Abstract
Interest in the bacteria responsible for the breakdown of lignocellulosic feedstuffs within the rumen has increased due to their potential utility in industrial applications. To date, most studies have focused on bacteria from domesticated ruminants. We have expanded the knowledge of the microbial ecology of ruminants by examining the bacterial populations found in the rumen of non-domesticated ruminants found in Canada. Next-generation sequencing of 16S rDNA was employed to characterize the liquid and solid-associated bacterial communities in the rumen of elk (Cervus canadensis), and white tailed deer (Odocoileus virginianus). Despite variability in the microbial populations between animals, principle component and weighted UniFrac analysis indicated that bacterial communities in the rumen of elk and white tail deer are distinct. Populations clustered according to individual host animal and not the association with liquid or solid phase of the rumen contents. In all instances, Bacteroidetes and Firmicutes were the dominant bacterial phyla, although the relative abundance of these differed among ruminant species and between phases of rumen digesta, respectively. In the elk samples Bacteroidetes were more predominant in the liquid phase whereas Firmicutes was the most prevalent phyla in the solid digesta (P = 1×10−5). There were also statistically significant differences in the abundance of OTUs classified as Fibrobacteres (P = 5×10−3) and Spirochaetes (P = 3×10−4) in the solid digesta of the elk samples. We identified a number of OTUs that were classified as phylotypes not previously observed in the rumen environment. Our results suggest that although the bacterial diversity in wild North American ruminants shows overall similarities to domesticated ruminants, we observed a number of OTUs not previously described. Previous studies primarily focusing on domesticated ruminants do not fully represent the microbial diversity of the rumen and studies focusing on non-domesticated ruminants should be expanded.
Introduction
Cellulose is a principal component of plant cell walls and the complete hydrolysis of this polymer requires the synergistic activity of a wide range of carbohydrate degrading enzymes. One of the best-characterized systems capable of effectively breaking down complex cellulolytic biomass is the rumen microbial community. In recent years, interest in plant cell-wall-degrading microbes and enzymes has increased due to the numerous potential industrial applications of these organisms and the proteins they express. In ruminants, digestion of the ingested plant biomass takes place under anaerobic conditions in the rumen. This anaerobic digestion chamber is inhabited by a diverse community of bacteria, archaea, protozoa and fungi that maintain a symbiotic relationship with the host, with bacteria playing the primary role in biomass degradation. These bacteria produce an array of enzymes that break down the lignocellulosic material. The resulting sugars are fermented into volatile fatty acids and used by the ruminant host for energy [1]. Considerable effort has been put forth into understanding the complex biology of this microbial ecosystem, including the application of metagenomics [2], metatranscriptomics [3], and genomic studies of major polysaccharide-degrading bacteria, as well as characterization of the enzymes they produce [4].
The rumen microbiome has been extensively studied, as the composition of this community can have a great impact on rumen function/dysfunction [4], [5], [6]. Until recently, the rumen microbiome was primarily studied using culture-based or classical molecular techniques (i.e., DGGE and ribosomal RNA clone libraries, respectively). Recent advances in next-generation sequencing have enhanced our ability to study this extremely complex environment. A recent examination of rumen microbe species in Genbank suggests that despite the extensive efforts that have gone into examining the microbial diversity of the rumen, there is still a great deal that we have yet to find out about this extraordinary environment [7]. To date, most studies have focused on domesticated ruminants, however, it is well known that the composition of the rumen microbial community varies depending on diet and ruminant species [7], [8], [9], [10]. There has been some effort to examine the microbial diversity of wild ruminants, but these studies have primarily examined a small number of clones using traditional 16S rDNA cloning techniques [8], [11], [12]. More recently, next generation sequencing approaches have been used to examine the bacterial diversity in the rumen of bovine [13], [14] the North American moose [15] and the Norwegian reindeer [16]. The diets of domesticated ruminants usually consist of high-quality forages or concentrates (e.g. hays, silages, or grain concentrates), whereas the diets of wild ruminants is more varied, depending on the nature of the browse and forage available for consumption at a given point in time within the environment. Given the different feeding strategies that are utilized by wild and domesticated ruminants, one would expect that the microbial populations in these hosts should be distinct. This is supported by a recent transcriptomic analysis of the rumen of musk-ox, which found a diverse array of genes coding for highly novel fibrolytic enzymes expressed in these animals [3] as well as the metagenomic analysis of the rumen microbiome of Norwegian reindeer [16].
We have conducted a study to examine the diversity of the rumen bacterial communities found in a number of wild Canadian ruminants using next-generation sequencing. Pyrosequencing of the V1-V3 region of the rRNA gene has been used to identify the prokaryotic diversity found in elk (Cervus canadensis), and white tailed deer (Odocoileus virginianus). Additionally, we have examined the differences between the bacterial communities associated with the solid and liquid fractions of rumen contents in these animals.
Materials and methods
Ethics Statement
Rumen samples from white tail deer were obtained, with permission, from animals harvested by licensed hunters during the open rifle fall hunting season in Sothern Alberta, during the course of their regular hunting activities. Rumen samples from elk were collected with permission at a licensed abattoir (Ft. Macleod, Alberta) immediately after animals were harvested. No animals were killed specifically for this study.
