All eukaryotic cells divide a finite number of times, yet the mechanistic basis of replicative aging remains unclear. Here, Tyler and colleagues investigate the effects of aging on chromatin structure and DNA instability in budding yeast. The use of spike-in controls reveals a global reduction in nucleosome occupancy during aging. Histone loss during aging leads to transcriptional induction of all yeast genes. Furthermore, the authors demonstrate elevated levels of DNA damage, retrotransposition, large-scale chromosome rearrangement, and translocation during aging.
Keywords: replicative aging, histone occupancy, gene expression, DNA rearrangement
Abstract
All eukaryotic cells divide a finite number of times, although the mechanistic basis of this replicative aging remains unclear. Replicative aging is accompanied by a reduction in histone protein levels, and this is a cause of aging in budding yeast. Here we show that nucleosome occupancy decreased by 50% across the whole genome during replicative aging using spike-in controlled micrococcal nuclease digestion followed by sequencing. Furthermore, nucleosomes became less well positioned or moved to sequences predicted to better accommodate histone octamers. The loss of histones during aging led to transcriptional induction of all yeast genes. Genes that are normally repressed by promoter nucleosomes were most induced, accompanied by preferential nucleosome loss from their promoters. We also found elevated levels of DNA strand breaks, mitochondrial DNA transfer to the nuclear genome, large-scale chromosomal alterations, translocations, and retrotransposition during aging.
Aging is the single highest risk factor for the majority of human malignancies. Despite the obvious importance of the aging process, there are still huge gaps in our knowledge of the biological changes that lead to aging and the molecular causes of these changes. The conservation of the fundamental mechanisms of aging and life span determination across eukaryotes enables the study of aging in the single-celled eukaryote budding yeast (Longo et al. 2012). Determining the molecular basis of the mechanisms that determine the replicative life span in yeast should help facilitate the development of therapeutic regimens to extend life span and delay the onset of age-associated disease in humans.
The packaging of the eukaryotic genome together with histones to form chromatin plays a critical role in regulating the activities of the genome. There is clear evidence that the presence of a histone octamer over any particular DNA sequence has a profound effect on the accessibility and therefore the genome stability and transcriptional and replicational potential of that particular DNA sequence (Bell et al. 2011). We demonstrated previously that the bulk amount of the histones decreases dramatically during replicative aging in budding yeast as a consequence of reduced protein synthesis of the histones (Feser et al. 2010). Yeast mutants that overexpress histones have an extended replicative life span, demonstrating that the loss of histones during aging is a cause of replicative aging (Feser et al. 2010). This reduction in histone level during aging is unlikely to be yeast-specific given that human fibroblasts display reduced synthesis of new histones upon successive cell divisions (O'Sullivan et al. 2010). Similarly, a profound loss of histones is also observed in senescent human cells both in vivo and in vitro (Ivanov et al. 2013).
Contradictory to the global increase in gene expression that one would expect to result from the significant degree of nucleosome loss during yeast aging, previous analyses of gene expression during yeast aging report similar numbers of genes with increased and decreased transcription (Lin et al. 2001; Koc et al. 2004; Lesur and Campbell 2004; Yiu et al. 2008). To reconcile these differences, we used improved experimental profiling and analysis approaches to examine where nucleosomes are lost from the yeast genome during aging and whether this loss correlates with increased transcription at these loci. Our study revealed that all of the changes in gene expression during aging are in fact increases and that there were no decreases. Moreover, we found that global nucleosome loss from the genome is the cause of the genome-wide increase in gene expression during aging. We also demonstrate elevated levels of DNA damage, retrotransposition, large-scale chromosome rearrangement, and translocation during aging.
Results
Global reduction in nucleosome occupancy and fuzzier nucleosome positioning during aging
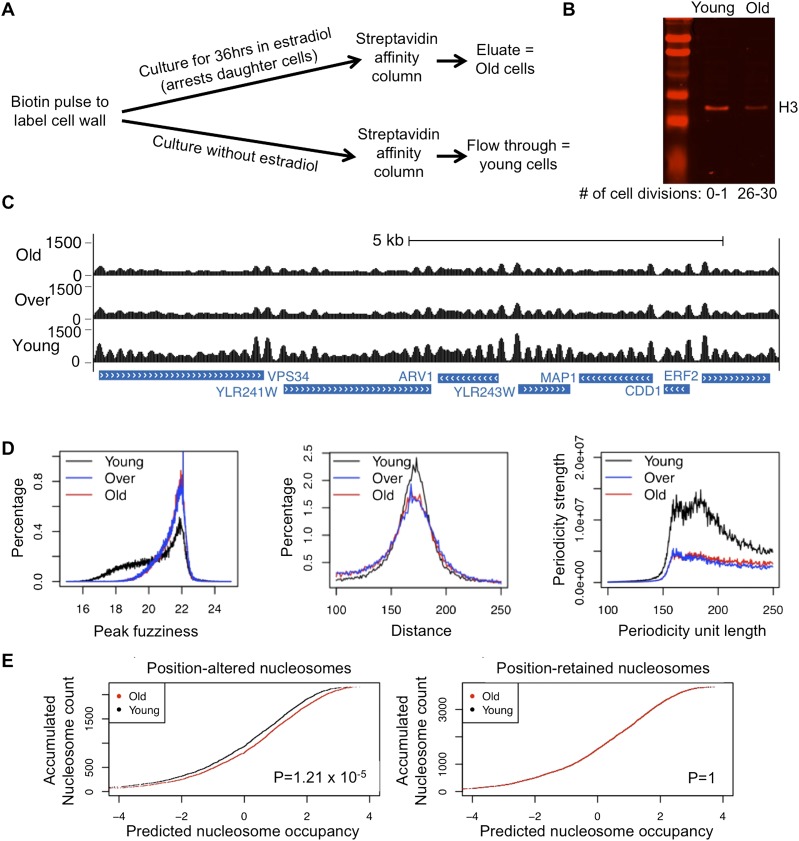
Replicative aging in budding yeast is an asymmetric process that affects mother cells, but not daughter cells, with the median replicative life span of ∼25 cell divisions and a maximum of 45 divisions. A major technical challenge in the study of replicative aging in yeast has been the difficulty in obtaining large numbers of old mother cells. For example, only one yeast cell per 100 million cells in the population has reached (or is older than) the median replicative age. To circumvent this problem, Gottschling's laboratory (Lindstrom and Gottschling 2009) created a “mother enrichment program” (MEP), which allows estradiol-inducible cell cycle arrest of only the daughter yeast cells. We used the MEP in concert with biotin affinity isolation (Smeal et al. 1996) to efficiently isolate the mother cells after 36 h of growth in estradiol-containing medium (Fig. 1A), which we found to be the time it takes for ∼85% of the mother cells to complete their replicative life span (Supplemental Fig. S1A). It is important to note that the yeast are still alive at this time point (Supplemental Fig. S1B) and have completed a median of 25 cell divisions (Supplemental Fig. S1C,D). Young cells were isolated via their failure to bind to the streptavidin resin from log phase cultures grown in parallel in the absence of estradiol in otherwise identical media. We verified that the MEP yeast strains used in this study, like the strains in our previous study (Feser et al. 2010), displayed a reduced H3 histone level during aging (Fig. 1B).
