Hair follicles (HFs) undergo cyclical periods of growth, which are fueled by stem cells (SCs). By conditionally targeting Sox9 in adult HF-SCs, Kadaja et al. found that SOX9 is essential for stemness and HF downgrowth. Numerous SOX9-sensitive targets encode extracellular factors, most notably enhancers of Activin/pSMAD2 signaling. Compromising Activin signaling recapitulates SOX9-dependent defects, and Activin partially rescues them. These findings reveal roles for SOX9 in regulating adult HF-SC maintenance and a role for SCs in coordinating their own behavior through non-cell-autonomous signaling within the niche.
Keywords: SOX9, hair follicle, stem cells, skin, Activin
Abstract
Hair follicles (HFs) undergo cyclical periods of growth, which are fueled by stem cells (SCs) at the base of the resting follicle. HF-SC formation occurs during HF development and requires transcription factor SOX9. Whether and how SOX9 functions in HF-SC maintenance remain unknown. By conditionally targeting Sox9 in adult HF-SCs, we show that SOX9 is essential for maintaining them. SOX9-deficient HF-SCs still transition from quiescence to proliferation and launch the subsequent hair cycle. However, once activated, bulge HF-SCs begin to differentiate into epidermal cells, which naturally lack SOX9. In addition, as HF-SC numbers dwindle, outer root sheath production is not sustained, and HF downgrowth arrests prematurely. Probing the mechanism, we used RNA sequencing (RNA-seq) to identify SOX9-dependent transcriptional changes and chromatin immunoprecipitation (ChIP) and deep sequencing (ChIP-seq) to identify SOX9-bound genes in HF-SCs. Intriguingly, a large cohort of SOX9-sensitive targets encode extracellular factors, most notably enhancers of Activin/pSMAD2 signaling. Moreover, compromising Activin signaling recapitulates SOX9-dependent defects, and Activin partially rescues them. Overall, our findings reveal roles for SOX9 in regulating adult HF-SC maintenance and suppressing epidermal differentiation in the niche. In addition, our studies expose a role for SCs in coordinating their own behavior in part through non-cell-autonomous signaling within the niche.
The renewal of interfollicular epidermis (IFE) is a constant process fueled by proliferation of cells in the basal epithelial layer. Epidermal differentiation can be easily followed by the unique spatiotemporal expression pattern of keratins and other structural proteins. Like all skin epithelial cells that maintain contact with basement membrane, basal undifferentiated keratinocytes express keratin 5 (K5), K14, and hemidesmosomal integrins α6 and β4 (Fuchs 2007). When basal cells start to differentiate, they lose contact with basement membrane and move into suprabasal spinous layers, a process associated with down-regulation of basal markers and induction of K1 and K10 (Fuchs and Green 1980). As cells continue upward, granular layer cells begin to express filaggrin and loricrin, components of terminally differentiated, outer stratum corneum layers that provide the skin barrier (Fuchs 2007).
In contrast, hair follicles (HFs) undergo synchronous, cyclical rounds of tissue growth (anagen), degeneration (catagen), and rest (telogen). During telogen, resident HF stem cells (SCs) can remain quiescent for weeks, enabling them to retain nucleotides and histones incorporated when in an active state. Such pulse-chase experiments traced the residence of HF-SCs to a niche known as the “bulge” (Cotsarelis et al. 1990; Tumbar et al. 2004). HF-SCs are typified by a group of transcription factors, including SOX9, NFATc1, TCF3/4, LHX2, and TBX1, which are not expressed in IFE or in the short-lived, transit-amplifying HF-SC progeny, called the matrix (Blanpain et al. 2004; Morris et al. 2004; Tumbar et al. 2004).
Upon activation, HF-SCs at the base of the bulge—a region now referred to as the hair germ (HG)—divide rapidly and generate a pool of matrix TA cells. Several days after HG activation, HF-SCs in the bulge proliferate briefly to self-renew and form a trail of cells, which forms the upper outer root sheath (ORS) of HFs (Greco et al. 2009; Rompolas et al. 2013). As the matrix and ORS grow downward, the dermal papilla (DP) is pushed further away from the bulge and upper ORS, prompting HF-SCs to return to quiescence. Matrix cells envelop and maintain contact with DP, enabling them to proliferate several times before they differentiate upward to form the hair and its channel or inner root sheath (IRS). The lower ORS remains proliferative and fuels replenishment of the matrix. As the destructive phase ensues and the matrix and lower ORS become apoptotic, spared ORS cells form a new SC niche: Upper ORS cells that divided the fewest times become the new bulge, those with slightly more activity become the new HG, and those that proliferated the most differentiate to form an inner bulge layer of terminally differentiated cells that secrete inhibitory BMP6 and FGF18 signals (Hsu et al. 2011). This keeps the HF in telogen until sufficient activating signals accumulate to again tip the balance and start a new hair cycle.
Bulge SCs, ORS cells, and HG express K5/K14 and α6β4, but only bulge SCs express CD34 (Trempus et al. 2003). Molecularly, HG is more similar to bulge than matrix and is thought to consist of “primed,” short-term HF-SCs (Greco et al. 2009). Matrix cells exhibit reduced expression of HF-SC markers. Upon terminal differentiation, each lineage induces expression of distinct keratins. Most notable are the group of hair keratins expressed by the cortex and cuticle of the hair shaft and K6 expressed by the hair shaft medulla and the companion layer that separates ORS from IRS.
SOX9 is intriguing in that it regulates the formation of many cell types, tissues, and organs, including not only HFs but also chondrocytes, testis, heart, lung, pancreas, bile duct, retina, sweat gland, and central nervous system (Vidal et al. 2005; Nowak et al. 2008; Lu et al. 2012; for review, see Lefebvre et al. 2007; Sarkar and Hochedlinger 2013). When Sox9 is conditionally ablated in embryonic skin, epidermal differentiation in the IFE and upper HF proceed unchecked, consistent with their natural lack of this factor, but HF-SCs do not form, and hair growth is arrested (Vidal et al. 2005; Nowak et al. 2008). Since no HF-SCs are generated in Sox9 mutant mice, it is not known whether and how this transcription factor regulates the behavior of HF-SCs once they are established.
In this study, we explore SOX9's function in adult HF-SCs and provide compelling evidence that SOX9 is essential for their maintenance but does not play a critical role in HF lineage differentiation or affect HF-SC activation at telogen → anagen. Rather, upon activation, SOX9-deficient HF-SCs lose HF stemness and differentiate along the epidermal lineage. This leads to premature arrest of HF downgrowth, which is dependent on bulge and upper ORS (normally SOX9-positive) but not TA matrix and hair shaft/IRS (normally SOX9-negative) lineages. Using genome-wide RNA sequencing (RNA-seq) profiling of purified wild-type and SOX9-deficient HF-SCs coupled with immunofluorescence and biochemical analyses, we further show that SOX9-deficient bulge cells lose HF-SC characteristics. Using SOX9 chromatin immunoprecipitation (ChIP) in vivo and deep sequencing (ChIP-seq) on adult HF-SCs purified directly from their native niche, we show that SOX9's targets include genes encoding key HF-SC transcription factors as well as extracellular factors. Activin signaling genes are particularly affected, and gene ablation and our rescue experiments show that SOX9 functions at least partially through regulating this pathway. Overall, our results reveal new functions and mechanisms for SOX9 and suggest that through Activin signaling, HF-SCs are able to suppress epidermal differentiation, sustain ORS production, and return to quiescence in early anagen.
Results
SOX9-expressing bulge HF-SCs contribute to all HF lineages
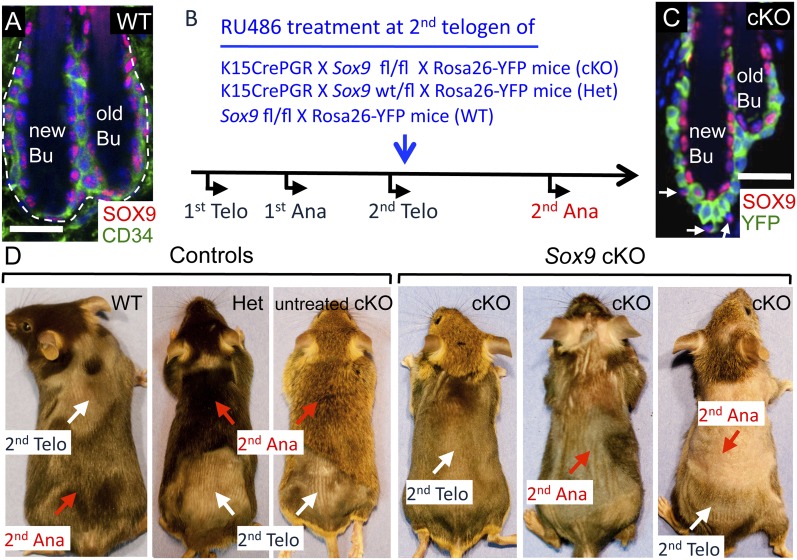
In telogen, SOX9 is detected in CD34+ HF-SCs, in HG cells, and in the terminally differentiated K6+ inner layer of the bulge (Fig. 1A; Supplemental Fig. 1A). After anagen induction, SOX9 expression in the downgrowing HF becomes restricted to early bulge progeny of upper ORS, known to maintain HF-SC stemness (Supplemental Figs. 1B, 2B). Indeed, when subjected to lineage tracing with Sox9-CreER and Rosa26-fl-stop-flYFP (R26YFP) mice, SOX9-expressing YFP+ cells gave rise to all adult hair lineages (Supplemental Fig. 1C). Prior to tamoxifen, no YFP+ cells were detected, documenting the specificity of Cre activation.
