Pre-mRNA 3′ end formation is coupled to transcription via the RNA Pol II C-terminal domain (CTD). However, how transcription and pre-mRNA maturation are coordinated in humans is poorly understood. Here, West and colleagues show that Pol II pausing promotes Ser2p by Cdk12, which recruits CPA factor CstF77 and is required for optimal 3′ end processing. This study delineates a reciprocal relationship between early steps in poly(A) site processing and Pol II Ser2p that ensures efficient pre-mRNA 3′ end formation in human cells.
Keywords: RNA polymerase II, C-terminal domain, cleavage and polyadenylation, pausing, Ser2, Cdk12
Abstract
3′ end formation of pre-mRNAs is coupled to their transcription via the C-terminal domain (CTD) of RNA polymerase II (Pol II). Nearly all protein-coding transcripts are matured by cleavage and polyadenylation (CPA), which is frequently misregulated in disease. Understanding how transcription is coordinated with CPA in human cells is therefore very important. We found that the CTD is heavily phosphorylated on Ser2 (Ser2p) at poly(A) (pA) signals coincident with recruitment of the CstF77 CPA factor. Depletion of the Ser2 kinase Cdk12 impairs Ser2p, CstF77 recruitment, and CPA, strongly suggesting that the processes are linked, as they are in budding yeast. Importantly, we additionally show that the high Ser2p signals at the 3′ end depend on pA signal function. Down-regulation of CPA results in the loss of a 3′ Ser2p peak, whereas a new peak is formed when CPA is induced de novo. Finally, high Ser2p signals are generated by Pol II pausing, which is a well-known feature of pA site recognition. Thus, a reciprocal relationship between early steps in pA site processing and Ser2p ensures efficient 3′ end formation.
Most eukaryotic pre-mRNAs are matured at their 3′ end by cleavage and polyadenylation (CPA) (Proudfoot 2012). During CPA, several multiprotein complexes (CPSF, CstF, CFI, and CFII) assemble onto the poly(A) (pA) signal within the pre-mRNA, with many additional factors also implicated (Zhao et al. 1999; Shi et al. 2009). A pA signal usually consists of an AAUAAA hexamer followed by a U-rich or G/U-rich element, although variants of the hexamer motif are also functional (Tian and Graber 2012). Pre-mRNA cleavage occurs between these two elements and is performed by CPSF73 (Mandel et al. 2006). Following cleavage, the upstream product is polyadenylated, and the 3′ product is degraded (Kim et al. 2004; Gromak et al. 2006). The only pre-mRNA transcripts not polyadenylated are those that code for replication-dependent histones. However, these transcripts use many of the same factors for cleavage of their 3′ ends, including CPSF73 as the endonuclease (Dominski et al. 2005; Kolev and Steitz 2005).
CPA is closely coupled to transcription via the C-terminal domain (CTD) of RNA polymerase II (Pol II), which is composed of repeats of the Tyr–Ser–Pro–Thr–Ser–Pro–Ser heptapeptide (McCracken et al. 1997). A dynamic binding platform for RNA biogenesis factors is created by cis/trans isomerization of prolines and phosphorylation of the other amino acids (Buratowski 2009; Eick and Geyer 2013). Of these modifications, Ser2 phosphorylation (Ser2p) is most strongly linked to 3′ end formation. In budding yeast, the Ser2 kinase Ctk1 is required for the cotranscriptional recruitment of several 3′ end processing factors to Pol II (Ahn et al. 2004). In humans, Cdk9, Brd4, and Cdk12 can phosphorylate CTD on Ser2, but Cdk12 is the proposed ortholog of Ctk1 (Peterlin and Price 2006; Bartkowiak et al. 2010; Devaiah et al. 2012). Similar to Ctk1, Cdk12 does not generally affect transcription, although a subset of genes is regulated (Blazek et al. 2011). Whether Cdk12 performs a function analogous to Ctk1 in coordinating 3′ end formation and transcription has not been tested. CTD phosphorylation status is also influenced by phosphatases, of which Fcp1 acts on Ser2p (Cho et al. 2001).
Although the importance of Ser2p for 3′ end processing is well established in yeast, the relationship between these two events is less well understood in mammals. However, Pol II lacking in Ser2 does not support efficient CPA in human cells (Gu et al. 2012). Moreover, Pol II that is most heavily phosphorylated on Ser2 often occurs as a peak at the pA site (Rahl et al. 2010; Brookes et al. 2012; Grosso et al. 2012; Hintermair et al. 2012). Global chromatin immunoprecipitation (ChIP) as well as analysis of individual genes demonstrate that this correlates with sites of Pol II pausing and is most frequently found on short highly expressed genes with the canonical AAUAAA motif (Glover-Cutter et al. 2008; Grosso et al. 2012). Pausing at pA sites is well characterized and mediated by capture of the emergent AAUAAA hexamer by CPSF bound to the body of Pol II (Nag et al. 2007). Pol II pausing can also occur over sequences downstream from the pA site, where it promotes transcriptional termination and 3′ end formation (Eggermont and Proudfoot 1993; Gromak et al. 2006; West and Proudfoot 2009).
The processes involved in recognizing pA sites are likely to be very relevant in the study of diseases such as cancer, in which there are widespread changes in pA signal usage (Mayr and Bartel 2009). When this is considered with its near-ubiquitous role in mRNA biogenesis, it is important to understand it. A lot of research has focused on elucidating the function of CTD modification in RNA processing, but little is known about whether the CTD code is influenced by pre-mRNA processing events for their own benefit. We describe here reciprocal coupling between 3′ end processing and Ser2p. This mechanism involves Pol II pausing that promotes Ser2p by Cdk12, which serves to recruit CstF77 and is required for optimal 3′ end processing.
Results
Ser2p levels are increased beyond the MYC gene pA site
We chose the human MYC gene as a model to analyze the coupling between 3′ end formation and transcription (Fig. 1A). It is highly expressed and was previously demonstrated to be a robust model to analyze Pol II association and processing factor recruitment (Glover-Cutter et al. 2008). ChIP was used to detect total Pol II (N20) or three of its well-characterized derivatives phosphorylated on CTD Ser5, Ser2, or Ser7 (Fig. 1B). It is important to note that we used antibodies that are the most specific available to these CTD states (Chapman et al. 2007; Hintermair et al. 2012). All antibodies gave substantially more signal than an IgG control (Supplemental Fig. 1). Pol II is most concentrated at the promoter, with an additional pause at the 3′ end. Ser5p and Ser7p signals were also highest at the beginning of the gene correlative with functions at the promoter and in 5′ capping (Komarnitsky et al. 2000; Schroeder et al. 2000; Schwer and Shuman 2011). Ser2p was low at the 5′ end of the gene but showed a significant peak beyond the pA signal. This peak of Ser2p at or shortly after pA signals has been observed previously on the MYC gene and many other genes in yeast and mammals (Glover-Cutter et al. 2008; Rahl et al. 2010; Preker et al. 2011; Bataille et al. 2012; Brookes et al. 2012; Grosso et al. 2012; Hintermair et al. 2012). While correlative with a function in CPA, why and how Ser2p is enriched at this position is not established in humans.
Figure 1.
(A) Diagram of the human MYC gene with exons (black boxes) and the pA site indicated. Regions assayed by ChIP are underlined and labeled, and a scale marker is shown. (B) ChIP experiment assaying the distribution of Pol II (N20), Ser5p, Ser2p, and Ser7p across the MYC gene. For each antibody, values are normalized to the maximum value (see the Materials and Methods for details). (NTS) A negative control region. (C) ChIP experiment assaying the distribution of CPSF30 and CstF77 across the MYC gene. For each antibody, values are normalized to the maximum value. (NTS) A negative control region. (D) Native ChIP analyzing Ser2p across the MYC gene. Values are normalized to the maximum value. (NTS) A negative control region. All error bars represent standard deviation from at least three biological replicates.