Figure 1.
(A) Schematic diagram of the purification of old and young cells. (B) Old cells isolated using the MEP were analyzed for histone H3 protein levels in comparison with young cells. Equivalent numbers of old and young cells (strain ZHY2) were analyzed by Western blotting. The number of cell divisions was determined by counting bud scars after calcofluor staining. (C) MNase-seq data for a typical region of the genome. “Over” refers to an old sample with overexpressed H3/H4. (D) Distribution of nucleosome fuzziness scores, neighboring distances, and occupancy periodicities. Periodicity was calculated based on the power spectrum density analysis algorithm (Chen et al. 2008, 2010). (E) Accumulated nucleosome count plotted relative to sequence-predicted nucleosome occupancy. (Left) Plot for nucleosomes with positions shifted up to 100 bp and not <50 bp in old cells relative to young cells. (Right) Plot for nucleosomes whose positions are the same in old and young cells; therefore, the curves for the old and young cells completely overlap with each other. P-values are calculated based on KS test between the distributions of nucleosome occupancy values in old and young cells.
In order to map the age-related changes in nucleosome occupancy and positioning, we performed micrococcal nuclease (MNase) digestion analysis of chromatin from old and young cells followed by paired-end Illumina sequencing (MNase-seq) of the mononucleosomal DNA fragments (Supplemental Fig. S2A). We verified that cross-linking efficiency was equivalent in the young and old yeast (Supplemental Fig. S2B). Given that we had shown previously that overexpression of H3 and H4 can extend life span (Feser et al. 2010), we also isolated old mothers from yeast overexpressing H3 and H4 for the same analysis, termed “over.” The young, old, and “over” samples were grown in the identical growth media containing 2% raffinose and 0.5% galactose for all experiments. The MNase-seq analysis was performed in three independent experiments using DANPOS for data analysis (Chen et al. 2013b). Our study was unique in including a spike-in normalization control to enable the detection of global differences in nucleosome organization per cell between the different samples (Supplemental Fig. S3A). The power of the spike-in control is apparent from the distinct nucleosome occupancy in the young and old chromatin (Fig. 1C) in comparison with the unnormalized data that show similar nucleosome occupancy between young and old chromatin (Supplemental Fig. S3B). The DANPOS analyses of nucleosome organization revealed several general features (Fig. 1C). First, the occupancy of histones was reduced ∼50% during aging and was restored to 60% of that observed in young cells by overexpressing histones. Single-cell analysis of GFP-tagged H4 expressed from the endogenous promoter indicated that most of the old population had reduced histone levels (Supplemental Fig. S4), as opposed to half of the old cells having normal histone content, and half of the old cells having no histones (which would be lethal). Second, histones in the old cell population occupy virtually all (97%) of the different possible nucleosome positions that were observed in the young cells regardless of the predicted strength of the DNA sequence for assembling nucleosomes. Third, by plotting fuzziness score distribution of a composite of all nucleosomes, we observed generally greater fuzziness scores in old cells relative to young cells, indicating that nucleosomes generally become fuzzier during aging (Fig. 1D, left). Young cells also showed a more uniform spacing between nucleosome units, with more nucleosomes displaying a 165-base-pair (bp) spacing distance (Fig. 1D, middle). Finally, the strength of nucleosome occupancy periodicity, which reflects the tendency of nucleosome positioning events to recur at certain distances along the DNA, was greatly reduced during aging (Fig. 1D, right). The increased fuzziness and reduced periodicity strength indicates that the stringency of nucleosome positioning is reduced in old cells, and reflects that, in the absence of steric hindrance from adjacent nucleosomes, histone octamers are able to adopt a wider range of positions on the DNA.
Although most nucleosome positions were maintained within the population during aging, 3.04% (2158 out of 70,934 nucleosomes) moved 50–100 bp from their original positions. These position-altered nucleosomes were preferentially depleted from promoters. This is due to the fact that nucleosomes tend to be well-positioned and hard to slide in promoter regions, whereas nucleosome positions are more flexible in promoter-distal gene bodies. For example, 18.55% and 48.44% of total nucleosomes fell within promoters (−350 to 50 bp) and promoter-distal gene bodies (from the transcription start site [TSS] 450 bp downstream from to 150 bp upstream of the transcription termination site [TTS]), respectively, while the percentage is only 10.80% and as many as 68.98% for the position-altered nucleosomes (Fisher's exact test P-value 1.3 × 10−43). Next, we analyzed whether the position-altered nucleosomes were moving toward DNA sequences that are predicted to better favor nucleosome occupancy compared with the original sequence occupied by the nucleosome in young cells (Kaplan et al. 2009) and found this to be the case: For the 2158 position-altered nucleosomes, their predicted occupancies are higher at positions in old cells relative to positions in young cells (Fig. 1E, left). Meanwhile, the changes in occupancy and fuzziness during aging were not influenced by the predicted propensity of the underlying DNA sequence to form nucleosomes (data not shown). As such, we conclude that for some nucleosomes, the lack of steric hindrance from adjacent nucleosomes allows histone octamers to move to DNA sequences that are more favorable for nucleosome formation during aging.