Figure 1.
Loss of SOX9 in adult HF-SCs causes deficient hair coat recovery. (A) Immunofluorescence of SOX9 and CD34 in the telogen HFs. (B) Strategy to knock out Sox9 in adult HF-SCs. (C) YFP and SOX9 immunofluorescence in telogen cKO HFs 2 wk after targeting. SOX9 is gone in HF-SCs, but K6+ inner bulge cells and YFP− cells in the CD34 layer (arrows) still express SOX9. (D) Hair coat recovery of Sox9 wild-type (WT), Het, and cKO mice after RU486 treatment at the beginning of second telogen (postnatal days 60–74 [P60–P74]). Untreated Sox9fl/fl × K15-CrePGR × R26YFP mice were also used as a control. Bars, 30 μm. The white dashed line denotes the epidermal–dermal border, and the blue channel is DAPI staining. (Ana) Anagen, (Bu) bulge, (Telo) telogen.
Equally important, YFP+ HF-SCs persisted long-term and were still fueling new hair regeneration 8 mo later. These results extended prior SOX9 studies on developing skin (Vidal et al. 2005; Nowak et al. 2008) and confirmed that SOX9+ cells within the adult bulge display the characteristics of HF-SCs.
Sox9 ablation in adult HF-SCs compromises hair coat regeneration
To address whether SOX9 is essential for SC maintenance and/or hair cycling, we bypassed SOX9's essential role in HF-SC establishment and conditionally targeted Sox9 ablation in established adult HF-SCs. For this purpose, we used the Keratin15 (K15) promoter to selectively drive expression of CrePGR in HF-SCs within the bulge and HG (Morris et al. 2004). By breeding to Sox9fl/fl × R26YFP mice, Sox9 ablation could be monitored by YFP (Fig. 1B). We administered RU486 in the extended second telogen to activate CrePGR and specifically ablate Sox9 in bulge HF-SCs (CD34+) and HG cells (CD34−) (Fig. 1C; Supplemental Fig. 2A,B). YFP− bulge cells included a few CD34+, K15+ HF-SCs but were mostly terminally differentiated K6+, K15− inner bulge cells (Hsu et al. 2011). Based on quantitative RNA PCR (qPCR) of mRNA from HF-SCs purified by fluorescence-activated cell sorting (FACS), Sox9 levels were decreased ∼80%–95% in YFP+ HF-SCs compared with heterozygous (Het) controls (Supplemental Fig. 2C). SOX9 expression in YFP− HF-SCs remained high, as expected.
Loss of SOX9 in HF-SCs did not result in an overt phenotype as long as HFs remained in telogen. Two weeks after RU486 treatment, percentages of YFP+ Sox9-null HF-SCs in skin were comparable with those of YFP+ Sox9 Het (control) HF-SCs. In addition, throughout telogen, Sox9-null HF-SCs remained quiescent like control HF-SCs (Supplemental Fig. 2D,E). As judged by shaving the skin and monitoring skin darkening, HFs lacking SOX9 in their SCs were able to enter into anagen (Fig. 1D). The transition from pink (telogen) to black (anagen) skin is a reflection of melanin production by differentiating melanocytes, which occurs in anagen and leads to melanin transfer to concomitantly differentiating hair shaft precursors (Müller-Röver et al. 2001).
After HFs transitioned from telogen to anagen, Sox9 conditional knockout (cKO) mice began to show deficient hair coat recovery with patchy and incomplete hair growth (Fig. 1D). Untreated Sox9 fl/fl;K15-CrePGR R26YFP mice showed normal hair coat recovery, as did RU486-treated Sox9 Het and wild-type controls (shown in Fig. 1D).
Sox9 cKO HFs launch the hair cycle normally, but once activated, their HF-SCs lose stemness
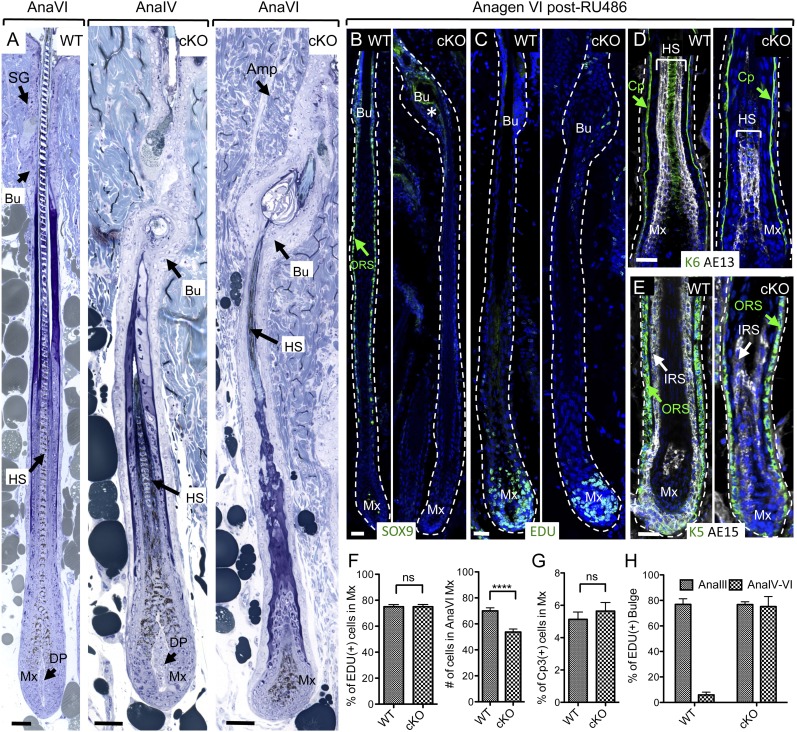
Following Sox9 ablation, mutant HF-SCs generated a seemingly normal early anagen-phase HF and hair shaft. Closer inspection, however, revealed that by Anagen IV, the bulge region showed signs of deformation, and HF downgrowth had markedly slowed. Moreover, by full anagen (Anagen VI), the hair shaft and IRS, which had formed normally at earlier stages, now showed signs of increased deformation (Fig. 2A,B; Supplemental Fig. 3A–C). Since the morphology of early anagen cKO HFs was normal and since Sox9 expression is normally silenced after wild-type ORS cells reach the TA matrix compartment, the later stage distortions of cKO HF/shaft morphology were likely secondary due to the physical barrier imposed by an increasingly aberrant bulge. Consistent with this notion, SOX9-deficient matrix cells were still proliferative (Fig. 2C) and gave rise to all HF lineages, including the K6+ companion layer, hair keratin+ (mAb AE13+) cells of cortex and cuticle, and trichohyalin+ (mAb AE15+) cells of IRS and hair shaft medulla (Fig. 2D,E).
Figure 2.
Sox9 ablation in adult HF-SCs results in bulge deformation and an anagen-dependent failure to return to quiescence. (A) Semithin sections of Sox9 wild-type (WT) and cKO HFs in the first anagen phase of the hair cycle following RU486 treatment in previous telogen. Note that (1) cKO HFs are shorter, and (2) IRS/hair shaft perturbations are subsequent to those of the bulge. (B) Anti-SOX9 immunofluorescence. (C) EdU incorporation (24-h pulse) in HF-SCs in first (full) Anagen VI following RU486. (D,E) K6 in the companion layer (Cp), hair keratins (AE13) in the hair shaft (HS) cortex and cuticle, and trichohyalin (AE15) in the IRS and HS medulla are still expressed in Sox9 cKO HFs. (F) Quantifications of EdU incorporation and total matrix cell number of Anagen VI HFs post-RU486. N = 3 mice for each genotype. (G) Quantification of activated caspase 3 in the matrix of Anagen VI HFs post-RU486. N = 3 mice for each genotype. (H) Quantifications of EdU incorporation in the bulge at the indicated time points. N = 4 mice for each genotype. Data are mean ± SEM. (****) P < 0.0001. Bars, 30 μm. White dashed lines denote the epidermal–dermal border, and the blue channel is DAPI staining. Asterisk indicates the nonspecific staining of SOX9 Ab. (Amp) Arrector pili muscle; (Bu) bulge; (DP) dermal papilla; (IRS) inner root sheath; (ORS) outer root sheath; (SG) sebaceous gland; (Mx) matrix; (ns) not significant.
