Abstract
To discover candidate biomarkers for diagnosis and detection of human laryngeal carcinoma and explore possible mechanisms of this cancer carcinogenesis, two-dimensional strong cation-exchange/reversed-phase nano-scale liquid chromatography/mass spectrometry analysis was used to identify differentially expressed proteins between the laryngeal carcinoma tissue and the adjacent normal tissue. As a result, 281 proteins with significant difference in expression were identified, and four differential proteins, Profilin-1 (PFN1), Nucleolin (NCL), Cytosolic non-specific dipeptidase (CNDP2) and Mimecan (OGN) with different subcellular localization were selectively validated. Semiquantitative RT-PCR and Western blotting were performed to detect the expression of the four proteins employing a large collection of human laryngeal carcinoma tissues, and the results validated the differentially expressed proteins identified by the proteomics. Furthermore, we knocked down PFN1 in immortalized human laryngeal squamous cell line Hep-2 cells and then the proliferation and metastasis of these transfected cells were measured. The results showed that PFN1 silencing inhibited the proliferation and affected the migration ability of Hep-2 cells, providing some new insights into the pathogenesis of PFN1 in laryngeal carcinoma. Altogether, our present data first time show that PFN1, NCL, CNDP2 and OGN are novel potential biomarkers for diagnosis and therapeutic targets for laryngeal carcinoma, and PFN1 is involved in the metastasis of laryngeal carcinoma.
Introduction
Laryngeal carcinoma, one of the most common types of cancer in the head and neck, accounts for 2.4% of new malignancies worldwide every year [1], [2]. This cancer is mainly squamous cell carcinoma, reflecting its origin from the squamous cells [3]. In addition, it is approved that laryngeal carcinoma may spread by direct extension to adjacent structures, and frequently distant metastasis to the lung [4], [5]. Up to now, most patients of laryngeal cancer could retain laryngeal function after the therapy if the disease was detected at an early stage. But unfortunately, the fact is that the disease is often diagnosed at advanced stages because of the lack of reliable, early diagnostic biomarkers. Therefore, identification of biomarkers for early detection and prognosis is important and may in turn lead to more effective treatments using multiplex technologies.
Proteomics, a study of the complete protein complements of the cell, is the integration of biochemical, genetics, and proteomics data in the detection of biomarkers for early detection of cancers [6]–[8]. Proteomics is currently considered to be a powerful tool for global evaluation of protein expression, and has been widely applied. It has been suggested that analysis of the cancer proteome can be beneficial to understand not only the association between protein alterations and malignancy, but also the effect of molecular intracellular mislocalization in tumour initiation [9]. Consistently, the development of increasingly high-throughput and sensitive mass spectroscopy-based proteomic techniques provides new opportunities to examine the physiology and pathophysiology of many biological samples. The two dimensional liquid chromatography tandem MS (2D LC-MS/MS) analysis is emerging as one of the more powerful quantitative proteomics methodologies in the search for tumour biomarkers [10], [11]. For instance, in previous study the authors used 2D LC-MS/MS to identify 100 differentially expressed proteins from rheumatoid arthritis patients, and concluded that up-regulation of vasculature development related proteins and down-regulation of redox-related proteins in fibroblast-like synoviocytes were predominant factors that may contribute to the pathogenesis of rheumatoid arthritis [12]. Moreover, using LC-MS/MS, Moon et al efficiently quantified the proteins of balding and non-balding dermal papilla cells (DPCs) from patients, and 128 up-regulated and 12 down-regulated proteins among 690 distinct proteins were identified in balding DPCs compared to non-balding DPCs [13].
A number of studies using proteomics based on surface-enhanced laser desorption/ionization time-of-flight MS have identified the differential serum proteins in laryngeal carcinoma, leading to discovery of potential biomarkers for diagnosis or prognosis [14], [15]. Although some proteomic studies on laryngeal carcinoma tissue have been reported [16]–[18], there are no clinically established biomarkers available for early detection and therapeutic targets of this cancer. Therefore, to obtain more information, in the present study, 2D LC-MS/MS was performed to identify the differential proteins between laryngeal carcinoma tissue and corresponding adjacent noncancerous tissue, and then the bioinformatics analyses, including gene ontology (GO) analysis, and protein network analysis of different proteins were conducted. Subsequently, values of the four differential proteins (PFN1, NCL, CNDP2 and OGN) with expressional alterations were selectively validated by semiquantitative RT-PCR and Western blotting. Furthermore, we first time show that PFN1, NCL, CNDP2 and OGN may be potential diagnostic and therapeutic targets for laryngeal carcinoma, and demonstrate that PFN1 is involved in the migration of human squamous cells.
Methods
Patients
Thirty-four laryngeal carcinoma tissues and corresponding adjacent noncancerous tissues were obtained from 34 patients who underwent surgical resection in Shanghai Changzheng Hospital, in accordance with approved human subject guidelines approved by the Scientific and Ethical Committee of Second Military Medical University. And an informed consent form was signed by the participants to proceed with the protocol research. All patients undergone resection and were not treated with neoadjuvant chemotherapy or radiotherapy. Two specimens were obtained from each patient, one from the centre of the tumor and the other of similar mass from remote areas (>1 cm) adjacent to the cancerous regions. All these samples were taken by experienced surgeons and examined by experienced pathologists, frozen immediately in liquid nitrogen, and then frozen at −80°C until use. The clinical details of the patients are shown in Table 1.
Table 1. Clinical characteristics of the patients.
| Characteristic | No. of patients (%) |
| Number of samples | N = 34 |
| Gender | |
| Male | 32/34(94.12) |
| Female | 2/34(5.88) |
| Age (years) | |
| Mean | 61.2±7.4 |
| Range | 38–75 |
| Clinical stage | |
| I | 8/34(23.53) |
| II | 6/34(17.65) |
| III | 11/34(32.35) |
| IV | 9/34(26.47) |
| Tumor location | |
| Glottic | 19/34(55.88) |
| Supraglottic | 11/34(32.35) |
| Subglottic | 2/34(5.88) |
| Transglottic | 2/34(5.88) |
Protein sample preparation
Samples collected from ten cancer tissues and the corresponding adjacent noncancerous tissues groups were pooled, respectively. 2 mg samples were ground in liquid nitrogen. One milliliter of lysis buffer (7 M urea, 2 M thiourea, 1x Protease Inhibitor Cocktail (Roche Ltd. Basel, Switzerland)) was added to sample, followed by sonication on ice and centrifugation at 13 000 rpm for 15 min at 4°C. The supernatant was stored in small aliquots at −80°C and the protein concentration was determined using a modified Bradford method.
2D-LC-MS/MS
One hundred micrograms of protein were reduced with 1 mM DTT for 45 min at 60°C, and carbamidomethylated with 5 mM iodoacetamide for 45 min at room temperature in the dark. Alkylated proteins were diluted four times with deionized water, and then digested with sequencing grade modified trypsin (Promega) overnight. The protease/protein ratio was 1: 50. The resulting peptide mixture was acidified with TFA to pH = 3, and then was desalted using a 1.3 ml C18 solid phase extraction column (Sep-Pak Cartridge) (Waters Corpoation, Milford, USA). The peptides were dried using a vacuum centrifuge and then resuspended with loading buffer (5 mM Ammonium formate containing 5% acetonitrile, pH 3.0), separated and analyzed by two-dimensional (2D) strong cation-exchange (SCX)/reversed-phase (RP) nano-scale liquid chromatography/mass spectrometry (2D-nanoLC/MS). The experiments were performed on a Nano Aquity UPLC system (Waters Corporation, Milford, USA) connected to an LTQ Orbitrap XL mass spectrometer (Thermo Electron Corp., Bremen, Germany) equipped with an online nano-electrospray ion source (Michrom Bioresources, Auburn, USA).
A 180 µm×2.4 cm SCX column (Waters Corporation, Milford, USA), which was packed with a 5 µm Poly Sulfoethyl Aspartamide (PolyLC, Columbia, MD, USA) was used for the first dimension. To recover hydrophobic peptides still retained on the SCX column after a conventional salt step gradient, a RP step gradient from 5% to 50% acetonitrile (ACN) was applied to the SCX column. A 15 µl plug was injected each time to form the step gradients. At last, 1 M Ammonium formate (NH4FA) was used to clean the SCX colum once. The plugs were loaded onto the SCX column with a loading buffer at a 15 µl/min flow rate for 6 min. A 15 µl peptide sample was loaded onto the SCX column before the gradient plugs were injected. The eluted peptides were captured by a trap column (Waters) while salts were diverted to waste. The trap column (2 cm x 180 µm) was packed with a 5 µm Symmetry C18 material (Waters). The RP analytical column (15 cm x 100 µm) was packed with a 1.7 µm Bridged Ethyl Hybrid (BEH) C18 material (Waters), and was used for the second dimension separation.
The peptides on the RP analytical column were eluted with a three-step linear gradient. Starting from 5% B to 40% B in 40 min (A: water with 0.1% formic acid; B: ACN with 0.1% formic acid), increased to 80% B in 3 min, and then to 5% B in 2 min. The column was re-equilibrated at initial conditions for 15 min. The column flow rate was maintained at 500 nl/min and column temperature was maintained at 35°C. The electrospray voltage of 1.9 kV versus the inlet of the mass spectrometer was used.
LTQ Orbitrap XL mass spectrometer was operated in the data-dependent mode to switch automatically between MS and MS/MS acquisition. Survey full-scan MS spectra with two microscans (m/z 300–1800) were acquired in the Obitrap with a mass resolution of 60,000 at m/z 400, followed by ten sequential LTQ-MS/MS scans. Dynamic exclusion was used with two repeat counts, 10 s repeat duration, and 90 s exclusion duration. For MS/MS, precursor ions were activated using 35% normalized collision energy at the default activation q of 0.25.