All genes are induced during replicative aging
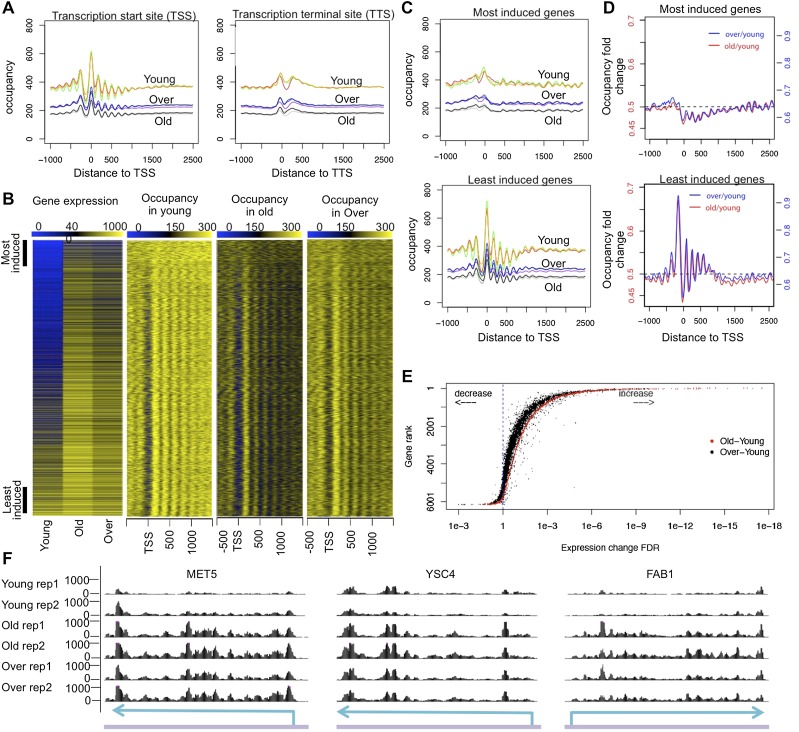
Most genes have a characteristic nucleosome-depleted region (NDR) just upstream of the TSS and over the TTS, flanked by a nucleosome-rich region containing highly occupied and well-phased nucleosomes, with the positioning of distal nucleosomes becoming increasingly less well defined (Lee et al. 2007; Mavrich et al. 2008). During aging, we found that this characteristic pattern of nucleosome depletion flanked by nucleosome phasing at the TSS and TTS was still present but weak (Fig. 2A). We predicted that the decay of the nucleosome phasing at the TTS and TSS, coupled with the globally reduced nucleosome occupancy over the yeast genome in old cells, would lead to profound changes in gene expression during aging given that loss of nucleosomes from the promoters activates model yeast genes even in the absence of the activators (Han and Grunstein 1988; Adkins and Tyler 2006). Therefore, we performed RNA sequencing (RNA-seq) analysis during the aging process using spike-in controls to allow normalization to cell number and detection of global expression changes per cell between the samples (Supplemental Fig. S3A; Supplemental Table S1). Figure 2B shows all genes in the yeast genome ranked from those with the most highly induced gene expression during aging (top) to those genes with the least induced gene expression during aging (bottom). The RNA-seq experiment during aging was performed three independent times, and the order of the gene ranking was not significantly changed between the three experiments (data not shown). These experiments indicate that expression of all genes in the yeast genome is induced upon loss of histones during aging even in the absence of the transcriptional activators. The gradation from yellow at the bottom to blue at the top in the young samples demonstrates that the genes whose expression changes most are genes that are repressed in young cells (Fig. 2B, blue), while those that are induced the least are highly expressed (Fig. 2B, yellow) in young cells (Fig. 2B). Given that all previous analyses of gene expression in yeast aging had concluded that roughly equivalent numbers of genes were induced and repressed during aging (Lin et al. 2001; Koc et al. 2004; Lesur and Campbell 2004; Yiu et al. 2008), while our analysis indicated that all genes are induced, we performed validation to show that those genes that are least induced during aging are still significantly induced (Fig. 2B; Supplemental Fig. S5). As such, the previous reports of repression of gene expression during aging were likely due to lack of normalization of the microarray data to cell number (Loven et al. 2012).
Figure 2.
(A) Nucleosome occupancy normalized based on spike-in control for all genes plotted relative to the TSS and TTS. All three replicates are plotted for each cell group. (B) Gene expression changes during aging are plotted from the RNA-seq analysis. Nucleosome occupancies for the same genes are shown on the right. Genes are ranked by the false discovery rate (FDR) of expression difference between old and young cells, with most induced genes on the top and least induced genes at the bottom. (C) Nucleosome occupancy plotted relative to the TSS; the top and bottom 500 genes ranked by expression up-regulation during aging are defined as most induced and least induced gene groups, respectively, and are plotted separately. (D) The fold change of nucleosome occupancy plotted relative to the TSS. Independent scales on the Y-axis are used to plot for old–young and over–young fold change. Horizontal dashed line indicates average fold change over all yeast genes. (E) Rank of gene expression change during aging plotted relative to the FDR value of gene expression change during aging (red) and during aging in the presence of H3/H4 overexpression (black). (F) RNA-seq data for three typical genes: MET5, YSC4, and FAB1. “Over” refers to an old sample with overexpressed H3/H4. Sky-blue arrows over a purple line indicate gene locus on genomic DNA, respectively.
Genes most induced during aging are normally repressed by nucleosome positioning over the promoter
Next, we compared the nucleosome occupancy flanking the TSS for all of the yeast genes in the same rank order (Fig. 2B). This analysis shows that the most highly transcribed genes in young cells have the most apparent NDR and clearest nucleosome phasing flanking the NDR (Fig. 2B,C), while genes that are transcribed at the lowest levels in young cells do not have a NDR and lack the phased flanking nucleosomes. It is also apparent that all genes show nucleosome loss by ∼50% during aging regardless of whether they are in the most induced or least induced group (Fig. 2B,C). However, the most induced genes show stronger nucleosome loss in TSS-proximal regions relative to TSS-distal regions, with occupancy fold change between the old and young cells ranging from 0.45 at the TSS to 0.5 at 2.5 kb downstream from the TSS, while the least induced genes show a reverse trend, with fold change ranging from 0.68 near the TSS to 0.5 at 2.5kb downstream from the TSS (Fig. 2D). The NDR in the least induced genes showed a decrease of occupancy by only 32.80% on average (Fig. 2D). These data suggest that repression of the most induced genes in young cells is a consequence of the fuzzy positioning of nucleosomes, which may block the accessibility of promoters to the transcription machinery, while the high degree of induction of these genes in old cells is due to loss of nucleosomes over the promoter. To provide further confirmation that the global induction of gene expression during aging is a consequence of loss of nucleosomes, we examined RNA changes upon overexpression of histones H3/H4, which partially restores the nucleosome occupancy (Fig. 2B). This analysis shows that overexpression of H3/H4 partially reversed the changes in gene expression (Fig. 2E,F). We conclude that the global loss of nucleosomes from the genome during aging results in global induction of gene expression. Furthermore, we conclude that the extent of induction during aging is dependent on the particular arrangement of the nucleosomes on the promoter, with genes that have a clear NDR and phased flanking nucleosomes being least induced, while genes with fuzzy nucleosome positioning and no NDR are most highly induced.