We did notice that cells expressing HF differentiation markers diminished as anagen progressed. EdU labeling and quantification revealed that even though percentages of labeled TA matrix cells were normal, their total numbers were also significantly reduced in the Sox9 cKO HFs (Fig. 2F). This was not attributable to apoptosis, as activated Caspase3 labeling was still very low in the mutant matrix (Fig. 2G). Pinpointing the defect further, we showed that SOX9-deficient HFs entered anagen with kinetics comparable with their wild-type counterparts, indicating that HG progenitors at the bulge base still seemed to be responding normally to their niche cues. Moreover, at Anagen III, when wild-type bulge HF-SCs self-renew (Hsu et al. 2011), Sox9 cKO bulge HF-SCs behaved normally. Surprisingly however, once activated to self-renew, SOX9-deficient bulge HF-SCs failed to return to quiescence (Fig. 2C, H). Soon thereafter, HF downgrowth was arrested.
Taken together, these results were all consistent with the view that SOX9 is essential for maintaining the bulge HF-SCs and upper ORS, and, without SOX9, these normally SOX9+ cells are unable to fuel the normally SOX9− TA matrix and differentiating hair cells.
Within the bulge niche, activated Sox9 cKO HF-SCs fail to repress epidermal differentiation
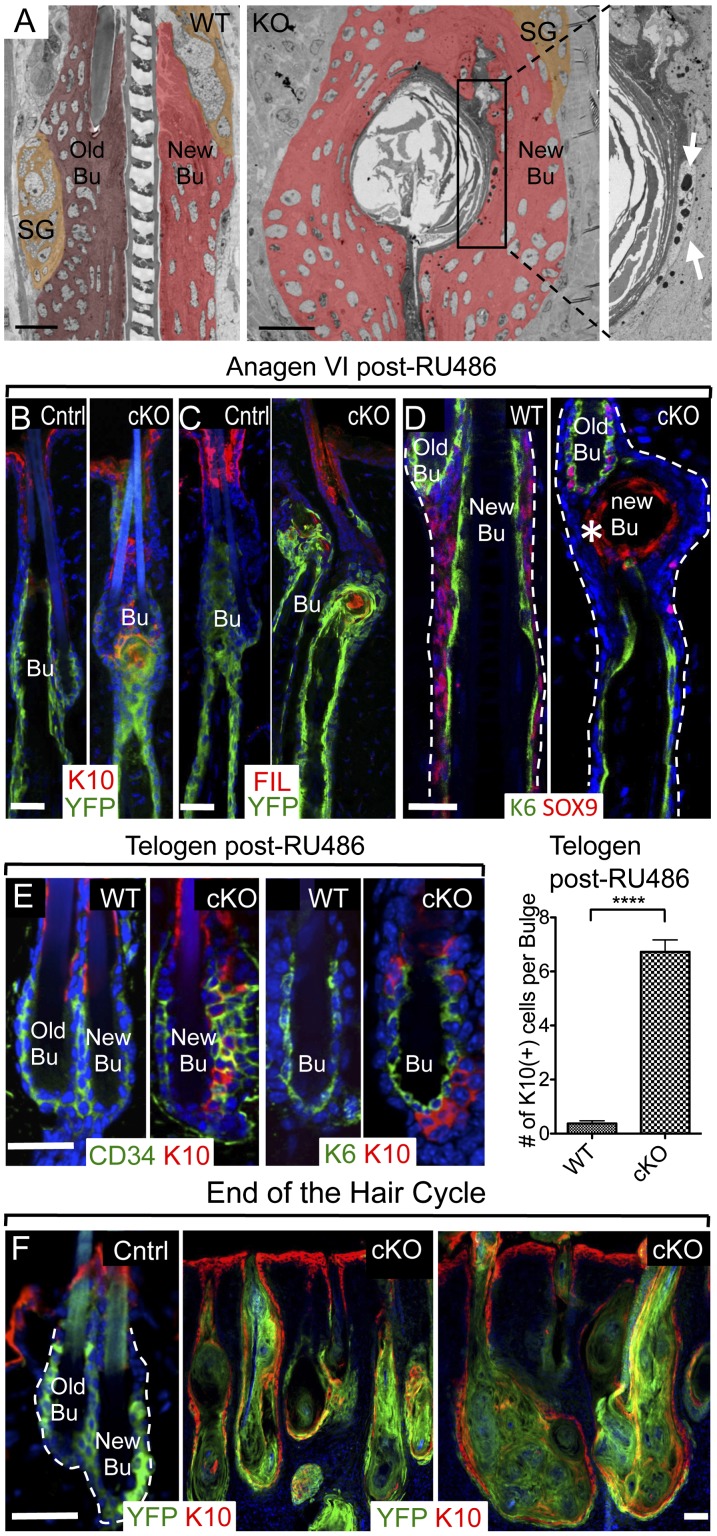
We returned to early anagen phase, where the first morphological consequences of Sox9 ablation in the bulge had emerged, which were soon followed by distortions in normally SOX9− hair cell populations. Closer inspection revealed the presence of keratinized pearls within the early SOX9-deficient bulge (Fig. 3A). Immunofluorescence microscopy further revealed the presence of K10 and filaggrin, indicative of terminal differentiation (Fig. 3B,C). The aberrant IFE differentiation appeared to be confined to the niche, as when Sox9 cKO HFs entered into anagen, YFP+ Sox9 cKO progeny along the ORS showed no signs of K10 and filaggrin immunostaining.
Figure 3.
Sox9 cKO HF-SCs differentiate into IFE cells. (A) Ultrastructure of the SC niche of wild-type (WT) and Sox9 cKO bulge of Anagen VI HFs after RU486 treatment. The new bulge (Bu), deformed at this stage, is marked by red pseudocolor; dark brown labels the old bulge, and light brown indicates the sebaceous gland. The boxed area, magnified at right, reveals keratinized pearls and keratohyalin granules (arrows) within the cKO bulge. (B) K10. (C) Immunostaining for filaggrin (FIL). K10 reveals atypical labeling of epidermal markers in the cKO bulge. (D) Immunolabeling shows that the K6+ inner bulge layer is not targeted and remains intact during the first anagen following Sox9 ablation in HF-SCs. Dashed lines denote the epidermal–dermal border. (E) Representative images of K10 immunostainings of telogen end-phase HFs treated with RU486 3 mo earlier at telogen start. Quantifications are at right. N = 3 mice for each genotype. Data are mean ± SEM. (****) P < 0.0001. (F) Depletion of SOX9 in adult HF-SCs leads eventually to the formation of K10-positive cysts. Blue channel is DAPI staining. (Cntrl) Control. Bars, 30 μm.
Intriguingly, while the actively cycling (newest) cKO bulge was undergoing these gross morphological and molecular changes, the old cKO bulge, which is not reused after its initial hair cycle (Hsu et al. 2011), remained quiescent and appeared to be largely intact morphologically (Supplemental Fig. 3A). It also displayed the characteristic SOX9+K6+ inner bulge layer, which still anchored to the old club hair (Fig. 3D). These findings further underscored the phenotypic dependency on hair cycle activation.
Spontaneous epidermal differentiation first appeared in the telogen bulge and was marked by a few K10+ cells within the CD34+ HF-SC layer (Fig. 3E). At this early stage, Sox9 cKO HF-SCs still exhibited molecular signs of stemness (Supplemental Fig. 2A). Soon after anagen entry, however, CD34 and other markers of stemness began to wane (Fig. 3E), and by the end of the first hair cycle in which Sox9 had been depleted, HFs were reduced to epidermal cysts (Fig. 3F).
Taken together, we placed the major SOX9 defect downstream from HF-SC survival and anagen onset but at the time (mid-anagen) when activated bulge HF-SCs normally return to quiescence. Moreover, our data showed that in SOX9's absence, activated HG progenitors still commit to HF differentiation programs, but activated bulge HF-SCs fail to both return to quiescence and also suppress epidermal differentiation within the niche.
Wnt and Notch signaling still occur in Sox9 cKO HFs
K10+ cysts have also been described when Notch signaling is compromised in HFs (Demehri and Kopan 2009; Hu et al. 2010; Lin et al. 2011). However, although signs of active Notch signaling, including the nuclear Notch intracellular domain (NICD) and downstream NICD target gene Hes1, were detected in the lower bulge/HG of telogen-phase HFs, they showed no significant differences between wild-type and cKO HFs (Supplemental Fig. 4A,B). Thus, changes in Notch signaling did not seem to account for the phenotype that we observed.
K10+ cysts have been also reported in HFs expressing a LEF1 mutant that lacks its β-catenin-binding domain and hence cannot be transactivated in response to Wnt signaling (Niemann et al. 2002). However, in the absence of SOX9, Wnt signaling still occurred, as judged by nuclear β-catenin in the activated HG cells and LEF1 in the emerging matrix cells (Supplemental Fig. 4C,D). Additionally, mRNA levels of Wnt targets seemed to be unaltered at the time point when phenotypic changes in Sox9 cKO telogen HFs were already evident (Supplemental Fig. 4E). mRNA levels of Tcf3 and Tcf4 were reduced somewhat but not enough to suggest that Wnt is responsible for the changes that we observed in the Sox9 cKO bulge.
Finally, inflammation can also lead to hair loss and cyst formation. Flow cytometry analysis, however, did not reveal any signs of immune cell infiltration in Sox9 cKO skin (Supplemental Fig. 4F).
