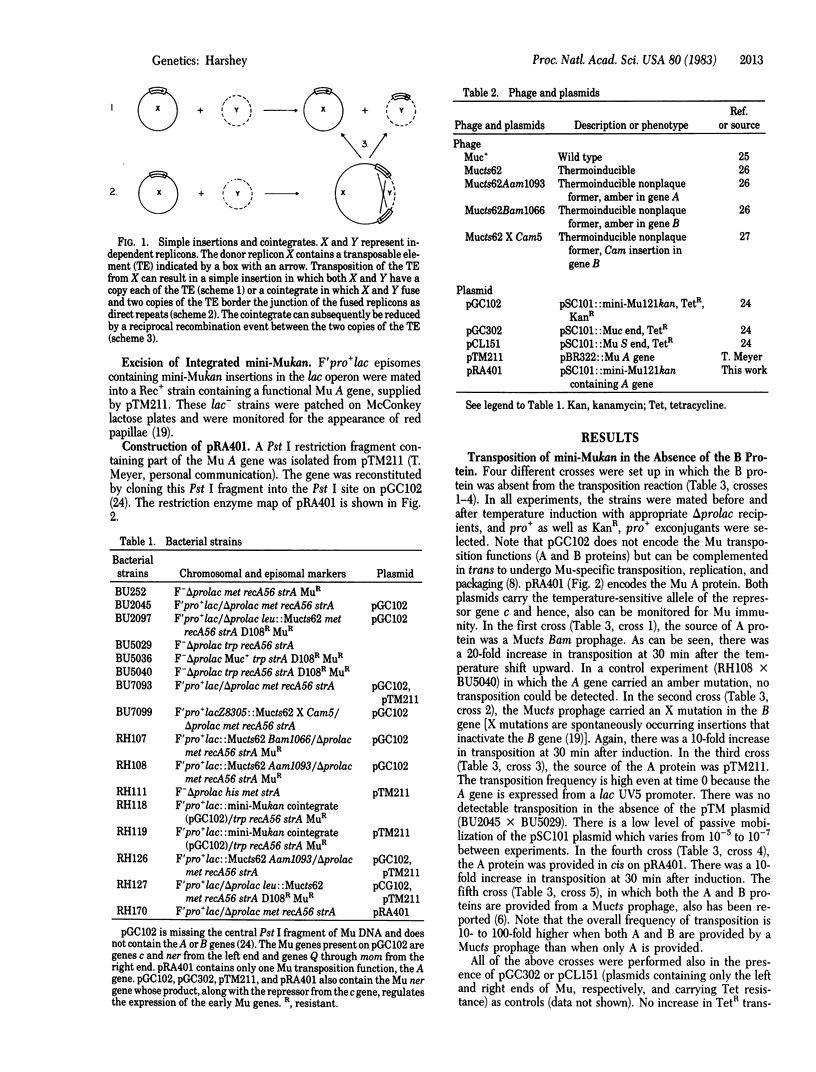
Abstract
Bacteriophage Mu is a self-contained mobile unit encoding functions that mediate its movement. There appear to be two alternate pathways for Mu DNA transposition that differ with respect to the end products they generate. During the lytic cycle of phage Mu growth the end products of transposition are predominantly cointegrates in an experimental system in which the induced Mu prophage is located on pSC101, a low-copy-number plasmid. On the other hand, Mu insertions into the host genome during lysogenization contain Mu DNA as simple insertions. Two Mu functions, encoded by the A and B genes, are required for Mu DNA transposition during its lytic growth. However, during lysogeny the product of gene B is not required for integration of Mu DNA. Evidence is presented here which shows that in the absence of the B gene product the majority of transposition events are simple insertions. This is in striking contrast to the situation in which the majority of the products are cointegrates in the presence of both A and B gene products. Additional evidence also suggests that these simple insertions do not arise through the resolution of cointegrate structures.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B. Mu insertion duplicates a 5 base pair sequence at the host inserted site. Cell. 1979 Jan;16(1):123–129. doi: 10.1016/0092-8674(79)90193-4. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I., Froshauer S. Insertion of a transposon for chloramphenicol resistance into bacteriophage Mu. Gene. 1978 Jul;3(4):303–314. doi: 10.1016/0378-1119(78)90040-9. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I. Reversal of mutator phage Mu integration. J Mol Biol. 1975 Jul 25;96(1):87–99. doi: 10.1016/0022-2836(75)90183-7. [DOI] [PubMed] [Google Scholar]
- Chaconas G., Harshey R. M., Bukhari A. I. Association of Mu-containing plasmids with the Escherichia coli chromosome upon prophage induction. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1778–1782. doi: 10.1073/pnas.77.4.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G., Harshey R. M., Sarvetnick N., Bukhari A. I. Mechanism of bacteriophage Mu DNA transposition. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):311–322. doi: 10.1101/sqb.1981.045.01.043. [DOI] [PubMed] [Google Scholar]
- Chaconas G., Harshey R. M., Sarvetnick N., Bukhari A. I. Predominant end-products of prophage Mu DNA transposition during the lytic cycle are replicon fusions. J Mol Biol. 1981 Aug 15;150(3):341–359. doi: 10.1016/0022-2836(81)90551-9. [DOI] [PubMed] [Google Scholar]
- Chaconas G., de Bruijn F. J., Casadaban M. J., Lupski J. R., Kwoh T. J., Harshey R. M., DuBow M. S., Bukhari A. I. In vitro and in vivo manipulations of bacteriophage Mu DNA: cloning of Mu ends and construction of mini-Mu's carrying selectable markers. Gene. 1981 Jan-Feb;13(1):37–46. doi: 10.1016/0378-1119(81)90041-x. [DOI] [PubMed] [Google Scholar]
- Coelho A., Leach D., Maynard-Smith S., Symonds N. Transposition studies using a ColE1 derivative carrying bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):323–328. doi: 10.1101/sqb.1981.045.01.045. [DOI] [PubMed] [Google Scholar]
- Faelen M., Huisman O., Toussaint A. Involvement of phage Mu-1 early functions in Mu-mediated chromosomal rearrangements. Nature. 1978 Feb 9;271(5645):580–582. doi: 10.1038/271580a0. [DOI] [PubMed] [Google Scholar]
- Faelen M., Resibois A., Toussaint A. Mini-mu: an insertion element derived from temperate phage mu-1. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1169–1177. doi: 10.1101/sqb.1979.043.01.132. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A. Isolation of conditional defective mutants of temperate phage Mu-1 and deletion mapping of the Mu-1 prophage. Virology. 1973 Jul;54(1):117–124. doi: 10.1016/0042-6822(73)90121-9. [DOI] [PubMed] [Google Scholar]