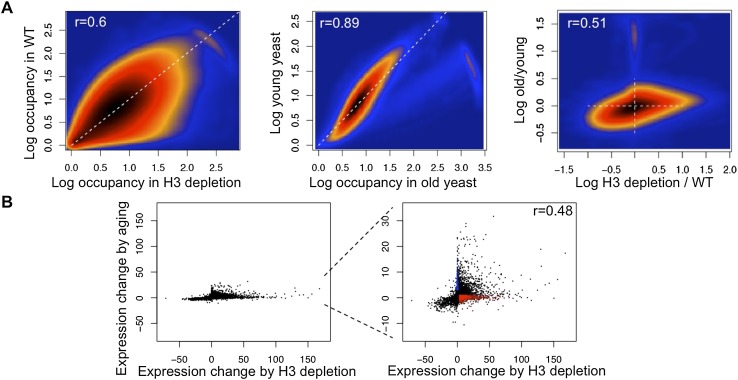
To verify that the changes in nucleosome occupancy and gene expression that we observed were indeed a consequence of the reduced histone content during aging, we compared our data with a recent analysis performed 3 h after transcriptional repression of histone H3 (Gossett and Lieb 2012). This histone depletion analysis specifically asked where the parental histones end up on the newly replicated DNA after a single round of replication in the absence of newly synthesized histone H3. In contrast, our study is asking where nucleosomes are formed as a result of the physiologically relevant progressive loss of all core histones that occurs through numerous cell divisions during aging. Because all previous nucleosome positioning analyses omitted spike-in controls for normalization, comparison was only possible when we used our data normalized without spike-in. Accordingly, we found that the changes in nucleosome occupancy and gene expression during H3 depletion and aging were related (Fig. 3A,B; Supplemental Fig. S6A). However, the expression of numerous genes changed during aging but not during H3 depletion (Fig. 3B), suggesting that these may be programmed aging events that are caused not by the sudden depletion of newly synthesized histones but rather the gradual and progressive loss of histones during aging. Furthermore, the histone H3 depletion led to a loss of numerous nucleosome positions (Gossett and Lieb 2012) that was not apparent during aging.
Figure 3.
(A) Comparison of nucleosome occupancy changes during histone H3 transcriptional repression (left) and aging (middle) and comparing aging with histone H3 transcriptional repression (right). (r) Pearson correlation coefficient. (B) Comparison of gene expression changes during aging and histone H3 transcriptional repression.
We also identified several novel genes that are induced during aging (Supplemental Table S2). For example, the region between 268,400 bp and 268,700 bp on chromosome VI is induced ∼100-fold during aging from the Watson DNA strand only (Supplemental Fig. S6B). This transcript does not encode an apparent ORF and is not polyadenylated (Supplemental Fig. S6C), suggesting that it is a cryptic transcript.
Genes most induced during aging have TATA-containing promoters predicted to better form nucleosomes
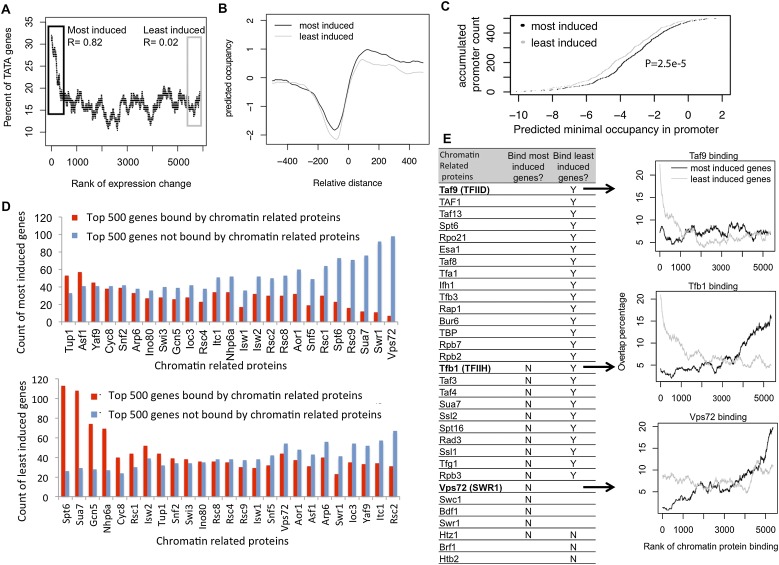
To examine the mechanistic basis for why the genes that are most induced during aging have fuzzier nucleosome positioning relative to the TSS and lack a NDR at the promoter, we looked for correlations with particular basal promoter DNA elements. We found that the 500 most highly induced genes were specifically enriched in TATA-containing promoters (Fig. 4A; Basehoar et al. 2004). This was not the case with the 500 least induced genes. Next, we compared the predicted nucleosome formation ability of the promoter regions (Kaplan et al. 2009) of the most and least induced genes. As expected, genes that were highly induced during aging tend to have promoter sequences that are predicted to be more likely to form nucleosomes, as compared with the genes whose expression is least induced (Fig. 4B,C). To examine whether the factors that occupy the promoter determine to what extent genes are induced, we analyzed the occupancy levels of chromatin-related proteins in young cells (Fig. 4D,E; Venters et al. 2011). It is apparent that the genes that are most induced are normally (in the young state) depleted of chromatin-related proteins that promote gene expression, such as the nucleosome remodeler SWR1, which incorporates the transcription-dependent H2AZ histone variant (Fig. 4D, top; Mizuguchi et al. 2004). Meanwhile, the genes most induced were enriched in repressive chromatin-related proteins, such as the histone chaperone Asf1, which assembles chromatin at the histone gene promoters (Fillingham et al. 2009), and the repressor Tup1, which stabilizes nucleosomes at actively repressed genes (Rizzo et al. 2011; Chen et al. 2013a). In contrast, the genes whose expression was least induced during aging (because they are already expressed at a close to maximal level in young cells) were enriched in the histone acetyltransferase Gcn5 and TFIIB (Sua7) (Fig. 4D, bottom). Interestingly, despite Spt6 being a histone chaperone that assembles chromatin onto promoters during repression (Adkins and Tyler 2006), Spt6 was enriched on the genes whose expression was least induced during aging and was depleted from the genes that are highly induced during aging. This presumably reflects the reported role of Spt6 in promoting transcription (Zhang et al. 2008).
Figure 4.
(A) The percentage of genes with TATA boxes in each of 500 genes ranked according to expression change during aging is shown. (R) Spearman's rank correlation coefficient. (B) The sequence-predicted nucleosome occupancy for the 500 most induced and the 500 least induced genes during aging, plotted relative to the TSS. (C) Accumulated count of promoters plotted relative to predicted minimal nucleosome occupancy. P-value is calculated based on KS test of the occupancy value distribution. (D) The 500 most and least induced genes during aging, ranked by chromatin remodeler occupancy, are defined as bound and not bound, respectively. (E, left) A list of chromatin-binding proteins that bind (Y) or do not bind (N) to the most induced or least induced genes during aging. We retrieved the top and bottom 500 genes bound by each protein and calculated their percentage of overlap with the most induced genes; if the percentage exceeded 15%, we then defined the protein as binding most induced genes. (Right, top) Genes were ranked by Taf9-binding intensity from highest to lowest and divided into groups, each containing 500 genes. For each group, the percentage of genes most induced or least induced during aging was calculated. The percentage for each group was then plotted relative to the Taf9-binding rank for each group. Similar plots were drawn for Tfb1 (middle) and Vps72 (bottom).
