Abstract
Hepatitis C virus (HCV) infection is the leading indication for liver transplantation and a major cause of graft failure. This study investigated whether cyclosporin A (CsA), a widely used immunosuppressant for organ transplantation, inhibits full cycle HCV replication and restores type I interferon (IFN) signaling pathway in human hepatocytes. CsA treatment of hepatocytes before, during, and after HCV infection significantly inhibited full cycle viral replication, which is evidenced by decreased expression of HCV RNA, protein, and infectious viruses in human hepatocytes. The suppression of HCV replication by CsA was associated with elevated levels of endogenous IFN-α in infected hepatocytes. Although CsA had little effect on IFN-α signaling pathway in uninfected hepatocytes, CsA treatment of HCV-infected hepatocytes specifically upregulated the expression of IFN regulatory factor-1 and inhibited the expression of suppressor of cytokine signaling-1 and protein inhibitor of activated signal transducers and activators of transcription-x, the primary negative regulators of IFN signaling pathway. These findings provide additional evidence to support the development of CsA-based prevention/treatment of HCV infection for transplant recipients.
Keywords: hepatitis C virus, cyclosporin A, innate immunity, interferon, organ transplantation
Organ transplantation has been accepted worldwide as an effective therapy for end-stage organ dysfunction, such as heart, liver, and kidney failure. However, transplant recipients show a high incidence of viral infections due to the administration of immunosuppressive agents. One common viral infection among patients undergoing immunosuppressive treatment after transplantation is hepatitis C virus (HCV) infection, the major cause of chronic hepatitis, liver cirrhosis, and carcinoma. In the United States, end-stage liver disease due to HCV infection has become the leading indication for liver transplantation. This is particularly relevant to patients who receive liver transplantation, as these individuals have high HCV infection rates before and after transplantation (1). It has been reported that serum HCV RNA levels are increased with corticosteroids treatment and immunosuppression (2, 3). HCV infection is an important cause of mortality in patients after organ transplantation, and there is a strong association between HCV infection and graft dysfunction, especially with the long-term result (4, 5). Therefore, treatment of HCV infection is extremely important for the long-term survival of transplant recipients. However, the strategy of anti-HCV therapy in transplant recipients is still controversial, because interferon-alpha (IFN-α), the currently most used anti-HCV agent, has been related to an increased risk of allograft dysfunction and loss (6, 7). In addition, IFN-α-based therapy is effective for <50% of people infected with HCV.
Cyclosporin A (CsA) is a widely used immunosuppressant for people who receive organ transplantation. It was recently recognized that CsA is a promising antiviral agent because of its potential ability to suppress HCV replication both in vivo and in vitro (8–10). CsA-mediated anti-HCV activity is not a result of its immunosuppressive effect (9–12). Studies from several groups have shown the anti-HCV activity of CsA is caused by its ability to bind cyclophilins (11–13). These studies evaluated HCV replication in either subgenomic or genome-length replicon systems (11, 14). The HCV replicon systems, however, only support continuous HCV RNA genome replication without producing infectious virus particles. Therefore, there is still a need to determine the impact of CsA treatment on HCV infection of human hepatocytes. Utilizing an HCV JFH-1 infectious cell system that fully recapitulates viral entry, replication, and viral production (15–17), we examined whether CsA has the ability to directly inhibit full cycle HCV replication and restore intracellular innate immunity in human hepatocytes. We also explored, at the cellular and molecular levels, the mechanisms of action of CsA on HCV and endogenous IFN-α in human hepatocytes. We were particularly interested in IFN-α, as IFN-α plays a key role in control of HCV infection. Our earlier study demonstrated that HCV has the ability to suppress intracellular IFN-α expression in human hepatocytes (18). Thus, it is of importance to determine whether CsA treatment of HCV-infected hepatocytes can result in the recovery of IFN-α-mediated intracellular innate immunity that is suppressed by the virus.
Materials and methods
Reagent
CsA was purchased from Sigma-Aldrich Inc. (St. Louis, Missouri, USA). Rabbit anti-HCV nonstructural protein 5A (NS5A) antibody was obtained from Chiron Company (Emeryville, California, USA). The second antibody (Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin [Ig]G) used for immunofluorescence assay was purchased from Invitrogen (Carlsbad, California, USA).
Hepatocytes and HCV infection
Huh7 cells were kindly provided by Dr Charles Rice (The Rockefeller University, New York, New York, USA). Huh7 cells were maintained in Dulbecco’s-modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS), 10 mM Hepes, 100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine at 5% carbon dioxide. For all experiments, functionally endotoxin-free media and reagents were used. The cell viability was measured by cell proliferation assay. To generate infectious HCV JFH-1, in vitro transcribed genomic JFH-1 RNA was transfected into Huh7 cells, as previously described (16). Cell culture supernatant collected at day 10 post transfection was centrifuged and passed through 0.22 μm filter. The cell-free media were then used as virus stocks, which were aliquoted and stored at −80°C. HCV RNA quantification was measured by the real-time reverse transcripition polymerase chain reaction (RT-PCR) assay, as previously described (19, 20).