Sample collection
Samples of rumen contents were removed from the rumen immediately after death and transported in an airtight container to a field lab within 30 min. Rumen samples from elk were collected at an abattoir immediately after animals were harvested. The elk were non-domesticated farmed animals that grazed on native prairie forage on farms in southern Alberta and southwestern Saskatchewan. Total rumen contents from elk (n = 15) and deer (n = 3) were transferred to a heavy walled 250 mL beaker and the solid and liquid phases were separated using a Bodum coffee filter plunger (http://www.bodum.com/ca/en-us/). Rumen contents were separated to evaluate differences in the microbial community associated with the solid and liquid digesta. Subsamples (5 mL) of liquid phase and ∼5 g of rumen solids were collected and frozen in liquid nitrogen and subsequently stored at −80°C.
DNA extraction
Frozen samples were ground in a Retsch RM 100 Mortar Grinder with the addition of 30 mL of 100 mM Tris-HCl pH 8.0, 500 mM EDTA pH 8.0, 1.5 M NaCl, 1 mg/mL Proteinase K in the presence of liquid nitrogen for 5 min. Following grinding, the samples were incubated at 50°C for 40 min, combined with 3 mL 2% SDS and incubated at 65°C for another 45 min. The lysate was centrifuged at 19,200×g for 10 min at room temperature to pellet debris. The lysate supernatant was combined 1∶1 (v/v) with 65°C 2% agarose, then poured into 90 mm square petri plates. The agarose containing the embedded DNA was equilibrated 3 times over 24 h against 30 volumes of TE (10 mM Tris pH 8.0, 1 mM EDTA pH 8.0) buffer and subsequently stored at 4°C. Large molecular weight DNA was eluted from the agarose using the “Freeze squeeze” method [17]. The DNA concentration was determined using the Quant-iT PicoGreen dsDNA assay kit, according to the manufacturers protocol (Life Technologies). Sequencing of the 16S rRNA genes was performed by Molecular Research LP, where pyrosequencing was carried out using the bTEFAP FLX massively parallel method [18]. A 100 ng DNA aliquot was used as a template in a 50 µL PCR reaction. The 16S rRNA gene universal bacterial primers 27F 5′-AGAGTTTGATCMTGGCTCAG-3′ [19] and 519R 5′-AGRGTTTGATCMTGGCTCAG-3′ [20] were used to obtain a 450 bp amplicon. The pyrosequencing targeted the V1-V3 hypervariable region of the 16s rRNA, using the procedure of Dowd et al., 2008. The metadata and sequence reads are available at the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession number PRJEB4222 and run accession numbers ERR318187-ERR318224.
Sequence analysis
Mothur v1.25 [21] was used for sequence analysis, OTU detection, taxonomic assignment and phylogenetic analysis. Weighted UniFrac calculations [22] and principle component analysis, using the Jaccard index, were carried out with Mothur to compare bacterial populations among different samples. The sequence data was first subjected to stringent quality control. This involved the use of pyronoise/flowgram noise reduction within Mothur, removal of all sequences shorter than 200 bp, sequences with more than 1 mismatch in the barcode region, 2 mismatches in the primer sequence or those that had homopolymers >9. The remaining sequences were aligned using the Silva bacterial database [23], which contained additional 16s sequences from an in-house database of rumen bacteria. After the alignment, the ends of the sequence were optimized using the optimize = end command in Mothur. Chimeric sequences were detected and removed using the sequence collection (UCHIME) as its own reference dataset [24]. Sequences were then subsampled to obtain a uniform number of sequences per sample for all subsequent analyses. A distance matrix was constructed using with Mothur at phylogenetic distances of 0.03 (species), 0.07 (genus) and 0.25 (phylum), respectively, to define OTUs. Mothur was also used to calculate sequence coverage, species diversity using inverse Simpson and Shannon-Weiner indices (Table 1), to create a cladogram based on differences in microbial communities using the Jaccard index, and to define the core microbiome in samples. Student t-tests were carried out to examine the significance of differences in the abundance of OTUs in the samples. Differences with a P-value <0.05 were determined to be statistically significant. Taxonomic identity of the OTUs belonging to the core microbiome was identified using ARB, with the same Silva bacterial database used for the sequence alignment.
Table 1. Sequence coverage, number of OTUs and richness of rumen samples included in this study at 93% similarity.
| Coverage (%) | Observed OTUs | Inverse Simpson | Shannon | |
| White Tailed Deer | ||||
| WTD1S | 86.7 | 685 | 63.8 | 5.32 |
| WTD1L | 90.0 | 531 | 30.0 | 4.62 |
| WTD2S | 84.0 | 859 | 86.1 | 5.80 |
| WTD2L | 85.3 | 720 | 36.6 | 5.25 |
| WTD3S | 84.3 | 818 | 111.0 | 5.75 |
| WTD3L | 85.5 | 823 | 155.2 | 5.91 |
Bold samples correspond to rumen solids and italic samples correspond to rumen liquids. The moose sample was mixed and contained both rumen liquid and solid. WTD – white tail deer and E – elk. Samples ending with S or L correspond to solid or liquid phases, respectively.