Identification of novel genes that influence the aging process
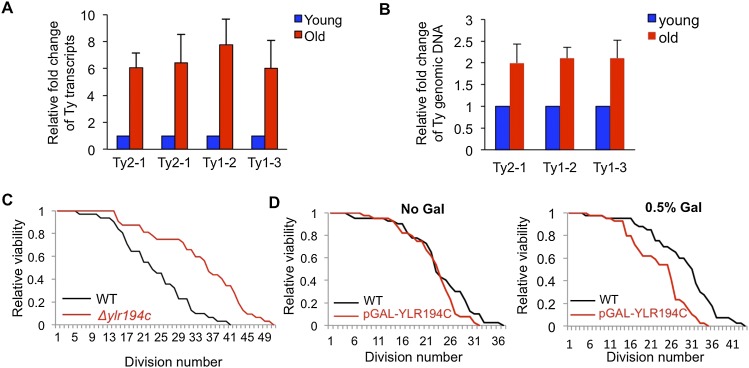
The genes whose expression was least induced during aging were significantly enriched in genes involved in protein synthesis (Supplemental Fig. S6D), contrary to previous reports citing that these genes are down-regulated during aging (Lesur and Campbell 2004). Meanwhile, the genes that were most highly induced during aging were significantly enriched in genes that fall into the gene ontology categories consistent with a function during meiosis and retrotransposition (Supplemental Fig. S6D). The yeast genome includes ∼330 retrotransposon insertions that fall within five distinct families, including the Ty1–Ty5 elements (Kim et al. 1998). Ty elements are normally expressed at a very low level; however, we found a striking increase in Ty element transcription during aging (Supplemental Table 1) that was confirmed by RT–PCR analysis (Fig. 5A) and was partially reversed by histone overexpression (Fig. 2B,E). Because the production of Ty RNA is the template for reverse transcription into the DNA precursor for integration into the genome via retrotransposition (Boeke et al. 1985), we determined whether there are increased levels of Ty DNA in the genome of old yeast as a consequence of increased Ty transcription. Quantitative PCR demonstrated that there is approximately a doubling of the retrotransposon content of the genome in old cells as compared with young cells (Fig. 5B), indicating that the aging process is accompanied by increased retrotransposition as a consequence of the loss of histones.
Figure 5.
(A) Real-time RT–PCR analysis of the fold increase in the expression of the indicated Ty elements during aging. Average and standard deviation of three independent experiments are plotted. (B) Real-time PCR analysis of the fold increase in the genomic DNA content of the indicated Ty elements during aging. Average and standard deviation of three independent experiments are plotted. (C). Replicative life span of isogenic yeast strains BY4741 (wild type [WT]; N = 31) and a strain deleted for YLR194C (ylr194cΔ; N = 32). (D) Replicative life span of isogenic yeast strains wild type (WT; BY4741) and pGAL-YLR194c grown on raffinose (No Gal; left panel) or 0.5% Gal medium (right panel; N = 40) to induce expression of Ylr194c.
To examine whether the nucleosome loss-mediated induction of specific genes during aging limits the number of further cell divisions, we examined the consequence of overexpression of multiple genes on replicative life span. YLR194C, which encodes a glycosylphosphatidylinositol (GPI)-anchored protein (Terashima et al. 2000), is 14-fold induced during aging at the RNA level and is also significantly increased at the protein level (data not shown). We found that deletion of YLR194C extended life span, and its forced overexpression significantly shortened life span (Fig. 5C,D). As such, it appears likely that the overexpression of YLR194C during aging limits the replicative potential of the old cells.
Mitochondrial DNA (mtDNA) and a segment of the right arm of chromosome XII are amplified during aging, correlating with increased DNA damage and translocations involving the rDNA and mtDNA
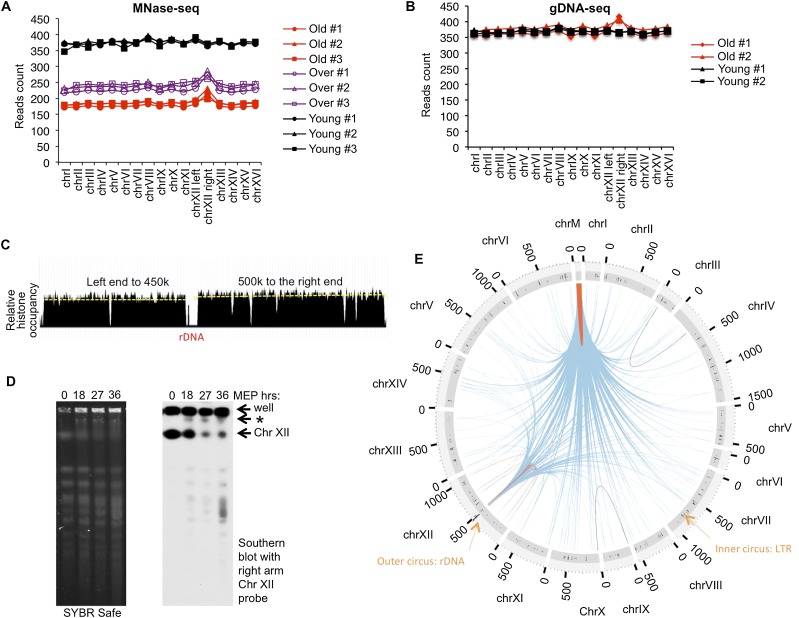
The reduction in nucleosome occupancy over each chromosome during aging was similar for all chromosomes, except for a region on chromosome XII spanning from the rDNA locus to the right end of the chromosome. This region had a nucleosome occupancy ∼15% higher than the rest of the genome in the old but not the young cells (Fig. 6A–C) that was also apparent in the genomic DNA sequencing (data not shown). To gain molecular proof of amplification of the segment of chromosome XII distal to the rDNA, we performed pulsed-field gel electrophoresis (PFGE) of intact chromosomes isolated from yeast at various times during aging. While most of the chromosomes become more faint and more smeared during aging, presumably reflecting increased DNA damage, a new band larger than chromosome XII appears during aging that hybridizes to a probe specific to genomic DNA centromere-distal to the rDNA locus (Fig. 6D). Notably, we confirmed that the increase in the copy number of the rDNA during aging is not sufficient to cause the appearance of this new chromosomal band (data not shown). We also confirmed that the increase in DNA breaks during aging is not due to increased apoptosis (Supplemental Fig. S1E). As such, it would appear that the right segment of chromosome XII distal to the rDNA is amplified or translocated onto another chromosome during aging in ∼15% of the cells in the population.
Figure 6.