We also performed ChIP to monitor the recruitment of the CPA factors CPSF30 and CstF77 to the MYC gene (Fig. 1C). CPSF30 was detected at all positions, which is consistent with the idea that CPSF may join Pol II at the promoter (Dantonel et al. 1997; Nag et al. 2007; Glover-Cutter et al. 2008). In contrast, CstF77 showed enrichment beyond the pA signal, consistent with previous reports that it is recruited at the end of the transcription cycle (Glover-Cutter et al. 2008; Davidson and West 2013). This experiment suggests that some members of the CPA machinery associate early with Pol II. However, complete assembly of a functional CPA complex does not occur until after the pA signal has been transcribed and is coincident with the highest Ser2p signal.
The 3′ peak of Ser2p could be explained by enhanced phosphorylation at those positions. Alternatively, Ser2p might already be present at upstream regions but in a state that is masked from detection. These possibilities are important to distinguish in order to properly interpret the ChIP data. To do so, we performed native ChIP on the MYC gene in the presence of high salt and SDS. We predicted that this would unmask any Ser2p that is bound by factors or obscured within CTD folding. Consistently, CstF77 was undetectable under these conditions (Supplemental Fig. 2). However, Ser2p should remain detectable, as Pol II is very tightly bound to DNA, aided by its so-called clamp region (Gnatt et al. 2001). To test this, we performed the experiment using the Ser2p antibody (Fig. 1D). Significantly, the Ser2p profile was very similar to that obtained by standard ChIP. To confirm the peak at the pA site, an extra primer pair was used spanning this region (pA). This experiment strongly suggests that Ser2p is increased beyond the pA site rather than being obscured before it.
Cdk12 activity is important for Ser2p and 3′ end formation
In budding yeast, most Ser2p is achieved by Ctk1 function and is required for the recruitment of CPA factors to genes (Ahn et al. 2004). Although CTD lacking Ser2 cannot efficiently recruit the CPA factor Pcf11 in humans (Gu et al. 2012), the kinase involved has not been investigated. Recently, Cdk12 was shown to promote Ser2p in metazoans and is the proposed ortholog of Ctk1 (Bartkowiak et al. 2010; Blazek et al. 2011). We used RNAi to deplete Cdk12 from HeLa cells so that we could study its function in Ser2p and 3′ end formation.
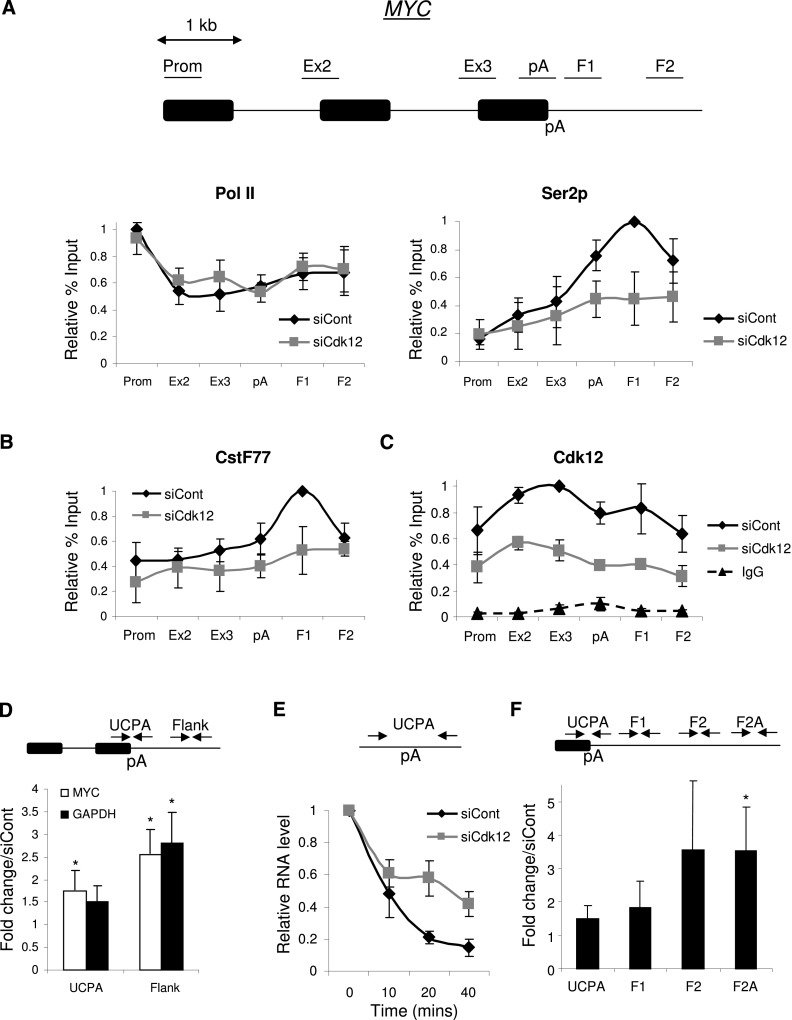
Following confirmation of successful Cdk12 depletion (Supplemental Fig. 3A), we performed ChIP on control and Cdk12 knockdown cells using the N20 and Ser2p and antibodies (Fig. 2A). Little difference in the Pol II profiles was evident, but we observed a reduction in Ser2p signals after the pA signal. This suggests that Cdk12 plays an important role in establishing the Ser2p profile across genes. Similarly, the CstF77 enrichment beyond the pA site was reduced upon depletion of Cdk12 (Fig. 2B). The recruitment of CPSF30, which has been proposed to bind to the body of Pol II in addition to the CTD, was unaffected by Cdk12 depletion as measured by ChIP (Supplemental Fig. 3B). These experiments confirm that Cdk12 phosphorylates Ser2p and suggest that this is important to recruit some 3′ end formation factors; namely, CstF77. This mechanism is analogous to that in budding yeast (Ahn et al. 2004). Finally, we analyzed the recruitment of Cdk12 to the MYC gene by ChIP (Fig. 2C). Cdk12 was present over all regions, and the signal was specific because it was reduced following treatment with Cdk12 siRNA. Cdk12 is not enriched over the pA site in comparison with upstream regions, suggesting that the high Ser2p at this position depends on modulation of Cdk12 activity.
Figure 2.
(A) ChIP experiment assaying the level and distribution of Pol II (N20) and Ser2p across the MYC gene in control or Cdk12-depleted cells. Values are normalized to the maximal value obtained in control siRNA-treated cells. (B) ChIP experiment assaying the level and distribution of CstF77 across the MYC gene in control or Cdk12-depleted cells. Values are normalized to the maximal value obtained in control siRNA-treated cells. (C) ChIP experiment assaying the level and distribution of Cdk12 across the MYC gene in control or Cdk12-depleted cells. IgG control values are also shown. Values are normalized to the maximal value obtained in control siRNA-treated cells. (D) Real-time PCR analysis of RNA not yet pA-cleaved (UCPA) and readthrough (Flank) RNA from the MYC and GAPDH genes in control and Cdk12-depleted samples. The diagram shows the final intron, pA site, and 3′ flank. Quantitation shows a fold change in Cdk12-depleted cells relative to control cells, normalized to terminal intron exon junction RNA. (*) P < 0.05. (E) Act D time-course analysis of non-pA-cleaved (UCPA) RNA in control or Cdk12-depleted cells. RNA levels were normalized to those present at time 0. (F) 4thio-UTP nuclear run-on (NRO) analysis of Myc transcriptional termination in control and Cdk12-depleted cells. The graph shows the fold change in cells depleted of Cdk12 compared with control cells. Signals were normalized to those from the intron 2–exon 3 junction. (*) P < 0.05. All error bars represent standard deviation from at least three biological replicates.