HF-SCs that lose SOX9 show decreased expression of HF-SC signature genes
Delving deeper into how SOX9 loss leads to a HF-to-IFE lineage fate switch, we transcriptionally profiled late telogen-phase HF-SCs (YFP+/ScaI−/CD34+/α6hi) purified from Sox9 cKO and Het mice treated 2 wk earlier in telogen with RU486 (Supplemental Fig. 5A). We chose this timing because typifying stemness markers were still expressed by SOX9-deficient HF-SCs, but early molecular changes were detectable (Supplemental Fig. 2).
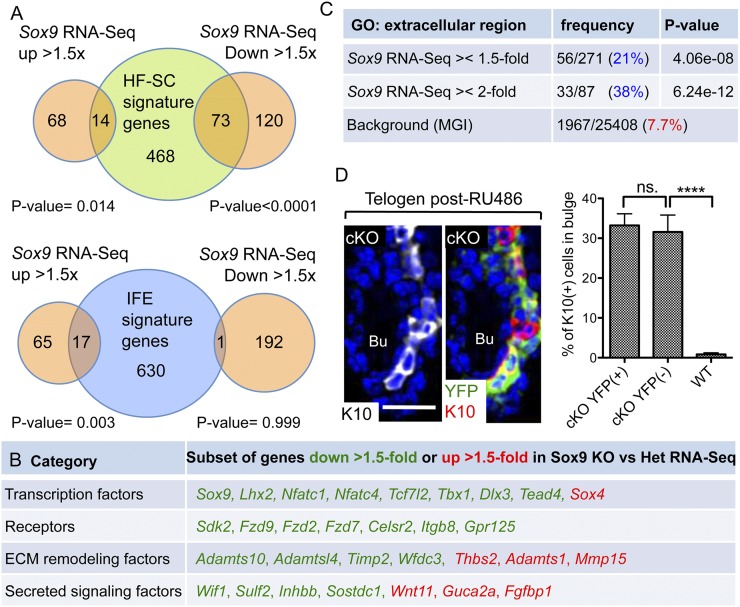
mRNAs isolated from these two samples were subjected to deep sequencing and bioinformatics analyses. Two-hundred-seventy-five protein coding genes were differentially expressed by ≥1.5-fold (P-value < 0.05) in Sox9-null versus Het HF-SCs. Interestingly, mRNAs that were down-regulated by ≥1.5× in Sox9-null HF-SCs overlapped significantly with not only the HF-SC signature but also the HF lineage, while mRNAs up-regulated by ≥1.5-fold in Sox9-null HF-SCs overlapped most significantly with mRNAs enriched in basal IFE cells (Fig. 4A; Supplemental Fig. 6B; Supplemental Table 2). The switch included repression of a cohort of HF lineage transcriptional regulators, including HF-SC genes Lhx2, Sox9, Tcf7l2, Tbx1, and Nfatc1 (Fig. 4B; Supplemental Fig. 6A; Supplemental Table 2). Additionally, transcriptional regulators for HF differentiation were transcribed at even lower levels than normal. This shift away from HF-SC and toward the IFE signature was consistent with the IFE differentiation observed within the Sox9 cKO HF bulge.
Figure 4.
Changes in gene expression in HF-SCs lacking SOX9. (A) Transcriptional profile of SOX9-deficient HF-SCs is shifted away from the normal HF-SC “signature” toward the basal IFE “signature.” The top diagram shows overlap of genes differentially expressed by ≥1.5× in Sox9 cKO versus Het HF-SCs (Sox9 RNA-seq), and genes were enriched ≥2× in telogen HF-SCs versus basal IFE (HF-SC signature genes). The bottom diagram shows overlap of Sox9 RNA-seq and genes enriched ≥2× in IFE versus telogen HF-SCs (IFE signature genes). (B) Table shows examples of genes ≥1.5-fold down-regulated (green) or ≥1.5-fold up-regulated (red) in Sox9 RNA-seq. (C) Gene ontology (GO) analyses of genes differentially expressed by ≥1.5× or ≥2× in Sox9 cKO versus Het HF-SCs denotes the “extracellular region” category as the most significantly enriched compared with the 7.6% frequency in the Mouse Genome Informatics (MGI) database. (D) K10 and YFP immunofluorescence of telogen Sox9 cKO bulge prior to hair cycling. (Blue) DAPI. At right, the percentage of K10+ cells within YFP+ and YFP− HF-SCs of the mosaic Sox9 cKO bulge (YFP is in 60%–80% of HF-SCs) and wild-type (WT) bulge. N = 3 mice for each genotype. Data are mean ± SEM. (****) P < 0.0001. Bar, 30 μm.
Gene ontology analyses revealed that many SOX9-dependent changes in gene expression were associated with secreted proteins (Fig. 4C). This was noteworthy, since the best-known SOX9 transcriptional targets of chondrocytes include extracellular matrix (ECM) genes such as Col2a1 (Lefebvre et al. 1997). To test the possibility that the SOX9-dependent phenotype may at least in part be rooted in cell-nonautonomous defects, we quantified K10 expression in YFP+ Sox9− and YFP− Sox9+ HF-SC subpopulations from Sox9-cKO × R26YFP mice. As shown in Figure 4D, K10 was detected in both bulge populations irrespective of whether SOX9 was present. In contrast, K10 was not seen in the bulge of wild-type HFs (Fig. 4D, shown in the graph). Together, these findings suggest that within the bulge niche, Sox9-null HF-SCs secrete factors that induce IFE differentiation in themselves and their neighboring bulge cells.
SOX9 functions primarily as an activator of gene expression in adult HF-SCs
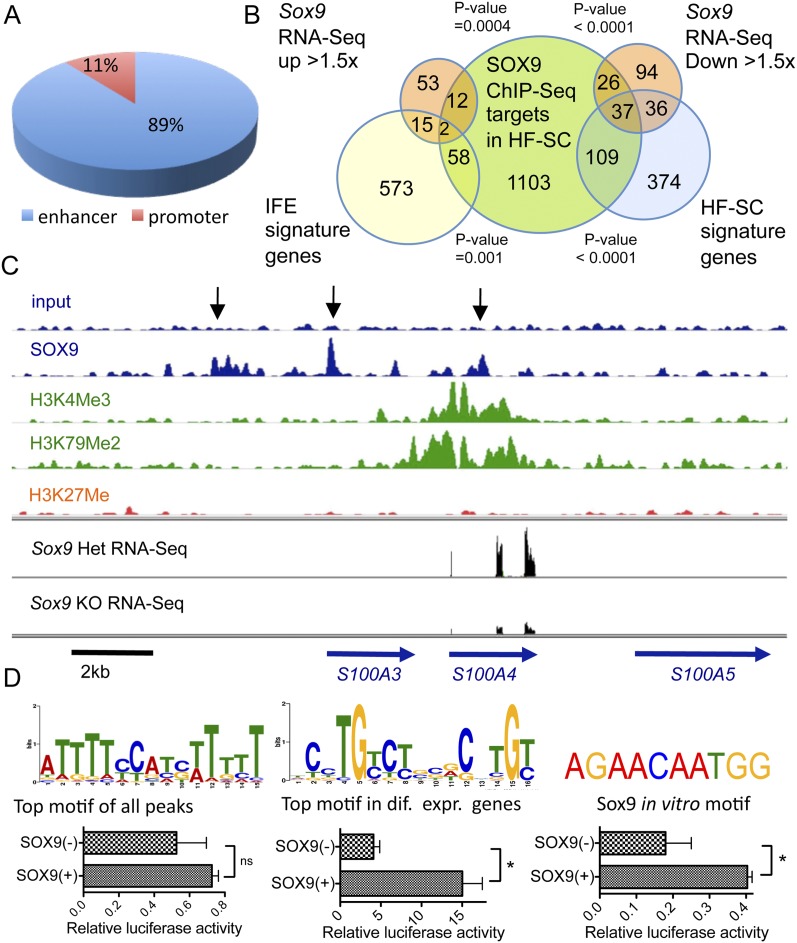
Second telogen HF-SCs could be FACS-purified in sufficient quantities to conduct SOX9 ChIP-seq in vivo (Supplemental Fig. 5B). One-thousand-three-hundred-eighty-four genes contained SOX9-binding sites in either the promoter (±2 kb of transcription start sites) or “enhancer” (−50 kb upstream of and +5 kb downstream from the gene body) (Fig. 5A; Supplemental Table 3). This list included Nfatc1, Lhx2, and Tcf7l2 (TCF4), previously shown to be critical for maintaining HF-SCs (Merrill et al. 2001; Nguyen et al. 2006, 2009; Rhee et al. 2006; Horsley et al. 2008; Folgueras et al. 2013). In this regard, SOX9 resembled SOX2, which, in embryonic SCs, binds to genes encoding pluripotency regulators (Boyer et al. 2005; Chen et al. 2008).
Figure 5.
