression of genes involved in multiple pathways in the lungs that were predicted to belong to the functional categories of phase I genes, Nrf2-regulated antioxidant and phase II genes, phase III detoxification genes, and other immune/inflammatory response genes. This suggested that the gene expression data corresponded to significant bronchoalveolar inflammation as well as enhanced oxidative stress and increased apoptosis as determined by immunohistochemical staining.60 Mathematical modeling techniques were not employed in the correlation-based analysis that was used in this earlier study. In other studies that explored CS-induced gene expression changes in A/J mice,61,62 gene expression data and other biological endpoints were also treated as unrelated variables. By contrast, in the present study, we included Bayesian network inference methods that can identify cause-and-effect relationships and predict mechanistic interaction networks, providing a strikingly different view of biological systems. Inferred molecular networks are typically interrogated to generate specific hypotheses and explore disease biology in a rigorous and systematic way.35,37,41
For the REFS™ approach, the use of multiple biological endpoints from the same animal would be optimal; however, this is not always possible. In the current study for example, to avoid physical irritation that might interfere with or influence gene expression changes, the lungs destined for gene expression analysis were not the ones subjected to bronchoalveolar lavage (BAL). As a consequence, the samples used to generate gene expression data were not from the same animals as those used to measure MMP-9 and TIMP-1 expression and MMP activity. Although this could be a source of mismatch in the Bayesian network building, because the mice can be considered to be genetically identical, we used a bootstrap technique to match the samples used for gene expression analysis and those used for other measured endpoints.
Molecular interaction networks
We developed an ensemble of networks for the A/J mouse model with different CS dose and exposure times. This model successfully recovered the measured experimental data on gene expression and four measured COPD-relevant endpoints. Based on the in silico 10-fold knockdown of gene expression in this ensemble of networks, 33 key molecular drivers for the four selected COPD-relevant endpoints were identified; the majority of these were associated with inflammation or COPD directly. These observations suggest that genes identified using this ensemble of networks represent promising key molecular drivers underlying the development of COPD and, as such, warrant further investigation.
BioModel™ predicted Psmd8, a 26S proteasome non-ATPase regulatory subunit 8, to be the master regulator of the molecular interaction networks. Interestingly, PSMD8 was reported to be more highly expressed in sedentary COPD patients than in sedentary controls,63 underlining a role for this molecule in COPD. Furthermore, Psmd8 was predicted to regulate Fuca1 and Ctsz in the network. BioModel™ also uncovered biologically valid cause-and-effect relationships but no direct interactions. The relationship between PSMD8 and CTSZ can be explained, for example, via ubiquitin. Both the proteasomal subunit and CTSZ directly bind ubiquitin.63 Pmsd8 binds to the ubiquitin protein ligase TRAF6, which interacts further with the ubiquitin specific peptidase USP21, which itself was shown to bind directly with FUCA1.64 FUCA1 is a lysosomal enzyme involved in the degradation of fucose-containing glycoproteins and glycolipids. In our network model, Fuca1 and Ctsz were predicted to interact and we propose that this interaction is likely to be via ubiquitin. In addition, the genes Cyba and Gusb, further downstream in the network, can be linked through ubiquitin.65,66
Various studies have shown that ubiquitin expression is up-regulated in the peripheral muscle of patients with COPD.67,68 Our data combined with literature searches hint that ubiquitin plays an as yet unexplored role in the development of COPD. Because our study focused on parenchymal lung tissue, it remains to be determined whether the up-regulation of ubiquitin in COPD is a general factor for disease development.
In summary, the successful application of REFS™ to this data set from the A/J mouse COPD inhalation study demonstrates that REFS™ is a powerful method for the integrative analysis of diverse data types. We believe that this study provides new insights into the mechanisms of smoking-induced emphysema. In addition, we show that BioModel™ can be leveraged to reduce the number of animals required for in vivo investigations since the effects of intermediate exposure time points can be simulated.
Materials and Methods
Ethics statement
All experimental procedures were in conformity with the American Association for Laboratory Animal Science Policy on the Humane Care and Use of Laboratory Animals (American Association for Laboratory Animal Science, 1996) in an AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care International)-accredited facility and were approved by the Institutional Animal Care and Use Committee (IACUC, Leuven, Belgium).
CS generation and animal exposure
Mice were exposed to CS from the reference cigarette 3R4F.69 Mainstream smoke was diluted with conditioned fresh air to reach a target concentration of 750 μg/l total particulate matter (TPM).
Female A/J mice (The Jackson Laboratory, Bar Harbor, ME, USA) were housed under controlled conditions in standard laboratory cages. All in vivo experimental protocols were approved by the local Ethics Committee and complied with strict governmental and international guidelines on animal experimentation. Mice (2–3 months old) were whole-body exposed for 2 or 4 h per day for 5 days/week for a period of 1 day to 5 months. A 30-min fresh-air period was included between the first and second hour of daily exposure. Between the second and third, and third and fourth hours of exposure, there were 60-min periods of fresh-air exposure. The exposure period began with an adaptation period: TPM concentrations were 125 μg/l on study days 1 and 2, 250 μg/l on study days 3 and 4, 375 μg/l on study days 7 and 8, 500 μg/l on study days 9 and 10, and 625 μg/l on study days 11 and 14. From study day 15, the target concentration was 750 μg/l. All animals received the same adaptation schedule except for those that were exposed for seven days. The adaptation phase for the 7-day time point was: TPM concentrations of 125 μg/l on days 1 and 2, 250 μg/l on day 3, 375 μg/l on day 4, 500 μg/l on day 5, 625 μg/l on day 6, and 750 μg/l on day 7. The inhalation protocol for the 7-day time point was different because the animals had to be given time to adapt progressively for animal welfare reasons as they were to be exposed for up to 5 months. Control animals were exposed to filtered, conditioned fresh air with the same exposure conditions as those for the 4-h CS-exposed group of mice. The smoke and air-exposed animals in the different groups were sacrificed after 1 day, 7 days, 1 month and 5 months. In addition, smoking cessation was modeled by 5 months of CS exposure followed by 2 months of no smoke exposure. This group was sacrificed after 7 months.