The 2D-LC-MS/MS experiment was repeat three times for cancer sample and corresponding adjacent noncancerous sample, respectively.
Peptide sequencing and data analysis
All MS/MS spectrums were identified by using SEQUEST [v.28 (revision 12), Thermo Electron Corp.] against the human UniProtKB/Swiss-Prot database (Release 2011_12_14, with 20249 entries), as previously described [12]. To reduce false positive identification results, a decoy database containing the reverse sequences was appended to the database. The searching parameters were set up as follows: full trypsin cleavage with two missed cleavage was considered, the variable modification was oxidation of methionine, the peptide mass tolerance was 20 ppm, and the fragment ion tolerance was 1 Da. Trans Proteomic Pipeline software (revision 4.0)(Institute of Systems Biology, Seattle, WA) was then utilized to identify proteins based upon corresponding peptide sequences with ≥95% confidence. The peptides results were filtered by Peptide Prophet with a p-value over 0.90 and a Protein Prophet probability of 0.95 was used for the protein identification results. Employing the APEX tool to quantified the protein abundances, the abundances estimated by normalizing for the measured total protein concentration. The false positive rate of less than 1% was set for all peptide identifications.
Bioinformatics analysis
The original data were derived from analysis using APEX software. Differentially expressed proteins were screened using the 2-sample t-test (P<0.05) and fold change (>1.5 or <0.667) method. All expression values of the differentially expressed proteins were first converted to a log form and then input as hierarchical clustering algorithms, where the Euclidean distance was used for distance and average for linkage for GO analysis. Differentially expressed genes were mapped to the appropriate GO database to calculate the number of genes at each node, using EASE software. The differentially expressed genes were classified according to bp (biologic process), cc (cellular component), and mf (molecular function) independently. In protein network analysis, interactions between genes in the range of the genomes analyzed were analyzed by downloading the pathway data in KEGG, MIPS, PubMed, MINT, Human Protein Reference Database (HPRD), BioGRID, Database of Interacting Proteins (DIP), and Reactome, using the BIND software package. Interrelationships between genes that had been reported in the literature were analyzed by co-citation calculation. The established gene network was able to directly reflect the interrelationships between genes at an overall level as well as the stability of the gene regulatory network.
Cell line and culture
The human laryngeal carcinoma cell line Hep-2 was obtained from the cell bank of the Shanghai Institute of Cell Biology (Shanghai, China). The cells were maintained in RPMI 1640 upplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin sulphate, and 1 mM sodium pyruvate at 37°C in 5% CO2.
siRNAs preparation and transfection
The siRNAs were chemically synthesised by Shanghai GenePharma Co., Ltd.. The siRNA sequences for PFN1 were previously described [19]: siRNA-PFN1: 5′- AGA AGG UGU CCA CGG UGG UUU -3′ (forward) and 5′- ACC ACC GUG GAC ACC UUC UUU -3′ (reverse). The negative control siRNAs were designed as follows: 5′-UAG CGA CUA AAC ACA UCA AUU-3′ (forward) and 5′-UUG AUG UGU UUA GUC GCU AUU-3′ (reverse). According to the manufacturer's specifications, the transfections of siRNA were carried out with Lipo2000 (Invitrogen) in 6-well plates. Until reached 50–70% confluence, the Hep-2 cells were transfected with 20 nM of siRNA for 6–12 h, and then replaced with the regular growth media. And cells were cultured for another 24–72 h before performing the experiments.
Semiquantitative RT-PCR
The total RNA was isolated from frozen tissues, and cells were extracted using TRIzol reagent (Takara). Two microgram of total RNA was used for cDNA synthesis using the RevertAidtm First Strand cDNA Synthesis Kit #1622 (Fermentas) according to the manufacturer's instructions. The primer sequences and the expected sizes of PCR products were as follows: PFN1, 5′-ATC GAC AAC CTC ATG GCG GAC G-3′(forward) and 5′-TTG CCA ACC AGG ACA CCC ACC T-3′(reverse) (140 bp); NCL, 5′-GAA AGC GTT GGA ACT CAC-3′(forward) and 5′-AAG TGT TCT CGC ATC TCG-3′(reverse) (103 bp); CNDP2, 5′-AAC TCA GGC CCT CCC TCT GTT GT-3′(forward) and 5′-GCT CCA GGA AGT GAC TGC GGC-3′(reverse) (146 bp); OGN, 5′- GTT GAC ATT GAT GCT GTA CCA CCC-3′(forward) and 5′-GCT TGG GAG GAA GAA CTG GA-3′(reverse) (241 bp). GAPDH, 5′-CAA GGT CAT CCA TGA CAA CTT TG-3′ (forward) and 5′-GTC CAC CAC CCT GTT GCT GTA G-3′(reverse) (496 bp). The PCR conditions used for the amplification were as follows: 94°C for 5 min, then 30 cycles of 94°C for 20 s, 55–60°C for 20 s, and 72°C for 30 s, followed by 72°C for 10 min. The RT-PCR products were analysed on a 1% agarose gel and visualised with ethidium bromide staining. The GAPDH gene was used as a positive control to assess the cDNA quality.
Cell proliferation assay
Cells (1×104/ml) were plated in 96-well plates. At 24, 48, and 72 h post-transfection with PFN1 siRNA, the cell viability was determined by cell counting kit-8 (CCK-8) assay (Dojindo) according to the manufacture's protocol.
Transwell assay
Transwell assay was performed using polycarbonate transwell filters (Corning, 8 µm) as previously described [20]. Briefly, at 12 h posttransfection, a sample of 0.8×105 cells were suspended in medium containing 1% FBS and added to the upper chamber. And the bottom chambers were filled with culture medium containing 20% FBS. After incubation for 24 h, the cells on the upper surface of the well were removed, and the cells on the lower surface were fixed in cold methanol and stained with 0.4% crystal violet (Sigma). For each experiment, the number of transmigrated cells in five random fields on the underside of the filter was counted and photographed, and three independent filters were analysed.
Western blotting
Whole-cell lysates were prepared from human tissue specimens and treated cells. For Western blotting analysis, equal amounts of proteins were separated using SDS-PAGE and transferred to a nitrocellulose membrane and then incubated with monoclonal antibody anti-PFN1 (Epitomics), monoclonal antibody anti-NCL (Santa Cruz), polyclonal antibody anti-CNDP2 (Proteintech), polyclonal antibody anti-OGN (Abgent), or monoclonal antibody anti-GAPDH (Bioworld) at 4°C overnight. The immunocomplexes were visualised using a horseradish peroxidase-conjugated antibody followed by a chemoluminescence reagent (Millipore) and detected on photographic film.
Statistical analysis
The data was expressed as the mean ± SD. All calculations were performed with SPSS version 11.7. The statistical analyses were performed with Student's t-test and analysis of variance. Multiple groups comparison in other assays was performed by one-way ANOVA.
All P values were two tailed, and <0.05 was considered statistically significant.
Results
Screening for differentially expressed proteins
Using APEX software, the original data were analyzed. Three independent experiments were performed in the laryngeal carcinoma (C) and the corresponding adjacent noncancerous (P) samples pools, respectively. According to the stringent criteria of having >1 unique peptide per protein present and a false discovery rate of ≤5%, 1,738 proteins were identified from the two sample pools. Following the statistical Student's 2-sample t-test analysis and the Fold change (C/P) methods, 141 proteins were significantly up-regulated using the criteria of P<0.05 and fold change >1.5, and 140 proteins were significantly down-regulated by P<0.05 and fold change <0.667 (Table 2). The expression values of the expressed proteins with significant difference were first converted to a log form and then input as hierarchical cluster algorithms. The results are shown in Figure 1.
Table 2. Differentially expressed proteins screened out compared the laryngeal carcinoma tissues (C) with the corresponding adjacent noncancerous tissues (P).