We next wanted to establish whether impaired Ser2p and CstF77 recruitment was important for overall 3′ end processing efficiency. We isolated total RNA from control and Cdk12-depleted cells and reverse-transcribed it with random hexamers. cDNA was then real-time PCR-amplified with primers spanning the MYC and GAPDH gene pA site and downstream flanking regions such that only unprocessed RNA was detected (Fig. 2D). More of these unprocessed transcripts were detected following Cdk12 depletion, indicating that 3′ end processing is more efficient when coupled to Ser2p by Cdk12. To gain a greater insight into this defect, we analyzed the rate of Myc pA site processing (Fig. 2E). This was done by monitoring non-pA-cleaved pre-mRNA following transcription inhibition by Actinomycin D (Act D) using primers spanning the pA cleavage site. In this experiment, cleavage at the pA site is predicted to cause a progressive loss of this product. When compared with control cells, Cdk12 depletion reduced the rate at which this species was lost, which is consistent with slower pA cleavage. A modest reduction in polyadenylated Myc RNA was also seen (Supplemental Fig. 4A,B). Importantly, exosome depletion did not alter the rate at which non-pA-cleaved Myc RNA was depleted, confirming that its loss is due to processing rather than degradation of the precursor (Supplemental Fig. 4C). Moreover, the rate of Myc splicing was unaffected by Cdk12 depletion, arguing that 3′ end formation is particularly sensitive to the presence of the protein (Supplemental Fig. 4E).
Defects in 3′ end processing reduce the efficiency of transcriptional termination (Whitelaw and Proudfoot 1986; Dye and Proudfoot 2001; Gromak et al. 2006). To test whether this was also a consequence of Cdk12 depletion, we performed a nuclear run-on (NRO) analysis of Myc transcripts (Fig. 2F). NRO detects RNA only from actively transcribing Pol II, and signals observed are reflective of the number of such polymerases at a given position on the template. Nuclei from control and Cdk12-depleted cells were incubated with 4-thio UTP to label nascent transcripts, which were subsequently biotinylated and purified before analysis by quantitative RT–PCR. We analyzed RNA over the pA site (UCPA) as well as over three downstream readthrough regions (F1, F2, and F2A). Importantly, we observed significantly more readthrough RNA over F2A in Cdk12-depleted cells as compared with control cells, confirming that full levels of Cdk12 are required for efficient Pol II termination in this case.
A functional pA site promotes high levels of Ser2p
Although our study focuses on a few model genes, a peak of Ser2p at the pA site is a common and evolutionarily conserved feature of protein-coding genes (Rahl et al. 2010; Bataille et al. 2012; Brookes et al. 2012; Grosso et al. 2012; Hintermair et al. 2012). Moreover, on Caenorhabditis elegans operons, Ser2p peaks are found at each internal 3′ end processing signal even though only a single pre-mRNA precursor is synthesized (Garrido-Lecca and Blumenthal 2010). These data argue that an aspect of CPA is involved in generating these high Ser2p signals to augment 3′ end formation. If this is the case, then mutation of a pA signal should disrupt the accumulation of Ser2p at the 3′ end of a gene.
To test this hypothesis, we used two HEK293 cell lines containing single-copy integrations of either the human β-globin gene (βWT) or a version with a mutated pA site (βpAm) (Davidson and West 2013). Expression of both genes was driven by a tetracycline-inducible CMV promoter (Fig. 3A). We performed ChIP on these β-globin genes to detect Pol II or its Ser2p derivative (Fig. 3B). Results were plotted to reflect the Ser2p levels normalized to Pol II levels. In the case of βWT, the proportion of Ser2p increased toward the 3′ end of the β-globin gene, similar to the MYC gene. In the case of βpAm, Ser2p density was highest at the 5′ end and had a generally flatter profile. Why the signal for Ser2p is slightly elevated at the 5′ end for βpAm is not clear but could reflect the disruption in processing. However, another ChIP experiment on integrated β-globin genes with normal and mutated pA sites gave a similar result, indicating that a pA site mutation disrupts the Ser2p pattern (Mapendano et al. 2010).
Figure 3.
(A) Diagram of βWT and βpAm genes integrated into HEK293 cells. The tetracycline-inducible CMV promoter (arrow), exons (black boxes), and pA site are indicated. Regions assayed by ChIP are underlined. (B) ChIP across βWT and βpAm to detect the relative ratio of Ser2p as compared with the Pol II (N20) signal. Note that the signal differential between Ex1 and 3′B is significant (P < 0.05) for βWT but not for βpAm. (C) 3′ RACE detection of PCPA of NR3C1 pre-mRNA in total RNA from control and U1 AMO-treated cells. The top panel shows the PCPA product, and the bottom panel shows product from the intronless TAF7 gene that acts as a loading control. The diagram shows the NR3C1 gene and a zoomed region showing primer pairs to detect PCPA within intron 2. (D) ChIP across the 5′ portion of the NR3C1 gene to detect the distribution and level of Pol II, Ser5p, Ser7p, and Ser2p in control or U1 AMO-treated cells. The diagram shows the 5′ portion of the NR3C1 gene, and amplicons are indicated and underlined. For each antibody, values are normalized to the maximal value obtained in control AMO-treated cells. All error bars represent standard deviation from at least three biological replicates.
We next performed the reciprocal experiment by asking whether de novo CPA would result in a corresponding peak of Ser2p. To test this hypothesis, we took advantage of the recently described function of U1 snRNA in inhibiting premature CPA (PCPA) (Kaida et al. 2010; Berg et al. 2012). A prominent example of this occurs in the NR3C1 pre-mRNA, which is subject to PCPA upon inhibition of U1 by an antisense morpholino (AMO). PCPA is rapidly induced following U1 inhibition, and an abundant product can be observed after 3 h (Davidson and West 2013). This was confirmed by 3′ RACE performed on cells treated with control or U1 AMOs (Fig. 3C).
To see whether this PCPA event affected CTD phosphorylation, we performed ChIP analysis on NR3C1 with N20, Ser5p, Ser2p, and Ser7p antibodies in control and U1 AMO-treated cells (Fig. 3D). Primers were used to analyze recruitment at positions between the promoter and 2 kb beyond the PCPA site. The overall Pol II profile was similar in both conditions over the regions assayed, but termination of transcription eventually occurred at downstream positions (Supplemental Fig. 5). There was little change in either Ser5p or Ser7p in U1 AMO samples compared with control treatment, showing that PCPA does not influence these modifications. Strikingly, the main difference between control and U1 AMO treatments was a dramatic increase in the Ser2p signal beyond the PCPA site in U1 AMO samples compared with the control. Importantly, a U2 AMO that inhibits splicing but does not promote PCPA did not affect the level of Ser2p (Supplemental Fig. 6). Thus, activation of a pA site induces high Ser2p, providing more support for a function of CPA in establishing the pattern of Ser2p across genes and, in particular, in promoting high levels of Ser2p near functional pA sites.