The mice in both the low- and high-dose groups were dissected 1–2 h after their last exposure. The lungs were retrieved and shock frozen immediately after dissection and stored at −80°C.
BALF collection protocol and endpoints analysis
BALF was collected from 10 mice per group for the determination of MMP-9 and TIMP-1 expression, and MMP activity. In brief, mice were exsanguinated under deep pentobarbital anesthesia. The trachea was cannulated and the lungs lavaged with 1 ml of Ca2+- and Mg2+-free Dulbecco’s phosphate-buffered saline warmed to 37°C. After centrifuging the lavage fluid (4°C, 5 min, 300 × g), the supernatant was aliquoted and stored at −80°C. MMP-9 and TIMP-1 levels were determined by multi-analyte profiling using high-density, quantitative immunoassays (Rules-Based Medicine Inc., Austin, TX, USA). MMP activity was determined as gelatinolytic activity using a fluorescence-labeled gelatin (EnzChek®Gelatinase/Collagenase Assay Kit; Invitrogen, Karlsruhe, Germany) according to the manufacturer’s instructions.
Microarray data generation
For microarray analysis, the parenchymal tissue from frozen lungs was prepared by laser capture micro-dissection and RNA was isolated using an RNeasy Micro Kit (Qiagen, Hilden, Germany). RNA was further processed using a GeneChip® 3′ IVT Express Kit. (Affymetrix, High Wycombe, UK). The biotin-labeled, fragmented RNA was hybridized against Affymetrix Mouse Genome 430 2.0 Arrays. The acquired Affymetrix CEL files that passed the quality check were used for further data processing and were submitted to ArrayExpress (E-MTAB-1426).
Microarray data processing and feature selection
Data processing
CEL files were processed using an in-house pipeline. GC Robust Multi-array Average (GCRMA) background correction, quantile normalization, and median polish summarization were performed to generate microarray expression values using affy, GCRMA, and affyPLM R packages.70–72
Low signal and unannotated probe sets
Pre-filtering improves the reliability of Affymetrix GeneChip results when used to analyze gene expression in complex tissues. In this study, probe sets were filtered out when the 95% quantile of the log2 expression value was <7, or when they did not belong to any gene.73–75
Redundant probe sets
Affymetrix GeneChip microarray designs often include multiple probe sets per gene or transcript unit. Probe sets that measure the same biomolecule tend to exhibit highly correlated behaviors across gene expression measurements. One probe set was selected from each group of correlated probe sets within a gene, thereby eliminating redundant variables. Two probe sets were considered to be correlated if the Pearson correlation coefficient was ≥0.6. The probe set that was differentially expressed in the largest number of comparisons between the experimental groups was selected. A probe set was defined as differentially expressed between two experimental conditions if the Benjamini-Hochberg corrected moderated t-test P-value was ≤0.01.76 When two probe sets within a gene were differentially expressed in an equal number of comparisons, the probe set that was closest to the 3′ end of the gene was selected. The genomic coordinates for each probe set were extracted from the NetAffx annotation file for Mouse 430 2 Array Version 31. If multiple alignments were associated with a given probe set, the alignment that matched the chromosome and strand for the gene was assigned to the probe set. The chromosome strand for the gene was extracted from the Ensembl mouse genome. Probe sets that corresponded to genes with no Ensembl ID were ignored. For each probe set, the start and stop positions from the selected alignment were stored. Based on the selection process described above, 1,988 redundant probe sets were removed.
Non-informative probe sets
Linear models were used to assess the covariance between endpoints and probe set expression measurements. To construct the linear models, each expression data point was matched with the group median value of the endpoint variable. Equation (1) describes the linear models (in R notation) and experimental conditions that were used for the individual endpoints.
| (1) |
Here, E is a continuous variable represented by the group median endpoint values; T is time modeled as a discrete categorical variable; and GE is the probe set gene expression values modeled as a continuous variable. The resulting slopes of the regression lines were tested for a significant difference from zero. A probe set was considered to significantly covary with the endpoint if the Benjamini-Hochberg corrected P-value for any time point was ≤0.01. Gene expression values per time point were normalized relative to sham by subtracting the median sham expression value within the same time point before fitting the linear models. In total, 10,643 probe sets were selected at this stage.
Calculation of total smoke exposure and transformation of endpoint variables
The animals in the study were exposed to a specific amount of particulate matter per hour each day. The total smoke exposure was calculated at a given time point based on the exposure schedule for each experimental group. The low-exposure groups were exposed to smoke for 2 h daily and the high-exposure groups were exposed to smoke for 4 h daily. Before REFS™ modeling, total smoke exposure values were logarithmically transformed. Table S1 displays the calculated total smoke exposure values and their logarithmic transformation.