Results
Pyrosequencing of elk and white tailed deer rumen samples
Pyro-sequencing of the V1-V3 region of the 16s rDNA gene in all of the elk and white tailed deer samples resulted in a total of 178,912 sequences being identified, 81,266 of which were unique. After applying quality control, 158,513 total and 60,399 unique sequences were retained. Sequences were subsampled (n = 2,862) to ensure a consistent and equal number of sequences from each sample were used in all comparisons and calculations. Representative rarefaction curves for the samples analyzed in this study can be found in Figure S1. The elk samples have a higher number of OTUs in both rumen phases compared to deer (Student t-test, P = 0.006) and species richness as estimated by both Inverse Simpson and Shannon indices was higher in elk compared to deer (P = 0.009 and 0.04, respectively). The sequence coverage, observed OTUs, and species richness, as estimated using the Inverse Simpson and Shannon indices, are shown in Table 1.
Bacterial community composition in wild ruminants
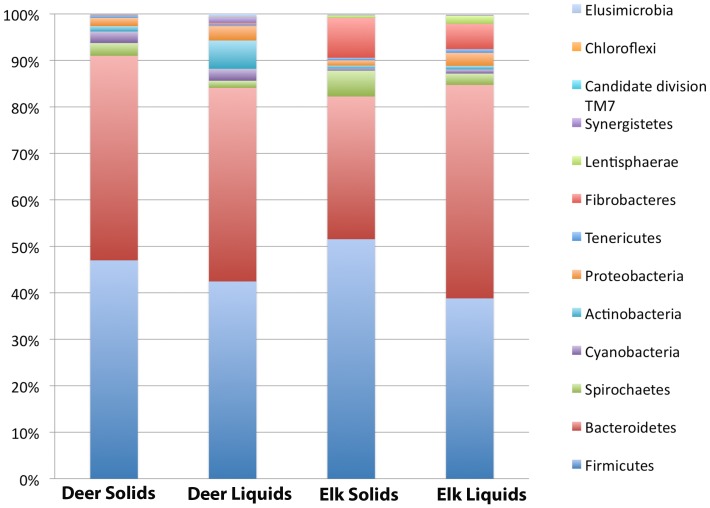
As in all other rumen environments examined to date, Bacteroidetes and Firmicutes were the predominant bacterial phyla in all samples, however, the proportion of sequences in each phylum varied in all samples examined (Fig. 1). At the class level, Clostridia and Bacteroidia were the most prevalent in both elk and deer. We observed a significant number of sequences belonging to Fibrobacteria in all elk samples, while these were almost completely absent in deer samples (P = <1×10−7). In total, 83 orders, 141 families, and 327 bacterial genera were identified when the samples across all animals.
Figure 1. Phylum level comparison of rumen microbiome associated with solid and liquid rumen phase from white tailed deer (n = 3) and elk (n = 15).
Values shown represent the percent of sequences assigned to a particular phylum averaged over all samples of the same type.
Prevotella made up 19.8% and 11.6% of the sequences in the solid phase and 27.5% and 18.2% of the sequences in the liquid phase of white tailed deer and elk, respectively. This indicates that Prevotella is more abundant in the liquid phase of the rumen compared to the solid phase in both white tail deer and elk (P = 0.001). Sequences assigned to the uncultured rumen bacterial cluster RFN8-YE57 were observed in all wild ruminants in this study, but were most abundant in elk (14.1%) and were more prevalent in rumen solids than liquids (P = 0.005).
Quinella ovalis made up 5.4% of the sequences in white tailed deer, but only 0.1% in elk. The abundance of Quinella sequences also varied between the solid and liquid phase and were approximately 2-fold higher in the liquid phase of the deer samples, however this trend was not found to be statistically significant (P = 0.48). Another member of the Veillonellaceae family, a species related to the genus Anaeromusa, was identified at similar levels in the solid and liquid phase rumen contents from white tailed deer (2.1%) but was almost absent from elk (<0.1%). An OTU that was identified as being similar to the genus Saccharofermentans and clustered with a number of uncultured rumen bacteria, was also abundant in the elk samples corresponding to 2.1 and 2.8% of the sequences in the liquid and solid phases, respectively but was absent in the deer. An OTU classified as Geosporobacter represented 1.2% of the sequences in both the liquid and solid phase in the deer but was <0.1% of the sequences in Elk. Ruminococcus was found in all animals at levels ranging from 1–2% in both the liquid and solid phases of digesta.
One of the most interesting observations was the high variability in the prevalence of Fibrobacter. A high percentage of the sequences in elk samples were classified as Fibrobacter however this phyla was found in very low levels in most of the deer samples. Comparing individual animals revealed high variability in the abundance of this genus. Within the elk samples, Fibrobacter was more abundant in rumen solids then rumen liquids (P = 1×10−5) ranging from 4.2% to 14.1% in the solid and 3.1% and 8.8% in the liquid phase of rumen contents. Within the white tailed deer samples one animal showed higher levels of F. succinogenes associated with the solid phase (0.8%).