(A) Average MNase-seq read coverage on each chromosome or chromosome segment. (B) Average genome sequencing read coverage on each chromosome or chromosome segment in the case of chromosome XII. (C) Nucleosome occupancy across the entire chromosome XII. (D) Analysis of intact yeast chromosomes by PFGE at different time points during aging. (Left panel) SYBR safe staining pattern of the chromosomes. (Right panel) Southern hybridization using a chromosome XII right arm-specific (distal to the rDNA locus) probe. During aging, a longer chromosome appears (labeled with an asterisk), along with apparent fragmentation of the chromosomes. (E) Circos plot of chromosomal translocations that were significantly increased (P < 1 × −3) in old cells relative to young cells. The rDNA locus is indicated by a black bar in the outer ligh-gray circus, and the LTR loci are marked on the inner dark-gray circus, as are the Ty elements. Links shown in blue indicate translocations connecting from either the rDNA region or the mtDNA to other chromosomes. Black links indicate other interchromosome translocation. Red links indicate intrachromosome translocation.
From our analysis of the genomic DNA sequences, we were able to measure the rate of translocations globally during aging. Both interchromosome and intrachromosome translocations happen with higher frequency on the mtDNA and the rDNA locus but not in other regions (Fig. 6E). From the genomic DNA sequences, we sequenced >20,000 translocation junctions and defined 210 pairs of genomic regions that show a significant increase in translocation frequency during aging (Supplemental Table S3). Strikingly, 207 of them involved the rDNA locus and/or the mitochondrial genome, and only 14 classes represent intrachromosomal translocations.
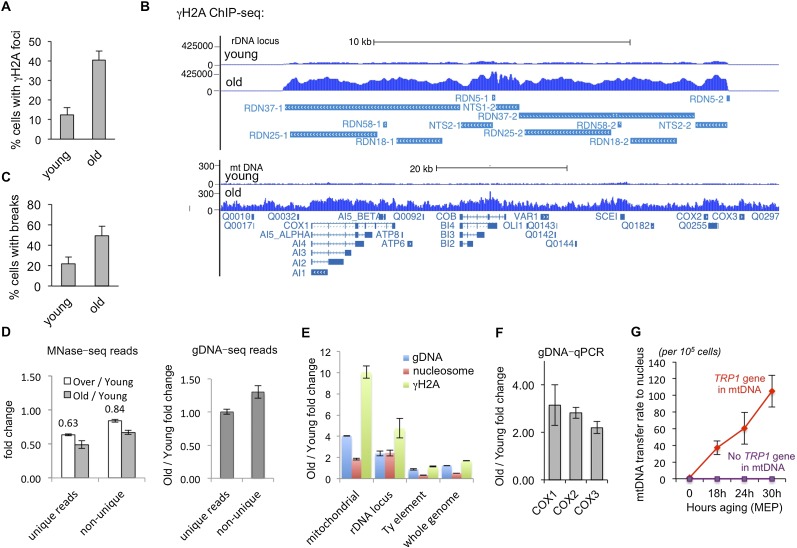
Given that chromosomal translocations are driven by dsDNA breaks, we asked whether DNA damage was more abundant in old cells. Indeed, we found that the number of cells with detectable foci of phosphorylated H2A (γH2A), which is an early DNA damage response, increased more than threefold during aging (Fig. 7A). Spike-in controlled chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) analysis of the abundance of γH2A during aging demonstrated it to be fivefold enriched at the rDNA locus in old cells (Fig. 7B). This suggests that the rDNA locus is suffering increased DNA damage during aging and is consistent with the rDNA being involved in the elevated frequency of translocations that occur during aging (Fig. 6E). Moreover, γH2A was also 10-fold enriched throughout the mtDNA in old cells (Fig. 7B). Given that the mtDNA within mitochondria is not packaged into chromatin, the enrichment of γH2A on the mtDNA is consistent with our observation of numerous translocations between the mtDNA and nuclear chromosomes in old cells (Fig. 6E). To examine directly whether there was an increase in DNA breaks per se during aging, we measured incorporation by terminal deoxynucleotidyl transferase (TdT) of dUTP-biotin onto 3′ DNA ends. Using this assay, the proportion of cells with DNA breaks increased significantly during aging (Fig. 7C). These data clearly demonstrate that the amount of DNA damage is elevated in the old cell population, consistent with the elevated rate of chromosomal translocations (Fig. 6E).
Figure 7.
(A) Quantitation of the percentage of young and old cells with detectable γH2A foci, as measured by immunofluorescence. Error bars represent standard deviation of three independently isolated sets of old and young cells. (B) ChIP-seq of γH2A occupancy in the rDNA and mtDNA of young and old cells. The data shown are not normalized to histone content or DNA content. (C) Quantitation of the percentage of young and old cells with detectable incorporation of biotin dUTP onto DNA ends by TdT, as detected by immunofluorescence. Error bars represent standard deviation of three independently isolated sets of old and young cells. (D) Fold change of unique reads or nonunique reads in each sequenced sample during aging. Error bars indicate standard deviation among replicates. (E) Fold change of rDNA reads, mtDNA reads, reads on Ty elements, or all the other reads in the genome sequencing, MNase-seq, and γH2A-ChIP-seq. Error bars indicate standard deviation among replicates. (F) Validation of increased mtDNA content in aged cells by quantitative PCR. Average and standard deviation of three independent experiments are shown, normalized to a spike in control added according to equivalent cell number. (G) Functional assay of transfer of mtDNA into the nucleus in a strain carrying a TRP1 gene in the mitochondrial genome (red) and a strain lacking the TRP1 gene (purple), as determined by frequency of colony formation on −TRP plates. Shown are the average and standard deviation of three independent experiments performed at the indicated times during aging using the MEP.
We noted that the level of nonunique sequencing reads in the genomic DNA sequencing increased by 1.3-fold in the old cell relative to the young cell; however, nonunique reads in the MNase-seq decreased by 0.67-fold (Fig. 7D). Accordingly, we found that the increase in the nonunique reads was partly due to the increase in the Ty sequences noted above (Figs. 5B, 7E) and a threefold increase in sequences from the rDNA (Fig. 7E), which is consistent with the elevated level of extrachromosomal ribosomal DNA circles (ERCs) that are produced from the rDNA by recombination and retained in the mother cells (Sinclair and Guarente 1997). We also noted a previously unreported increase in the mtDNA content of aging cells (Fig. 7E), which was confirmed by quantitative PCR analysis of the mitochondrially encoded COX1, COX2, and COX3 genes (Fig. 7F). Upon removal of the mtDNA, Ty, and rDNA sequence reads from the genomic DNA sequencing, it is clear that young and old cells have globally equivalent genomic DNA contents (Fig. 7E), and this was validated by quantitative PCR analysis (data not shown). Importantly, the enrichment of γH2A on the rDNA and mtDNA is more significant than the enrichment of the nucleosomes and genomic DNA at these loci during aging (Fig. 7E), suggesting that DNA damage is indeed more abundant at these loci in old cells. We propose that the increase in mtDNA content, coupled with the translocation of mtDNA onto nuclear chromosomes (Fig. 6F) and increased γH2A on mtDNA (Fig. 7B), represents nuclear transfer of the mtDNA during the repair of double-strand breaks (Ricchetti et al. 1999) during aging. To functionally test whether there is an increase in mtDNA transfer to the nuclear genome during aging, we used a strain in which the TRP1 gene had been inserted into the mtDNA (Thorsness and Fox 1993). The TRP1 gene is not transcribed by the mitochondrial RNA polymerase but is transcribed by RNA polymerase II upon its release from the mtDNA. Using this assay, we found that the rate of transfer of the TRP1 gene from the mtDNA to the nucleus is increased greatly during replicative aging (Fig. 7G), consistent with the preponderance of translocations involving the mtDNA during aging (Fig. 6E).