The distributions of the endpoint variables were assessed for normality. MMP-9 expression and MMP activity data followed a log-normal distribution. The values of the MMP-9 and MMP activity variables were logarithmically transformed before REFS™ analysis and feature selection. Zero values were replaced by the logarithm of the minimum value for each variable.
Missing lung weight values for the low-dose CS group at 5 months were imputed based on the assumption of normality. A quadratic model was fitted for the lung weight values across the study timeline. This was used to predict the average lung weight for low smoke exposure at 5 months; the logarithm of smoke exposure was calculated as 5.16, and the corresponding lung weight average was predicted to be 0.23 g. Lung weight values were imputed by sampling from the normal distribution, N(0.23,σ). The standard deviation was estimated from lung weight measurements in the high-dose CS group at 5 months (Fig. S2).
Linking endpoint data with gene expression
Because of biological assay implementation challenges, endpoint measurements and gene expression profiles were obtained between mice. To match the samples for which phenotypic endpoints were measured to those for which gene expressions were profiled, bootstrap was leveraged. Covariation and error terms of linear models linking endpoint variables and gene expression were estimated conservatively as mixtures of estimates from 10 different data frames. Each data frame was constructed by bootstrap sampling of the endpoint values within an experimental group. Samples were then matched with the microarray data. Endpoints were sampled without replacement for every experimental group except MMP activity in the low-dose CS group at 5 months. MMP activity in this group was sampled with replacement because there were fewer endpoint measurements than microarray data points. Values for a given endpoint were sampled independently of other endpoints, except when multiple endpoints were measured in the same animal where values for all measured endpoints were matched to a gene expression profile within the same experimental group.
Constructing BioModel™: REFS™ methodology
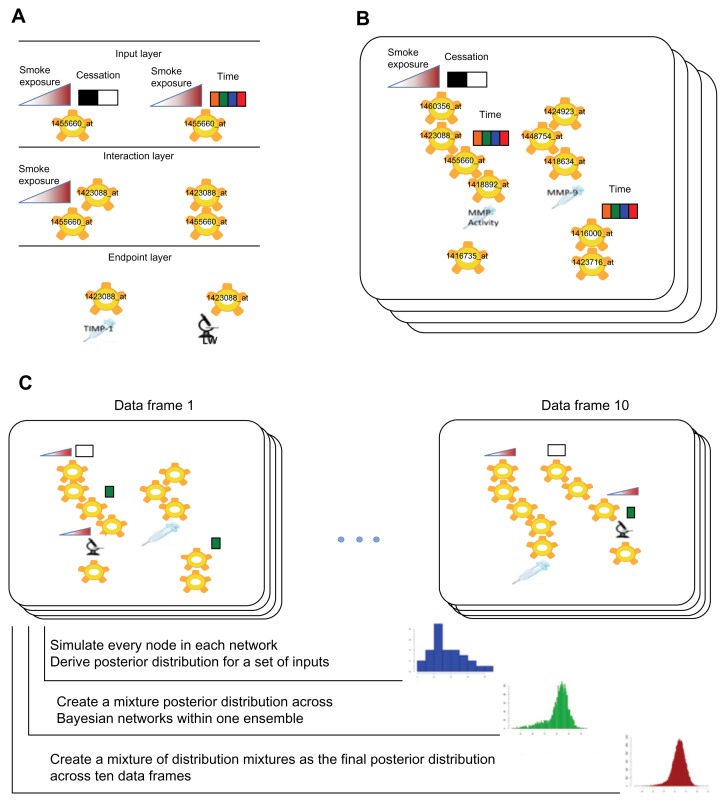
A multivariate system of random variables where each variable is either discrete (time, smoke exposure cessation) or continuous (smoke exposure, gene expression, endpoints) may be characterized probabilistically using a joint probability distribution function. The explicit formulation of a joint probability distribution requires the estimation of a large number of parameters. However, a joint probability distribution may often be factorized into a product of local conditional probability distributions (Fig. 6A). This approach yields a framework where each particular factorization and choice of parameters is a distinct probabilistic model of the process that created the observed experimental data.41 Learning models from a data set determine which factorizations of the data joint distribution are most likely given the observation, and what are the likely values for the parameters.
Figure 6.
Schematic representation of REFS™ data analysis steps and model simulation workflow. A. The network fragment enumeration step. Local linear regression models are evaluated against observed data. B. Ensemble of Bayesian networks sampled by the Metropolis Monte-Carlo sampling algorithm from the space of all possible networks. C. The simulations workflow. Specific values are set for the study input variables: CS total exposure, CS exposure time, and CS cessation. Posterior distributions from individual networks are combined to create the posterior mixture distribution across 100 networks and 10 data frames. LW: lung weight. The colored time boxes indicate multiple time points.
Each factorization of the joint probability function is represented by a unique DAG with a vertex for each random variable and directed edges between vertices to represent the dependencies between variables functionalized under local conditional distributions. In addition to the graph, the model also specifies distributions for the parameters of local conditional distributions. The likelihood function gives the posterior distribution of the parameter values around the maximum likelihood estimate.
To determine which factorizations are likely given the data, a Bayesian framework is used to compute the posterior probability of the model P(M|D) from Bayes’ Theorem (equation 2).
| (2) |
Here, P(D) is the probability of the observed data and P(M) is the prior probability of the model. Each local conditional model is scored using the Schwartz’s Bayesian Information Criterion approximated to the above posterior probability. The total DAG score is defined as the sum of the local model scores. Linear regression, sigmoidal regression, and ANCOVA models between probe sets, endpoints, smoke exposure and cessation, and time factors were exhaustively scored.