Comparison of bacterial communities between ruminants
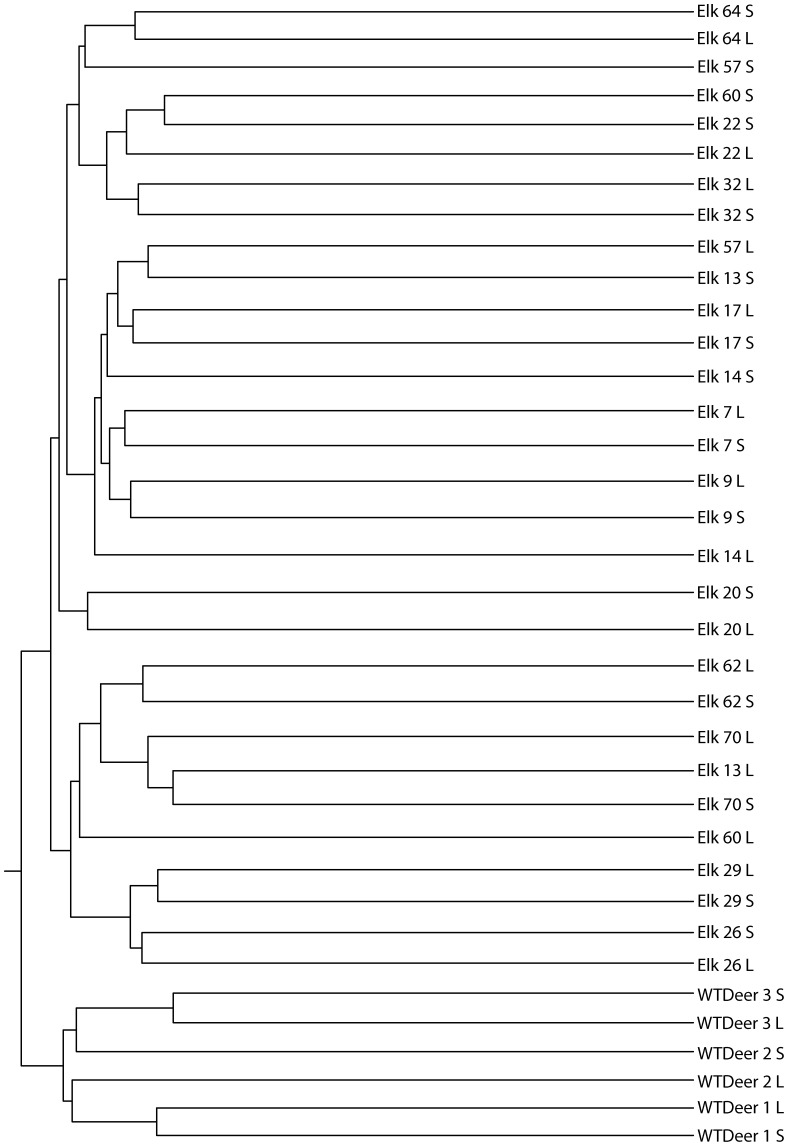
Clustering of the samples based on a Jaccard similarity plot at 97% identity clearly showed that the deer samples clustered separately from the elk samples (Fig. 2). Furthermore, the samples generally clustered together based on the individual animal species and not on the basis of the digestive fraction (i.e. liquid versus solid). This is true of all the deer samples and 8 of the 15 elk samples. The elk samples form two large clades however there are no defining characteristics of these samples that explain this clustering. With the exception of samples 52, 53, 55, and 59, the elk samples did not cluster based on the location of the animal prior to harvest in southern Alberta and southwestern Saskatchewan.
Figure 2. Microbial composition of wild ruminants assessed using Jaccard analysis of OTUs at 97% identity.
Samples are labeled animal elk or white tailed deer - animal number - S (rumen solids) or L (rumen liquids).
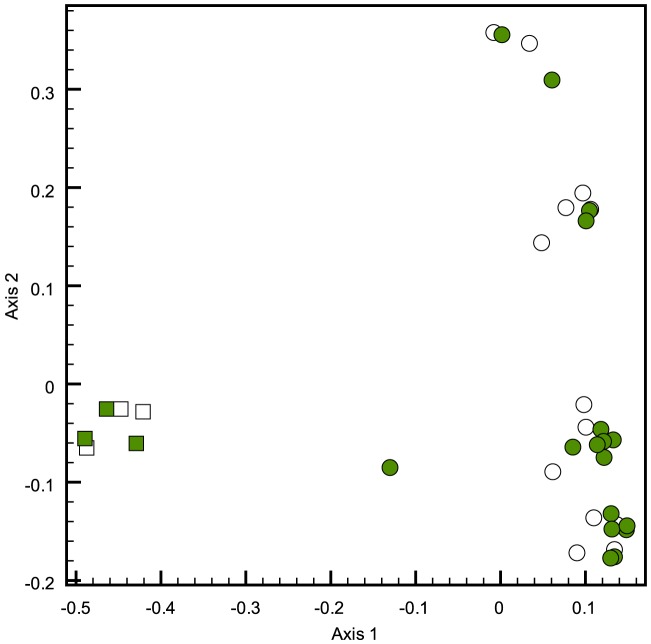
Weighted UniFrac analysis was carried out on the samples to quantify the differences in the microbial communities in the samples examined (Table 2). The microbial community associated with the white tail deer was significantly different (P<0.001) from the microbial communities associate with all elk samples. A comparison of the microbial communities in the liquid and solid rumen digesta showed that in some individuals the community varied significantly whereas in others it did not differ (Table 2). In particular, the liquid and solid phases in the deer sample did not show statistically significant differences in composition, however the difference in the Elk samples was significant. Principle component analysis showed distinct clustering of the communities based on ruminant species (Fig. 3). Elk samples showed a high degree of variability in community composition.