CPSF73 depletion reduces the level of Ser2p on transcribed genes
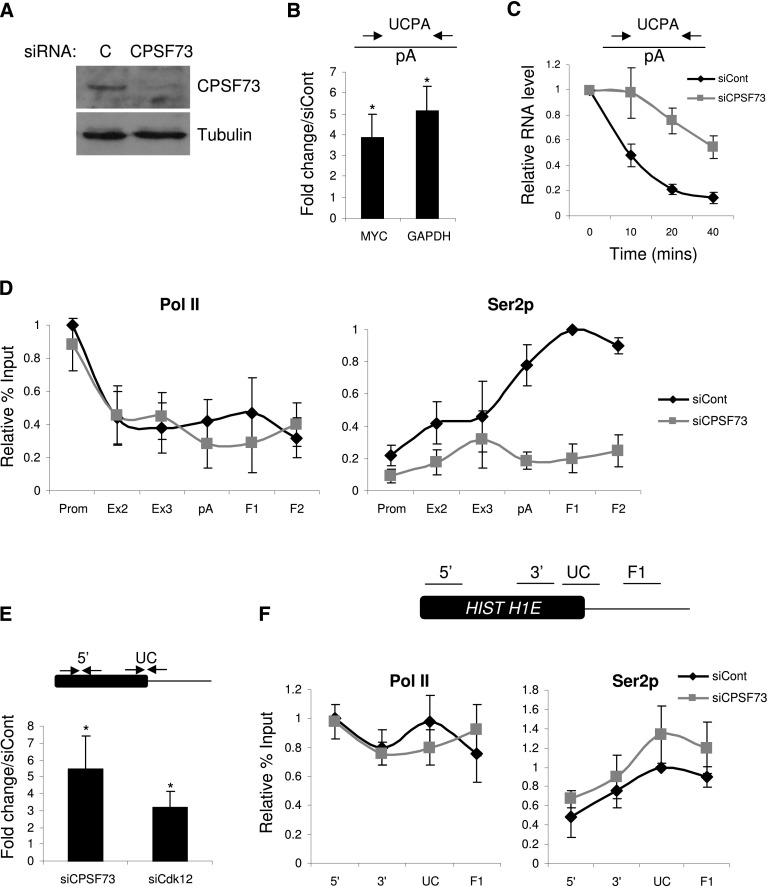
To see whether trans-acting factors also mediate CPA-dependent Ser2p, we used RNAi to deplete the pA endonuclease CPSF73 from HeLa cells. After confirming successful protein knockdown (Fig. 4A), we tested that the depletion was sufficient to observe a 3′ end processing defect. Total RNA was isolated from control and CPSF73-depleted cells and reverse-transcribed with random hexamers. Next, primers spanning MYC or GAPDH gene pA sites were used to perform real-time PCR and detect transcripts on which CPA had not yet occurred (Fig. 4B). CPSF73 depletion resulted in a threefold to fourfold increase in the level of these species, establishing that protein levels were sufficiently low to inhibit CPA. Consistently, Act D time-course analysis revealed a substantial reduction in the rate at which Myc transcripts were pA-cleaved (Fig. 4C), and fewer polyadenylated Myc transcripts were produced following CPSF73 depletion (Supplemental Fig. 7A,B).
Figure 4.
(A) Western blotting of protein extracts from cells treated with control or CPSF73 siRNAs. The top panel shows CPSF73 protein, and the bottom panel shows Tubulin loading control. (B) Real-time PCR analysis of RNA not yet pA-cleaved (UCPA) from the MYC and GAPDH genes in control and CPSF73-depleted samples. Quantitation shows a fold change in CPSF73-depleted cells relative to control cells normalized to the terminal intron–exon junction. (*) P < 0.05. (C) Act D time-course analysis of non-pA-cleaved (UCPA) RNA in control or CPSF73-depleted cells. RNA levels were normalized to those present at time 0. (D) ChIP experiment assaying the level and distribution of Pol II (N20) and Ser2p across the MYC gene in control or CPSF73-depleted cells. Values are normalized to the maximal value obtained in control siRNA-treated cells. (E) Real-time PCR analysis of histone H1E RNA 3′ end processing in control, CPSF73-depleted, or Cdk12-depleted samples. The primer pairs used are indicated on the accompanying diagram. Quantitation shows a fold change relative to control cells normalized to signal over the Histone gene body (5′) (*) P < 0.05. (F) ChIP experiment assaying the level and distribution of Pol II (N20) and Ser2p across the histone H1E gene in control or CPSF73-depleted cells. Values are normalized to the maximal value obtained in control siRNA-treated cells. The diagram shows the positions of amplicons. All error bars represent standard deviation from at least three biological replicates.
Next, we tested the effect that reduced CPA has on Pol II loading and Ser2p. ChIP was performed on both control and CPSF73-depleted cells using the N20 and Ser2p antibodies (Fig. 4D). CPSF73 RNAi caused little change in Pol II occupancy, as determined by the N20 signal. However, its depletion resulted in a more dramatic effect on Ser2p, which was reduced at all positions tested but in particular over the region beyond the pA site. The same was true for GAPDH and ACTB genes (Supplemental Fig. 7C). Finally, Cdk12 protein levels were equivalent in control and CPSF73-depleted cells, arguing that its indirect depletion was not responsible (Supplemental Fig. 7D). These data provide further evidence that efficient 3′ end formation is important to establish high levels of Ser2p by implicating CPSF73 in the process.
Histone RNA processing defects are not associated with changes in Ser2p level
CPSF73 catalysis is also a crucial part of histone pre-mRNA processing, although these RNAs are not polyadenylated (Dominski et al. 2005; Kolev and Steitz 2005). We were interested in whether there was also reciprocal coupling between 3′ end formation of histone transcripts and Ser2p or the effect was pA site-specific. Using histone H1E RNA as a model, we sought to confirm the role of CPSF73 in histone RNA processing in vivo and test for involvement of Cdk12. We performed quantitative RT–PCR on total RNA from control, CPSF73-depleted, and Cdk12-depleted cells using primers spanning the site of 3′ end formation (Fig. 4E). We observed a substantial processing defect in CPSF73-depleted cells, in line with its anticipated role in histone RNA maturation. Impaired processing was also observed in Cdk12-depleted samples, indicating that Ser2p may also be important.
To investigate whether 3′ end processing of histone RNA by CPSF73 was involved in promoting Ser2p, we performed ChIP on control and CPSF73-depleted cells using Pol II and Ser2p antibodies (Fig. 4F). However, there was little difference in the level or distribution of Pol II or its Ser2p derivative in both conditions. Thus, 3′ end processing of histone RNA and CPSF73 catalysis per se does not promote elevated Ser2p levels in a way that is observable by ChIP. It should be noted that Ser2p levels are much lower over Histone H1E as compared with MYC, which may reflect the distinct mechanisms of 3′ end formation (Supplemental Fig. 8).
CPA does not impact on Ser2p levels
We next wanted to determine the step in CPA that promotes Ser2p. For this purpose, we subdivided CPA into three stages: Pol II pausing and pA site recognition, pA site cleavage, and polyadenylation. Polyadenylation was tested first, as it distinguishes pA site processing (which we found promotes Ser2p) from histone RNA processing (which we found does not). Accordingly, we compared Pol II and Ser2p ChIP signals on the MYC gene in control cells or cells treated with cordycepin (CDY), which inhibits polyadenylation (Fig. 5A). As previously documented (Anamika et al. 2012), CDY caused a mild reduction of Pol II signal at the promoter; however, the level and distribution of Ser2p was not significantly changed. Thus, polyadenylation does not positively impact on Ser2p.
Figure 5.
(A) ChIP analysis of the level and distribution of Pol II and Ser2p in cells treated with DMSO or the polyadenylation inhibitor CDY. Values are normalized to the maximal value obtained in DMSO-treated cells. (B) RNA-IP experiment using Pol II (N20) and Ser2p antibodies detecting unspliced Myc intron 1 (Ex1–In1) and intron 2 (Ex2–In2) as well as non-pA-cleaved (UCPA) and flank (F1) RNA. For each antibody, values are normalized to the maximal value obtained. (C) Western blot analysis of CstF77 (top panel) and PABPN1 (loading control; bottom panel) in cells treated with control and CstF77 siRNAs. (D) Real-time PCR analysis of RNA not yet pA-cleaved (UCPA) from the MYC and GAPDH genes in control and CstF77-depleted samples. Quantitation shows a fold change in CstF77-depleted cells relative to control cells, normalized to the terminal intron–exon junction. (*) P < 0.05. (E) Act D time-course analysis of non-pA-cleaved (UCPA) RNA in control or CstF77-depleted cells. RNA levels are relative to those present at time 0. (F) ChIP experiment assaying the level and distribution of Pol II (N20) and Ser2p across the MYC gene in control or CstF77-depleted cells. Values are normalized to the maximal value obtained in control siRNA-treated cells. All error bars represent standard deviation from at least three biological replicates.