Even with such local model restrictions, the space of all possible graphs is too large to be explored by an exhaustive search. Instead, the Metropolis method was used to generate samples from an equilibrium Boltzmann distribution of candidate structures.77 Each step in a Metropolis Markov Chain corresponds to local transformations such as adding or deleting network fragments. To accelerate convergence, simulated annealing was applied with a decreasing annealing temperature. In the REFS™ implementation, the temperature was stopped at T = 1 because sampling at this temperature corresponds directly to sampling from the posterior distribution, P(M|D). As illustrated in Figure 6B, 100 network models were sampled for each of the 10 data frames generated by the bootstrap-driven match of gene expression data and endpoint measurements. The final BioModel™ of probabilistic gene interactions and their effects on study endpoints in A/J mice was a mixture of 10 REFS™ ensembles.
Model simulations
Stochastic simulations of a probabilistic model allow predictions about individual variables to be made under different conditions. These conditions can be perturbations of other variables in the model and/or different values of the input factors. Monte Carlo simulations were used to generate posterior probabilities for gene expression and endpoint variables using sampling parameters and error terms from their respective posterior distributions. A typical simulation routine sweeps the network iteratively and generates samples of variables with ‘parents’ that have already acquired a value in previous iterations, until all variables have values. One full sweep produces one sample per network in an ensemble (Fig. 6C, blue histogram); the sweep is then repeated multiple times per network for 100 networks within one ensemble (Fig. 6C, green histogram); the final posterior distribution is therefore a mixture of 1,000 samples (100 networks across 10 ensembles) as depicted in Figure 6C, red histogram. Interventions such as knockdown of gene transcript expression can be simulated in silico by reducing the expression level of that gene by a pre-specified value. For example, a 10-fold knockdown was performed by subtracting log210 from the baseline gene expression level. A network sweep is then performed to estimate the knockdown effect by sampling the posterior distribution of all other variables in the model.
Modeling experimental conditions
Model performance was assessed by simulating the experimental conditions and correlating model predictions with the observed data. In simulations, only the factor variables were set to their experimental values. These factor variables were CS exposure, time, and cessation status. Other variables in each network of the ensemble were propagated from factor variables until all variables had been assigned a calculated value. Pearson’s correlation coefficient between the observed and REFS™-simulated data was calculated for every endpoint and probe set.
Identification of molecular drivers of study endpoints
Baseline and gene expression knockdown simulations of BioModel™ were performed to identify key molecular drivers for the study endpoints. Simulations were performed for four different study conditions: 1 month, no CS exposure; 1 month, high CS exposure; 5 months, high CS exposure; 5 months high CS exposure plus 2 months of no CS exposure (cessation). For each experimental condition, baseline transcript expression and endpoint values were calculated by simulation with BioModel™. BioModel™ was used to find a subset of gene expression variables that are upstream and therefore causal of a particular endpoint in the model. A probe set was said to be causal of an endpoint if it was upstream of this endpoint in at least one DAG. Causal probe sets were found for every endpoint variable across 100 DAGs and 10 data frames in the model, resulting in 1,118 from a set union. The following algorithm was used to assess whether any of the causal relationships were likely to be observed in a similarly sized validation study: A causal probe set was knocked down 10-fold in the model and the posterior distribution of the corresponding endpoint was estimated by creating a mixture posterior distribution across 100 networks for each data frame (Fig. 6C, green histogram). The next objective was to estimate the chance of observing a significant difference in the endpoint measurement if samples were to be drawn from the baseline posterior distribution or the posterior distribution under the gene expression perturbation. A number of samples equal to the study size (101 animals) were drawn from the baseline endpoint posterior distribution and the perturbed endpoint posterior distribution. The t-test statistic for the difference of the means and its P-value was calculated from the drawn samples. This sampling procedure was repeated 100 times and the average P-value was recorded. Average P-values for all causal probe sets were corrected for multiple testing using the Benjamin-Hochberg correction. At the end of this workflow, 10 P-values were calculated for every causal probe set; these corresponded to the 10 data frames used in the model development. A causal probe set was defined as a significant molecular driver of the endpoint if the second smallest adjusted P-value was ≤0.05. This threshold detected a substantial perturbation effect and also guaranteed that a significant difference between endpoint measurements was achieved in at least two of the 10 ensembles created.
Simulation-based inference from interaction networks
Network inference from a REFS™ model can be done by examining the topology of Bayesian networks in the ensemble and visualizing network fragments or edges based on a fragment or edge frequency threshold.41 However, it is important to note that the presence of topological links does not always translate into a sizeable effect on a network dependent variable. In addition, model topology in itself accounts for all experimental conditions and therefore will not reveal insights specific to a particular experimental arm or explain differences between experimental groups. Thus, a more robust approach to network inference in REFS™ models, or any other network inference model, would be to use simulations of variable perturbations.
To generate a molecular interaction network of significant drivers for a particular experimental group, the input variables time, CS exposure, and CS-exposure cessation were set to the experimental group values. Each significant driver probe set identified by REFS™ was down-regulated by 10-fold in the model. Changes in downstream network variables were evaluated. If a downstream variable changed by ≥ two-fold, then the relationship between the perturbed and downstream variables was recorded as a parent—child relationship. The fold change was calculated between the means of perturbed and baseline posterior distributions mixed across all ensembles and networks. To aid in the visualization of the final network, direct links between gene expression variables were omitted when an indirect link existed.