Immunofluorescence staining
Immunofluorescence evaluation of HCV NS5A protein expression in HCV-infected hepatocytes was carried out as previously described (19). Briefly, after wash with 1x phosphate-buffered saline (PBS), the cells were fixed with PBS containing 4% paraformaldehyde and 4% sucrose for 20 min, permeabilized with PBS containing 0.5% triton for 10 min, washed 3 times with PBS, and pretreated with a blocking solution for 30 min. Cells were then incubated with rabbit anti-HCV NS5A antibody (1:100) for 1 h, and subsequently incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (1:200). After being washed 3 times with PBS, cells were viewed under a fluorescence microscope (Olympus America Inc., Center Valley, Pennsylvania USA).
Titration of extracellular and intracellular HCV JFH-1
The infectivity titer of HCV JFH-1was determined on Huh7 cells by the end-point dilution and immunofluorescence assay as previously described (17). For titration of extracellular virus, the infected cell culture supernatant was serially diluted 10-fold in DMEM-10%FCS and used to infect naïve Huh7 cells (104 cells/well) in 96-well plates. HCV infectivity was examined 72 h post infection by immunofluorescence assay (17). The viral titer is expressed as focus-forming units per milliliter of supernatant (FFU/mL). For titration of intracellular infectious virus, HCV JFH-1-infected cells were washed once with PBS and subsequently incubated with trypsin-EDTA (Invitrogen, Carlsbad, California, USA) for 5 min at 37°C. Cells were resuspended in DMEM-10%FCS and collected by centrifugation at 1500 rpm for 3 min. The cell pellets were resuspended in DMEM-10%FCS and were lysed by 4 freeze-thaw cycles in dry ice and a 37°C water bath, respectively. Cell debris was pelleted by centrifugation for 5 min at 4000 rpm. The supernatants were collected and used for HCV infectivity titration (17, 21).
Quantitative real-time RT-PCR
Total cellular RNA (1 μg) extracted from Huh7 cells was subjected to reverse transcription using the reverse transcription system from Promega (Madison, Wısconsin, USA). The real-time RT-PCR for the HCV RNA quantification was performed as previously described (22, 23). The PCR amplified products were analyzed using the software MyiQ provided with the thermocycler (Icycler iQ real-time PCR detection system, Bio-Rad Laboratories, Hercules, California, USA). The levels of glyceraldehyde-3-phosphate dehydrogenase (GADPH) mRNA were used as an endogenous reference to normalize the quantities of target mRNA. To examine the amount of HCV RNA in culture supernatant, HCV RNA was extracted from 200 μL of supernatant by TRI-Reagent-BD (Molecular Research Center Inc., Cincinnati, Ohio, USA). Extracted RNA was then purified by an RNeasy Mini Kit and treated with RNase-free DNase digestion (Qiagen, Valencia, California, USA). Copy numbers of HCV RNA were determined by the real-time RT-PCR (19, 23). The special oligonucleotide primers used in this study are listed in Table 1. The oligonucleotide primers were synthesized by Integrated DNA Technologies Inc. (Coralville, Iowa, USA).
Table 1.
Primers used for real-time reverse-transcription polymerase chain reaction assay
| Primer
|
Orientation
|
Sequence
|
|---|---|---|
| GAPDH | Sense | 5′-GGTGGTCTCCTCTGACTTCAACA-3′ |
| Antisense | 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ | |
| HCV | Sense | 5′-RAYCACTCCCCTGTGAGGACC-3′ |
| Antisense | 5′-TGRTGCACGGTCTACGAGACCTC-3′ | |
| IFN-α | Sense | 5′-TTTCTCCTGCCTGAAGGACAG-3′ |
| Antisense | 5′-GCTCATGATTTCTGCTCTGACA-3′ | |
| IFN-β | Sense | 5′-GCCGCATTGACCATCTATGAGA-3′ |
| Antisense | 5′-GAGATCTTCAGTTTCGGAGGTAAC-3′ | |
| SR-BI | Sense | 5′-GGTCCCTGTCATCTGCCAA-3′ |
| Antisense | 5′-CTCCTTATCCTTTGAGCCCTTT-3′ | |
| Occludin | Sense | 5′-AAGCAAGTGAAGGGATCTGC-3′ |