Discussion
By using spike-in controls for normalization, we were able to show that nucleosome occupancy is decreased by ∼50% across the whole genome during aging, leading to transcriptional up-regulation of every gene in the yeast genome. The fuzzier nucleosome positioning observed during aging indicates that steric hindrance from adjacent nucleosomes contributes to the stringency of nucleosome positioning. From the highly touted influence of the DNA sequence in positioning nucleosomes, one might have expected that only sequences with highly predicted nucleosome occupancy would be assembled into nucleosomes in old cells. In contrast, we found that most nucleosome positions in the genome are still represented in the population of aged, naturally histone-depleted cells. We also show that the reduced expression of genes during aging previously reported (Lesur and Campbell 2004) was likely due to lack of normalization of the data, requiring a re-evaluation of the accepted changes in gene expression during aging. For example, the ribosomal protein genes have been reported to be repressed during aging (Lesur and Campbell 2004; Yiu et al. 2008). However, we show that that is clearly not the case, as we found that ribosomal protein genes and all other yeast genes are in fact induced during aging.
It is quite striking that we found the rDNA to be the point of demarcation of the amplification of the segment of the right arm of chromosome XII in the old cells. The rDNA has received attention in the aging field as the locus that produces ERCs as a result of recombination that has been suggested to be a cause of aging (Sinclair and Guarente 1997). Meanwhile others have proposed that rDNA instability per se, not ERC formation, is the cause of aging (Kobayashi 2008). Recently, it was shown that increased replicative age induces mitotic recombination in the ribosomal RNA gene cluster in daughter cells (Lindstrom et al. 2011), leading to loss of heterozygosity of the right segment of chromosome XII at a rate of up to 11 events per 1000 divisions, measured genetically (Lindstrom et al. 2011). Our work is the first time that the amplification during aging of the right segment of chromosome XII has been reported in mother cells, and the high frequency of this genomic rearrangement during aging is unprecedented, occurring in 15% of the population of mothers.
Given the experimental difficulty with isolating old yeast cells, no one had previously examined whether levels of DNA damage increase during replicative aging. Using the MEP, we show that DNA damage foci and DNA breaks significantly increase during aging. The regions with increased DNA damage during aging include the rDNA locus, which is likely the driving force behind the previously proposed rDNA instability, ERC formation, and loss of heterozygosity during aging as well as the amplification of the right segment of chromosome XII that we observed in 15% of old cells. Furthermore, we uncovered evidence for DNA damage on chromatinized mtDNA and elevated levels of chromosomal translocations during aging, particularly involving the rDNA locus and the mtDNA. Indeed, mtDNA has previously been observed to be transferred into the nuclear genome during the process of double-strand break repair (Ricchetti et al. 1999). It seems likely that the elevated levels of dsDNA damage that we observed during aging drive the translocations and nuclear transfer of mtDNA, accounting for the fourfold increase in the mtDNA content of old cells.
Retrotransposons from yeast to humans are normally silenced by a repressive chromatin structure. Accordingly, we observed derepression of the yeast Ty retrotransposons during aging concomitant with the loss of nucleosomes during aging and partly reversed by overexpression of the histones. This increased Ty transcription led to increased levels of Ty DNA during aging presumably due to retrotransposition. Retrotransposition itself is mutagenic, can disrupt gene function and expression by providing novel promoters, and results in additional identical DNA sequences that mediate illegitimate recombination and translocations between chromosomes. Derepression of retrotransposons due to chromatin changes also is a feature of aged human cells at least in vitro because aged human adult stem cells show increased expression of the Alu retrotransposons (Wang et al. 2011). Very recently, replicatively senescent human cells have been shown to have increased expression of the Alu and Line1 retrotransposons that correlated with a more open chromatin structure as well as increased retrotransposition (De Cecco et al. 2013).
The fact that there are increased levels of (1) amplification of the right segment of chromosome XII, (2) Ty retrotransposons, (3) mtDNA, and (4) translocations in old cells indicates that these genetic changes are not effectively passed on to the daughter cells. Given that these genetic changes were observed in populations of old cells, where 85% of the cells have completed their replicative life span, the most plausible explanation is that the cells that have these increased genetic changes are not effective at producing daughter cells. It would be interesting to determine in the future whether these genetic changes contribute to the replicative age clock that prevents further cell divisions. It would be also interesting to investigate how much of the increased genomic instability during aging results from genotoxic stress versus chromatin changes. Finally, this work presents candidate factors for future studies toward determining the mechanism of aging and developing novel strategies to achieve longevity.
Materials and methods
Yeast strains and growth
All strains are derivatives of Saccharomyces cerevisiae S288c (BY4741), and genotypes are provided in the Supplemental Material. One-step PCR-mediated gene replacement and epitope tagging were carried out using standard techniques. Cells were cultured in YEPD (1% yeast extract, 2% peptone, 2% glucose) unless otherwise indicated. Cells were grown exponentially overnight to mid-log phase before the initiation of all experiments. For overexpression of histones H3/H4 under the control of the pGAL1/10 divergent promoters, cells were cultured in YEP plus the indicated carbon source. Replicative life span assays were performed on YEPD with 2% glucose or YEP with 2% raffinose and 0.5%, 1%, or 2% galactose (YEPRG) as indicated. Plasmids expressing histones were maintained by growth in synthetic complete medium lacking the appropriate amino acid and supplemented with raffinose and galactose for strains carrying galactose-inducible promoter fusions.
Purification of old and young cells
For isolating old cells, we used the MEP as previously described (Lindstrom and Gottschling 2009) with slight modifications described in the Supplemental Material. The identical medium was used for the young, old, and “over” samples for all experiments in order to ensure that changes in chromatin or gene expression were not due to differences in sugar source.
Life span measurement by micromanipulation
Replicative life span of virgin mother cells was determined as previously described with minor modifications (Kennedy et al. 1994). Cells were streaked onto YEPD plates, and a single colony was selected for life span analysis. Cells were incubated nightly at 12°C to impede division. Yeast were initially grown on YEP + 2% glycerol plates to eliminate yeast cells lacking mitochondria.