We next asked whether pA site cleavage promotes Ser2p. This is not a trivial issue to address because separating pA site recognition from cleavage is very difficult. However, we reasoned that if pA cleavage were a prerequisite for high Ser2p levels, then non-pA-cleaved RNA would only be found in association with Pol II that was not heavily phosphorylated on Ser2. This would be in contrast to the situation by ChIP, where Ser2p peaks at the pA site. To test this, we performed RNA immunoprecipitation (RNA-IP) of Myc transcripts with the Pol II and Ser2p antibodies (Fig. 5B). We analyzed unspliced intron 1 (Ex1–In1), unspliced intron 2 (Ex2–In2), non-pA-cleaved (UCPA), and 3′ flanking region (F1) transcripts. Similar amounts of each RNA were precipitated with the Pol II antibody, but when the Ser2p antibody was used, UCPA and F1 transcripts were immunoprecipitated more efficiently than those upstream, consistent with the ChIP data. If high Ser2p occurred only after pA site cleavage, it might have been expected that no Ser2p peak would be seen by RIP or that, if it was, UCPA transcripts would not be associated with it. This result therefore suggests that high Ser2p can occur on Pol II associated with transcripts not yet pA-cleaved. In support of this, direct visualization of transcription by other groups found that most Pol II present beyond pA sites is associated with RNA that was yet to be cleaved (Osheim et al. 1999, 2002).
As a further assessment of the role of pA site cleavage in Ser2p, we used RNAi to deplete cells of CstF77 (Fig. 5C). This factor is important for pA site cleavage, but our findings suggest that it is most efficiently recruited after Ser2 becomes highly phosphorylated. We first tested whether our depletion was sufficient to impair cleavage of the MYC and GAPDH gene pA sites using RT–PCR on RNA from control and CstF77-depleted cells (Fig. 5D). Uncleaved pA sites from both genes accumulated twofold to threefold following CstF77 depletion, confirming its function in CPA. This was accompanied by a reduced rate of Myc pA cleavage as measured by Act D time-course analysis (Fig. 5E) and a reduction in the number of polyadenylated Myc transcripts (Supplemental Fig. 9). Finally, we performed ChIP under both conditions to detect Pol II and Ser2p over the MYC gene (Fig. 5F). Importantly, even though CstF77 depletion impaired pA site cleavage, it did not affect Ser2p levels, indicating that the two processes can be uncoupled. These data further argue that Ser2p is promoted by a step prior to pA site cleavage.
Early events in 3′ end formation promote Ser2p
We next considered the possibility that pausing over the pA site might be responsible, since the 3′ peak of Ser2p that is observed genome-wide is coincident with a peak of paused Pol II (Grosso et al. 2012). Furthermore, there is little or no Pol II pausing on histone genes and no reciprocal coupling of histone H1E RNA processing and Ser2p, which is present at low levels (Fig. 4F; Supplemental Fig. 8; Anamika et al. 2012).
It has been shown that binding of CPSF30 by nonstructural protein 1A (NS1A) from influenza virus abrogates AAUAAA-dependent Pol II pausing (Nag et al. 2007). NS1A binds to CPSF30 and prevents the interaction of CPSF complexes with pre-mRNA (Nemeroff et al. 1998). We made two HEK293 cell lines containing a stably integrated single copy of the wild-type NS1A gene or a mutant version encoding NS1A that cannot bind CPSF30. Transcription of both genes was driven by a tetracycline-inducible promoter to allow rapid and coordinated expression (Supplemental Fig. 10A). This system is advantageous over RNAi, since CPSF30 activity and 3′ end formation are more rapidly inhibited, and the mutant that does not bind to CPSF30 is a robust control.
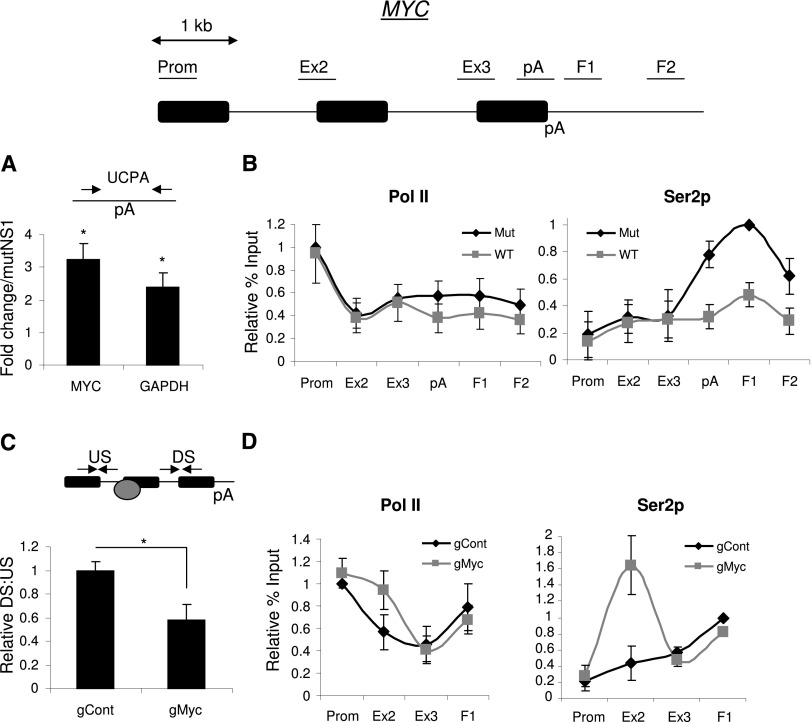
To test the system, total RNA was isolated from wild-type and mutant NS1A cells induced with tetracycline. Following random hexamer-primed reverse transcription, uncleaved Myc and GAPDH pA sites were quantitated by real-time PCR (Fig. 6A). Twofold to threefold more of these species were present in cells expressing wild-type NS1A when compared with those expressing mutant NS1A, confirming that inhibition of CPSF30 impairs CPA. Consistently, wild-type NS1A also impaired the recruitment of CPSF30 to the MYC gene (Supplemental Fig. 10B). We next tested the effects of NS1A on the level and distribution of Pol II and its Ser2p derivative (Fig. 6B). Results for Pol II were similar in both cell lines, but Ser2p levels were depleted at the 3′ end of MYC in cells expressing wild-type NS1A as compared with mutant NS1A. This experiment provides further support for a role of CPSF and Pol II pausing in promoting high levels of Ser2p at 3′ ends of genes. CstF77 recruitment was also reduced by expression of wild-type NS1A (Supplemental Fig. 10B).
Figure 6.
(A) Real-time PCR analysis of Myc or GAPDH RNA that is not cleaved at the pA site (UCPA) in cells expressing wild-type or mutant NS1A. Quantitation shows a fold change in wild-type NS1A cells relative to mutant NS1A cells normalized to the terminal intron–exon junction. (*) P < 0.05. (B) ChIP experiment assaying the level and distribution of Pol II (N20) and Ser2p across the MYC gene in wild-type (WT) or mutant (Mut) NS1A-expressing cells. Values are normalized to the maximal value obtained in mutant NS1A cells. Primer pairs used are underlined on the accompanying diagram. (C) RT–PCR determination of Pol II pausing at the Myc gRNA. Levels of RNA from upstream (US) of or downstream (DS) from the gRNA-binding site were compared and plotted. The graph shows the DS:US ratio in cells expressing control or Myc-specific gRNA. The diagram shows primers and the position to which the Cas9:gRNA complex is targeted (gray circle). (*) P < 0.05. (D) ChIP analysis of the level and distribution of Pol II (N20) and Ser2p across the MYC gene in cells transfected with a dCas9 expression construct in addition to a plasmid expressing control or MYC-specific gRNA. Values are normalized to the maximal value obtained in control gRNA transfected cells. All error bars represent standard deviation from at least three biological replicates.