| Antisense | 5′-GGGGTTATGGTCCAAAGTCA-3′ | |
| LDLR | Sense | 5′-ACTGGTGTGAGAGGACCACC-3′ |
| Antisense | 5′-CAAAGGAAGACGAGGAGCAC-3′ | |
| CD81 | Sense | 5′-CGCCAAGGATGTGAAGCAGTTC-3′ |
| Antisense | 5′-TCCCGGAGAAGAGGTCATCGAT-3′ | |
| CLDN1 | Sense | 5′-GGCAGATCCAGTGCAAAGTC-3′ |
| Antisense | 5′-TCTTCTGCACCTCATCGTCTT-3′ | |
| IRF-1 | Sense | 5′-TGAAGCTACAACAGATGAGG-3′ |
| Antisense | 5′-AGTAGGTACCCCTTCCCATC-3′ | |
| IRF-3 | Sense | 5′-ACCAGCCGTGGACCAAGAG-3′ |
| Antisense | 5′-TACCAAGGCCCTGAGGCAC-3′ | |
| IRF-5 | Sense | 5′-AAGCCGATCCGGCCAA-3′ |
| Antisense | 5′-GGAAGTCCCGGCTCTTGTTAA-3′ | |
| IRF-7 | Sense | 5′-TGGTCCTGGTGAAGCTGGAA-3′ |
| Antisense | 5′-GATGTCGTCATAGAGGCTGTTGG-3′ | |
| SOCS-1 | Sense | 5′-GACGCCTGCGGATTCTACTG-3′ |
| Antisense | 5′-GGCCATCTTCACGCTAAGGG-3′ | |
| SOCS-2 | Sense | 5′-TGCAAGGATAAGCGGACAGG-3′ |
| Antisense | 5′-CAGAGATGCTGCAGAGATGG-3′ | |
| SOCS-3 | Sense | 5′-TGCGCCTCAAGACCTTCAGC-3′ |
| Antisense | 5′-GATGCGCAGGTTCTTGGTCC-3′ | |
| PIAS-1 | Sense | 5′-TCCCACCCAATCTTTGTGTG-3′ |
| Antisense | 5′-GCCGCATTTTACCAAGTGGA-3′ | |
| PIAS-3 | Sense | 5′-TGCTGGCCGGAACAAGAGTG-3′ |
| Antisense | 5′-AGGGGGCAAAGAGAGAAGGG-3′ | |
| PIAS-x | Sense | 5′-TCTTCTGACGAAGAGGAAGACC-3′ |
| Antisense | 5′-TCAGAAGATGTTCCAAGCTTCA-3′ | |
| PIAS-y | Sense | 5′-GAGAAGAAGCCCACCTGGA-3′ |
| Antisense | 5′-ACACTCGCTCAGGATCTTCG-3′ |
GAPDH, glyceraldehyde-3 -phosphate dehydrogenase; HCV, hepatitis C virus; IFN, interferon; IRF, interferon regulatory factor; SOCS, suppressor of cytokine signaling; PIAS, protein inhibitors of activated STAT; STAT, signal transducers and activators of transcription.
Quantitative real-time RT-PCR of microRNA (miRNA)
Total cellular RNA, including miRNA, was extracted from cells using miRNeasy Mini Kit (Qiagen). Total RNA (1 μg) was reverse-transcribed with miScript Reverse Transcription Kit from Qiagen. The real-time RT-PCR for the quantification of a subset of miRNAs (miR-122, miR-196a, miR-296, miR-351, miR-431, and miR-448) was carried out with miScript Primer Assays and miScript SYBR Green PCR Kit (Qiagen).
Statistical analysis
Where appropriate, data were expressed as mean ± standard deviation of triplicate cultures. Statistical significance was assessed by Student t-test. Statistical analyses were performed with Graphpad Instat Statistical Software. Statistical significance was defined as P<0.05.
Results
CsA suppresses HCV JFH-1 replication
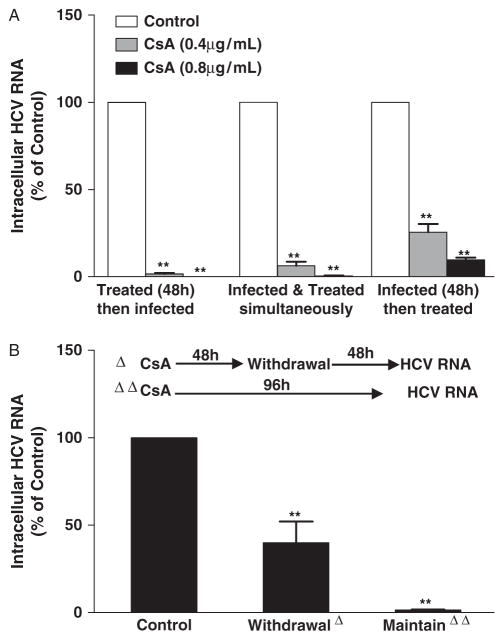
We first examined whether CsA has cytotoxicity on human hepatocytes. CsA, at the concentrations of <3 μg/mL, had little cytotoxicity in hepatocytes. To assess the effect of CsA on HCV JFH-1 replication and infectivity, Huh7 cells were infected with HCV JFH-1 for 72 h, and then treated with CsA at various concentrations for 48 h. CsA, in a dose-and time-dependent manner, suppressed HCV JFH-1 replication in hepatocytes, which was evidenced by decreased expression of both intracellular and extracellular HCV RNA (Fig. 1A, B, and F) as well as infectious virus (Fig. 1C and D). The inhibition of HCV JFH-1 replication by CsA treatment was confirmed by the experiments examining NS5A protein expression in hepatocytes treated with or without CsA (Fig. 1E). To further determine the protective effect of CsA on hepatocytes, we treated hepatocytes with CsA under 3 different conditions. CsA treatment of hepatocytes before, during, and after HCV infection significantly suppressed HCV expression (Fig. 2A). In order to determine whether CsA-mediated inhibition of HCV replication could be maintained after the withdrawal of CsA treatment, we removed CsA from HCV-infected cultures. As indicated in Figure 2B, the withdrawal of CsA resulted in the rebound of HCV replication. In contrast, CsA remaining in HCV-infected cultures prevented virus from rebounding (Fig. 2B).
Fig. 1.