Table 2. Weighted-UniFrac comparison of microbial communities found in rumen samples based on Jaccard analysis of 97% similarity.
| Samples compared | P value |
| WT deer Sol – WT deer Liq | 0.53 |
| WT deer Sol – Elk Sol | <0.001 |
| WT deer Sol – Elk Liq | <0.001 |
| WT deer Liq – Elk Sol | <0.001-0.42 |
| WT deer Liq – Elk Liq | <0.001-0.42 |
| Elk Sol – Elk Sol | <0.001-0.09 |
| Elk Liq – Elk Liq | <0.001-0.30 |
| Elk Sol – Elk Liq | <0.001-0.32 |
The P-value is a measure of the significance with which the microbial communities compared differ. P-values of <0.001 are highly significant, 0.001–0.01 are significant, 0.01–0.05 are marginally significant, 0.05–0.1 are suggestive and >0.1 is not significant. P-value ranges indicate the upper and lower limits of the calculation when multiple samples were compared.
Figure 3. Principle component analysis of 16S profiles from rumen contents collected from elk (Solid-blue, Liquid-Black), white tailed deer (Solid-Red, Liquid-Green).
Individual variation in rumen bacteria
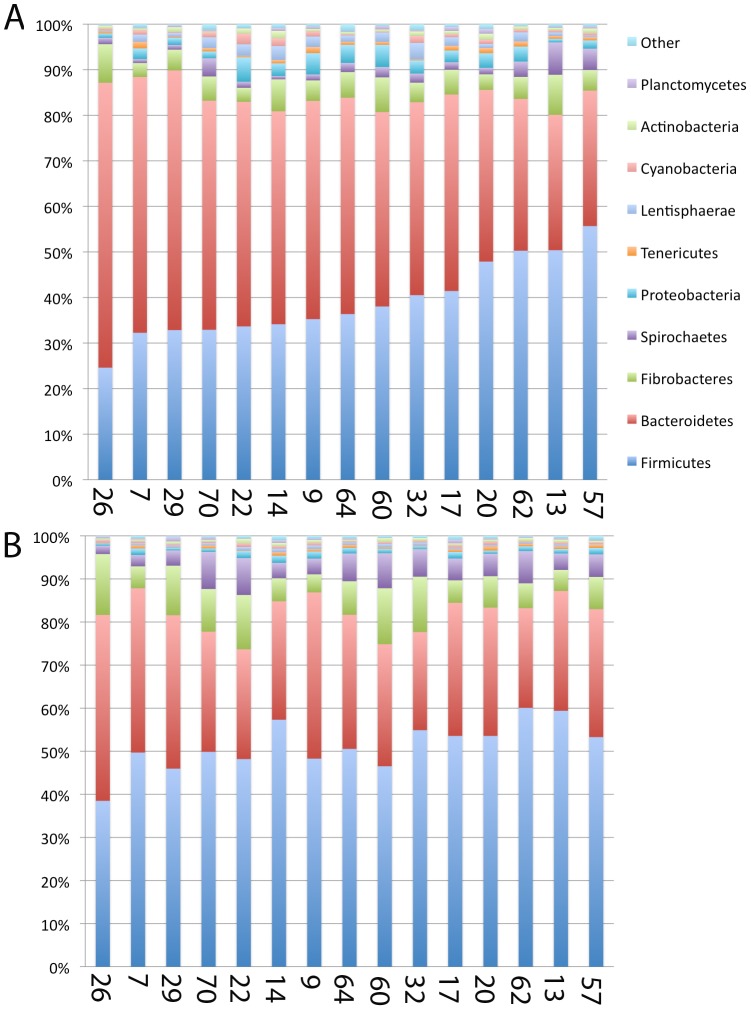
Examination of samples from the elk provides a view of bacterial diversity within the same ruminant species over a larger number of individuals (n = 15). A comparison of the sequences classified at the phylum level showed that in all cases the majority of the bacteria belonged to Firmicutes and Bacteriodes (Fig. 4). In the elk samples, Firmicutes was found to be more abundant in the solid rumen contents (P = 1×10−5) whereas Bacteriodes was found at higher levels in the liquid fraction (P = 1×10−5). Fibrobacteres accounted for a large percentage of the sequences in elk with a higher percentage in the solid than the liquid fraction (P = 5×10−3). The Spirochaetes made up a larger fraction of the phyla in the solid phase, relative to the liquid phase (P = 3×10−4). Comparison of the distribution of bacterial phyla in all individuals revealed that in all cases Firmicutes, Bacteroidetes and Fibrobacteres were the dominant phyla, however the relative proportion was highly variable among individual hosts (Fig. 4). Deeper phylogenetic classification of the reads revealed that the majority of elk samples, both liquid and solid, were dominated by Prevotella ruminocola, an uncultured Prevotella species, RFN8-YE57 and Fibrobacter succinogenes. Although these genera tended to account for a large proportion of those identified, the relative amounts in each sample were highly variable. Animal 20 was unique in that it was the only individual that had significant levels of Q. ovalis and Heliobacillus within its bacterial community. These sequences were found at higher levels in the liquid phase of this animal.