| Uniprot ID | Identified Proteins | Gene name | Fold change (C/P) | T test |
| P29508 | Serpin B3 | SERPINB3 | 15.30 | 0.00677 |
| P53634 | Dipeptidyl-peptidase 1 | CTSC | 13.70 | 0.00474 |
| P02792 | Ferritin light chain | FTL | 9.91 | 0.01259 |
| P04899 | Guanine nucleotide-binding protein G(i), alpha-2 subunit | GNAI2 | 9.77 | 0.00089 |
| Q15181 | Inorganic pyrophosphatase | PPA1 | 8.51 | 0.01315 |
| P19971 | Thymidine phosphorylase | TYMP | 8.06 | 0.00101 |
| P40227 | T-complex protein 1 subunit zeta | CCT6A | 7.86 | 0.00698 |
| P59998 | Actin-related protein 2/3 complex subunit 4 | ARPC4 | 7.64 | 0.00176 |
| P50552 | Vasodilator-stimulated phosphoprotein | VASP | 7.25 | 0.01336 |
| P13797 | Plastin-3 | PLS3 | 6.52 | 0.00256 |
| P09467 | Fructose-1,6-bisphosphatase 1 | FBP1 | 6.37 | 0.04326 |
| O00299 | Chloride intracellular channel protein 1 | CLIC1 | 5.91 | 0.00516 |
| P52895 | Aldo-keto reductase family 1 member C2 | AKR1C2 | 5.82 | 0.0004 |
| O15533 | Tapasin | TAPBP | 5.81 | 0.01439 |
| Q16630 | Cleavage and polyadenylation specificity factor subunit 6 | CPSF6 | 5.76 | 0.04131 |
| P54578 | Ubiquitin carboxyl-terminal hydrolase 14 | USP14 | 5.47 | 0.01414 |
| P07737 | Profilin-1 | PFN1 | 5.20 | 0.00692 |
| P37837 | Transaldolase | TALDO1 | 5.12 | 0.0124 |
| P31939 | Bifunctional purine biosynthesis protein PURH | ATIC | 5.05 | 0.01149 |
| P35637 | RNA-binding protein FUS | FUS | 5.02 | 0.01108 |
| P47929 | Galectin-7 | LGALS7 | 4.92 | 0.00664 |
| O00764 | Pyridoxal kinase | PDXK | 4.91 | 0.0044 |
| P23141 | Liver carboxylesterase 1 | CES1 | 4.86 | 0.01626 |
| A6NIZ1 | Ras-related protein Rap-1b | RAP1B | 4.78 | 0.00595 |
| P42224 | Signal transducer and activator of transcription 1-alpha/beta | STAT1 | 4.60 | 0.00168 |
| P55209 | Nucleosome assembly protein 1-like 1 | NAP1L1 | 4.41 | 0.02318 |
| O14979 | Heterogeneous nuclear ribonucleoprotein D-like | HNRPDL | 4.34 | 0.03791 |
| P11413 | Glucose-6-phosphate 1-dehydrogenase | G6PD | 4.30 | 0.01466 |
| P58107 | Epiplakin | EPPK1 | 4.30 | 0.0157 |
| Q96AB3 | Isochorismatase domain-containing protein 2, mitochondrial | ISOC2 | 4.26 | 0.01074 |
| P28838 | Cytosol aminopeptidase | LAP3 | 4.25 | 0.01448 |
| O75569 | Interferon-inducible double stranded RNA-dependent protein kinase activator A | PRKRA | 4.21 | 0.04359 |
| Q12874 | Splicing factor 3A subunit 3 | SF3A3 | 4.18 | 0.01359 |
| Q96HE7 | ERO1-like protein alpha | ERO1L | 4.09 | 0.02022 |
| O60664 | Mannose-6-phosphate receptor-binding protein 1 | M6PRBP1 | 3.91 | 0.00369 |
| P99999 | Cytochrome c | CYCS | 3.91 | 0.00451 |
| P19338 | Nucleolin | NCL | 3.90 | 0.02815 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | IDH1 | 3.81 | 0.01901 |
| P23246 | Splicing factor, proline- and glutamine-rich | SFPQ | 3.67 | 0.00085 |
| Q01518 | Adenylyl cyclase-associated protein 1 | CAP1 | 3.61 | 0.01283 |
| Q07666 | KH domain-containing, RNA-binding, signal transduction-associated protein 1 | KHDRBS1 | 3.55 | 0.0109 |
| Q96KP4 | Cytosolic non-specific dipeptidase | CNDP2 | 3.50 | 0.01266 |
| P30043 | Flavin reductase | BLVRB | 3.46 | 0.02879 |
| P05164 | Myeloperoxidase | MPO | 3.44 | 0.00316 |
| P36871 | Phosphoglucomutase-1 | PGM1 | 3.44 | 0.03173 |
| P50995 | Annexin A11 | ANXA11 | 3.43 | 0.03113 |
| P02786 | Transferrin receptor protein 1 | TFRC | 3.40 | 0.04239 |
| Q16658 | Fascin | FSCN1 | 3.36 | 0.01824 |
| P63104 | 14-3-3 protein zeta/delta | YWHAZ | 3.35 | 0.01256 |
| P00491 | Purine nucleoside phosphorylase | NP | 3.32 | 0.00487 |
| Q9UNM6 | 26S proteasome non-ATPase regulatory subunit 13 | PSMD13 | 3.28 | 0.04739 |
| P68104 | Putative elongation factor 1-alpha-like 3 | EEF1AL3 | 3.28 | 0.00105 |
| Q13347 | Eukaryotic translation initiation factor 3 subunit I | EIF3I | 3.20 | 0.01014 |
| Q96FQ6 | Protein S100-A16 | S100A16 | 3.17 | 0.01151 |
| P52565 | Rho GDP-dissociation inhibitor 1 | ARHGDIA | 3.15 | 0.00299 |
| Q99715 | Collagen alpha-1(XII) chain | COL12A1 | 3.12 | 0.02148 |
| P29401 | Transketolase | TKT | 3.09 | 0.00809 |
| P53582 | Methionine aminopeptidase 1 | METAP1 | 3.08 | 0.00304 |
| P52209 | 6-phosphogluconate dehydrogenase, decarboxylating | PGD | 2.96 | 0.03408 |
| P17096 | High mobility group protein HMG-I/HMG-Y | HMGA1 | 2.95 | 0.02965 |
| P13693 | Translationally-controlled tumor protein | TPT1 | 2.88 | 0.02137 |
| Q01469 | Fatty acid-binding protein, epidermal | FABP5 | 2.86 | 0.03051 |
| Q14974 | Importin subunit beta-1 | KPNB1 | 2.84 | 0.01796 |
| P31949 | Protein S100-A11 | S100A11 | 2.82 | 0.01872 |
| P02144 | Myoglobin | MB | 2.81 | 0.01049 |
| Q16543 | Hsp90 co-chaperone Cdc37 | CDC37 | 2.77 | 0.01228 |
| P50914 | 60S ribosomal protein L14 | RPL14 | 2.75 | 0.00198 |
| Q9Y490 | Talin-1 | TLN1 | 2.67 | 0.0068 |
| P06733 | Alpha-enolase | ENO1 | 2.64 | 0.01238 |
| P24821 | Tenascin | TNC | 2.63 | 0.01489 |
| P37802 | Transgelin-2 | TAGLN2 | 2.57 | 0.03438 |
| P61160 | Actin-related protein 2 | ACTR2 | 2.57 | 0.00239 |
| P30838 | Aldehyde dehydrogenase, dimeric NADP-preferring | ALDH3A1 | 2.56 | 0.03337 |
| P13010 | ATP-dependent DNA helicase 2 subunit 2 | XRCC5 | 2.55 | 0.00755 |
| P30085 | UMP-CMP kinase | CMPK1 | 2.55 | 0.0115 |
| Q9UN86 | Ras GTPase-activating protein-binding protein 2 | G3BP2 | 2.55 | 0.01204 |
| P18206 | Vinculin | VCL | 2.53 | 0.02193 |
| Q92616 | Translational activator GCN1 | GCN1L1 | 2.53 | 0.02057 |
| P05198 | Eukaryotic translation initiation factor 2 subunit 1 | EIF2S1 | 2.50 | 0.03971 |
| O00303 | Eukaryotic translation initiation factor 3 subunit F | EIF3F | 2.49 | 0.01838 |
| P09110 | 3-ketoacyl-CoA thiolase, peroxisomal | ACAA1 | 2.49 | 0.00319 |
| P09651 | Heterogeneous nuclear ribonucleoprotein A1 | HNRNPA1 | 2.49 | 0.04643 |
| P63241 | Eukaryotic translation initiation factor 5A-1 | EIF5A | 2.48 | 0.00699 |
| P16401 | Histone H1.5 | HIST1H1B | 2.47 | 0.02978 |
| P60842 | Eukaryotic initiation factor 4A-I | EIF4A1 | 2.47 | 0.02891 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 2.43 | 0.01183 |
| P07195 | L-lactate dehydrogenase B chain | LDHB | 2.41 | 0.0067 |
| Q12931 | Heat shock protein 75 kDa, mitochondrial | TRAP1 | 2.40 | 0.00724 |
| Q99832 | T-complex protein 1 subunit eta | CCT7 | 2.40 | 0.00158 |
| P18669 | Phosphoglycerate mutase 1 | PGAM1 | 2.39 | 0.02256 |
| Q07960 | Rho GTPase-activating protein 1 | ARHGAP1 | 2.37 | 0.00799 |
| P60174 | Triosephosphate isomerase | TPI1 | 2.36 | 0.00203 |
| P13639 | Elongation factor 2 | EEF2 | 2.35 | 0.00292 |
| P26641 | Elongation factor 1-gamma | EEF1G | 2.34 | 0.00212 |
| Q86VP6 | Cullin-associated NEDD8-dissociated protein 1 | CAND1 | 2.29 | 0.00207 |