Effect of cyclosporin A (CsA) on hepatitis C virus (HCV) replication. (A–E) Dose-dependent effect of CsA on HCV replication. HCV JFH-1-infected Huh7 cells were treated with CsA at indicated concentrations at 72 h post infection. Cell lysates and culture supernatants were collected at 48 h post treatment for the HCV RNA analysis and HCV infectivity assay. The levels of HCV RNA (A, B) and the titers of infectious virus (C, D) were determined by the real-time reverse transcription polymerase chain reaction (RT-PCR) and serial dilution/immunofluorescence analyses, respectively. The levels of intracellular HCV RNA (A), with normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, are expressed as % of control (without CsA treatment, which is defined as 100). The levels of extracellular HCV RNA (B) are expressed as viral RNA copies/mL. The intracellular (C) or extracellular (D) HCV infectivity is expressed as focus-forming units per milliliter (FFU/mL) of cell lysate or supernatant, respectively. HCV nonstructrual protein 5A (NS5A) protein expression (E) was determined by immunofluorescence staining with antibody against NS5A (green). The cell nuclei were stained by Hoechst (blue). One representative experiment is shown (magnification × 200). (F) Tıme course effect of CsA on HCV replication. HCV JFH-1-infected Huh7 cells were treated with CsA (0.8 μg/mL) at 72 h post infection. At different time points, total cellular RNA extracted from the cell cultures was subjected to the real-time RT-PCR for HCV and GAPDH RNA quantification. The levels of intracellular HCV RNA, with normalization to GAPDH mRNA, are expressed as % of control (without CsA treatment, which is defined as 100). The results (A–D, F) shown are mean ± standard deviation of triplicate cultures, representative of 3 experiments (*P<0.05, **P<0.01).
Fig. 2.
Effect of cyclosporin A (CsA) on hepatitis C virus (HCV) infection and replication under different conditions. (A) Effect of different CsA treatment conditions on HCV JFH-1 infection of human hepatocytes. Huh7 cells were either treated with CsA first for 48 h and then infected with HCV JFH-1 for 72 h, or treated with CsA and infected with HCV JFH-1 simultaneously and then cultured for 72 h, or infected with HCV JFH-1 first for 48 h and then treated with CsA for 72 h. Total cellular RNA extracted from the cell cultures was subjected to the real-time reverse transcription polymerase chain reaction (RT-PCR) assay for HCV and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA quantification. The levels of intracellular HCV RNA, with normalization to GAPDH mRNA, are expressed as % of control (without CsA treatment, which is defined as 100). (B) Effect of CsA treatment withdrawal on HCV replication. HCV JFH-1-infected Huh7 cells (at 72 h post infection) were treated with CsA (0.8 μg/mL) for 48 h, the cells were then cultured for additional 48 h in the presence or absence of CsA (0.8 μg/mL). Total cellular RNA extracted from the cell cultures was subjected to the real-time RT-PCR for HCV and GAPDH RNA quantification. The levels of intracellular HCV RNA, with normalization to GAPDH mRNA, are expressed as % of control (without CsA treatment, which is defined as 100). The data shown are the mean ± standard deviation of triplicate culture, representative of 3 experiments (**P<0.01).
CsA selectively restores IFN-α expression in HCV-infected hepatocytes
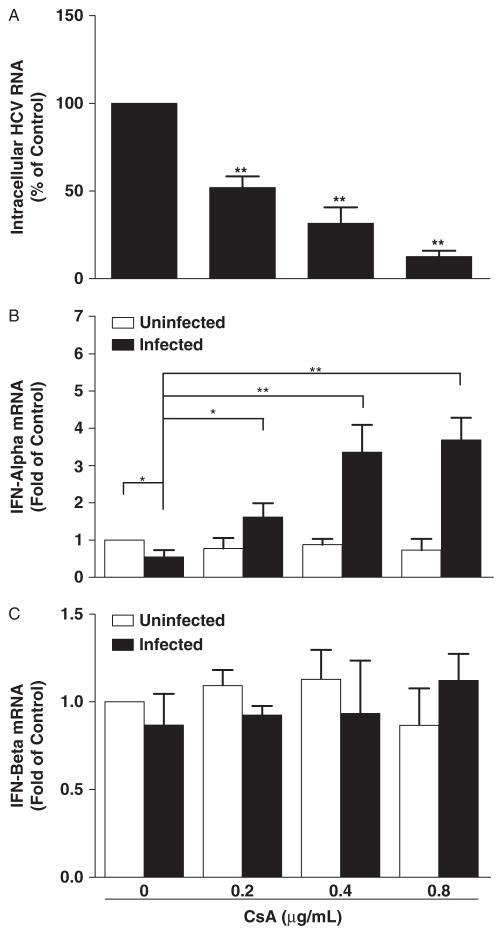
It has been documented that HCV has the ability to suppress the type I IFN pathway (18, 24, 25). We thus determined whether CsA could restore the expression of IFN-α or -β in HCV-infected hepatocytes. As shown in Figure 3B, HCV infection reduced IFN-α expression in hepatocytes. However, when HCV JFH-1-infected cells were treated with CsA, IFN-α expression was significantly increased (Fig. 3B). In contrast, CsA had little effect on IFN-β expression in HCV-infected hepatocytes (Fig. 3C). In addition, CsA treatment of uninfected hepatocytes had little effect on the expression of either IFN-α or IFN-β (Fig. 3B and C). The effect of CsA on IFN-α expression was negatively correlated with HCV RNA expression in hepatocytes (Fig. 3A and B).