Figure 4. Diversity of bacterial phyla found in the Liquid (A) and Solid (B) fraction of rumen contents collected from 15 elk.
Samples are arranged from the lowest to highest level of Firmicutes based on the percentage of total reads assigned to that phyla. Numbers along the x-axis indicate the animal identification number each sample was obtained from. The percentage of total reads (x-axis) assigned to each phylum is plotted for all 15 samples (y-axis).
Core microbiome Analysis
The nature of the core microbiome was determined for all samples originating from white tailed deer and elk. Taxa that were found to be present in all of the samples in the analysis were deemed to be members of a core microbiome. The results of this analysis are shown in Table 3. There were 8 taxa from three phyla that were shared among all animals examined in this study. Analysis of the core microbiome of the elk rumen identified 12 conserved taxa, including F. succinogenes, the vadinHA42 cluster of Ruminococcaceae, Treponema byrantii, and an uncultured member of the genus Saccharofermentans. The core microbiome of the white tailed deer samples showed 11 shared taxa in the solid phase and 10 in the liquid phase, respectively, including Q. ovalis in the liquid and an uncultured member of the BS11 gut group in the solid fraction of digesta.
Table 3. Taxa identified within the core microbiome of the rumen of elk, white tailed deer.
| All animals | Elk solid phase | Elk liquid phase | Deer solid phase | Deer liquid phase |
| Lachnospiraceae Wet75 sp1 | Lachnospiraceae Wet75 sp1 | Lachnospiraceae Wet75 sp1 | Lachnospiraceae Wet75 sp1 | Lachnospiraceae Wet75 sp1 |
| Uncultured Prevotella | Uncultured Prevotella | Uncultured Prevotella | Uncultured Prevotella | Uncultured Prevotella |
| Unclassified Synergistetes | Unclassified Synergistetes | Unclassified Synergistetes | Unclassified Synergistetes | Unclassified Synergistetes |
| Lachnospiraceae Wet75 sp2 | Lachnospiraceae Wet75 sp2 | Lachnospiraceae Wet75 sp2 | Lachnospiraceae Wet75 sp2 | Lachnospiraceae Wet75 sp2 |
| Prevotella ruminicola | Prevotella Ruminicola | Prevotella ruminicola | Prevotella ruminicola | Prevotella ruminicola |
| RFN8-YE57 | RFN8-YE57 | RFN8-YE57 | RFN8-YE57 | RFN8-YE57 |
| Incertia sedis Clos-spor | Incertia sedis Clos-spor | Incertia sedis Clos-spor | Incertia sedis Clos-spor | Incertia sedis Clos-spor |
| RC9 gut group | RC9 gut group | RC9 gut group | RC9 gut group | RC9 gut group |
| VadinHA42 | VadinHA42 | VadinHA42 | Quinella ovalis | |
| Treponema bryantii | Uncultured Para-prevotella | Uncultured Para-prevotella | BS11 gut group | |
| Fibrobacter succinogenes | Fibrobacter succinogenes | BS11 gut group | Prevotella byrantii | |
| Uncultured Saccharofermentans | Prevotella byrantii | Prevotella byrantii | ||
| Phyla represented | ||||
| Firmicutes, Bacteroidetes Synergistetes | Firmicutes, Bacteroidetes, Spirochaetes, Fibrobacteres Synergistetes | Firmicutes, Bacteroidetes, Fibrobacteres Synergistetes | Firmicutes, Bacteroidetes Synergistetes | Firmicutes, Bacteroidetes Synergistetes |
Discussion
Our results show that the bacterial communities found in the rumen of a number of wild ruminants are significantly different from each other. Furthermore, examination of multiple individuals shows that, although the phyla present within the rumen of a particular host species are generally conserved, the relative proportion of these microbes varies. These assertions are supported by distance-based clustering, using the Jaccard similarity index and principal component analysis, as well as weighted UniFrac calculations. We also compared the composition of the bacterial communities found within the liquid and solid phases of the rumen and found distinct differences in the composition of these communities in elk, but not deer. This agrees with the results of a comparison of the bacterial population in the liquid and solid rumen phases of cattle [5]. As is commonly seen in studies of the rumen microbiome, there was a high degree of variability in community composition between individuals. The deer samples were highly ruminated, having a thick, porridge-like consistency and were difficult to separate into liquid and solid fractions, which may contribute to the observed results.