Supplementary Data
The four endpoints measured after exposure to high and low CS doses at different time points. MMP activity was determined as gelatinolytic activity (μU/ml) in BALF. MMP-9 and TIMP-1 were measured by multiplexed immunoassays. *indicates a statistically significant difference between the mean of the group beneath it and the mean of the sham group at the same time point; # indicates a statistically significant difference between the mean of the group beneath it and the mean of the corresponding group at the previous time point. The Wilcoxon rank sum test calculated the significance level as 5%. Bars represent the mean ± SEM. ND, not determined.
Linear and quadratic models of lung weight (g) across logarithmically transformed smoke exposure (TPM × h/l) conditions. Filled circles indicate the predicted average lung weight at 5 months obtained using linear (red line) and quadratic (blue line) regression models.
Table S1.
Total CS exposure calculated for each experimental group on linear and logarithmic scales.
| EXPERIMENTAL GROUP | SMOKE EXPOSURE | SMOKE EXPOSURE (LOG10 SCALE) |
|---|---|---|
| 1 day low | 250 | 2.397940009 |
| 7 days low | 5500 | 3.740362689 |
| 1 month low | 24000 | 4.380211242 |
| 5 months low | 144000 | 5.158362492 |
| 1 day high | 500 | 2.698970004 |
| 7 days high | 11000 | 4.041392685 |
| 1 month high | 48000 | 4.681241237 |
| 5 months high | 288000 | 5.459392488 |
Acknowledgments
We are grateful to Sam Ansari for submitting the gene expression data to ArrayExpress (E-MTAB-1426).
Footnotes
COMPETING INTERESTS: Yang Xiang, Ulrike Kogel, Stephan Gebel, Michael J. Peck, Manuel C. Peitsch, and Julia Hoeng are employees of Philip Morris International. Viatcheslav R. Akmaev is an employee of Berg Biosystems and was an employee of GNS Healthcare.
Author Contributions
Conceived and designed the experiments: UK, SG, MJP, MCP, JH. Performed the experiments: UK, SG, MCP, JH. Analyzed the data: VRA, YX, MJP, JH. Contributed reagents/materials/analysis tools: YX, UK, SG, MCP, VRA, MJP, JH. Wrote the paper: YX, UK, SG, MJP, MCP, VRA, JH. All authors reviewed and approved of the final manuscript.
DISCLOSURES AND ETHICS
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
FUNDING: This work was funded by Philip Morris International.
REFERENCES
- 1.Barnes PJ. Chronic obstructive pulmonary disease. N Engl J Med. 2000 Jul 27;343(4):269–80. doi: 10.1056/NEJM200007273430407. [DOI] [PubMed] [Google Scholar]
- 2.Rabe KF, Hurd S, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007 Sep 15;176(6):532–55. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 3.Spurzem JR, Rennard SI. Pathogenesis of COPD. Semin Respir Crit Care Med. 2005 Apr;26(2):142–53. doi: 10.1055/s-2005-869535. [DOI] [PubMed] [Google Scholar]
- 4.Mannino DM, Watt G, Hole D, et al. The natural history of chronic obstructive pulmonary disease. Eur Respir J. 2006 Mar;27(3):627–43. doi: 10.1183/09031936.06.00024605. [DOI] [PubMed] [Google Scholar]
- 5.Buist AS, McBurnie MA, Vollmer WM, et al. International variation in the prevalence of COPD (the BOLD Study): a population-based prevalence study. Lancet. 2007 Sep 1;370(9589):741–50. doi: 10.1016/S0140-6736(07)61377-4. [DOI] [PubMed] [Google Scholar]
- 6.Kim KH, Pandey SK, Kabir E, Susaya J, Brown RJ. The modern paradox of unregulated cooking activities and indoor air quality. J Hazard Mater. Nov 15;195:1–10. doi: 10.1016/j.jhazmat.2011.08.037. [DOI] [PubMed] [Google Scholar]
- 7.Rennard SI, Vestbo J. COPD: the dangerous underestimate of 15% Lancet. 2006 Apr 15;367(9518):1216–9. doi: 10.1016/S0140-6736(06)68516-4. [DOI] [PubMed] [Google Scholar]
- 8.Wright JL, Churg A. Animal models of cigarette smoke-induced chronic obstructive pulmonary disease. Expert Rev Respir Med. 2010 Dec;4(6):723–34. doi: 10.1586/ers.10.68. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro SD. The pathogenesis of emphysema: the elastase:antielastase hypothesis 30 years later. Proc Assoc Am Physicians. 1995 Oct;107(3):346–52. [PubMed] [Google Scholar]
- 10.Ohnishi K, Takagi M, Kurokawa Y, Satomi S, Konttinen YT. Matrix metalloproteinase-mediated extracellular matrix protein degradation in human pulmonary emphysema. Lab Invest. 1998 Sep;78(9):1077–87. [PubMed] [Google Scholar]
- 11.Finlay GA, O’Driscoll LR, Russell KJ, et al. Matrix metalloproteinase expression and production by alveolar macrophages in emphysema. Am J Respir Crit Care Med. 1997 Jul;156(1):240–7. doi: 10.1164/ajrccm.156.1.9612018. [DOI] [PubMed] [Google Scholar]
- 12.Imai K, Dalal SS, Chen ES, et al. Human collagenase (matrix metalloproteinase-1) expression in the lungs of patients with emphysema. Am J Respir Crit Care Med. 2001 Mar;163(3 Pt 1):786–91. doi: 10.1164/ajrccm.163.3.2001073. [DOI] [PubMed] [Google Scholar]
- 13.Segura-Valdez L, Pardo A, Gaxiola M, Uhal BD, Becerril C, Selman M. Upregulation of gelatinases A and B, collagenases 1 and 2, and increased parenchymal cell death in COPD. Chest. 2000 Mar;117(3):684–94. doi: 10.1378/chest.117.3.684. [DOI] [PubMed] [Google Scholar]