SOX9 functions primarily as a transcriptional activator in adult HF-SCs. (A) Distribution of SOX9 peaks between promoters (−2 kb to +2 kb of transcription start sites) and “enhancers” (−50 kb of transcription start sites, gene body, +5 kb downstream from the gene) of SOX9-bound genes. (B) Diagram showing overlap of in vivo SOX9-bound targets (ChIP-seq) and mRNAs differentially expressed (RNA-seq) in Sox9 cKO versus Het HF-SCs. Overlaps represent the number of genes bound by SOX9 and whose expression is up-regulated or down-regulated by ≥1.5-fold in Sox9 cKO. Overlaps with HF-SC signature and IFE signature genes are also shown. (C) ChIP-seq profiles of the S100A4 genomic locus in wild-type (WT) telogen HF-SCs. Input represents the sequencing profile of genomic DNA used in ChIP pull-downs. Black arrows indicate locations of DNA fragments enriched in SOX9 ChIP. The SOX9 ChIP-seq profile is also compared with published data of histone modifications in telogen HF-SCs (Lien et al. 2011): H3K4me3 Ab pulls down promoter regions, H3K79me2 marks actively transcribed DNA, and the H3K27me3 label is enriched in polycomb-repressed chromatin. Below is an RNA-seq profile of the S100A4 locus in Sox9 cKO and Het HF-SCs. The locations of RNA-seq fragments match with exons in the S100A4 gene. All tracks are set to the same scale. The lengths of genes and direction of transcription are indicated by blue arrows. (D) SOX9-binding motifs (from left): the most enriched motif within SOX9 peaks in HF-SCs, the motif enriched most within SOX9 peaks associated with differentially expressed genes in Sox9 cKO versus Het, and the previously reported SOX9 in vitro motif (Mertin et al. 1999). Below, relative luciferase activities driven by SOX9 motifs were used as an enhancer. Assays were performed in 293FT cells (SOX9-negative) in the presence of a SOX9 expression vector or empty vector. All results were normalized to luciferase activity when firefly luciferase reporter with minimal CMV was used and to the Renilla luciferase control reporter driven by the HSV TK promoter. Experiments for each construct were performed in triplicate in duplicate wells. Data are mean ± SEM. (ns) Not significant; (*) P < 0.05.
Comparison of SOX9 ChIP-seq data with our list of genes ≥1.5-fold differentially expressed in Sox9 cKO versus wild-type HF-SCs produced a short list of 77 putative SOX9-regulated HF-SC genes (Supplemental Table 2C). Notably, the overlap with ChIP-seq data was significantly higher with genes whose expression was decreased in the absence of Sox9, suggesting that SOX9 functions primarily as a transcriptional activator in HF-SCs. Thus, whereas 33% of down-regulated genes bound SOX9 (P-value < 0.0001), only 17% of up-regulated genes did so (P-value = 0.0004) (Fig. 5B).
Fifty-nine percent of putative SOX9 targets that were down-regulated ≥1.5-fold in Sox9 cKO HF-SCs were also enriched in wild-type HF-SCs compared with IFE progenitors. This included Wwp2, which was previously identified by ChIP-qPCR as a direct downstream SOX9 target in chondrocytes (Nakamura et al. 2011). Further insights into SOX9's role as a transactivator came from comparing our SOX9 ChIP-seq profile with activating and repressive histone methylation marks on HF-SC chromatin (Supplemental Table 3; Lien et al. 2011). Figure 5C shows these profiles for S100A4, a representative HF-SC signature gene that bound SOX9. As illustrated, at least three distinct SOX9 peaks were identified within and upstream of the gene body, whereas histone H3 trimethylation of lysine residue 4 (H3K4me3), analogous to the H3K79me2 mark, characteristic of transcriptional elongation (Lien et al. 2011), was prevalent over the S100A4 gene body. Conversely, S100A4 lacked the repressive Ezh2/polycomb complex mark H3K27me3. These marks agreed well with our S100A4 RNA-seq profiles, where transcription of all three exons of S100A4 was repressed upon loss of SOX9 in HF-SCs (Fig. 5C).
We carried out unbiased sequence motif searches first within the 600 SOX9 peaks with the strongest ChIP-seq signals and then within peaks of genes whose expression changed by >1.5× upon Sox9 ablation. The resultant top two motifs suggested a low sequence specificity of SOX9 binding in HF-SCs (Fig. 5D). Moreover, even the motif underlying the strongest peaks in HF-SC chromatin diverged at several sites from the SOX9 DNA-binding motif that emerged from in vitro studies in which recombinant SOX9 was exposed to a mixture of random oligonucleotides (Mertin et al. 1999). Similar differences have been noted previously, first for the 44 putative targets bound by SOX9 in ChIP hybridizations of 80 selected chondrocyte promoters (Oh et al. 2010) and then for the 109 putative targets identified in a genome-wide SOX9 ChIP-on-ChIP of embryonic testis chromatin (Bhandari et al. 2012). That said, none of these consensus sites were identical, further pointing to the weak specificity of SOX9 for its in vivo targets. Low binding specificity is emerging as a general feature of Sox proteins (Lefebvre et al. 1998; Kamachi et al. 1999).
Chromatin binding does not necessarily imply transcription factor regulation. To address whether SOX9 has the capacity to regulate the SOX9-binding motifs that we identified from our in vivo SOX9 ChIP-seq study, we compared the relative activities of enhancers composed of 3× multimerized motifs identified from either in vitro oligonucleotide analyses (Mertin et al. 1999), genome-wide analyses of the strongest SOX9 peaks, or analyses of the SOX9 peaks from the 103 genes that exhibited differential expression upon Sox9 ablation in HF-SCs. To evaluate SOX9 dependency and enhancer activity, we transfected our expression vectors with or without an additional SOX9 expression vector (controlled for equal DNA levels).
Each enhancer showed some SOX9-dependent activity (Fig. 5D). However, when compared relative to the Renilla luciferase internal control, the most potent enhancer and the one with greatest sensitivity to SOX9 (>15×) was that constructed from the in vivo motif that was present specifically in genes that also showed SOX9 transcriptional sensitivity in HF-SCs (shown in Fig. 5D). These results show that when multimerized, this SOX9-binding motif is sufficient on its own to act as an enhancer. That said, the weaker activity of the enhancer composed of in vitro optimized SOX9-binding sites, coupled with the relative lack of specificity of the in vivo SOX9 motifs, raises the possibility that the SOX9-dependent regulation conferred by in vivo SOX9 motifs may be complex and involve associated factors. Although beyond the scope of the present study, our findings are consistent with accumulating evidence suggesting that Sox factors do not act alone in their function (for review, see Kamachi and Kondoh 2013).
SOX9 is a positive regulator of genes involved in Activin/TGFβ signaling
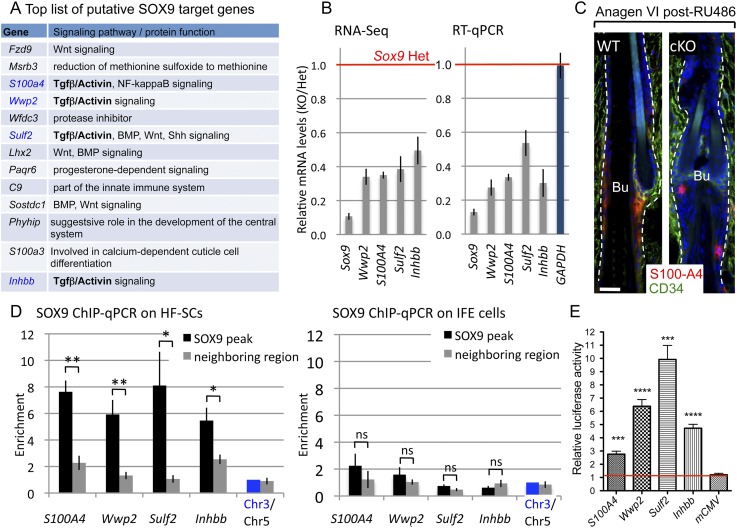
To gain further insights into the pathways preferentially regulated by SOX9, we focused on highly expressed HF-SC signature genes (FPKM >4 in Sox9 Het RNA-seq) that were strongly bound by SOX9 in their promoter/enhancer and also down-regulated in Sox9 cKO HF-SCs more than twofold in both RNA-seq replicates. Among this short list of 13 genes were Wwp2, S100A4, Sulf2, and Inhbb, previously reported to enhance TGFβ/Activin signaling (Fig. 6A; Matsuura et al. 2010; Soond and Chantry 2011; Zheng et al. 2013). qRT–PCR on independently purified HF-SCs confirmed that their mRNAs were down-regulated in cKO HF-SCs (Fig. 6B). GAPDH and Ppib were internal mRNA loading controls. Bulge-specific reduction of S100A4 was further confirmed by immunolabeling (Fig. 6C).
Figure 6.