| P38606 | V-type proton ATPase catalytic subunit A | ATP6V1A | 2.26 | 0.03249 |
| Q15691 | Microtubule-associated protein RP/EB family member 1 | MAPRE1 | 2.23 | 0.03292 |
| P62244 | 40S ribosomal protein S15a | RPS15A | 2.19 | 0.01589 |
| P69905 | Hemoglobin subunit alpha | HBA1 | 2.19 | 0.00953 |
| Q07021 | Complement component 1 Q subcomponent-binding protein, mitochondrial | C1QBP | 2.17 | 0.03131 |
| P02751 | Fibronectin | FN1 | 2.16 | 0.01678 |
| P09211 | Glutathione S-transferase P | GSTP1 | 2.15 | 0.02025 |
| P23381 | Tryptophanyl-tRNA synthetase, cytoplasmic | WARS | 2.13 | 0.00901 |
| P26599 | Polypyrimidine tract-binding protein 1 | PTBP1 | 2.13 | 0.03262 |
| P29692 | Elongation factor 1-delta | EEF1D | 2.13 | 0.0437 |
| P14618 | Pyruvate kinase isozymes M1/M2 | PKM2 | 2.13 | 0.00143 |
| P11940 | Polyadenylate-binding protein 1 | PABPC1 | 2.12 | 0.02495 |
| P13667 | Protein disulfide-isomerase A4 | PDIA4 | 2.11 | 0.01605 |
| P13796 | Plastin-2 | LCP1 | 2.11 | 0.00619 |
| P09960 | Leukotriene A-4 hydrolase | LTA4H | 2.10 | 0.01662 |
| Q15149 | Plectin-1 | PLEC1 | 2.07 | 0.00424 |
| Q13011 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | ECH1 | 2.06 | 0.01475 |
| P04632 | Calpain small subunit 1 | CAPNS1 | 2.06 | 0.03241 |
| P00918 | Carbonic anhydrase 2 | CA2 | 2.05 | 0.0467 |
| P40616 | ADP-ribosylation factor-like protein 1 | ARL1 | 2.02 | 0.01116 |
| P40939 | Trifunctional enzyme subunit alpha, mitochondrial | HADHA | 2.01 | 0.00595 |
| P23528 | Cofilin-1 | CFL1 | 2.01 | 0.02903 |
| Q8NBS9 | Thioredoxin domain-containing protein 5 | TXNDC5 | 1.93 | 0.00024 |
| P52566 | Rho GDP-dissociation inhibitor 2 | ARHGDIB | 1.92 | 0.04385 |
| Q14103 | Heterogeneous nuclear ribonucleoprotein D0 | HNRNPD | 1.92 | 0.03422 |
| Q08211 | ATP-dependent RNA helicase A | DHX9 | 1.91 | 0.01272 |
| Q8TE68 | Epidermal growth factor receptor kinase substrate 8-like protein 1 | EPS8L1 | 1.91 | 0.02329 |
| O15372 | Eukaryotic translation initiation factor 3 subunit H | EIF3H | 1.85 | 0.01438 |
| P07858 | Cathepsin B | CTSB | 1.84 | 0.00487 |
| P17655 | Calpain-2 catalytic subunit | CAPN2 | 1.84 | 0.03102 |
| P10809 | 60 kDa heat shock protein, mitochondrial | HSPD1 | 1.82 | 0.02513 |
| P51970 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | NDUFA8 | 1.82 | 0.04023 |
| P14625 | Endoplasmin | HSP90B1 | 1.82 | 0.0194 |
| P30153 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | PPP2R1A | 1.81 | 0.0417 |
| O75955 | Flotillin-1 | FLOT1 | 1.78 | 0.00694 |
| Q02878 | 60S ribosomal protein L6 | RPL6 | 1.78 | 0.01691 |
| O75083 | WD repeat-containing protein 1 | WDR1 | 1.75 | 0.01675 |
| P12268 | Inosine-5′-monophosphate dehydrogenase 2 | IMPDH2 | 1.75 | 0.0418 |
| P24158 | Myeloblastin | PRTN3 | 1.74 | 0.01096 |
| Q7KZF4 | Staphylococcal nuclease domain-containing protein 1 | SND1 | 1.73 | 0.00169 |
| P00558 | Phosphoglycerate kinase 1 | PGK1 | 1.72 | 0.00095 |
| P60660 | Myosin light polypeptide 6 | MYL6 | 1.70 | 0.02066 |
| Q13200 | 26S proteasome non-ATPase regulatory subunit 2 | PSMD2 | 1.69 | 0.04337 |
| P61019 | Ras-related protein Rab-2A | RAB2A | 1.65 | 0.0369 |
| Q92597 | Protein NDRG1 | NDRG1 | 1.61 | 0.00689 |
| P02768 | Serum albumin | ALB | 1.57 | 0.00957 |
| O00231 | 26S proteasome non-ATPase regulatory subunit 11 | PSMD11 | 1.56 | 0.02677 |
| Q9P2E9 | Ribosome-binding protein 1 | RRBP1 | 0.66 | 0.02565 |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein | HSPG2 | 0.65 | 0.02063 |
| P06576 | ATP synthase subunit beta, mitochondrial | ATP5B | 0.65 | 0.04632 |
| Q9ULV4 | Coronin-1C | CORO1C | 0.58 | 0.04756 |
| P29966 | Myristoylated alanine-rich C-kinase substrate | MARCKS | 0.58 | 0.01746 |
| P62861 | 40S ribosomal protein S30 | FAU | 0.57 | 0.00947 |
| P06396 | Gelsolin | GSN | 0.56 | 0.04161 |
| P02652 | Apolipoprotein A-II | APOA2 | 0.56 | 0.04 |
| P02763 | Alpha-1-acid glycoprotein 1 | ORM1 | 0.55 | 0.00937 |
| P31146 | Coronin-1A | CORO1A | 0.55 | 0.034 |
| P11047 | Laminin subunit gamma-1 | LAMC1 | 0.54 | 0.02215 |
| P20700 | Lamin-B1 | LMNB1 | 0.53 | 0.03269 |
| P10412 | Histone H1.2 | HIST1H1C | 0.53 | 0.01299 |
| P14866 | Heterogeneous nuclear ribonucleoprotein L | HNRNPL | 0.53 | 0.00906 |
| P09622 | Dihydrolipoyl dehydrogenase, mitochondrial | DLD | 0.53 | 0.00322 |
| P14923 | Junction plakoglobin | JUP | 0.52 | 0.01465 |
| O95994 | Anterior gradient protein 2 homolog | AGR2 | 0.52 | 0.04294 |
| P49748 | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADVL | 0.52 | 0.03597 |
| P08603 | Complement factor H | CFH | 0.52 | 0.01851 |
| Q16891 | Mitochondrial inner membrane protein | IMMT | 0.51 | 0.03273 |
| P05091 | Aldehyde dehydrogenase, mitochondrial | ALDH2 | 0.51 | 0.02974 |
| P00505 | Aspartate aminotransferase, mitochondrial | GOT2 | 0.50 | 0.00699 |
| Q13813 | Spectrin alpha chain, brain | SPTAN1 | 0.50 | 0.00471 |
| P11216 | Glycogen phosphorylase, brain form | PYGB | 0.50 | 0.02901 |
| Q6YN16 | Hydroxysteroid dehydrogenase-like protein 2 | HSDL2 | 0.50 | 0.00185 |
| P10155 | 60 kDa SS-A/Ro ribonucleoprotein | TROVE2 | 0.50 | 0.03947 |
| P30084 | Enoyl-CoA hydratase, mitochondrial | ECHS1 | 0.50 | 0.0033 |
| P19823 | Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2 | 0.49 | 0.02225 |
| P30048 | Thioredoxin-dependent peroxide reductase, mitochondrial | PRDX3 | 0.49 | 0.01532 |
| P08727 | Keratin, type I cytoskeletal 19 | KRT19 | 0.49 | 0.01893 |
| Q9UHG3 | Prenylcysteine oxidase 1 | PCYOX1 | 0.49 | 0.02023 |
| P27635 | 60S ribosomal protein L10 | RPL10 | 0.49 | 0.02615 |
| P27824 | Calnexin | CANX | 0.49 | 0.04302 |
| Q15582 | Transforming growth factor-beta-induced protein ig-h3 | TGFBI | 0.49 | 0.00143 |
| P01024 | Complement C3 | C3 | 0.49 | 0.00256 |
| P00488 | Coagulation factor XIII A chain | F13A1 | 0.49 | 0.01016 |
| P02747 | Complement C1q subcomponent subunit C | C1QC | 0.48 | 0.04694 |
| Q01082 | Spectrin beta chain, brain 1 | SPTBN1 | 0.48 | 0.01722 |
| P39656 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase 48 kDa subunit | DDOST | 0.48 | 0.04726 |
| P04080 | Cystatin-B | CSTB | 0.47 | 0.04055 |
| P01023 | Alpha-2-macroglobulin | A2M | 0.47 | 0.02189 |
| Q9NSE4 | Isoleucyl-tRNA synthetase, mitochondrial | IARS2 | 0.46 | 0.04337 |
| P36269 | Gamma-glutamyltransferase 5 | GGT5 | 0.46 | 0.00259 |
| P21810 | Biglycan | BGN | 0.46 | 0.00989 |
| P31040 | Succinate dehydrogenase flavoprotein subunit, mitochondrial | SDHA | 0.45 | 0.00591 |
| P02788 | Lactotransferrin | LTF | 0.45 | 0.02393 |
| P62158 | Calmodulin | CALM1 | 0.44 | 0.01293 |
| P01857 | Ig gamma-1 chain C region | IGHG1 | 0.43 | 0.00036 |
| P22695 | Cytochrome b-c1 complex subunit 2, mitochondrial | UQCRC2 | 0.43 | 0.00169 |