- 14.Demedts IK, Morel-Montero A, Lebecque S, et al. Elevated MMP-12 protein levels in induced sputum from patients with COPD. Thorax. 2006 Mar;61(3):196–201. doi: 10.1136/thx.2005.042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molet S, Belleguic C, Lena H, et al. Increase in macrophage elastase (MMP-12) in lungs from patients with chronic obstructive pulmonary disease. Inflamm Res. 2005 Jan;54(1):31–6. doi: 10.1007/s00011-004-1319-4. [DOI] [PubMed] [Google Scholar]
- 16.Grumelli S, Corry DB, Song LZ, et al. An immune basis for lung parenchymal destruction in chronic obstructive pulmonary disease and emphysema. PLoS Med. 2004 Oct;1(1):e8. doi: 10.1371/journal.pmed.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuder RM, Yoshida T, Arap W, Pasqualini R, Petrache I. State of the art. Cellular and molecular mechanisms of alveolar destruction in emphysema: an evolutionary perspective. Proc Am Thorac Soc. 2006 Aug;3(6):503–10. doi: 10.1513/pats.200603-054MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churg A, Wright JL. Proteases and Emphysema. Curr Opin Pulm Med. 2005;11(2):153–9. doi: 10.1097/01.mcp.0000149592.51761.e3. [DOI] [PubMed] [Google Scholar]
- 19.MacNee W. Pulmonary and systemic oxidant/antioxidant imbalance in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2005;2(1):50–60. doi: 10.1513/pats.200411-056SF. [DOI] [PubMed] [Google Scholar]
- 20.Laurell CB, Eriksson S. The electrophoretic alpha1-globulin pattern of serum in alpha1-antitrypsin deficiency. COPD. 2013 Mar;10(Suppl 1):3–8. doi: 10.3109/15412555.2013.771956. [DOI] [PubMed] [Google Scholar]
- 21.Barnes PJ. Mediators of chronic obstructive pulmonary disease. Pharmacol Rev. 2004 Dec;56(4):515–48. doi: 10.1124/pr.56.4.2. [DOI] [PubMed] [Google Scholar]
- 22.Cawston T, Carrere S, Catterall J, et al. Matrix metalloproteinases and TIMPs: properties and implications for the treatment of chronic obstructive pulmonary disease. Novartis Found Symp. 2001;234:205–18. doi: 10.1002/0470868678.ch13. discussion 218–28. [DOI] [PubMed] [Google Scholar]
- 23.Demedts IK, Brusselle GG, Bracke KR, Vermaelen KY, Pauwels RA. Matrix metalloproteinases in asthma and COPD. Curr Opin Pharmacol. 2005 Jun;5(3):257–63. doi: 10.1016/j.coph.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Betsuyaku T, Nishimura M, Takeyabu K, et al. Neutrophil granule proteins in bronchoalveolar lavage fluid from subjects with subclinical emphysema. Am J Respir Crit Care Med. 1999 Jun;159(6):1985–91. doi: 10.1164/ajrccm.159.6.9809043. [DOI] [PubMed] [Google Scholar]
- 25.Lanone S, Zheng T, Zhu Z, et al. Overlapping and enzyme-specific contributions of matrix metalloproteinases-9 and -12 in IL-13-induced inflammation and remodeling. J Clin Invest. 2002 Aug;110(4):463–74. doi: 10.1172/JCI14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Churg A, Wang R, Wang X, Onnervik PO, Thim K, Wright JL. Effect of an MMP-9/MMP-12 inhibitor on smoke-induced emphysema and airway remodelling in guinea pigs. Thorax. 2007 Aug;62(8):706–13. doi: 10.1136/thx.2006.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.March TH, Wilder JA, Esparza DC, et al. Modulators of cigarette smoke-induced pulmonary emphysema in A/J mice. Toxicol Sci. 2006 Aug;92(2):545–59. doi: 10.1093/toxsci/kfl016. [DOI] [PubMed] [Google Scholar]
- 28.Seagrave J, Barr EB, March TH, Nikula KJ. Effects of cigarette smoke exposure and cessation on inflammatory cells and matrix metalloproteinase activity in mice. Exp Lung Res. 2004 Jan-Feb;30(1):1–15. doi: 10.1080/01902140490252858. [DOI] [PubMed] [Google Scholar]
- 29.Vlahos R, Bozinovski S, Jones JE, et al. Differential protease, innate immunity, and NF-kappaB induction profiles during lung inflammation induced by subchronic cigarette smoke exposure in mice. Am J Physiol Lung Cell Mol Physiol. 2006 May;290(5):L931–45. doi: 10.1152/ajplung.00201.2005. [DOI] [PubMed] [Google Scholar]
- 30.Beeh KM, Beier J, Kornmann O, Buhl R. Sputum matrix metalloproteinase-9, tissue inhibitor of metalloprotinease-1, and their molar ratio in patients with chronic obstructive pulmonary disease, idiopathic pulmonary fibrosis and healthy subjects. Respir Med. 2003 Jun;97(6):634–9. doi: 10.1053/rmed.2003.1493. [DOI] [PubMed] [Google Scholar]
- 31.Kang MJ, Oh YM, Lee JC, et al. Lung matrix metalloproteinase-9 correlates with cigarette smoking and obstruction of airflow. J Korean Med Sci. 2003 Dec;18(6):821–7. doi: 10.3346/jkms.2003.18.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margolin A, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang Y, Talikka M, Belcastro V, et al. Divergence Weighted Independence Graphs for the Exploratory Analysis of Biological Expression Data. Journal of Health and Medical Informatics. 2011:S2–001. [Google Scholar]
- 34.Marbach D, Prill RJ, Schaffter T, Mattiussi C, Floreano D, Stolovitzky G. Revealing strengths and weaknesses of methods for gene network inference. Proceedings of the National Academy of Sciences. 2010;107(14):6286. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pe’er D, Regev A, Elidan G, Friedman N. Inferring subnetworks from perturbed expression profiles. Bioinformatics. 2001;17(Suppl 1):S215–24. doi: 10.1093/bioinformatics/17.suppl_1.s215. [DOI] [PubMed] [Google Scholar]