- Giphart-Gassler M., Reeve J., van de Putte P. Polypeptides encoded by the early region of bacteriophage Mu synthesized in minicells of Escherichia coli. J Mol Biol. 1981 Jan 5;145(1):165–191. doi: 10.1016/0022-2836(81)90339-9. [DOI] [PubMed] [Google Scholar]
- Harshey R. M., Bukhari A. I. A mechanism of DNA transposition. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1090–1094. doi: 10.1073/pnas.78.2.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey R. M., McKay R., Bukhari A. I. DNA intermediates in transposition of phage Mu. Cell. 1982 Jun;29(2):561–571. doi: 10.1016/0092-8674(82)90172-6. [DOI] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- Howe M. M., Schumm J. W. Transposition of bacteriophage Mu: properties of lambda phages containing both ends of Mu. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):337–346. doi: 10.1101/sqb.1981.045.01.047. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Kamp D. Nucleotide sequences of the attachment sites of bacteriophage Mu DNA. Nature. 1979 Jul 19;280(5719):247–250. doi: 10.1038/280247a0. [DOI] [PubMed] [Google Scholar]
- Kamp D., Kahmann R. Two pathways in bacteriophage Mu transposition? Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):329–336. doi: 10.1101/sqb.1981.045.01.046. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Morita C., Heffron F. Transposon Tn3 encodes a site-specific recombination system: identification of essential sequences, genes, and actual site of recombination. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4041–4045. doi: 10.1073/pnas.78.7.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer P. J., Cohen S. N. Effect of temperature on translocation frequency of the Tn3 element. J Bacteriol. 1979 Aug;139(2):515–519. doi: 10.1128/jb.139.2.515-519.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebart J. C., Ghelardini P., Paolozzi L. Conservative integration of bacteriophage Mu DNA into pBR322 plasmid. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4362–4366. doi: 10.1073/pnas.79.14.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungquist E., Bukhari A. I. State of prophage Mu DNA upon induction. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3143–3147. doi: 10.1073/pnas.74.8.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. I., Kikuchi A., Nash H. A., Weisberg R. A., Friedman D. I. Site-specific recombination of bacteriophage lambda: the role of host gene products. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1121–1126. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- Pato M. L., Reich C. Instability of transposase activity: evidence from bacteriophage mu DNA replication. Cell. 1982 May;29(1):219–225. doi: 10.1016/0092-8674(82)90106-4. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starlinger P. IS elements and transposons. Plasmid. 1980 May;3(3):241–259. doi: 10.1016/0147-619x(80)90039-6. [DOI] [PubMed] [Google Scholar]
- TAYLOR A. L. BACTERIOPHAGE-INDUCED MUTATION IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1963 Dec;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner B., Pato M., Toussaint A., Faelen M. Replication of mini-Mu prophage DNA. Virology. 1981 Aug;113(1):379–387. doi: 10.1016/0042-6822(81)90163-x. [DOI] [PubMed] [Google Scholar]
- Westmaas G. C., van der Maas W. L., van de Putte P. Defective prophages of bacteriophage Mu. Mol Gen Genet. 1976 Apr 23;145(1):81–87. doi: 10.1007/BF00331561. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Gassler M., Stevens W. F., van de Putte P. On the control of transcription of bacteriophage Mu. Mol Gen Genet. 1974;131(2):85–96. doi: 10.1007/BF00266145. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Lotterman B. Kinetics of Mu DNA synthesis. Mol Gen Genet. 1977 Mar 7;151(2):169–174. doi: 10.1007/BF00338691. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Giphart-Gassler M., Goosen N., Goosen T., van Leerdam E. Regulation of integration and replication functions of bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):347–353. doi: 10.1101/sqb.1981.045.01.048. [DOI] [PubMed] [Google Scholar]