SOX9 is a positive regulator of genes involved in Activin/TGFβ signaling. (A) Top list of putative SOX9 targets that were highly expressed in adult HF-SCs (FPKM >5 in Het RNA-seq), strongly influenced by SOX9 loss (down-regulated by twofold or more in cKO versus Het), and associated with a strong SOX9 peak in ChIP-seq (peak height ≥13). (B) RT-qPCR verification of RNA-seq data. Independent FACS-purified samples were analyzed for putative SOX9 target genes involved in TGFβ/Activin signaling. For RT-qPCR, all results were normalized to Ppib expression; GAPDH was used as an internal control. Data are mean ± SEM. N = 4 mice per genotype. (C) S100A4 immunofluorescence in Sox9 cKO and wild-type (WT) HFs in the first full anagen after targeting. White lines denote the epidermal–dermal border. (Blue) DAPI. Bar, 30 μm. (D) Independent SOX9 ChIP-qPCR on FACS-purified HF-SCs (SOX9-positive) and IFE (SOX9-negative) cells. For each gene, primers were designed to amplify region where the SOX9 peak was detected (black bars) and neighbor regions 4–8 kb away from the SOX9 peak (gray bars). Note that enrichment of DNA fragments was specific to HF-SCs and regions underneath the peaks, validating the specificity of SOX9 Ab and binding. Results were normalized to the intergenic Chr3 region; the intergenic Chr5 region was used as a control. Experiments were done in triplicate. Data are mean ± SEM. (*) P < 0.05; (**) P < 0.01. (E) Relative luciferase activities driven by sequences (300–600 bp) encompassing SOX9 ChIP-seq binding sites. Results are presented as the enhancement of luciferase activity in the presence of a SOX9 expression vector compared with an empty expression vector. All results are normalized to the luciferase activity of Renilla luciferase control reporter vector. Experiments for each construct were performed in four replicates in duplicate wells. Data are mean ± SEM. (***) P < 0.001; (****) P < 0.0001.
To further validate our SOX9 ChIP-seq data, we performed SOX9 ChIP-qPCR on HF-SCs (SOX9+) and IFE (SOX9−). These analyses confirmed that the enrichment obtained from SOX9 ChIP was SOX9-specific and did not occur when SOX9− cells were used in this assay (Fig. 6D). Enrichment of DNA fragments in SOX9 ChIP-qPCR was specific to regions where SOX9 ChIP-seq peaks were detected, as shown by analyses of neighboring chromatin regions and random chromatin regions lacking SOX9 peaks (shown in Fig. 6D).
To verify that SOX9 regulates the transcription of these genes, we carried out luciferase reporter assays in cultured cells with or without Sox9 expression vector. Relative to the control minimal CMV promoter, each 300- to 600-base-pair (bp) element encompassing SOX9-binding sites of either S100A4, Wwp2, Sulf2, or Inhbb displayed marked enhancer activity, elevating luciferase expression by as much as 10× in the presence of SOX9 (Fig. 6E). The effects were SOX9-dependent, as without SOX9, levels plummeted to that of the minimal promoter (Fig. 6E, red line). These data establish Wwp2, Inhbb, S100A4, and Sulf2 as bona fide targets of SOX9.
SOX9 maintains the identity of HF-SCs through Activin signaling
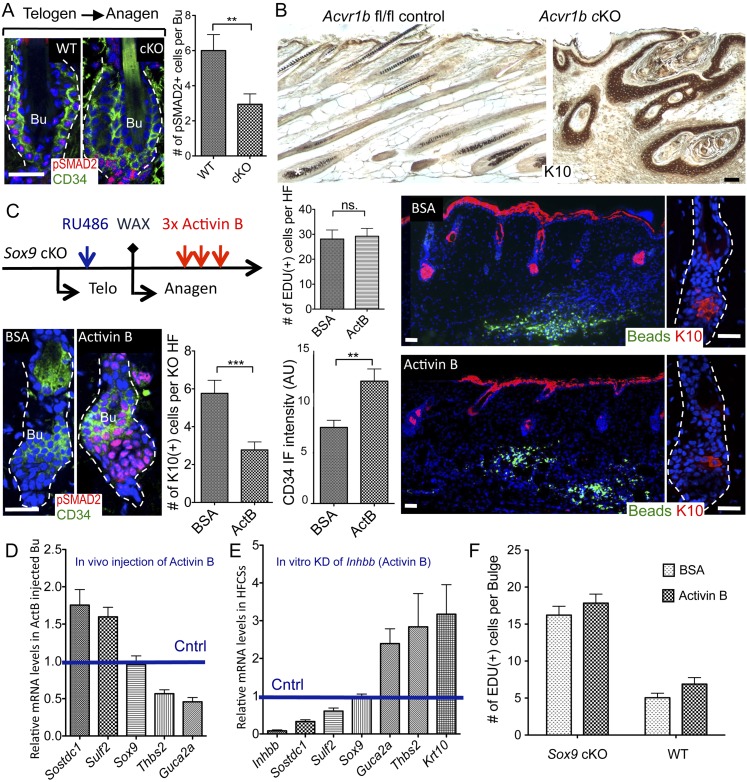
pSMAD2, the downstream effector of TGFβ/Activin signaling, is up-regulated at the telogen → anagen transition and aids in driving HF regeneration (Oshimori and Fuchs 2012). Since Wwp2, Inhbb, S100A4, and Sulf2 have been implicated as enhancers of TGFβ/Activin signaling, their down-regulation in Sox9 cKO HF-SCs suggested that SOX9 may be involved at least in part in regulating this pathway. Indeed, in comparison with the wild-type bulge at telogen → anagen, pSMAD2 staining was significantly reduced in Sox9 cKO bulge HF-SCs at the same stage (Fig. 7A).
Figure 7.
SOX9 maintains the identity of HF-SCs through Activin signaling. (A) pSMAD2 immunofluorescence of HFs during the first telogen–anagen transition after RU486. Quantifications are at right. N = 3 mice per genotype. Data are mean ± SEM. (**) P < 0.01. (B) K10 immunohistochemistry of cKO Acvr1b mice and fl/fl control (wild-type [WT]) mice. Asterisk indicates melanin. (C) Intradermally injected Activin B inhibits the trans-differentiation of HF-SCs to IFE caused by SOX9 loss. Mice treated with RU486 in second telogen were depilated to synchronize hair cycles and expedite the phenotype. Recombinant Activin B and BSA were injected intradermally into the same mouse at two different locations over the 3 d. Immunostaining for pSMAD2 and CD34 in Activin B and BSA-treated HFs are shown at left; K10 is shown at right. Quantifications of K10+ cells, EdU+ cells per HF, and the total fluorescence intensity of the CD34 label are shown in the middle. N = 4 mice. Data are mean ± SEM. (**) P < 0.01; (***) P < 0.001. White lines denote the epidermal–dermal border. (Blue) DAPI. Bar, 30 μm. (D) RT-qPCR quantification of mRNA levels of secreted factors in Activin B-treated bulge. HF-SCs were FACS-purified from Activin B and BSA injection sites independently from two Sox9 cKO mice. Data are mean ± SEM. (E) RT-qPCR quantification of mRNA levels of secreted factors in cultured wild-type HF-SCs, where Activin B. expression has been reduced by Inhbb shRNA. Data are mean ± SEM based on four independent experiments. (F) Quantifications of EdU+ cells in wild-type and Sox9 cKO bulge at Anagen IV, after treatment with BSA or Activin B. N = 2 mice. Data are mean ± SEM.
Mice whose skin epithelium lacks TGF-β receptor II (and hence TGFβ signaling) do not exhibit gross alterations during normal homeostasis (Guasch et al. 2007; Oshimori and Fuchs 2012). In contrast, when Activin receptor type 1B (Acvr1b) is conditionally targeted for ablation, HFs degenerate, resulting in large cysts with keratinaceous debris (Qiu et al. 2011). Given these phenotypic parallels, we focused on the hypothesis that SOX9 acts on genes of the Activin pathway to sustain stemness and block IFE differentiation.
Similar to Sox9 cKO HFs, cysts in Actvr1b cKO mice showed signs of IFE differentiation and K10 expression (Fig. 7B). To test whether Activin signaling is directly important for repressing IFE fate in the HF-SC niche, we asked whether Activin B, encoded by Inhbb, could rescue this defect. To this end, we injected RU486-treated Sox9 cKO mice in their second telogen with recombinant Activin B or BSA (control) along with fluorescent beads to mark intradermal injection sites. Since prominent signs of HF → IFE transition surface at telogen → anagen, we then waxed back skins to remove hair shafts and induce synchronized anagen.
As shown in Figure 7C, Activin B restored nuclear pSMAD2 and significantly reduced the number of K10+ Sox9 cKO HF-SCs. This decline in K10 was clearly specific to Activin B, as it tapered off further from the injection site (shown in Fig. 7C). Concomitantly, CD34 immunofluorescence intensity was increased in Activin-treated, Sox9 cKO HFs. While CD34 repression is a common sign of HF-SC proliferation, proliferation was not affected.
To further study the role of Activin in SOX9-mediated control of the bulge niche, we treated telogen Sox9 cKO HFs with Activin B and BSA, FACS-purified HF-SCs from injection sites, and measured their mRNA levels of secreted factors whose expression was altered by SOX9 loss. Interestingly, mRNA levels of HF-SC signature genes Sostdc1 and Sulf2 were increased, while levels of IFE signature genes Thbs2 and Guca2a were decreased after the Activin B injection (Fig. 7D). In a good correlation, Sostdc1 and Sulf2 expression was decreased, while Thbs2 and Guca2a were increased after knocking down Inhbb in cultured wild-type HF-SCs (Fig. 7E).