- 36.Friedman N, Koller D. Being Bayesian about network structure. A Bayesian approach to structure discovery in Bayesian networks. Machine learning. 2003;50(1):95–125. [Google Scholar]
- 37.Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005 Apr 22;308(5721):523–9. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 38.Chaibub Neto E, Ferrara CT, Attie AD, Yandell BS. Inferring causal phenotype networks from segregating populations. Genetics. 2008 Jun;179(2):1089–100. doi: 10.1534/genetics.107.085167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Zhu J, Lum PY, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008 Mar 27;452(7186):429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khalil I, Brewer MA, Neyarapally T, Runowicz CD. The potential of biologic network models in understanding the etiopathogenesis of ovarian cancer. Gynecol Oncol. 2010 Feb;116(2):282–5. doi: 10.1016/j.ygyno.2009.10.085. [DOI] [PubMed] [Google Scholar]
- 41.Xing H, McDonagh PD, Bienkowska J, et al. Causal modeling using network ensemble simulations of genetic and gene expression data predicts genes involved in rheumatoid arthritis. PLoS Comput Biol. 2011 Mar;7(3):e1001105. doi: 10.1371/journal.pcbi.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tetley TD. Inflammatory cells and chronic obstructive pulmonary disease. Curr Drug Targets Inflamm Allergy. 2005 Dec;4(6):607–18. doi: 10.2174/156801005774912824. [DOI] [PubMed] [Google Scholar]
- 43.Da Wei Huang BTS, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Haspel JA, Choi AM. Autophagy: a core cellular process with emerging links to pulmonary disease. Am J Respir Crit Care Med. 2011 Dec 1;184(11):1237–46. doi: 10.1164/rccm.201106-0966CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryter SW, Choi AM. Autophagy in the lung. Proc Am Thorac Soc. 2010 Feb;7(1):13–21. doi: 10.1513/pats.200909-101JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryter SW, Lee SJ, Choi AM. Autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. Expert Rev Respir Med. 2010 Oct;4(5):573–84. doi: 10.1586/ers.10.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gasque P. Complement: a unique innate immune sensor for danger signals. Mol Immunol. 2004 Nov;41(11):1089–98. doi: 10.1016/j.molimm.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 48.Schleimer RP. Innate immune responses and chronic obstructive pulmonary disease: “Terminator” or “Terminator 2”? Proc Am Thorac Soc. 2005;2(4):342–346. doi: 10.1513/pats.200504-030SR. discussion 371–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marc MM, Korosec P, Kosnik M, et al. Complement factors c3a, c4a, and c5a in chronic obstructive pulmonary disease and asthma. Am J Respir Cell Mol Biol. 2004 Aug;31(2):216–9. doi: 10.1165/rcmb.2003-0394OC. [DOI] [PubMed] [Google Scholar]
- 50.Kosmas EN, Zorpidou D, Vassilareas V, Roussou T, Michaelides S. Decreased C4 complement component serum levels correlate with the degree of emphysema in patients with chronic bronchitis. Chest. 1997 Aug;112(2):341–7. doi: 10.1378/chest.112.2.341. [DOI] [PubMed] [Google Scholar]
- 51.Vaisar T, Kassim SY, Gomez IG, et al. MMP-9 sheds the beta2 integrin subunit (CD18) from macrophages. Mol Cell Proteomics. 2009 May;8(5):1044–60. doi: 10.1074/mcp.M800449-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Chen L, Li Y, Guan XY. Overexpression of cathepsin Z contributes to tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. PLoS One. 2011;6(9):e24967. doi: 10.1371/journal.pone.0024967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies G, Sinnott M, Withers S. Comprehensive Biological Catalysis. Academic Press; London: 1997. [Google Scholar]
- 54.Koivunen A, Maisi P, Konttinen Y, Sandholm M. Gelatinolytic activity in tracheal aspirates of horses with chronic obstructive pulmonary disease. Acta veterinaria Scandinavica. 1997;38(1):17. doi: 10.1186/BF03548504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terpstra G, De Weger R, Wassink G, Kreuknit J, Huidekoper H. Changes in alveolar macrophage enzyme content and activity in smokers and patients with chronic obstructive lung disease. International journal of clinical pharmacology research. 1987;7(4):273. [PubMed] [Google Scholar]
- 56.Puljic R, Benediktus E, Plater-Zyberk C, et al. Lipopolysaccharide-induced lung inflammation is inhibited by neutralization of GM-CSF. European Journal of Pharmacology. 2007;557(2–3):230–5. doi: 10.1016/j.ejphar.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 57.Meng F, Francis H, Glaser S, et al. Role of stem cell factor and granulocyte-colony stimulating factor in remodeling during liver regeneration. Hepatology. 2011 doi: 10.1002/hep.24673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Cui D, Tong X, et al. The role of matrix metalloproteinases in extracellular matrix remodelling in chronic obstructive pulmonary disease rat models. Zhonghua Nei Ke Za Zhi. 2002 Jun;41(6):393–8. [PubMed] [Google Scholar]