| P46781 | 40S ribosomal protein S9 | RPS9 | 0.43 | 0.04449 |
| Q02218 | 2-oxoglutarate dehydrogenase E1 component, mitochondrial | OGDH | 0.43 | 0.01132 |
| P24539 | ATP synthase subunit b, mitochondrial | ATP5F1 | 0.42 | 0.01379 |
| P17931 | Galectin-3 | LGALS3 | 0.42 | 0.00253 |
| P01009 | Alpha-1-antitrypsin | SERPINA1 | 0.41 | 0.00246 |
| P00738 | Haptoglobin | HP | 0.41 | 0.0076 |
| P62280 | 40S ribosomal protein S11 | RPS11 | 0.41 | 0.04435 |
| P43304 | Glycerol-3-phosphate dehydrogenase, mitochondrial | GPD2 | 0.41 | 0.02182 |
| O60716 | Catenin delta-1 | CTNND1 | 0.41 | 0.01993 |
| Q96IU4 | Abhydrolase domain-containing protein 14B | ABHD14B | 0.40 | 0.04326 |
| Q14152 | Eukaryotic translation initiation factor 3 subunit A | EIF3A | 0.40 | 0.04596 |
| P51888 | Prolargin | PRELP | 0.40 | 0.0218 |
| P02511 | Alpha-crystallin B chain | CRYAB | 0.40 | 0.02841 |
| Q8NCW5 | Apolipoprotein A-I-binding protein | APOA1BP | 0.39 | 0.014 |
| P84098 | 60S ribosomal protein L19 | RPL19 | 0.39 | 0.01034 |
| O75306 | NADH dehydrogenase iron-sulfur protein 2, mitochondrial | NDUFS2 | 0.39 | 0.0012 |
| P07099 | Epoxide hydrolase 1 | EPHX1 | 0.39 | 0.01386 |
| P49755 | Transmembrane emp24 domain-containing protein 10 | TMED10 | 0.39 | 0.03179 |
| P60709 | Actin, cytoplasmic 2 | ACTG1 | 0.39 | 0.00425 |
| P00450 | Ceruloplasmin | CP | 0.38 | 0.01124 |
| P21796 | Voltage-dependent anion-selective channel protein 1 | VDAC1 | 0.38 | 0.02369 |
| P02545 | Lamin-A/C | LMNA | 0.38 | 0.00148 |
| P39059 | Collagen alpha-1(XV) chain | COL15A1 | 0.38 | 0.02395 |
| P63167 | Dynein light chain 1, cytoplasmic | DYNLL1 | 0.38 | 0.00646 |
| Q14134 | Tripartite motif-containing protein 29 | TRIM29 | 0.38 | 0.00844 |
| P51571 | Translocon-associated protein subunit delta | SSR4 | 0.37 | 0.02087 |
| Q9UN36 | Protein NDRG2 | NDRG2 | 0.37 | 0.02335 |
| P01834 | Ig kappa chain C region | IGKC | 0.37 | 0.03372 |
| Q02790 | FK506-binding protein 4 | FKBP4 | 0.37 | 0.01019 |
| P04217 | Alpha-1B-glycoprotein | A1BG | 0.36 | 0.02191 |
| Q9BS26 | Thioredoxin domain-containing protein 4 | TXNDC4 | 0.36 | 0.02078 |
| P30040 | Endoplasmic reticulum protein ERp29 | ERP29 | 0.35 | 0.0485 |
| P12532 | Creatine kinase, ubiquitous mitochondrial | CKMT1A | 0.35 | 0.02472 |
| Q08380 | Galectin-3-binding protein | LGALS3BP | 0.35 | 0.00749 |
| P30049 | ATP synthase subunit delta, mitochondrial | ATP5D | 0.33 | 0.01374 |
| P63244 | Guanine nucleotide-binding protein subunit beta-2-like 1 | GNB2L1 | 0.33 | 0.002 |
| P35232 | Prohibitin | PHB | 0.33 | 0.01226 |
| Q01081 | Splicing factor U2AF 35 kDa subunit | U2AF1 | 0.31 | 0.01422 |
| Q9UQ80 | Proliferation-associated protein 2G4 | PA2G4 | 0.31 | 0.01201 |
| P02730 | Band 3 anion transport protein | SLC4A1 | 0.30 | 0.01198 |
| P31930 | Cytochrome b-c1 complex subunit 1, mitochondrial | UQCRC1 | 0.29 | 0.00459 |
| O75367 | Core histone macro-H2A.1 | H2AFY | 0.29 | 0.00555 |
| P11177 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | PDHB | 0.29 | 0.04049 |
| P24752 | Acetyl-CoA acetyltransferase, mitochondrial | ACAT1 | 0.28 | 0.01184 |
| P08294 | Extracellular superoxide dismutase [Cu-Zn] | SOD3 | 0.27 | 0.04222 |
| Q05707 | Collagen alpha-1(XIV) chain | COL14A1 | 0.27 | 0.00088 |
| P62826 | GTP-binding nuclear protein Ran | RAN | 0.27 | 0.03551 |
| Q99623 | Prohibitin-2 | PHB2 | 0.27 | 0.00683 |
| P04844 | Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit 2 | RPN2 | 0.26 | 0.02265 |
| Q12907 | Vesicular integral-membrane protein VIP36 | LMAN2 | 0.26 | 0.02661 |
| P04181 | Ornithine aminotransferase, mitochondrial | OAT | 0.26 | 0.01802 |
| P62750 | 60S ribosomal protein L23a | RPL23A | 0.25 | 0.0077 |
| P06732 | Creatine kinase M-type | CKM | 0.25 | 0.02901 |
| P14927 | Cytochrome b-c1 complex subunit 7 | UQCRB | 0.25 | 0.04749 |
| P51884 | Lumican | LUM | 0.25 | 0.0166 |
| Q9UH99 | Protein unc-84 homolog B | UNC84B | 0.24 | 0.01415 |
| P00403 | Cytochrome c oxidase subunit 2 | MT-CO2 | 0.24 | 0.00441 |
| P02675 | Fibrinogen beta chain | FGB | 0.24 | 0.01004 |
| P49257 | Protein ERGIC-53 | LMAN1 | 0.23 | 0.00243 |
| Q03252 | Lamin-B2 | LMNB2 | 0.23 | 0.00382 |
| P58546 | Myotrophin | MTPN | 0.23 | 0.03507 |
| P32969 | 60S ribosomal protein L9 | RPL9 | 0.22 | 0.03767 |
| O95299 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | NDUFA10 | 0.21 | 0.02844 |
| Q16795 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | NDUFA9 | 0.21 | 0.01163 |
| P04843 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 | RPN1 | 0.20 | 0.00436 |
| Q8TDL5 | Long palate, lung and nasal epithelium carcinoma-associated protein 1 | LPLUNC1 | 0.20 | 0.02273 |
| P20774 | Mimecan | OGN | 0.20 | 0.03045 |
| P02679 | Fibrinogen gamma chain | FGG | 0.19 | 0.00129 |
| P48047 | ATP synthase subunit O, mitochondrial | ATP5O | 0.19 | 0.01449 |
| P62841 | 40S ribosomal protein S15 | RPS15 | 0.19 | 0.02324 |
| Q92817 | Envoplakin | EVPL | 0.19 | 0.00422 |
| P09493 | Tropomyosin alpha-1 chain | TPM1 | 0.19 | 0.01514 |
| P19652 | Alpha-1-acid glycoprotein 2 | ORM2 | 0.18 | 0.00543 |
| Q9UIJ7 | GTP:AMP phosphotransferase mitochondrial | AK3 | 0.18 | 0.01683 |
| P00367 | Glutamate dehydrogenase 1, mitochondrial | GLUD1 | 0.18 | 0.01563 |
| P10916 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | MYL2 | 0.18 | 0.00277 |
| P00387 | NADH-cytochrome b5 reductase 3 | CYB5R3 | 0.17 | 0.00535 |
| Q9UI09 | NADH dehydrogenase 1 alpha subcomplex subunit 12 | NDUFA12 | 0.17 | 0.00843 |
| P32322 | Pyrroline-5-carboxylate reductase 1, mitochondrial | PYCR1 | 0.17 | 0.00997 |
| P45880 | Voltage-dependent anion-selective channel protein 2 | VDAC2 | 0.17 | 0.00812 |
| Q9BSJ8 | Extended synaptotagmin-1 | FAM62A | 0.17 | 0.02282 |
| P12883 | Myosin-7 | MYH7 | 0.15 | 0.04314 |
| P07585 | Decorin | DCN | 0.15 | 0.00903 |
| P45378 | Troponin T, fast skeletal muscle | TNNT3 | 0.14 | 0.03654 |
| Q07954 | Prolow-density lipoprotein receptor-related protein 1 | LRP1 | 0.13 | 0.01165 |
| P61626 | Lysozyme C | LYZ | 0.13 | 0.00319 |
| P20618 | Proteasome subunit beta type-1 | PSMB1 | 0.11 | 0.03038 |
| Q96A32 | Myosin regulatory light chain 2, skeletal muscle isoform | MYLPF | 0.09 | 0.00292 |
| P01876 | Ig alpha-1 chain C region | IGHA1 | 0.07 | 0.00055 |
| Q9BXN1 | Asporin | ASPN | 0.07 | 0.00292 |
| P35749 | Myosin-11 | MYH11 | 0.06 | 0.03254 |
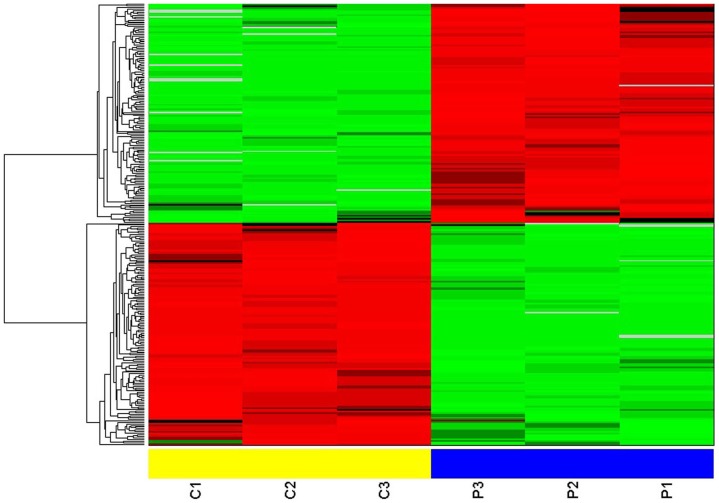
Figure 1. Hierarchical cluster analysis of the proteins expressed with statistically significant differences (P<0.05, and fold change >1.5 or <0.667) in cancer tissue and paracancerous normal tissue from patients with laryngeal carcinoma.