Once activated, Sox9 cKO HF-SCs cannot return to quiescence. To test whether restoring Activin signaling can restore this defect, we synchronized the hair cycle by waxing mice and then injected Activin B and BSA intradermally between 4 and 6 d after depilation; i.e., the time when proliferation of wild-type HF-SC starts to decrease. EdU labeling and quantifications showed that Activin B did not affect HF-SC proliferation in either wild-type or cKO mice (Fig. 7F).
Taken together, these findings were consistent with the non-cell-autonomous effects that we observed previously between SOX9 loss and IFE fate conversion and provide strong evidence that this link operates at least partially through Activin signaling. However, it also illustrates the multifaceted ways in which SOX9 regulates HF-SC behavior.
Discussion
Cell-autonomous and non-cell-autonomous functions for SOX9 in governing the SC niche
When Sox9 is targeted in development, HF-SCs never form, resulting in the arrest of SOX9-dependent HF morphogenesis but not SOX9-independent IFE (Vidal et al. 2005; Nowak et al. 2008). Since HF-SCs fail to develop without SOX9, SOX9's function as an HF-SC transcriptional regulator has remained unclear. This issue is also largely unexplored for most SOX9+ cell types that contribute to adult tissue homeostasis (Furuyama et al. 2011; Guo et al. 2012; Lu et al. 2012). We now address this issue for the HF. Our results show clearly that when Sox9 is ablated in adult HF-SCs, they are unable to maintain stemness. Moreover, by governing expression of a number of extracellular factors, SOX9 can exert both cell-autonomous and non-cell-autonomous effects on SC behavior. Indeed, one of the first signs of Sox9 ablation was that a few intact wild-type HF-SCs within the niche began to aberrantly differentiate along the IFE lineage.
Defects in SC behavior were manifested soon thereafter. While HFs transitioned from telogen → anagen and SOX9-deficient bulge HF-SCs launched self-renewal on cue in early anagen, they failed to return to quiescence in Anagen IV. Despite this activity, cKO HF-SCs could not sustain ORS or TA matrix pools, grinding hair lineage differentiation to a halt and underscoring the paucity of stemness within these proliferating Sox9-ablated bulge cells. Indeed, SOX9-deficient HF-SCs differentiated along the IFE lineage, fully exhausting the HF-SC pool in bulge and upper ORS, where SOX9 is normally expressed.
Additionally noteworthy was the paucity of HF differentiation defects even though HF-SC-mediated fueling of TA matrix progenitors was severely compromised by SOX9 loss. In wild-type HFs, SOX9 marks the HF-SCs in the bulge and the upper ORS but is down-regulated as HF lineages progress. In contrast, SOX4 and SOX6 are expressed in the HG, ORS, and matrix (Rendl et al. 2005; Greco et al. 2009; Lien et al. 2011), and SOX18 and SOX21 are expressed later in hair differentiation (Pennisi et al. 2000; Kiso et al. 2009). Whether these changes in gene expression compensate and contribute to the relative normalcy of Sox9-null HF-SC progeny is beyond the scope of the present study. However, in some other tissues, redundancy between SOX9 and some of its family members has been described (Lefebvre et al. 1998; Stolt et al. 2003).
Adult SCs reside in a discrete microenvironment, which plays a dominant role in regulating SC maintenance, self-renewal, activation, and differentiation. In the HF-SC niche, signals from the inner bulge layer, DP, dermal fibroblasts, and adipocyte precursor cells have all been shown to impact HF-SC quiescence and hair cycle activation (Botchkarev et al. 1999; Plikus et al. 2008; Rendl et al. 2008; Festa et al. 2011; Hsu et al. 2011). While these collective studies provide insights into how stemness is influenced by extrinsic signaling, few studies have focused on whether SCs themselves might express factors that impact their own and their neighbors' behavior and, if so, how.
After identifying genes whose expression had changed upon conditional ablation of Sox9 in adult HF-SCs, we realized that a remarkable number of these genes encode either secreted proteins or proteins with an extracellular domain. Moreover, in HFs containing a mixture of wild-type and Sox9 mutant HF-SCs, IFE differentiation was induced by not only the mutant but also the wild-type HF-SCs. These findings expose a hitherto unrecognized communication among HF-SCs within their native niche to maintain their own stemness. Moreover, our findings also illuminate the importance of SOX9 to niche microenvironment.
It was notable that many of SOX9's bona fide target genes encode a group of proteins known to enhance Activin/TGFβ signaling and that Activin B injections rescued the IFE differentiation defect in the Sox9 mutant niche. These findings underscore Inhbb, which encodes Activin B, as a physiologically significant SOX9 target. Moreover, given the striking differences between conditional loss of TGFβ and Activin receptor signaling with regard to their effects on the HF-SC niche, our findings further suggest that despite their shared ability to activate pSMAD2-mediated transcription, TGFβs and Activins have nonoverlapping functions that cannot be compensated for by the expression of each other. In this regard, it is intriguing that TGFβ2 is induced by HG at anagen onset, enhances pSMAD2 in the HG, and promotes HG proliferation by lowering BMP signaling (Oshimori and Fuchs 2012). In contrast, consistent with the conventional role of TGFβ family members in restricting keratinocyte proliferation, Sox9 deficiency (and reduced Activin/pSMAD2) was manifested in the activated bulge HF-SCs, which did not return to quiescence in mid-anagen.
Broader mechanistic issues for regulating SC behavior
The complexities that we unearthed in examining one SOX9 target, Inhbb, in this study make it unlikely that a few linear pathways will explain SOX9's governance of stemness. Further support for this notion stems from the fact that even though Lhx2 is one of the genes most markedly repressed upon loss of SOX9, its loss-of-function phenotype is quite distinct from that of Sox9. On the one hand, LHX2 and SOX9 orchestrate quiescence and stemness, and their loss leads to proliferation and loss of HF-SC markers and behavior. On the other hand, each has their own impact on lineage suppression, with LHX2 playing a more important role in sebocyte differentiation (Folgueras et al. 2013), and SOX9 playing a greater role in suppressing IFE differentiation. Together, these factors maintain stemness and direct HF-SCs along the correct lineage. That said, our studies show that SOX9 binds and regulates Lhx2 in HF-SCs, while LHX2 ChIP-seq did not reveal binding to Sox9 (Folgueras et al. 2013). This could account for the more severe, combined Activin B/LHX2-deficient-like phenotype of our Sox9 cKO mice.
In closing, we are left with the view that adult HF-SC behavior is determined by inputs from multiple niche cell types, including SCs themselves. This guarantees that the decision of when to make tissue and what lineage to follow happens in synchrony and only when a variety of cellular inputs from the tissue are met. Our findings might also be relevant to human epidermal cells, which, unlike their mouse counterparts, express SOX9, which, when overexpressed, causes reduced expression of differentiation-related genes (Shi et al. 2013).
Materials and methods
Mice and labeling experiments
Sox9fl/fl mice (Akiyama et al. 2002) were crossed with transgenic Krt15-CrePGR mice (Morris et al. 2004) and R26YFP mice (Srinivas et al. 2001). Cre-PGR was activated in 2-mo-old mice by RU486 treatment: 7-d intraperitoneal injection of 1% RU486 (VWR) in sesame oil (0.75mg/10g body weight per day; Sigma) together with 2-wk topical application of 4% RU486 in ethanol. Knock-in Sox9-creERT2 mice (Soeda et al. 2010) were crossed with R26YFP mice. CreER was activated by intraperitoneal injection of 2-mo-old mice with 2% tamoxifen (Sigma) in corn oil (1mg/10 g body weight per day for 2 d; Sigma). For EdU pulse experiments, mice received intraperitoneal injections of EdU (25 μg/g; Invitrogen) for 24 h (anagen HFs) or 10 d (telogen HFs). Depilation of mid-dorsal HFs was achieved on anesthetized mice with molten wax, which was peeled off after hardening. Recombinant human Activin B (500 ng; R&D Systems) or BSA control was repeatedly intradermally injected for 3 d with FluoSpheres (Invitrogen) into the back skin post-depilation. Postnatal day 50–60 (P50–P60) CD1 mice from Charles River Laboratories were used for SOX9 ChIP-seq and ChIP-qPCR experiments. Mice were housed and bred in the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited Comparative Bioscience Center (CBC) at The Rockefeller University in accordance with National Institutes of Health guidelines, and procedures were performed with Institutional Animal Care and Use Committee (IACUC)-approved protocols.
ChIP-seq assay and analysis
All materials, methods, and sequencing for ChIP-seq have been previously described (Lien et al. 2011). Independent immunoprecipitations were performed on FACS-sorted populations. Isolation of HF-SCs and IFE cells by FACS has been described elsewhere (Nowak and Fuchs 2009; Folgueras et al. 2013). Two replicates of SOX9 ChIP-seq were performed in HF-SCs, and one replicate was performed in IFE cells; 5 × 106 cells were used for each ChIP-seq run. Rabbit polyclonal anti-mouse SOX9 (Millipore, AB5535) was used for ChIP-seq. ChIP-seq reads were aligned to the mouse genome (mm9, build 37), and peaks were called with MACS software (Zhang et al. 2008). SOX9-bound targets were defined as mouse RefSeq genes with a peak at their promoters (−2 kb to +2 kb of transcription start sites) or enhancers (−50 kb of transcription start sites, gene body, +5 kb downstream from the gene). For the identification of SOX9 targets, only peaks that were specific to HF-SCs were called. The MEME software suite (Bailey et al. 2009) was applied to the sequences under the SOX9 peaks (200 bp around summit) for finding enriched motifs. ChIP-seq raw data will be submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo).