A number of studies from various ruminants all point to Bacteroides and Firmicutes being the dominant phyla in the rumen ecosystem [5], [8], [9], [11], [14]. Our results confirm this finding and show that even within wild ruminants the above are the dominant phyla and compose the foundation of the core microbiome of wild ruminants. There have been relatively few studies that have used high throughput next-generation based sequencing to examine the bacterial diversity in the rumen thus far. One such study that examined the bovine rumen found Prevotella to be most prevalent followed by Oscillibacter and Coprococcus [13]. Prevotella and Tannerella were over represented in the liquid phase and Butyrivibrio and Blautia in the solid phase of digesta [13]. Tannerella was not observed in any of our samples and Blautia was seen only in low abundance (<0.3%) in elk. Oscillibacter was only found in white tailed deer and was more prevalent in the solid phase than the liquid phase of digesta (0.5% versus 0.1% of sequences). Q. ovalis was found to account for a substantial proportion of the rumen bacteria in white tailed deer but not elk. Q. ovalis is a member of the Veillonellaceae family that plays a role in the fermentation of starch and sugar to propionate, a characteristic consistent with its prevalence in the liquid phase of rumen contents. Another OTU that was found in high abundance is the rumen microbe labeled RFN8-YE57. This bacterium was first identified in the forestomach of an eastern grey kangaroo and belongs to the Family of the Lachnospiracaeae, within Clostridial cluster XIVa. It is related to Butyrivibrio and Pseudobutyrivibrio, which are non-sporulating, non-motile gram-negative rods that produce acetate, succinate, and N-butyrate as fermentation products [25]. Little is known about the specific role of this bacterium in the rumen, however the high abundance of this organism suggests it is an important member of the rumen microbiome. Another abundant OTU found in the elk samples but not in the deer was classified as Victivallis. The only described species, Victivallis vadensis, is a gram-negative, strictly anaerobic coccum, that displays cellobiose-degrading activity and is syntrophic with the methanogenic archaea Methanospirillum hungatei [26]. This genus was first identified in a human fecal sample but has not been previously described in the rumen. An OTU with similarity to the genus Geosporobacter was observed at levels of ∼1% of the sequences identified in deer. The only characterized member of this genus was isolated from a deep aquifer and is a sporulating, non-motile, gram-positive, obligate anaerobe that can ferment a diverse range of carbohydrates to generate acetate, H2 and CO2 [27].
F. succinogenes was highly abundant and a member of the core microbiome in the rumen of elk, but was virtually absent from the deer samples. This may be due to dietary differences and the fact that deer are browsers, as opposed to elk, which are grazers. The Fibrobacter sequences in elk were more abundant in the solid (8.5%) as compared to the liquid (5.4%) fraction (P = 5×10−3), which is consistent with the role of this organism in fiber degradation. A number of studies have found that Fibrobacter is not consistently found in the rumen [5], [10], [11], [15], [28], suggesting that the distribution of this organism may be quite variable in the rumen environment and that this organism only plays an important role in fiber digestion in some ruminant species. The high non-specific nuclease content of this bacterium [29], its sensitivity to lysis, as well as difficulties in amplifying Fibrobacter DNA [30] or natural variation in the levels of this organism may play roles in the observed variability of Fibrobacter in the rumen [31].
Two recent studies have utilized next-generation sequencing to investigate the bacterial diversity in Moose [15] and Reindeer [16]. Ishaq and Wright (2012) used the phylochip approach to examine the bacterial diversity in moose and found that the most predominate phyla were identified as Bacteriodetes and Proteobacteria. This contrasts other studies, which consistently show that Firmicutes and Bacteriodetes make up the dominant phyla in the rumen and is likely a result of both real differences in the individual rumen microbiome of the moose, and differences in the technique that was used in this study. A draw back of using this microarray-based approach is that species identification is restricted to the probes present on the chip. Furthermore, these probes were not specifically developed to represent bacterial members of the rumen environment, something acknowledged by the authors. The other studies examining the rumen microbiome of non-domesticated wild ruminants have focused on the Norwegian reindeer [11], [12], [16]. Both metagenomic sequencing targeting the V1-V3 of the 16s rRNA gene [16] and sequencing of full length 16s rRNA clones [11], [12] have been used and both have identified a number of novel sequences. These sequences were suggested to represent novel species that have not been observed in domesticated ruminants [11], [16]. Interestingly, the sequences observed in the reindeer microbial community were not observed in the samples examined in this study. These studies provide additional support to the hypothesis that the rumen microbiome varies with host species and that efforts should be made to study under-represented ruminant species.
Conclusions
This work examined the bacterial communities found in elk and white tail deer. These hosts exhibit both similarities and differences to the rumen microbiome found in domesticated ruminants. Our results suggest that the current studies focusing primarily on domesticated bovine, sheep, and goats are not capturing the full diversity of microbes that are found within the rumen environment. A greater focus on examining the rumen microbiome of non-domesticated ruminants could help to identify novel microbes and enzymes of commercial interest.
Supporting Information
Representative rarefaction curves of wild ruminant samples. Curves represent the number of OTUs at 97% similarity level observed as a function of sequencing depth. For clarity, not all of the samples examined are displayed.
(TIF)
Acknowledgments
The technical support of Lyn Paterson is gratefully acknowledged.