Three independent experiments were performed in cancer tissue (C1, C2, C3) and paracancerous normal tissue (P1, P2, P3).
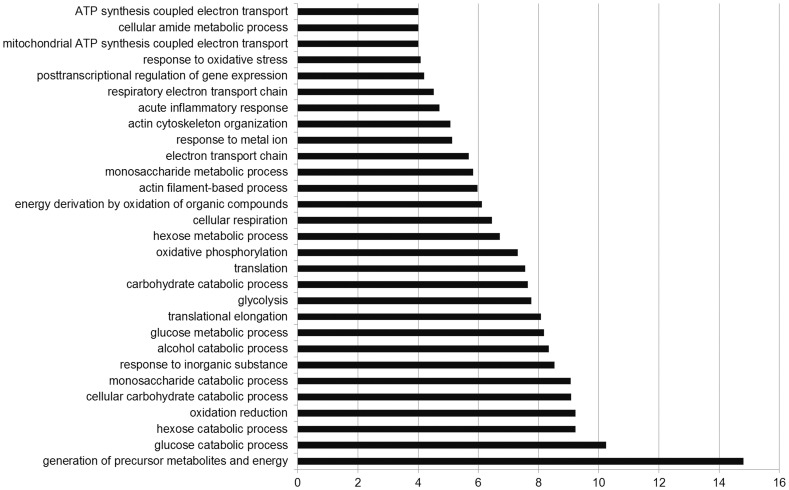
GO analysis of the proteins with significant difference in expression
To get more insight on the biological significance of the differentially expressed proteins in human laryngeal carcinoma, GO analysis was conducted on 281 differentially expressed proteins (Figure 2). According to biologic process analysis, it showed that each group was enriched with the proteins of different functions, suggesting that the differentially expressed proteins may play a distinctive role in human laryngeal carcinogenesis by these signaling pathways.
Figure 2. Gene ontology analysis of differentially expressed proteins classified according to biologic process.
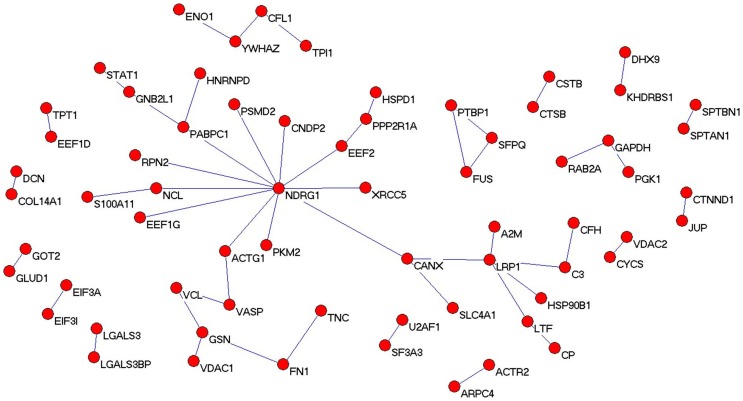
Analysis of the differential protein network
To identify the potential interrelationships between proteins expressed with significant difference, a protein–protein interaction network was built up with Pajek software. Consistently, the differential protein network was established by integrating three different types of interaction: 1) protein–protein interactions obtained in well established high-throughput experiments such as yeast 2-hybrid experiments; 2) gene interactions reported in the literature; and 3) protein interaction, gene regulation, and protein decoration. The results are shown in Figure 3. These proteins may have important roles in laryngeal carcinoma oncogenesis and progression, and their presence in this network diagram confirms the relevance of the differentially expressed proteins data set and their association to laryngeal carcinoma, in some way.
Figure 3. Protein network analysis.
The protein–protein interaction network of differential proteins is shown.
Validation of differentially expressed proteins indentified by proteomics
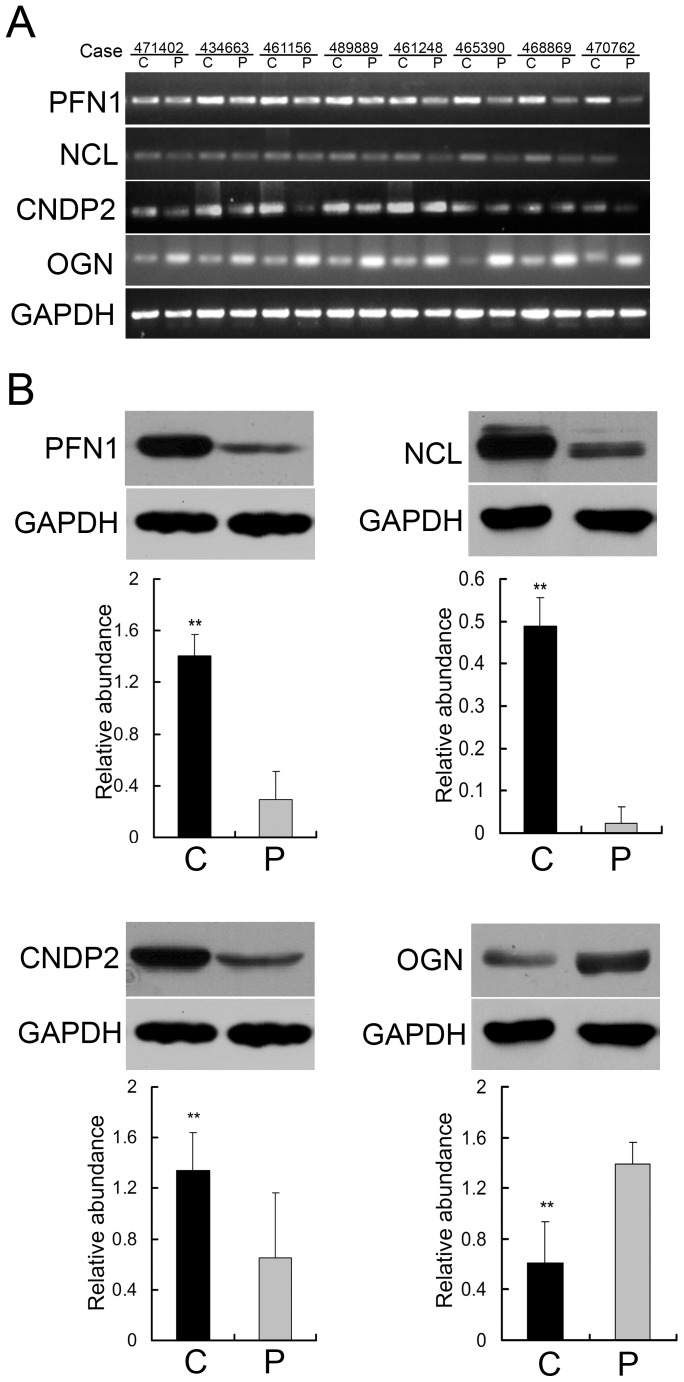
Next, semiquantitative RT-PCR was performed to detect the mRNA levels of PFN1, NCL, CNDP2 and OGN in 8 cases of paired laryngeal carcinoma tissues. As shown in Figure 4A, in most cases, PFN1, NCL and CNDP2 exerted an increased mRNA expression, while OGN displayed a decreased level in the carcinoma tissues compared with the adjacent normal tissue. The protein expression level of the four selected molecules was further investigated with 24 paired cases of laryngeal carcinoma and non-cancer tissue sections using Western blotting. And the results revealed that the expression of PFN1, NCL, and CNDP2 was elevated and that of OGN was reduced in laryngeal carcinoma tissue compared with the adjacent normal tissue (Figure 4B). Thus, these results validate the differentially expressed proteins indentified by the proteomics.
Figure 4. Validation of differentially expressed proteins in laryngeal carcinoma tissue and the adjacent normal tissue by semiquantitative RT-PCR and Western blotting.
(A) The representative image of mRNA levels of PFN1, NCL, CNDP2 and OGN between laryngeal carcinoma tissue and their corresponding normal tissue in 8 cases of tissues measured by semiquantitative RT-PCR. (B) The representative result of Western blotting show the expressions of PFN1, NCL, CNDP2 and OGN in the laryngeal carcinoma tissue and the adjacent normal tissue, respectively. Histograms are representative the relative abundance of proteins mean from 24 cases of tissues. (**P<0.01 by One-way ANOVA).
PFN1 silencing inhibits the proliferation and metastasis of the human laryngeal carcinoma Hep-2 cells
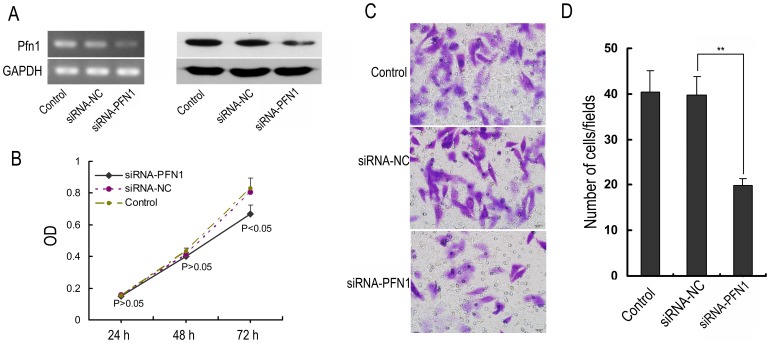
To know whether down-regulation of PFN1 is involved in laryngeal carcinoma carcinogenesis, Hep-2 cells were transfected with siRNA to specifically target PFN1 or negative control siRNA, and then the proliferation and metastasis of the transfected cells were measured. The transfection efficiency was confirmed by semiquantitative RT-PCR and Western blotting. The data showed that PFN1 expression in the siRNA-PFN1 group was reduced significantly at both the mRNA (24 h) and protein (48 h) levels when compared to the levels in the negative control siRNA and untreated control groups (Figure 5A). Next, the proliferation of siRNA-transfected Hep-2 cells was determined by CCK-8 assay. As shown in Figure 5B, within 48 hours, the percentages of viable cells were not significantly different in the PFN1 siRNA group when compared to the negative control siRNA and untreated control groups. And the cells of PFN1 siRNA group showed a significantly decreased proliferation at 72 h time points. Additionally, transwell assay was performed to further determine whether the downregulation of PFN1 could influence the migration ability of Hep-2 cells. As expectably, the numbers of cells in the siRNA-PFN1 group that migrated to the lower surfaces of the transwells were reduced in comparison to those in the negative control siRNA and untreated control groups (Figure 5C and D). These results indicate that PFN1 silencing affects the proliferation and migration ability of Hep-2 cells.