Pol II pausing is sufficient to promote Ser2p
The above data are consistent with a function for pA site pausing in promoting Ser2p. To validate this model, we sought to promote a single site-specific Pol II pause in a region where Ser2p was normally low and test whether it was increased as a consequence. We used the Streptococcus pyogenes type II CRISPR system that has recently proved useful for genome modification in human cells (Cong et al. 2013; Mali et al. 2013). Briefly, a guide RNA (gRNA) is used to target an endonuclease, Cas9, to a chosen part of the genome. If a catalytically dead Cas9 (dCas9) is expressed, the complex remains bound to the DNA, where it impedes Pol II progress (Gilbert et al. 2013; Qi et al. 2013). To target this complex to the MYC gene, we made a construct expressing a gRNA directed close to the intron 1 exon 2 boundary of the MYC gene, where Ser2p is lower than at the 3′ end of the gene. To test whether the MYC gRNA could restrict Pol II elongation, cells were transfected with a dCas9 expression plasmid and the MYC-specific or control gRNA constructs. Total RNA was isolated and reverse-transcribed before being real-time PCR-amplified with primers upstream of and downstream from the gRNA-binding site (Fig. 6C). We then calculated the ratio of downstream to upstream RNA. When compared with cells expressing the control gRNA, significantly less downstream RNA was recovered in cells expressing the MYC gRNA. This indicates that a proportion of Pol II is paused at the gRNA-binding site.
Finally, we performed ChIP on cells expressing dCas9 with either the control or MYC gRNA to detect Pol II or Ser2p (Fig. 6D). When compared with the control gRNA, the MYC gRNA caused a modest accumulation of Pol II at the end of intron 1, where otherwise the pattern was similar. This is consistent with the expected obstruction to Pol II elongation provided by gRNA:dCas9. It should be noted that not all MYC loci are likely to be bound by gRNA:dCas9 due to incomplete targeting evident in transfection-based genome-editing experiments (Wang et al. 2013). Importantly, however, the MYC gRNA induced a more substantial peak of Ser2p over this region. These experiments strongly support the idea that Pol II pausing can promote Ser2p.
Transcripts extending beyond the region of high Ser2p are inefficiently cleaved at the pA site
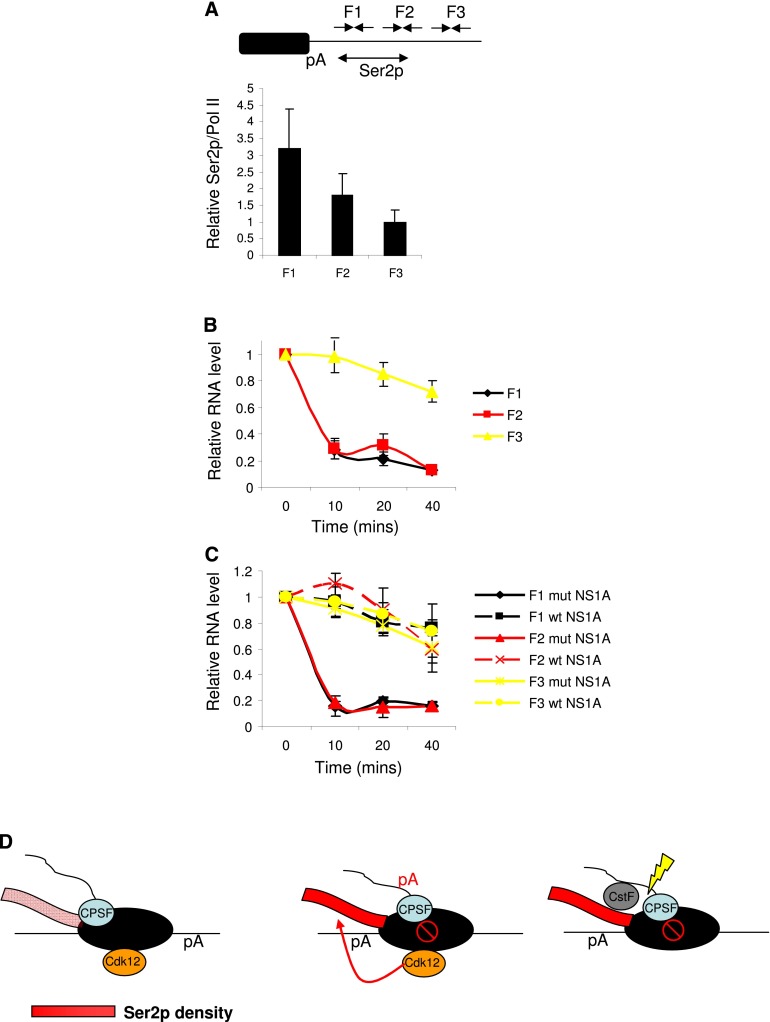
We finally wanted to test whether the reciprocal coupling that we describe is important for efficient CPA. Accordingly, the rate of 3′ end formation on transcripts from the region of high Ser2p was compared with that of downstream RNAs produced by Pol II that escapes pausing. Pol II over this downstream region (F3) displayed lower Ser2p than that over upstream F1 and F2 regions (Fig. 7A). To assay processing rates, we inhibited transcription using Act D and isolated total RNA samples at 10-min intervals over a 40-min period. Following reverse transcription with random hexamers, real-time PCR was used to quantitate RNA from F1, F2, and F3 (Fig. 7B). Both F1 and F2 transcripts were strongly depleted after only 10 min, which indicates that pA cleavage takes place very quickly. In contrast, F3 RNA remained stable throughout the time course, indicating that transcripts extending to this region were very inefficiently processed compared with those upstream.
Figure 7.
(A) Graph showing the relative ratio of Ser2p ChIP signal as compared with total Pol II (N20) over the F1, F2, and F3 regions. Note that Ser2p/N20 is lowest for the F3 region (values normalized to those for the Ex2 amplicon). The diagram shows the position of amplicons, and the region of high Ser2p is also indicated. (B) Act D time-course analysis of RNA from the F1, F2, or F3 regions of the MYC gene 3′ flank. For each product, values are plotted relative to those obtained at the 0 time point following normalization to U6 snRNA. (C) Act D time-course analysis of RNA from the F1, F2, and F3 regions of the MYC gene 3′ flank in cells expressing wild-type (wt) or mutant (mut) NS1A. For each product, values are plotted relative to those obtained at the 0 time point following normalization to U6 snRNA. (D) Model: Upstream of the pA signal, Ser2p is present at lower levels, and CPSF accompanies Pol II. Transcription of the pA site induces Pol II pausing (open red circle with slash) involving CPSF, and this triggers Ser2p by Cdk12. As a result, CstF77 is recruited, and CPA takes place efficiently. Ser2p density is indicated by a color gradient of pink (lower) to red (higher) on the CTD. All error bars represent standard deviation from at least three biological replicates.
If the loss of F1 and F2 signal is a consequence of CPA, it should be prevented when the process is impaired. To test this, we repeated the Act D time course but analyzed the decay of F1, F2, and F3 in cells expressing wild-type and mutant NS1A protein (Fig. 7C). The decay in mutant NS1A cells was rapid, as before; however, it was much slower in cells expressing the wild-type protein, which inhibits CPA. This result confirms that the low levels of F1 and F2 transcripts observed following Act D treatment were due to functional 3′ end processing. There was little difference between wild-type and mutant NS1A samples assayed for F3, since transcripts extending to this region are already poorly processed. In sum, these data strongly suggest that the mechanism that we describe promotes efficient CPA.