MNase digestion, genomic DNA isolation, library construction, sequencing, and data analysis
The same number of young, old, and histone H3/H4-overexpressing cells was collected, and the cells were cross-linked with formaldehyde followed by spheroplasting and MNase digestion, as detailed in the Supplemental Material. The same number of purified young and old cells was subjected to genomic DNA isolation. The masterpure yeast DNA purification kit (catalog nos. MPY80010 and MPY 80200) was used to isolate the genomic DNA. The libraries were generated using a KAPA library preparation kit (KK8200, Illumina series). A spike-in control that mimics the size of the mononucleosomal fragment was included during the library construction and was added according to the number of cells used to isolate the mononucleosomal fragments. Samples (two RNA-seq replicates, three MNase-seq replicates, and two genomic-seq replicates) were prepared for paired-end sequencing. All libraries were sequenced using the Illumina HiSeq2000 system. The data analysis is described at length in the Supplemental Material.
Real-time RT–PCR
For real-time PCR analysis of cDNAs, a Roche Applied Sciences LightCycler 480 was used with the LightCycler 480 SYBR Green I master mix kit (Roche, catalog no. 04887352001) according to the kit directions.
Quantitative Western blotting analysis
Total protein extract was isolated and boiled prior to loading gels. IRDye anti-rabbit secondary antibody that fluoresced at 700 nm was used.
PFGE analysis
Analyses of intact yeast chromosomes were performed using a CHEF DR II system (Bio-Rad) followed by Southern hybridization using a chromosome XII probe distal to the rDNA locus labeled by random prime labeling with 32P. Yeast cells were isolated at different time points as indicated using the MEP and were used to prepare agarose gel molds following the manufacturer's instructions (Bio-Rad). The electrophoretic conditions were 6 V/cm and 24 h at 14°C with a 50- to 150-sec pulse time and a 1% agarose gel.
Acknowledgments
We thank Dan Gottschling and Peter Thorsness for generously sharing yeast strains, Briana Dennehey and members of the Tyler laboratory for critically reading the manuscript, and Banafsheh Mirnikjoo for performing some of the preliminary studies. This work was supported by National Institutes of Health (NIH) grants GM64475 and CA95641, a University of Texas University Trust award, an M.D. Anderson Stars award and a Cancer Prevention Research Institute of Texas (CPRIT) Rising Star award (to J.K.T.), and CPRIT RP110471-C3, Department of Defense grant W81XWH-10-1-0501, and NIH R01HG007538 (to W.L.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.233221.113.
References
- Adkins MW, Tyler JK 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell 21: 405–416 [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Bell O, Tiwari VK, Thoma NH, Schubeler D 2011. Determinants and dynamics of genome accessibility. Nat Rev Genet 12: 554–564 [DOI] [PubMed] [Google Scholar]
- Boeke JD, Garfinkel DJ, Styles CA, Fink GR 1985. Ty elements transpose through an RNA intermediate. Cell 40: 491–500 [DOI] [PubMed] [Google Scholar]
- Chen K, Meng Q, Ma L, Liu Q, Tang P, Chiu C, Hu S, Yu J 2008. A novel DNA sequence periodicity decodes nucleosome positioning. Nucleic Acids Res 36: 6228–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wang L, Yang M, Liu J, Xin C, Hu S, Yu J 2010. Sequence signatures of nucleosome positioning in Caenorhabditis elegans. Genomics Proteomics Bioinformatics 8: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wilson MA, Hirsch C, Watson A, Liang S, Lu Y, Li W, Dent SY 2013a. Stabilization of the promoter nucleosomes in nucleosome-free regions by the yeast Cyc8-Tup1 corepressor. Genome Res 23: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, Li W 2013b. DANPOS: Dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res 23: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM 2013. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK 2010. Elevated histone expression promotes life span extension. Mol Cell 39: 724–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Kainth P, Lambert JP, van Bakel H, Tsui K, Pena-Castillo L, Nislow C, Figeys D, Hughes TR, Greenblatt J, et al. 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol Cell 35: 340–351 [DOI] [PubMed] [Google Scholar]
- Gossett AJ, Lieb JD 2012. In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae. PLoS Genet 8: e1002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Grunstein M 1988. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55: 1137–1145 [DOI] [PubMed] [Google Scholar]
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al. 2013. Lysosome-mediated processing of chromatin in senescence. J Cell Biol 202: 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR Jr, Guarente L 1994. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol 127: 1985–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF 1998. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 8: 464–478 [DOI] [PubMed] [Google Scholar]
- Kobayashi T 2008. A new role of the rDNA and nucleolus in the nucleus–rDNA instability maintains genome integrity. BioEssays 30: 267–272 [DOI] [PubMed] [Google Scholar]
- Koc A, Gasch AP, Rutherford JC, Kim HY, Gladyshev VN 2004. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc Natl Acad Sci 101: 7999–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Lesur I, Campbell JL 2004. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Mol Biol Cell 15: 1297–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Manchester JK, Gordon JI 2001. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J Biol Chem 276: 36000–36007 [DOI] [PubMed] [Google Scholar]
- Lindstrom DL, Gottschling DE 2009. The mother enrichment program: A genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics 183: 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Leverich CK, Henderson KA, Gottschling DE 2011. Replicative age induces mitotic recombination in the ribosomal RNA gene cluster of Saccharomyces cerevisiae. PLoS Genet 7: e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy B 2012. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab 16: 18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA 2012. Revisiting global gene expression analysis. Cell 151: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J 2010. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 17: 1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti M, Fairhead C, Dujon B 1999. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature 402: 96–100 [DOI] [PubMed] [Google Scholar]
- Rizzo JM, Mieczkowski PA, Buck MJ 2011. Tup1 stabilizes promoter nucleosome positioning and occupancy at transcriptionally plastic genes. Nucleic Acids Res 39: 8803–8819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Smeal T, Claus J, Kennedy B, Cole F, Guarente L 1996. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84: 633–642 [DOI] [PubMed] [Google Scholar]
- Terashima H, Yabuki N, Arisawa M, Hamada K, Kitada K 2000. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol Gen Genet 264: 64–74 [DOI] [PubMed] [Google Scholar]
- Thorsness PE, Fox TD 1993. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics 134: 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. 2011. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 41: 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, et al. 2011. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 10: 3016–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu G, McCord A, Wise A, Jindal R, Hardee J, Kuo A, Shimogawa MY, Cahoon L, Wu M, Kloke J, et al. 2008. Pathways change in expression during replicative aging in Saccharomyces cerevisiae. J Gerontol A Biol Sci Med Sci 63: 21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Fletcher AG, Cheung V, Winston F, Stargell LA 2008. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol Cell Biol 28: 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]