Figure 5. Effects of PFN1 silencing on the proliferation and metastasis of Hep-2 cells.
(A) The mRNA (24 h) and protein (48 h) expression of PFN1 after specific siRNA transfection in Hep-2 cells. The levels of mRNA and protein were determined by semiquantitative RT-PCR and Western blotting, respectively. (B) The cell viability of Hep-2 cells harvested 24, 48, and 72 h post-transfection after treatment with siPFN1. The optical density (OD) represents the proliferative characters of the treated cells. (C) The directed migratory capacities of Hep-2 cells after the siPFN1 transfected for 24 h were evaluated using a Transwell migration study. Images of cells on the undersurface of a filter are shown. Bar, 20 µm. (D) The number of cells per field in control and treated cells is shown. Values are the mean ± SD from three independent experiments. **P<0.01.
Discussion
Previous proteomics study using 2D-MS identified the differential proteins in laryngeal carcinoma tissue [17], few candidate proteins were detectable because of low resolution, and most of the differentially expressed proteins detected were high-abundance proteins. In addition, using SELDI-TOF-MS method to identify the proteomic shift in laryngeal carcinoma serum, Cheng et al and Liu group reported different findings and conclusions that few proteins were found to vary in concert, and the discrepancies might be due to their technical problems such as varying ability of mass spectrometry to identify a particular protein [14], [15]. Thus, successful application of proteomic technologies to biomedical and clinical research is leading to the discovery of disease-specific biomarkers for diagnosis and treatment monitoring, providing insight into the underlying pathologies and allowing identification of novel therapeutic targets [12]. In the current study, we used 2 chromatographic methods coupled with MS to detect differentially expressed proteins and thereby greatly raised the number of detectable proteins. The 2D LC-MS/MS analysis performed in this study led to the identification of 1738 proteins, among which 281 were differentially expressed with significance between the laryngeal carcinoma tissues and the corresponding adjacent noncancerous tissues. Of these, 141 proteins were upregulated, and the remaining 140 proteins were downregulated. To get more insight on the biological significance of the differentially expressed proteins in laryngeal carcinoma process, hierarchical cluster, gene ontology and protein network analysis were performed on 281 differential proteins. Stage-specific and coregulated expression profiles of the differentially expressed proteins were displayed in the hierarchical cluster analysis. GO analysis revealed that each functional group may play a distinctive role during laryngeal carcinoma carcinogenesis. Additionally, the network diagram confirmed the relevance of the differentially expressed proteins provided a handle by which to identify upstream activators and downstream effectors.
Considerable proteins, such as YWHAZ, S100-A11, glutathione S-transferase, alpha-enolase, flavin reductase, fascin, and carbonic anhydrase, have been reported to be associated with laryngeal carcinoma in previous proteomics studies but without clinical validation and in-depth functional research [17], [18]. However, in our present study, four of the altered expressed proteins with different subcellular localization, such as PFN1: extracellular, NCL: nucleolus, nucleus and cytoplasm, CNDP2: cytoplasm, and OGN: extracellular, have been observed to be differentially expressed in cancers from other origins but not previously in laryngeal carcinoma [21]–[24]. Meanwhile, these candidates have been proved to be involved in multiple cellular pathways related to carcinogenesis, including proliferation, differentiation, apoptosis, migration, and invasion. Thus, the expression of PFN1, NCL, CNDP2 and OGN were further investigated employing a large collection of human laryngeal carcinoma tissues. Noteworthy, the effects of PFN1 in the proliferation and migration of human squamous cells were also analysed.
Profilin-1(PFN1), as an important actin-binding protein and ubiquitously expressed profilin isoform, has been considered as an essential control element for actin polymerization by virtue of its ability to funnel actin monomers (G-actin) to the growing filament and interact with almost all major protein families, which involved in nucleation and/or elongation of actin filaments [25]. Deregulation of PFN1 has been reported in various adenocarcinomas (breast, pancreas, hepatic, and gastric), and indicating that the molecule may function as a tumor-suppressor gene [26]–[29]. Especially, PFN1 plays crucial roles in metastasis and carcinogenesis of mammary epithelial cells by regulating membrane protrusion, motility, and invasion [30]. To know whether down-regulation of PFN1 is involved in laryngeal carcinoma carcinogenesis, we knocked down PFN1 in human laryngeal carcinoma cells Hep-2, and then detected whether PFN1 knockdown decreased the proliferation and metastasis of Hep-2 cells. The data showed that PFN1 silencing inhibited the proliferation and affected the migration ability of Hep-2 cells, demonstrating that PFN1 plays an important role in human laryngeal carcinoma carcinogenesis. To our knowledge, this is the first report to establish a correlation between PFN1 down-regulation and carcinogenesis of human laryngeal carcinoma, and PFN1 as a potential biomarker for early detection of this cancer.
Nucleolin (NCL) is another protein found overexpressed in laryngeal carcinoma. As a multifunctional phosphoprotein, NCL has a bipartite nuclear localization signal sequence and binds RNA through its RNA recognition motifs [31]. It has been shown to be up-regulated in highly proliferative cells and regulated many aspects of DNA and RNA metabolism, chromatin structure, rRNA maturation, cytokinesis, nucleogenesis, cell proliferation and growth [32], [33]. Further, the expression of NCL was reported to be increased in pancreatic ductal adenocarcinoma and the overexpression of the protein was found in other human cancers such as gliomas, melanoma, and non-small cell lung cancer [34]–[37]. Similarly, our semiquantitative RT-PCR and Western blotting results confirmed on a larger series of specimens the increased expression of NCL in the laryngeal carcinoma, indicating the possibility that the overexpression of this protein is more specific to cancer.
Cytosolic non-specific dipeptidase 2 (CNDP2), also known as carboxypeptidase of glutamate-like (CPGL), is expressed in all human tissues [38]. Previously, Zhang et al observed that CNDP2 is downregulated in hepatocellular cancer and could inhibit the viability, colony formation, and invasion of hepatocellular carcinoma cells [23]. A recent report also demonstrated that the loss of CNDP2 functioned as a tumour suppressor gene in pancreatic cancer and that the loss of CNDP2 suppressed proliferation, induced G0/G1 accumulation, and inhibited the migration ability of a pancreatic cancer cell line [39]. However, not all tumours express a low CNDP2 level, and the molecular function of CNDP2 is largely unknown. Okamura et al showed through quantitative proteomic analysis that renal cell carcinoma tissues have a high level of CNDP2 expression [40]. Tripathi et al found that CNDP2 was up-regulated in breast cancer tissues compared with normal breast epithelium [41]. The discrepant expression of CNDP2 in different tumours may due to its tissue specificity. In consistent with the later researches, our proteomic investigation revealed the overexpression of CNDP2 in the laryngeal carcinoma, and providing the information that this protein might be an accessible biomarker for certain type of cancers.
Mimecan (OGN), a secretory protein, belongs to a family of small leucine-rich proteoglycans (SLRPs). The expression of OGN was absent in several cancer cell lines, implicating its potential role as a tumor suppressor gene in cancer biology, although its physiological function has not been fully elucidated [42]. Even though, various human diseases, such as primary open-angle glaucoma and pituitary tumors, have been reported to associate with the expression of OGN [43]. Concomitantly, the differential expression of this protein serves as an excellent pathological biomarker to distinguish non-small cell lung cancers from small cell lung cancers [44]. Here, our validation experiments demonstrated the significant down-expression of OGN in a large group of laryngeal carcinoma patients, hinting that OGN may be a potential tumour suppressor gene involved in laryngeal carcinoma initiation and progression.
Taken together, in this study, the use of 2D LC-MS/MS identified 281 significantly differentially expressed proteins in human laryngeal carcinoma, and four differential proteins (PFN1, NCL, CNDP2 and OGN) with expressional changes were selectively verified. It was showed that panel of the four proteins, or some of them, could serve as novel potential biomarkers for detection or therapeutic targets of human laryngeal carcinoma. Moreover, it was found that PFN1 knockdown decreased the metastasis of Hep-2 cells, demonstrating that PFN1 plays an important role in metastasis of laryngeal carcinoma. Thus, our findings reported here could have potential clinical value in diagnosis of human laryngeal carcinoma, and would provide some valuable information for further study of molecular mechanisms of this cancer.