Discussion
We investigated how transcription, CTD modification, and CPA are linked in human cells. We propose that a reciprocal relationship between Ser2p and pausing during CPA coordinates 3′ end formation of pre-mRNAs and transcription. We suggest that Pol II pausing, mediated by the pA signal and CPSF, promotes high levels of Ser2p by Cdk12. This augments the recruitment of CstF to form a functional CPA complex to execute 3′ end formation. This mechanism is advantageous for 3′ end formation, as pA site cleavage is inefficient if Pol II reads through the region of high Ser2p. Our model is presented in Figure 7D.
Our data indicate that the effect of CPA on Ser2p is mediated by Pol II pausing, which is a well-described consequence of pA signal transcription (Orozco et al. 2002; Nag et al. 2007; Grosso et al. 2012). We argue this for several reasons: First, the Ser2p signal increased over the pA signal by ChIP and RNA-IP experiments, showing that template-bound Pol II that is heavily phosphorylated on Ser2 is associated with non-pA-cleaved RNA. Second, we found that there is no effect of CPSF73 depletion on Ser2p on the histone H1E gene even though processing is impaired. This suggests that CPSF73 catalysis per se and therefore pre-mRNA cleavage are not involved in Ser2p. Consistently, it has been proposed that Pol II pausing is transient or absent on histone genes (Anamika et al. 2012). Third, we found that CstF77 depletion does not impact on Ser2p even though pA site cleavage is impaired. Fourth, site-specific Pol II pausing induced by dCas9 is associated with a Ser2p peak even though no pA signal is present. Finally, direct visualization of Pol II transcription by electron microscopy indicates that Pol II that is associated with pA-cleaved RNA is rarely seen either in Drosophila or on human minigenes in Xenopus (Osheim et al. 1999, 2002). This being the case, the majority of the ChIP signal beyond pA signals is likely to be contributed by Pol II that is associated with unprocessed pA sites.
Our native ChIP experiment strongly suggests that Ser2p is added beyond the pA site rather than being present upstream but masked by bound factors. However, it should be noted that CTD repeats with Tyr1p, Ser5p, or Ser7p can be poorly detected by this Ser2p antibody in vitro (Chapman et al. 2007; Hintermair et al. 2012). Therefore, a Ser2p peak at the 3′ end may result from the loss of one or more of these inhibitory marks instead of the gain of Ser2p. This would be reflected by a sharp pA signal-dependent reduction in Tyr1p, Ser5p, or Ser7p, but, to our knowledge, this has never been described in humans. Finally, the H5 antibody is often used to detect Ser2p but is less specific than the 3E10 antibody that we used, as it reacts strongest with CTD phosphorylated on Ser5 and Ser2 (Chapman et al. 2007). Even so, ChIP sequencing with H5 or 3E10 generates very similar average profiles, with peaks of signal at or beyond the pA signal (Rahl et al. 2010; Brookes et al. 2012; Grosso et al. 2012; Hintermair et al. 2012). As we propose, these combined data support a straightforward model in which Ser2p is increased after the pA site.
We showed that a transcriptional pause can act as a molecular trigger for Ser2p, and a matter for future study will be to understand how pausing leads to enhanced Cdk12 activity. The ChIP experiments in Figure 2 demonstrate that human Cdk12 is present across the MYC gene. This is very similar to what was observed with Drosophila Cdk12, which was detected broadly across several tested genes (Bartkowiak et al. 2010). Pausing might trigger Ser2p by localized recruitment of Cdk12 or by promoting its activity. Our results favor the latter because, although Cdk12 is present at the 3′ end of the MYC gene, there is not a striking peak. A further possibility is that Ser2p is repressed by a phosphatase that is inactivated in a 3′ end processing-dependent manner. We found this possibility unlikely given that most available evidence does not support inhibition of Ser2p phosphatase activity at gene ends (Bataille et al. 2012; Fuda et al. 2012). Finally, our own preliminary analysis found that Ser2p was not increased prematurely upon Fcp1 depletion (data not shown).
Our observation of reciprocal coupling between CPA-mediated Pol II pausing and CTD modification is an important conceptual advance in understanding cotranscriptional pre-mRNA processing. We found that early events in 3′ end formation—specifically, Pol II pausing—promote high levels of Ser2p. This modification is in turn required for the efficient recruitment of a CPA complex and subsequent 3′ end formation. Thus, a pA signal is at least partly responsible for generating the CTD state necessary for its processing. We speculate that analogous coupling might be used in other cases of cotranscriptional RNA maturation, in particular those requiring Ser2p. The mechanism that we propose parallels another case of reciprocal coupling between pre-mRNA maturation and the chromatin template whereby Histone modification and splicing influence one another (Luco et al. 2010; de Almeida et al. 2011; Kim et al. 2011; Bieberstein et al. 2012).
Materials and methods
Primers
A list of primers, AMO sequences, and siRNA targets is provided in Supplemental Table 1.
Plasmids
βWT and βpAm HEK293 cell lines were already described (Davidson and West 2013). For gRNA expression, the gRNA AAVS1-T1 plasmid (41817, Addgene) was modified by PCR to include sequences complementary to MYC (see Supplemental Table 1). dCas9 was made using the hCas9 D10A plasmid (41816, Addgene) as a template for site-directed mutagenesis whereby His841 was changed to Ala. To make the NS1A cell lines, pcDNA-NS1 (a kind gift from Ervin Fodor) was amplified with NS1F and NS1R. The product was then inserted into a vector prepared by amplifying pcDNA5 FRT/TO, containing human β-globin, with promR and βpAF primers. To make mutant NS1, this plasmid was amplified with NS1mF and NS1mR primers.
Cell culture
Cells were grown in DMEM supplemented with 10% fetal calf serum. Act D was used at 10 μg/mL. Electroporation of morpholinos was performed on a confluent 10-cm-diameter dish of cells using 10 μM AMO in 400 μL of DMEM using a 4-mm gap cuvette (960 μF, 280 V in a Bio-Rad gene pulser). RNA was isolated 3 h after electroporation. For transient transfection, 6 μg each of gRNA and dCas9 constructs were transfected using JetPrime (Polyplus), and experiments were performed 48 h later. For RNAi, 20% confluent 60-mm dishes were transfected with 18 μL of 2 μM siRNA and 5 μL of RNAiMAX (Life Technologies) and left for 72 h. The process was then repeated after replating of cells to 20% confluence. Stable cell line generation was with the Flp-IN system (Life Technologies) and is described elsewhere (Davidson and West 2013).
Antibodies
Antibodies used were Pol II (N20:sc899, Santa Cruz Biotechnology), Ser2p (3E10, ChromoTek), Ser5p (3E8, ChromoTek), Ser7p (4E12, ChromoTek), CstF77 (C0249, Sigma), CPSF73 (C2747, Sigma), CPSF30 (SAB4200170, Sigma; A301-584A, Bethyl Laboratories), Tubulin (T6557, Sigma), Cdk12 for Western (NB100-87011, Novus Biologics), PABPN1 (75855, Abcam), and Cdk12 for ChIP (57311, Abcam).
RNA analysis
Total RNA was isolated using Trizol (Life Technologies) and DNase-treated with Turbo DNase (Life Technologies). For real-time PCR analysis, 1 μg of RNA was reverse-transcribed using Inprom II (Promega). Parallel reactions were performed in the absence of reverse transcriptase. One-twentieth of the cDNA mix was used for real-time PCR using 5–10 pmol of forward and reverse primer and Brilliant III SYBR mix (Agilent Technologies) in a Qiagen Rotorgene machine. Differences were calculated using comparative quantitation.