Funding Statement
This study was supported through funding from Genome Alberta and the Agriculture and Agri-Food Canada Genomics Research and Development Initiative. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Russell JB, Rychlik JL (2001) Factors that alter rumen microbial ecology. Science 292: 1119–1122. [DOI] [PubMed] [Google Scholar]
- 2. Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, et al. (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331: 463–467. [DOI] [PubMed] [Google Scholar]
- 3. Qi M, Wang P, O'Toole N, Barboza PS, Ungerfeld E, et al. (2011) Snapshot of the eukaryotic gene expression in muskoxen rumen—a metatranscriptomic approach. PLoS One 6: e20521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krause DO, Denman SE, Mackie RI, Morrison M, Rae AL, et al. (2003) Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol Rev 27: 663–693. [DOI] [PubMed] [Google Scholar]
- 5. Kong Y, Teather R, Forster R (2010) Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol Ecol 74: 612–622. [DOI] [PubMed] [Google Scholar]
- 6. Hook SE, Steele MA, Northwood KS, Dijkstra J, France J, et al. (2011) Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol Ecol 78: 275–284. [DOI] [PubMed] [Google Scholar]
- 7. Kim M, Morrison M, Yu Z (2011) Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol Ecol 76: 49–63. [DOI] [PubMed] [Google Scholar]
- 8. Nelson KE, Zinder SH, Hance I, Burr P, Odongo D, et al. (2003) Phylogenetic analysis of the microbial populations in the wild herbivore gastrointestinal tract: insights into an unexplored niche. Environ Microbiol 5: 1212–1220. [DOI] [PubMed] [Google Scholar]
- 9. Pitta DW, Pinchak E, Dowd SE, Osterstock J, Gontcharova V, et al. (2010) Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb Ecol 59: 511–522. [DOI] [PubMed] [Google Scholar]
- 10. de Menezes AB, Lewis E, O'Donovan M, O'Neill BF, Clipson N, et al. (2011) Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol Ecol 78: 256–265. [DOI] [PubMed] [Google Scholar]
- 11. Sundset MA, Praesteng KE, Cann IK, Mathiesen SD, Mackie RI (2007) Novel rumen bacterial diversity in two geographically separated sub-species of reindeer. Microb Ecol 54: 424–438. [DOI] [PubMed] [Google Scholar]
- 12. Sundset MA, Edwards JE, Cheng YF, Senosiain RS, Fraile MN, et al. (2009) Molecular diversity of the rumen microbiome of Norwegian reindeer on natural summer pasture. Microb Ecol 57: 335–348. [DOI] [PubMed] [Google Scholar]
- 13. Fouts DE, Szpakowski S, Purushe J, Torralba M, Waterman RC, et al. (2012) Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS One 7: e48289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, et al. (2013) Simultaneous Amplicon Sequencing to Explore Co-Occurrence Patterns of Bacterial, Archaeal and Eukaryotic Microorganisms in Rumen Microbial Communities. PLoS One 8: e47879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishaq SL, Wright AD (2012) Insight into the bacterial gut microbiome of the North American moose (Alces alces). BMC Microbiol 12: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pope PB, Mackenzie AK, Gregor I, Smith W, Sundset MA, et al. (2012) Metagenomics of the Svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci. PLoS One 7: e38571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tautz D, Renz M (1983) An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem 132: 14–19. [DOI] [PubMed] [Google Scholar]
- 18. Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, et al. (2008) Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrant E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, NY: John Wiley and Sons. pp. 115–175.
- 20. Turner S, Pryer KM, Miao VP, Palmer JD (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46: 327–338. [DOI] [PubMed] [Google Scholar]
- 21. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ouwerkerk D, Klieve AV, Forster RJ, Templeton JM, Maguire AJ (2005) Characterization of culturable anaerobic bacteria from the forestomach of an eastern grey kangaroo, Macropus giganteus . Lett Appl Microbiol 41: 327–333. [DOI] [PubMed] [Google Scholar]
- 26. Zoetendal EG, Plugge CM, Akkermans AD, de Vos WM (2003) Victivallis vadensis gen. nov., sp. nov., a sugar-fermenting anaerobe from human faeces. Int J Syst Evol Microbiol 53: 211–215. [DOI] [PubMed] [Google Scholar]
- 27. Klouche N, Fardeau ML, Lascourreges JF, Cayol JL, Hacene H, et al. (2007) Geosporobacter subterraneus gen. nov., sp. nov., a spore-forming bacterium isolated from a deep subsurface aquifer. Int J Syst Evol Microbiol 57: 1757–1761. [DOI] [PubMed] [Google Scholar]
- 28. Zened A, Combes S, Cauquil L, Mariette J, Klopp C, et al. (2013) Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol Ecol 83: 504–514. [DOI] [PubMed] [Google Scholar]
- 29. Lee SF, Forsberg CW, Gibbins AM (1992) Type II DNA restriction-modification system and an endonuclease from the ruminal bacterium Fibrobacter succinogenes S85. J Bacteriol 174: 5275–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Larue R, Yu Z, Parisi VA, Egan AR, Morrison M (2005) Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ Microbiol 7: 530–543. [DOI] [PubMed] [Google Scholar]
- 31.Stewart CS, Flint HJ, Bryant MP (1997) The rumen bacteria. The Rumen Microbial Ecosystem. 2nd ed: Chapman & Hill. pp. 10–55.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative rarefaction curves of wild ruminant samples. Curves represent the number of OTUs at 97% similarity level observed as a function of sequencing depth. For clarity, not all of the samples examined are displayed.
(TIF)