Funding Statement
This study was supported by funds from the Shanghai Science and Technology Commission (13431900303, 13431900303); Shanghai Health and Family Planning Commission and Young Start-up Projects of Changzheng Hospital (2012CZQN07). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Parkin DM, Bray F, Ferlay J, Pisani P (2001) Estimating the world cancer burden: Globocan 2000. Int J Cancer 94: 153–156. [DOI] [PubMed] [Google Scholar]
- 2. Lefebvre JL (2006) Laryngeal preservation in head and neck cancer: multidisciplinary approach. Lancet Oncol 7: 747–755. [DOI] [PubMed] [Google Scholar]
- 3. Tomasino RM, Bazan V, Daniele E, Nuara R, Morello V, et al. (1996) Biological characterization of laryngeal squamous-cell carcinoma. Anticancer Res 16: 2257–2267. [PubMed] [Google Scholar]
- 4. Medina JE, Ferlito A, Robbins KT, Silver CE, Rodrigo JP, et al. (2011) Central compartment dissection in laryngeal cancer. Head Neck 33: 746–752. [DOI] [PubMed] [Google Scholar]
- 5. Zvrko E, Mikić A, Jancić S (2012) Relationship of E-cadherin with cervical lymph node metastasis in laryngeal cancer. Coll Antropol. 2012 Nov 36 Suppl 2119–24. [PubMed] [Google Scholar]
- 6. Herrmann PC, Liotta LA, Petricoin EF III (2001) Cancer proteomics: the state of the art. Dis Markers 17: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kavallaris M, Marshall GM (2005) Proteomics and disease: opportunities and challenges. Med J Aust 182: 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johann DJ Jr, McGuigan MD, Patel AR, Tomov S, Ross S, et al. (2004) Clinical proteomics and biomarker discovery. Ann N Y Acad Sci 1022: 295–305. [DOI] [PubMed] [Google Scholar]
- 9. Ma Y, Peng J, Liu W, Zhang P, Huang L, et al. (2009) Proteomics identification of desmin as a potential oncofetal diagnostic and prognostic biomarker in colorectal cancer. Mol Cell Proteomics 8: 1878–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeSouza LV, Grigull J, Ghanny S, Dubé V, Romaschin AD, et al. (2007) Endometrial carcinoma biomarker discovery and verification using differentially tagged clinical samples with multidimensional liquid chromatography and tandem mass spectrometry. Mol Cell Proteomics 6: 1170–1182. [DOI] [PubMed] [Google Scholar]
- 11. Zeng GQ, Zhang PF, Deng X, Yu FL, Li C, et al. (2012) Identification of candidate biomarkers for early detection of human lung squamous cell cancer by quantitative proteomics. Mol Cell Proteomics 11: M111.013946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang JG, Xu WD, Zhai WT, Li Y, Hu JW, et al. (2012) Disorders in angiogenesis and redox pathways are main factors contributing to the progression of rheumatoid arthritis: a comparative proteomics study. Arthritis Rheum 64: 993–1004. [DOI] [PubMed] [Google Scholar]
- 13. Moon PG, Kwack MH, Lee JE, Cho YE, Park JH, et al. (2013) Proteomic analysis of balding and non-balding mesenchyme-derived dermal papilla cells from androgenetic alopecia patients using on-line two-dimensional reversed phase-reversed phase LC-MS/MS. J Proteomics 85: 174–191. [DOI] [PubMed] [Google Scholar]
- 14. Liu C, Pan C, Wang H, Yong L (2011) Effect of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry on identifying biomarkers of laryngeal carcinoma. Tumour Biol 32: 1139–1145. [DOI] [PubMed] [Google Scholar]
- 15. Cheng L, Zhou L, Tao L, Zhang M, Cui J, et al. (2009) Preliminary study of proteomic shift from normal to premalignant laryngeal lesions and to laryngeal squamous cell carcinoma. Acta Otolaryngol 129: 774–778. [DOI] [PubMed] [Google Scholar]
- 16. Li ZB, Lehar M, Braga N, Westra W, Liu LH, et al. (2003) Study of human laryngeal muscle protein using two-dimensional electrophoresis and mass spectrometry. Proteomics 3: 1325–1334. [DOI] [PubMed] [Google Scholar]
- 17. Sewell DA, Yuan CX, Robertson E (2007) Proteomic signatures in laryngeal squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec 69: 77–84. [DOI] [PubMed] [Google Scholar]
- 18. Ralhan R, Desouza LV, Matta A, Chandra Tripathi S, Ghanny S, et al. (2008) Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol Cell Proteomics 7: 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding Z, Lambrechts A, Parepally M, Roy P (2006) Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J Cell Sci 119: 4127–4137. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Z, Zhang J, Miao L, Liu K, Yang S, et al. (2012) Interleukin-11 promotes the progress of gastric carcinoma via abnormally expressed versican. Int J Biol Sci 8: 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu N, Zhang W, Yang Y, Liang YL, Wang LY, et al. (2006) Profilin 1obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics 6: 6095–6106. [DOI] [PubMed] [Google Scholar]
- 22. Peng L, Liang J, Wang H, Song X, Rashid A, et al. (2010) High levels of nucleolar expression of nucleolin are associated with better prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin Cancer Res 16: 3734–3742. [DOI] [PubMed] [Google Scholar]
- 23. Zhang P, Chan DW, Zhu Y, Li JJ, Ng IO-L, et al. (2006) Identification of carboxypeptidase of glutamate like-B as a candidate suppressor in cell growth and metastasis in human hepatocellular carcinoma. Clinical cancer research 12: 6617–6625. [DOI] [PubMed] [Google Scholar]
- 24. Orr B, Riddick AC, Stewart GD, Anderson RA, Franco OE, et al. (2012) Identification of stromally expressed molecules in the prostate by tag-profiling of cancer-associated fibroblasts, normal fibroblasts and fetal prostate. Oncogene 31: 1130–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Witke W (2004) The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol 14: 461–469. [DOI] [PubMed] [Google Scholar]
- 26. Oien KA, Vass JK, Downie I, Fullarton G, Keith WN (2003) Profiling, comparison and validation of gene expression in gastric carcinoma and normal stomach. Oncogene 22: 4287–4300. [DOI] [PubMed] [Google Scholar]
- 27. Wu N, Zhang W, Yang Y, Liang YL, Wang LY, et al. (2006) Profilin 1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics 6: 6095–6106. [DOI] [PubMed] [Google Scholar]
- 28. Janke J, Schluter K, Jandrig B, Theile M, Kolble K, et al. (2000) Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin 1. J Exp Med 191: 1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, et al. (2006) Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics 5: 157–171. [DOI] [PubMed] [Google Scholar]
- 30. Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, et al. (2012) Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell 22: 615–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tajrishi MM, Tuteja R, Tuteja N (2011) Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol 4: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Angelov D, Bondarenko VA, Almagro S, Menoni H, Monge' lard F, et al. (2006) Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J 25: 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mi Y, Thomas SD, Xu X, Casson LK, Miller DM, et al. (2003) Apoptosis in leukemia cells is accompanied by alterations in the levels and localization of nucleolin. J Biol Chem 278: 8572–8579. [DOI] [PubMed] [Google Scholar]
- 34. Peng L, Liang J, Wang H, Song X, Rashid A, et al. (2010) High levels of nucleolar expression of nucleolin are associated with better prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin Cancer Res 16: 3734–3742. [DOI] [PubMed] [Google Scholar]
- 35. Galzio R, Rosati F, Benedetti E, Cristiano L, Aldi S, et al. (2012) Glycosilated nucleolin as marker for human gliomas. J Cell Biochem 113: 571–579. [DOI] [PubMed] [Google Scholar]
- 36. Mourmouras V, Cevenini G, Cosci E, Epistolato MC, Biagioli M, et al. (2009) Nucleolin protein expression in cutaneous melanocytic lesions. J Cutan Pathol 36: 637–646. [DOI] [PubMed] [Google Scholar]
- 37. Zhao H, Huang Y, Xue C, Chen Y, Hou X, et al. (2013) Prognostic significance of the combined score of endothelial expression of nucleolin and CD31 in surgically resected non-small cell lung cancer. PLoS One 8: e54674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Teufel M, Saudek V, Ledig JP, Bernhardt A, Boularand S, et al. (2003) Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J Biol Chem 278: 6521–6531. [DOI] [PubMed] [Google Scholar]
- 39. Lee JH, Giovannetti E, Hwang JH, Petrini I, Wang Q, et al. (2012) Loss of 18q22.3 involving the carboxypeptidase of glutamate-like gene is associated with poor prognosis in resected pancreatic cancer. Clin Cancer Res 18: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okamura N, Masuda T, Gotoh A, Shirakawa T, Terao S, et al. (2008) Quantitative proteomic analysis to discover potential diagnostic markers and therapeutic targets in human renal cell carcinoma. Proteomics 8: 3194–3203. [DOI] [PubMed] [Google Scholar]
- 41. Tripathi A, King C, De la Morenas A, Perry VK, Burke B, et al. (2008) Gene expression abnormalities in histologically normal breast epithelium of breast cancer patients. Int J Cancer 122: 1557–1566. [DOI] [PubMed] [Google Scholar]
- 42. Wang Y, Ma Y, Lü B, Xu E, Huang Q, et al. (2007) Differential expression of mimecan and thioredoxin domain-containing protein 5 in colorectal adenoma and cancer: a proteomic study. Exp Biol Med (Maywood) 232: 1152–1159. [DOI] [PubMed] [Google Scholar]
- 43. Hu SM, Li F, Yu HM, Li RY, Ma QY, et al. (2005) The mimecan gene expressed in human pituitary and regulated by pituitary transcription factor-1 as a marker for diagnosing pituitary tumors. J Clin Endocrinol Metab 90: 6657–6664. [DOI] [PubMed] [Google Scholar]
- 44. Zheng CX, Zhao SX, Wang P, Yu HM, Wang CF, et al. (2009) Different expression of mimecan as a marker for differential diagnosis between NSCLC and SCLC. Oncol Rep 22: 1057–1061. [DOI] [PubMed] [Google Scholar]