Fig. 3.
Effect of cyclosporin A (CsA) on type I interferon (IFN) expression. Hepatitis C virus (HCV) JFH-1-infected or uninfected Huh7 cells were treated with CsA at indicated concentrations at 72 h post infection. Total cellular RNA extracted from the cell cultures at 48 h post treatment was subjected to the real-time reverse transcription polymerase chain reaction assay for HCV (A), IFN-α (B), IFN-β (C), and glyceraldehyde-3-phosphate dehydrogenase RNA quantification. Intracellular HCV expression is expressed as HCV RNA levels relative (%) to control (without CsA treatment, which is defined as 100). IFN-α/β expression is expressed as mRNA levels relative (fold) to the control (uninfected Huh7 cells without CsA treatment, which is defined as 1). The data shown are the mean ± standard deviation of triplicate culture, representative of 3 experiments (*P<0.05, **P<0.01).
CsA modulates the expression of the regulators of IFN pathway
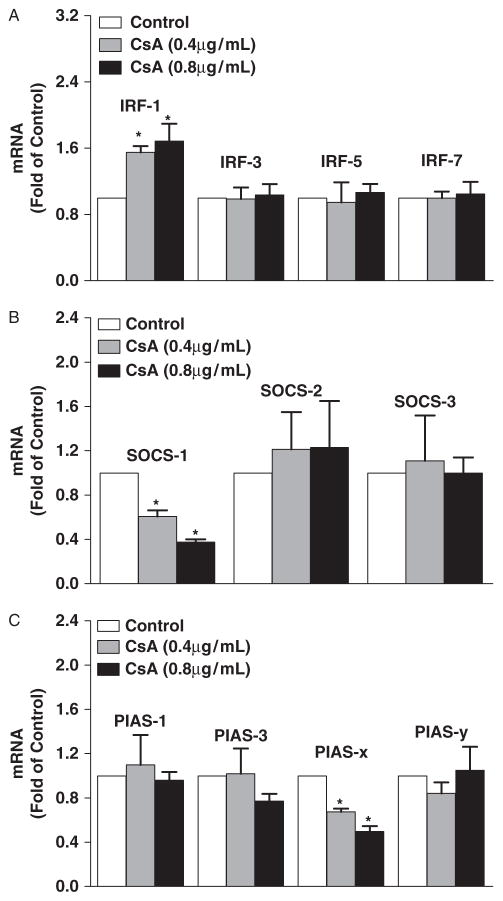
Type I IFN production is primarily controlled by multiple transcription factors. We thus examined whether CsA could modulate the expression of the key regulators of IFN pathway. As shown in Figure 4, although CsA had little effect on the expression of interferon regulatory factors (IRFs)-3, -5, and -7, CsA treatment induced the expression of IRF-1 (Fig. 4A). In addition, CsA treatment of HCV-infected hepatocytes decreased the expression of suppressor of cytokine signaling-1 (SOCS-1) and protein inhibitors of activated signal transducers and activators of transcription (PIAS-x), the 2 key negative regulators of IFN signaling pathway (Fig. 4B and C).
Fig. 4.
Effect of cyclosporin A (CsA) on the expression of the regulators of interferon (IFN) signaling pathway. Hepatitis C virus (HCV) JFH-1-infected Huh7 cells were treated with CsA at indicated concentrations for 24 h. Total cellular RNA extracted from the cell cultures was subjected to the real-time reverse transcription polymerase chain reaction assay for interferon regulatory factor (IRF)-1, -3, -5, -7 (A), suppressor of cytokine signaling (SOCS)-1, -2, -3 (B), protein inhibitors of activated STAT (PIAS)-1, 3, x, y (C), and glyceraldehyde-3-phosphate dehydrogenase mRNA quantification. The data are expressed as mRNA levels relative (fold) to the control (without CsA treatment, which is defined as 1). The data shown are the mean ± standard deviation of triplicate culture, representative of 3 experiments (*P<0.05).
CsA does not affect HCV entry receptors and miRNA expression in hepatocytes
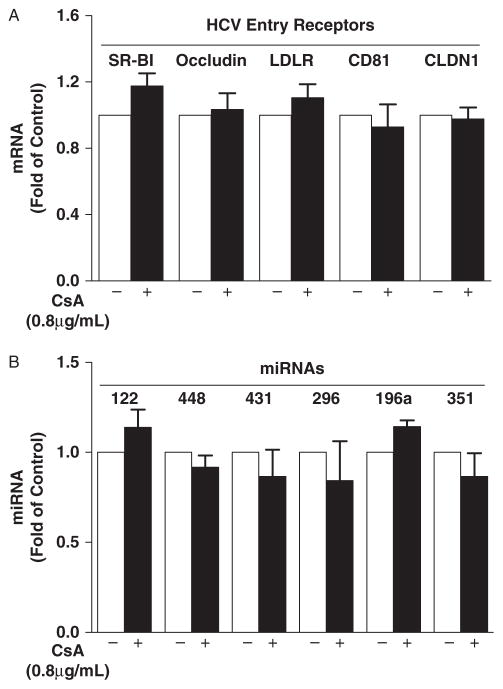
Several cell surface molecules have been proposed as HCV entry receptors or co-receptors. Suppression of the expression of these receptors could reduce HCV entry and expression in hepatocytes. We thus examined the expression of 5 known HCV entry receptors (SR-BI, Occludin, LDLR, CD81, and CLDN1) in CsA-treated Huh7 cells. As shown in Figure 5A, CsA had little effect on the expression of these HCV receptors. We also examined whether CsA treatment of hepatocytes modulates the expression of several intracellular miRNAs that have recently been identified to modulate HCV replication in hepatocytes (26, 27). As shown in Figure 5B, CsA treatment had little effect on the expression of miR-122, miR-448, miR-431, miR-296, miR-196a, and miR-351 in hepatocytes.