ChIP
In a confluent 10-cm dish, HeLa or HEK293 cells were cross-linked in 1% formaldehyde for 10 min at room temperature (4 × 60-mm dishes were used for RNAi experiments). Cross-links were quenched in 125 mM glycine, and cells were rinsed in PBS and then collected by centrifugation (500g for 5 min). Cells were sonicated in 400 μL of RIPA buffer (150 mM NaCl, 1% NP40, 0.5% DOC, 0.1% SDS, 50 mM Tris-Cl at pH 8, 5 mM EDTA at pH 8) for 30 sec on, 30 sec off for 12 min on high in a Bioruptor. Chromatin was centrifuged at 13,000g for 10 min, and supernatants added to 20 μL of protein A/G Dynabeads (Invitrogen) preincubated in 500 μL of RIPA buffer for 2 h with antibody (generally 2–4 μg per ChIP). No antibody controls were performed in parallel. Following overnight rotation at 4°C, the beads were washed twice in RIPA buffer, four times in wash buffer (500 mM NaCl, 1% NP40, 1% DOC, 100 mM Tris.Cl at pH 8.5), and twice in RIPA buffer. Immune complexes were eluted in 0.1 M NaHCO3/1%SDS (15 min of rotation at room temperature). Cross-links were reversed for 5 h at 65°C (250 mM NaCl, 1 μg RNase A). DNA was phenol-chloroform-extracted and ethanol-precipitated. Generally, 1/50th was used for each real-time PCR reaction.
RNA-IP
For RNA-IP, the same protocol was used as for ChIP except extracts were DNase-treated (30 min at room temperature) before immunoprecipitation as well as after elution (1 h at 37°C). Immunoprecipitations were performed in the presence of RNase inhibitor. Half of the elution was reverse-transcribed, and the other half was used for a parallel minus reverse transcriptase control.
Native high-salt ChIP
A 10-cm dish of HeLa cells was scraped in PBS and then lysed in 5 mL of HLB before underlayering with 1 mL of HLB + 10% sucrose. Nuclei were pelleted by spinning at 500g for 5 min. These were resuspended in 1 mL of MNase buffer, and chromatin was digested for 10 min at 37°C with 1 U of micrococcal nuclease (Sigma). Nuclei were then lysed in high-salt RIPA buffer (500 mM NaCl, 1% NP40, 0.5% DOC, 0.1% SDS, 50 mM Tris-Cl at pH 8, 5 mM EDTA at pH 8). Extracts were immunoprecipitated overnight in high-salt RIPA buffer in the presence of beads to which antibodies had been coupled. The next day, these were washed four times for 5 min in high-salt RIPA buffer and then once in RIPA buffer. Elution and analysis were performed as for standard ChIP.
ChIP quantitation
Our ChIP experiments often required data obtained from different antibodies to be plotted on the same graph. Moreover, as is common for ChIP, different batches of the same antibody frequently varied in their effectiveness. Importantly, however, the pattern of recruitment was always robust. To account for this, values were plotted as relative percentage of input, where we calculated the percentage of input for each experiment before maximum immunoprecipitation as a relative percent input of 1. Each plot displays the average and standard deviation of at least three experiments quantitated in this way.
4-thio UTP NRO
A detailed protocol for 4-thio UTP NRO is described in Davidson et al. ( 2012).
Acknowledgments
We thank Ervin Fodor (Oxford University) and Adolfo Garcia-Sastre (Mount Sinai School of Medicine) for the NS1A expression plasmid and NS1A antibody, respectively. We also thank Atlanta Cook, Gracjan Michlewski, and Marie-Joelle Schmidt for critically reading the manuscript, and David Tollervey for comments during the revision process. This work was supported by a Wellcome Trust Research Career Development award to S.W., and an EMBO long-term post-doctoral fellowship awarded to L.M. Grant numbers were 088499/Z/09/2 (to S.W.) and Centre core grant from the Wellcome Trust (092076). Funding for the open access charge was from The Wellcome Trust.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.231274.113.
Freely available online through the Genes & Development Open Access option.
References
- Ahn SH, Kim M, Buratowski S 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Anamika K, Gyenis A, Poidevin L, Poch O, Tora L 2012. RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts. PLoS ONE 7: e38769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL 2010. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24: 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F 2012. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 45: 158–170 [DOI] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberstein NI, Oesterreich FC, Straube K, Neugebauer KM 2012. First exon length controls active chromatin signatures and transcription. Cell Rep 2: 62–68 [DOI] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM 2011. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 25: 2158–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. 2012. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D 2007. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 15: 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389: 399–402 [DOI] [PubMed] [Google Scholar]
- Davidson L, West S 2013. Splicing-coupled 3′ end formation requires a terminal splice acceptor site, but not intron excision. Nucleic Acids Res 41: 7101–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L, Kerr A, West S 2012. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J 31: 2566–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. 2011. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 18: 977–983 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ 3rd, Singer DS 2012. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci 109: 6927–6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Purdy M, Wagner EJ, Marzluff WF 2005. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol 25: 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105: 669–681 [DOI] [PubMed] [Google Scholar]
- Eggermont J, Proudfoot NJ 1993. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J 12: 2539–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D, Geyer M 2013. The RNA polymerase II carboxy-terminal domain code. Chem Rev 113: 8456–8490 [DOI] [PubMed] [Google Scholar]
- Fuda NJ, Buckley MS, Wei W, Core LJ, Waters CT, Reinberg D, Lis JT 2012. Fcp1 dephosphorylation of the RNA polymerase II C-terminal domain is required for efficient transcription of heat shock genes. Mol Cell Biol 32: 3428–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Lecca A, Blumenthal T 2010. RNA polymerase II C-terminal domain phosphorylation patterns in Caenorhabditis elegans operons, polycistronic gene clusters with only one promoter. Mol Cell Biol 30: 3887–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL 2008. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15: 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD 2001. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science 292: 1876–1882 [DOI] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot NJ 2006. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol 26: 3986–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso AR, de Almeida SF, Braga J, Carmo-Fonseca M 2012. Dynamic transitions in RNA polymerase II density profiles during transcription termination. Genome Res 22: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Eick D, Bensaude O 2012. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res 41: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, et al. 2012. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J 31: 2784–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S 2004. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL 2011. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci 108: 13564–13569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev 19: 2583–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH 2010. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 40: 410–422 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Nag A, Narsinh K, Martinson HG 2007. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat Struct Mol Biol 14: 662–669 [DOI] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell 1: 991–1000 [DOI] [PubMed] [Google Scholar]
- Orozco IJ, Kim SJ, Martinson HG 2002. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J Biol Chem 277: 42899–42911 [DOI] [PubMed] [Google Scholar]
- Osheim YN, Proudfoot NJ, Beyer AL 1999. EM visualization of transcription by RNA polymerase II: Downstream termination requires a poly(A) signal but not transcript cleavage. Mol Cell 3: 379–387 [DOI] [PubMed] [Google Scholar]
- Osheim YN, Sikes ML, Beyer AL 2002. EM visualization of Pol II genes in Drosophila: Most genes terminate without prior 3′ end cleavage of nascent transcripts. Chromosoma 111: 1–12 [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Preker P, Almvig K, Christensen MS, Valen E, Mapendano CK, Sandelin A, Jensen TH 2011. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res 39: 7179–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ 2012. Ending the message: Poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 14: 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Shuman S 2011. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell 43: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, Frank J, Manley JL 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Graber JH 2012. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA 3: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ 2009. Transcriptional termination enhances protein expression in human cells. Mol Cell 33: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E, Proudfoot N 1986. α-Thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α2 globin gene. EMBO J 5: 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]