Statistics
To determine the significance between two groups, comparisons were made using unpaired two-tailed Student's t-test in Prism5 (GraphPad software). For all statistical tests, the 0.05 level of confidence was accepted for statistical significance.
Acknowledgments
We are grateful to G.H. Su for sharing the Acvr1b KO and wild-type tissue slides. We thank E. Ezratty, A.R. Folgueras, I. Matos, Y.-C. Hsu, N. Oshimori, W.-H. Lien, C.-Y. Chang, C.P. Lu, E. Heller, S.E. Williams, S. Beronja, S. Naik, and other Fuchs laboratory members for helpful discussions and critical reading of the manuscript. We thank D. Oristian and A. Aldeguer for expert assistance in the mouse facility; M. Nikolova, J. Racelis, S. Chai, E. Wong, and P. Janki for excellent technical assistance; the Rockefeller flow cytometry facility, headed by S. Mazel, for cell sorting; the Rockefeller Bioimaging facility, headed by A. North, for assistance with image acquisition; and the Rockefeller and Weil Cornell Medical School Genomics Resource Centers for their assistance with high-throughput sequencing. M.K. was supported by an Human Frontier Science Program Post-doctoral Fellowship. E.F. is a Howard Hughes Medical Institute Investigator. This work was supported by a grant from the National Institutes of Health (R01-AR31737).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.233247.113.
References
- Akiyama H, Chaboissier M-C, Martin JF, Schedl A, de Crombrugghe B 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16: 2813–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME suite: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari RK, Haque MM, Skinner MK 2012. Global genome analysis of the downstream binding targets of testis determining factor SRY and SOX9. PLoS ONE 7: e43380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E 2004. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118: 635–648 [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, Botchkareva NV, Roth W, Nakamura M, Chen LH, Herzog W, Lindner G, McMahon JA, Peters C, Lauster R, et al. 1999. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat Cell Biol 1: 158–164 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Vega VB, Ng H-H 2008. Transcriptional regulatory networks in embryonic stem cells. Cold Spring Harb Symp Quant Biol 73: 203–209 [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM 1990. Label-retaining cells reside in the bulge area of pilosebaceous unit: Implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61: 1329–1337 [DOI] [PubMed] [Google Scholar]
- Demehri S, Kopan R 2009. Notch signaling in bulge stem cells is not required for selection of hair follicle fate. Development 136: 891–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa E, Fretz J, Berry R, Schmidt B, Rodeheffer M, Horowitz M, Horsley V 2011. Adipocyte lineage cells contribute to the skin stem cell niche to drive hair cycling. Cell 146: 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueras AR, Guo X, Pasolli HA, Stokes N, Polak L, Zheng D, Fuchs E 2013. Architectural niche organization by LHX2 is linked to hair follicle stem cell function. Cell Stem Cell 13: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E 2007. Scratching the surface of skin development. Nature 445: 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Green H 1980. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell 19: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, et al. 2011. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet 43: 34–41 [DOI] [PubMed] [Google Scholar]
- Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Cruz-Racelis Dela J, Fuchs E 2009. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E 2007. Loss of TGFβ signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell 12: 313–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell G, et al. 2012. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148: 1015–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E 2008. NFATc1 balances quiescence and proliferation of skin stem cells. Cell 132: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-C, Pasolli HA, Fuchs E 2011. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Lefort K, Qiu W, Nguyen B-C, Rajaram RD, Castillo E, He F, Chen Y, Angel P, Brisken C, et al. 2010. Control of hair follicle cell fate by underlying mesenchyme through a CSL–Wnt5a–FoxN1 regulatory axis. Genes Dev 24: 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Kondoh H 2013. Sox proteins: Regulators of cell fate specification and differentiation. Development 140: 4129–4144 [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Cheah KS, Kondoh H 1999. Mechanism of regulatory target selection by the SOX high-mobility-group domain proteins as revealed by comparison of SOX1/2/3 and SOX9. Mol Cell Biol 19: 107–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiso M, Tanaka S, Saba R, Matsuda S, Shimizu A, Ohyama M, Okano HJ, Shiroishi T, Okano H, Saga Y 2009. The disruption of Sox21-mediated hair shaft cuticle differentiation causes cyclic alopecia in mice. Proc Natl Acad Sci 106: 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B 1997. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro α1(II) collagen gene. Mol Cell Biol 17: 2336–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B 1998. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J 17: 5718–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B 2007. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol 39: 2195–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien W-H, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E 2011. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 9: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-Y, Kao C-H, Lin KM-C, Kaartinen V, Yang L-T 2011. Notch signaling regulates late-stage epidermal differentiation and maintains postnatal hair cycle homeostasis. PLoS ONE 6: e15842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CP, Polak L, Rocha AS, Pasolli HA, Chen S-C, Sharma N, Blanpain C, Fuchs E 2012. Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell 150: 136–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura I, Lai C-Y, Chiang K-N 2010. Functional interaction between Smad3 and S100A4 (metastatin-1) for TGF-β-mediated cancer cell invasiveness. Biochem J 426: 327–335 [DOI] [PubMed] [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, Fuchs E 2001. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev 15: 1688–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertin S, McDowall SG, Harley VR 1999. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res 27: 1359–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G 2004. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol 22: 411–417 [DOI] [PubMed] [Google Scholar]
- Müller-Röver S, Handjiski B, van der Veen C, Eichmüller S, Foitzik K, McKay IA, Stenn KS, Paus R 2001. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol 117: 3–15 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Yamamoto K, He X, Otsuki B, Kim Y, Murao H, Soeda T, Tsumaki N, Deng JM, Zhang Z, et al. 2011. Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat Commun 2: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Rendl M, Fuchs E 2006. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 127: 171–183 [DOI] [PubMed] [Google Scholar]
- Nguyen H, Merrill BJ, Polak L, Nikolova M, Rendl M, Shaver TM, Pasolli HA, Fuchs E 2009. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet 41: 1068–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann C, Owens DM, Hülsken J, Birchmeier W, Watt FM 2002. Expression of ΔNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development 129: 95–109 [DOI] [PubMed] [Google Scholar]
- Nowak JA, Fuchs E 2009. Isolation and culture of epithelial stem cells. Methods Mol Biol 482: 215–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, Fuchs E 2008. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 3: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C-D, Maity SN, Lu J-F, Zhang J, Liang S, Coustry F, de Crombrugghe B, Yasuda H 2010. Identification of SOX9 interaction sites in the genome of chondrocytes. PLoS ONE 5: e10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E 2012. Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell 10: 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi D, Bowles J, Nagy A, Muscat G, Koopman P 2000. Mice null for sox18 are viable and display a mild coat defect. Mol Cell Biol 20: 9331–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, de la Cruz D, Baker RE, Maini PK, Maxson R, Chuong C-M 2008. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451: 340–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Li X, Tang H, Huang AS, Panteleyev AA, Owens DM, Su GH 2011. Conditional activin receptor type 1B (Acvr1b) knockout mice reveal hair loss abnormality. J Invest Dermatol 131: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M, Lewis L, Fuchs E 2005. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol 3: e331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M, Polak L, Fuchs E 2008. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev 22: 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H, Polak L, Fuchs E 2006. Lhx2 maintains stem cell character in hair follicles. Science 312: 1946–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Greco V 2013. Spatial organization within a niche as a determinant of stem-cell fate. Nature 502: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Hochedlinger K 2013. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Sohn K-C, Li Z, Choi D-K, Park YM, Kim J-H, Fan Y-M, Nam YH, Kim S, Im M, et al. 2013. Expression and functional role of Sox9 in human epidermal keratinocytes. PLoS ONE 8: e54355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda T, Deng JM, de Crombrugghe B, Behringer RR, Nakamura T, Akiyama H 2010. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis 48: 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soond SM, Chantry A 2011. Selective targeting of activating and inhibitory Smads by distinct WWP2 ubiquitin ligase isoforms differentially modulates TGFβ signalling and EMT. Oncogene 30: 2451–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier M-C, Schedl A, Wegner M 2003. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 17: 1677–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempus CS, Morris RJ, Bortner CD, Cotsarelis G, Faircloth RS, Reece JM, Tennant RW 2003. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol 120: 501–511 [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E 2004. Defining the epithelial stem cell niche in skin. Science 303: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal VPI, Chaboissier M-C, Lützkendorf S, Cotsarelis G, Mill P, Hui C-C, Ortonne N, Ortonne J-P, Schedl A 2005. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol 15: 1340–1351 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Gai X, Han S, Moser CD, Hu C, Shire AM, Floyd RA, Roberts LR 2013. The human sulfatase 2 inhibitor 2,4-disulfonylphenyl-tert-butylnitrone (OKN-007) has an antitumor effect in hepatocellular carcinoma mediated via suppression of TGFB1/SMAD2 and Hedgehog/GLI1 signaling. Genes Chromosomes Cancer 52: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]