Fig. 5.
Effect of cyclosporin A (CsA) on the expression of hepatitis C virus (HCV) entry receptors and HCV-related microRNAs (miRNAs). (A) Effect of CsA on HCV entry receptor expression. HCV JFH-1-infected Huh7 cells were treated with CsA at indicated concentration at 72 h post infection. Total cellular RNA was extracted from the cell cultures at 48 h post treatment and subjected to the real-time reverse transcription polymerase chain reaction (RT-PCR) assay for SR-BI, Occludin, LDLR, CD81, CLDN1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA quantification. (B) Effect of CsA on HCV-related miRNA expression. Total cellular RNA extracted from the cell cultures was subjected to the real-time RT-PCR for miRNA (miR-122, miR-448, miR-431, miR-296, miR-196a, and miR-351) and GAPDH RNA quantification. The HCV receptor and miRNA expression is expressed as RNA levels relative (fold) to control (without CsA treatment, which is defined as 1). The data shown are the mean ± standard deviation of triplicate culture, representative of 3 experiments.
Discussion
Using the recently established HCV JFH-1 system that recapitulates viral entry, replication, and production of infectious virus, we have evaluated the effect of CsA on HCV infection/replication. We have demonstrated that CsA treatment of human hepatocytes could inhibit full cycle HCV infection/replication, which was evidenced by decreased expression of HCV RNA, protein, and infectious virus in human hepatocytes. CsA treatment of human hepatocytes not only inhibited the levels of HCV RNA both intracellularly and extracellularly, but also suppressed the production of infectious viruses within cells and in culture supernatant (Fig. 1). Wıth the HCV JFH-1 system, we were able to examine whether CsA pretreatment could protect hepatocytes from HCV infection (Fig. 2). Inhibition of virus by CsA was also observed even after HCV infection had taken place (Fig. 2). Among 3 different conditions of CsA treatment, CsA pretreatment of hepatocytes demonstrated the optimal inhibition of HCV (Fig. 2), suggesting that pre-binding of cellular cyclophilins by CsA is important. This finding suggests an anti-HCV benefit of preadministration of CsA to transplant recipients. In addition, we showed that continued CsA treatment in HCV-infected cultures was necessary, as withdrawal of CsA from HCV-infected cultures resulted in viral rebound (Fig. 2B). This inhibitory effect of CsA on HCV replication was potent, as up to 90% of HCV inhibition was observed in the cells treated with CsA at the concentrations that are achievable clinically. This observation is highly significant, as high-dose CsA treatment can cause side effects in transplant recipients (28). It was reported that CsA treatment could induce significant oxidative stress in liver at concentrations of >10 μM, which is due to the cytotoxicity effect of CsA on hepatocytes (29). However, when CsA was used at concentrations of <10 μM, little cytotoxicity was observed (29). In this study, we used CsA at concentrations ranging from 0.05 to 1.6 μg/mL (<10 μM threshold) and did not find a cytotoxic effect on human hepatocytes.
The suppression of HCV replication by CsA was associated with elevated levels of endogenous IFN-α in hepatocytes. This CsA-mediated IFN-α induction could not be a result of its direct effect on IFN-α and/or IFN-α regulatory genes, as CsA treatment had little effect on IFN-αexpression in uninfected hepatocytes. It is likely that CsA, through the suppression of HCV and its protein expression, restores IFN-α expression in hepatocytes. This statement is supported by our earlier study (18) and by the reports of others (24, 25), showing that HCV and its proteins such as NS5A could inhibit IFN signaling pathway in hepatocytes. Because IFN-α plays a key role in host cell innate immunity against viral infections, including HCV, suppression of IFN-α expression could provide a favorable microenvironment for HCV infection/replication. Our further studies showed that CsA treatment not only restored the levels of endogenous IFN-α but also enhanced the expression of IFN-α in HCV JFH-1-infected hepatocytes. This finding, however, disagrees with the report that CsA does not activate IFN response genes in the HCV replicon system (11). This discrepancy may be due to the fact that 2 different cell systems (HCV replicon system vs. full cycle infectious model) were used by the 2 laboratories. It is known that the HCV subgenomic replicon system only supports continuous HCV RNA genome replication without producing infectious virus particles. In our study, we used the HCV JFH-1 infectious cell system that fully recapitulates viral entry, replication, and viral production. Nevertheless, future studies are necessary in order to determine the mechanism(s) involved in the CsA action on host cell innate immunity against HCV infection/replication.