- 59.Tetley TD. Antiprotease Therapy. In: al CMe, editor. Therapeutic Strategies in COPD. Oxford: Clinical Publishing; 2005. pp. 233–45. [Google Scholar]
- 60.Rangasamy T, Misra V, Zhen L, Tankersley CG, Tuder RM, Biswal S. Cigarette smoke-induced emphysema in A/J mice is associated with pulmonary oxidative stress, apoptosis of lung cells, and global alterations in gene expression. Am J Physiol Lung Cell Mol Physiol. 2009 Jun;296(6):L888–900. doi: 10.1152/ajplung.90369.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meireles SI, Esteves GH, Hirata R, Jr, et al. Early changes in gene expression induced by tobacco smoke: Evidence for the importance of estrogen within lung tissue. Cancer Prev Res (Phila) 2010 Jun;3(6):707–17. doi: 10.1158/1940-6207.CAPR-09-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Izzotti A, Cartiglia C, Longobardi M, et al. Gene expression in the lung of p53 mutant mice exposed to cigarette smoke. Cancer Res. 2004 Dec 1;64(23):8566–72. doi: 10.1158/0008-5472.CAN-04-1420. [DOI] [PubMed] [Google Scholar]
- 63.Radom-Aizik S, Kaminski N, Hayek S, Halkin H, Cooper DM, Ben-Dov I. Effects of exercise training on quadriceps muscle gene expression in chronic obstructive pulmonary disease. J Appl Physiol. 2007 May;102(5):1976–1984. doi: 10.1152/japplphysiol.00577.2006. [DOI] [PubMed] [Google Scholar]
- 64.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009 Jul 23;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagner SA, Beli P, Weinert BT, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011 Oct;10(10):M111 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim W, Bennett EJ, Huttlin EL, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011 Oct 21;44(2):325–40. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan YM, Jiang YH, Liu XJ. Ubquitin expression is up-regulated in periperal muscle in patients with chronic obstructive pulmonary disease. CHEST Journal. 2005;128(4_MeetingAbstracts):247S-a. [Google Scholar]
- 68.Debigare R, Cote CH, Maltais F. Ubiquitination and proteolysis in limb and respiratory muscles of patients with chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2010 Feb;7(1):84–90. doi: 10.1513/pats.200906-051JS. [DOI] [PubMed] [Google Scholar]
- 69.Davies H, Vaught A. The reference cigarette. Research cigarettes. 2003 [Google Scholar]
- 70.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 71.Wu C, Irizarry R, Macdonald J, Gentry J. gcrma: Background Adjustment Using Sequence Information. R package version. 2005;2.22.0 [Google Scholar]
- 72.Bolstad B, Collin F, Brettschneider J, et al. Quality assessment of Affymetrix GeneChip data. 2005 [Google Scholar]
- 73.Hulshizer R, Blalock EM. Post hoc pattern matching: assigning significance to statistically defined expression patterns in single channel microarray data. BMC Bioinformatics. 2007;8:240. doi: 10.1186/1471-2105-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004 Feb 17;101(7):2173–8. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Norris CM, Kadish I, Blalock EM, et al. Calcineurin triggers reactive/inflammatory processes in astrocytes and is upregulated in aging and Alzheimer’s models. J Neurosci. 2005 May 4;25(18):4649–58. doi: 10.1523/JNEUROSCI.0365-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smyth G. Limma: linear models for microarray data. 2005 [Google Scholar]
- 77.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic acids research. 2003;31(24):7280–301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The four endpoints measured after exposure to high and low CS doses at different time points. MMP activity was determined as gelatinolytic activity (μU/ml) in BALF. MMP-9 and TIMP-1 were measured by multiplexed immunoassays. *indicates a statistically significant difference between the mean of the group beneath it and the mean of the sham group at the same time point; # indicates a statistically significant difference between the mean of the group beneath it and the mean of the corresponding group at the previous time point. The Wilcoxon rank sum test calculated the significance level as 5%. Bars represent the mean ± SEM. ND, not determined.
Linear and quadratic models of lung weight (g) across logarithmically transformed smoke exposure (TPM × h/l) conditions. Filled circles indicate the predicted average lung weight at 5 months obtained using linear (red line) and quadratic (blue line) regression models.
Table S1.
Total CS exposure calculated for each experimental group on linear and logarithmic scales.
| EXPERIMENTAL GROUP | SMOKE EXPOSURE | SMOKE EXPOSURE (LOG10 SCALE) |
|---|---|---|
| 1 day low | 250 | 2.397940009 |
| 7 days low | 5500 | 3.740362689 |
| 1 month low | 24000 | 4.380211242 |
| 5 months low | 144000 | 5.158362492 |
| 1 day high | 500 | 2.698970004 |
| 7 days high | 11000 | 4.041392685 |
| 1 month high | 48000 | 4.681241237 |
| 5 months high | 288000 | 5.459392488 |