Clinically, the choice of regimens of immunosuppressant after organ transplantation may have significant impact on the outcome of HCV-related liver disease, as some of the immunosuppressants are known to interfere with anti-HCV therapy. For example, it was reported (30) that treatment with tacrolimus, a calcineurin inhibitor of serine/threonine protein phosphatase, can elicit resistance to IFN-α therapy by disturbance of tyrosine phosphorylation of signal transducers and activators of transcription-1 (STAT-1), a primary component of the IFN signaling cascade. Because CsA possesses the same action as tacrolimus (31), we examined whether CsA has the ability to affect the STAT-IFN pathway, modulating the IRFs. We found that IRF-1 expression was upregulated in CsA-treated and HCV-infected hepatocytes. IRF-1 is among the key positive regulators for IFN-α induction. To further explore the mechanisms involved in CsA-mediated enhancement of IFN-α expression, we determined whether CsA treatment inhibits the expression of negative regulators in the IFN signaling pathway. Two suppressor families, SOCSs and PIASs, have been identified as the negative regulators in the IFN signaling pathway (32, 33). Among the suppressor family members examined, the levels of SOCS-1 and PIAS-x were decreased in CsA-treated hepatocytes (Fig. 4B and C). These findings support the report that CsA can inhibit nuclear translocation of tyrosine phosphorylated STAT-1, resulting in the decrease of SOCS-1 and PIAS-x expression (30). Since SOCS-1 and PIAS-x are specific inhibitors of STAT-mediated gene activation (33, 34), suppression of these factors provides a sound mechanism for the CsA action on IFN-α.
In conclusion, our study provides compelling experiment evidence that CsA treatment of human hepatocytes inhibits full cycle HCV JFH-1 replication, leading to the restoration and enhancement of intracellular IFN-α expression. Although CsA is a potent immunosuppressant, it appears to be beneficial for people infected with HCV, given its ability to inhibit HCV replication and restore hepatocyte innate immunity. Because of the importance of host cell innate immunity in protecting transplant recipients from viral infections, it is of great interest to develop intervention strategies to reconstitute intracellular innate responses to control HCV replication. Thus, future studies are necessary for the development of CsA-based prevention/treatment of HCV infection for transplant recipients.
Acknowledgments
We thank Dr Charles Rice (The Rockefeller University, New York, New York, USA) and Dr Takija Wakita (Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan) for generously providing us with the Huh7 cell line and the plasmid pJFH-1, respectively. This study was supported by grants from the National Institutes of Health (DA 22177, DA 12815, and DA 27550 to Dr W.-Z Ho).
References
- 1.Fabrizi F, Poordad FF, Martin P. Hepatitis C infection and the patient with end-stage renal disease. Hepatology. 2002;36 (1):3–10. doi: 10.1053/jhep.2002.34613. [DOI] [PubMed] [Google Scholar]
- 2.Fong TL, Valinluck B, Govindarajan S, Charboneau F, Adkins RH, Redeker AG. Short-term prednisone therapy affects aminotransferase activity and hepatitis C virus RNA levels in chronic hepatitis C. Gastroenterology. 1994;107 (1):196–199. doi: 10.1016/0016-5085(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 3.Mas V, Alvarellos T, Chiurchiu C, Camps D, Massari P, de Boccardo G. Hepatitis C virus infection after renal transplantation: viral load and outcome. Transplant Proc. 2001;33 (1–2):1791–1793. doi: 10.1016/s0041-1345(00)02682-8. [DOI] [PubMed] [Google Scholar]
- 4.Fong TL, Hou L, Hutchinson IV, Cicciarelli JC, Cho YW. Impact of hepatitis C infection on outcomes after heart transplantation. Transplantation. 2009;88 (9):1137–1141. doi: 10.1097/TP.0b013e3181bd3e59. [DOI] [PubMed] [Google Scholar]
- 5.Haji SA, Starling RC, Avery RK, et al. Donor hepatitis-C seropositivity is an independent risk factor for the development of accelerated coronary vasculopathy and predicts outcome after cardiac transplantation. J Heart Lung Transplant. 2004;23 (3):277–283. doi: 10.1016/S1053-2498(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 6.Weclawiack H, Kamar N, Mehrenberger M, et al. Alpha-interferon therapy for chronic hepatitis C may induce acute allograft rejection in kidney transplant patients with failed allografts. Nephrol Dial Transplant. 2008;23 (3):1043–1047. doi: 10.1093/ndt/gfm678. [DOI] [PubMed] [Google Scholar]
- 7.Carbognin SJ, Solomon NM, Yeo FE, et al. Acute renal allograft rejection following pegylated IFN-alpha treatment for chronic HCV in a repeat allograft recipient on hemodialysis: a case report. Am J Transplant. 2006;6 (7):1746–1751. doi: 10.1111/j.1600-6143.2006.01374.x. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama H, Yoshinaga H, Tanaka T, et al. Effects of cyclosporin A on hepatitis C virus infection in bone marrow transplant patients. Bone Marrow Transplantation Team Bone Marrow Transplant. 1997;20 (11):993–995. doi: 10.1038/sj.bmt.1700996. [DOI] [PubMed] [Google Scholar]
- 9.Inoue K, Sekiyama K, Yamada M, Watanabe T, Yasuda H, Yoshiba M. Combined interferon alpha2b and cyclosporin A in the treatment of chronic hepatitis C: controlled trial. J Gastroenterol. 2003;38 (6):567–572. doi: 10.1007/s00535-002-1104-5. [DOI] [PubMed] [Google Scholar]
- 10.Inoue K, Yoshiba M. Interferon combined with cyclosporine treatment as an effective countermeasure against hepatitis C virus recurrence in liver transplant patients with end-stage hepatitis C virus related disease. Transplant Proc. 2005;37 (2):1233–1234. doi: 10.1016/j.transproceed.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa M, Sakamoto N, Tanabe Y, et al. Suppression of hepatitis C virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology. 2005;129 (3):1031–1041. doi: 10.1053/j.gastro.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 12.Yang F, Robotham JM, Nelson HB, Irsigler A, Kenworthy R, Tang H. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82 (11):5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciesek S, Steinmann E, Wedemeyer H, et al. Cyclosporine A inhibits hepatitis C virus nonstructural protein 2 through cyclophilin A. Hepatology. 2009;50 (5):1638–1645. doi: 10.1002/hep.23281. [DOI] [PubMed] [Google Scholar]
- 14.Nakagawa M, Sakamoto N, Enomoto N, et al. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem Biophys Res Commun. 2004;313 (1):42–47. doi: 10.1016/j.bbrc.2003.11.080. [DOI] [PubMed] [Google Scholar]
- 15.Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309 (5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 16.Wakita T, Pietschmann T, Kato T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11 (7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong J, Gastaminza P, Cheng G, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102 (26):9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang T, Lin RT, Li Y, et al. Hepatitis C virus inhibits intracellular interferon alpha expression in human hepatic cell lines. Hepatology. 2005;42 (4):819–827. doi: 10.1002/hep.20854. [DOI] [PubMed] [Google Scholar]
- 19.Ye L, Wang X, Wang S, et al. CD56 + T cells inhibit hepatitis C virus replication in human hepatocytes. Hepatology. 2009;49 (3):753–762. doi: 10.1002/hep.22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Ye L, Peng JS, et al. Morphine inhibits intrahepatic interferon-alpha expression and enhances complete hepatitis C virus replication. J Infect Dis. 2007;196 (5):719–730. doi: 10.1086/520093. [DOI] [PubMed] [Google Scholar]
- 21.Gastaminza P, Kapadia SB, Chisari FV. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J Virol. 2006;80 (22):11074–11081. doi: 10.1128/JVI.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Zhang T, Ho C, Orange JS, Douglas SD, Ho WZ. Natural killer cells inhibit hepatitis C virus expression. J Leukoc Biol. 2004;76 (6):1171–1179. doi: 10.1189/jlb.0604372. [DOI] [PubMed] [Google Scholar]
- 23.Yang JH, Lai JP, Douglas SD, Metzger D, Zhu XH, Ho WZ. Real-time RT-PCR for quantitation of hepatitis C virus RNA. J Virol Methods. 2002;102 (1–2):119–128. doi: 10.1016/s0166-0934(02)00007-1. [DOI] [PubMed] [Google Scholar]
- 24.Pawlotsky JM, Roudot-Thoraval F, Bastie A, et al. Factors affecting treatment responses to interferon-alpha in chronic hepatitis C. J Infect Dis. 1996;174 (1):1–7. doi: 10.1093/infdis/174.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Melen K, Fagerlund R, Nyqvist M, Keskinen P, Julkunen I. Expression of hepatitis C virus core protein inhibits interferon-induced nuclear import of STATs. J Med Virol. 2004;73 (4):536–547. doi: 10.1002/jmv.20123. [DOI] [PubMed] [Google Scholar]
- 26.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309 (5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen IM, Cheng G, Wieland S, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449 (7164):919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel K, McHutchison JG. Initial treatment for chronic hepatitis C: current therapies and their optimal dosing and duration. Cleve Clin J Med. 2004;71 (Suppl 3):S8–S12. doi: 10.3949/ccjm.71.suppl_3.s8. [DOI] [PubMed] [Google Scholar]
- 29.Wolf A, Trendelenburg CF, Diez-Fernandez C, et al. Cyclosporine A-induced oxidative stress in rat hepatocytes. J Pharmacol Exp Ther. 1997;280 (3):1328–1334. [PubMed] [Google Scholar]
- 30.Hirano K, Ichikawa T, Nakao K, et al. Differential effects of calcineurin inhibitors, tacrolimus and cyclosporin A, on interferon-induced antiviral protein in human hepatocyte cells. Liver Transpl. 2008;14 (3):292–298. doi: 10.1002/lt.21358. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398 (6724):256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 32.Vlotides G, Sorensen AS, Kopp F, et al. SOCS-1 and SOCS-3 inhibit IFN-alpha-induced expression of the antiviral proteins 2,5-OAS and MxA. Biochem Biophys Res Commun. 2004;320 (3):1007–1014. doi: 10.1016/j.bbrc.2004.06.051. [DOI] [PubMed] [Google Scholar]
- 33.Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006;16 (2):196–202. doi: 10.1038/sj.cr.7310027. [DOI] [PubMed] [Google Scholar]
- 34.Chung CD, Liao J, Liu B, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278 (5344):1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]