Abstract
This proof-of-concept study demonstrates the application of a novel nucleic acid detection platform to detect Clostridium difficile in subjects presenting with acute diarrheal symptoms. This method amplifies three genes associated with C. difficile infection, including genes and deletions (cdtB and tcdC) associated with hypervirulence attributed to the NAP1/027/BI strain. Amplification of DNA from the tcdB, tcdC, and cdtB genes was performed using a droplet-based sandwich platform with quantitative real-time PCR in microliter droplets to detect and identify the amplified fragments of DNA. The device and identification system are simple in design and can be integrated as a point-of-care test to help rapidly detect and identify C. difficile strains that pose significant health threats in hospitals and other health-care communities.
CME Accreditation Statement: This activity (“JMD 2014 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint sponsorship of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2014 CME Program in Molecular Diagnostics”) for a maximum of 48 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Clostridium difficile is an anaerobic, spore-forming, Gram-positive bacterium that colonizes the human colon and typically presents as an opportunistic infection after other colonic flora have been eradicated by commonly used antibiotics.1–3 C. difficile infection can cause severe diarrhea, pseudomembranous colitis, and toxic megacolon, and may require urgent colectomy or result in death.1–5 C. difficile infection has rapidly increased in incidence, severity, and mortality, particularly in the United States, Canada, and Europe, and C. difficile is the leading cause of nosocomial infectious mortality.4–7 In the United States, C. difficile infection was responsible for $4.8 billion in excess hospital costs in 2008 and an estimated 14,000 deaths from 2006 to 2007.8,9 The changing epidemiology of C. difficile has been marked by hospital outbreaks due to a hypervirulent strain, NAP1/027/BI, characterized by severe disease and increased morbidity and mortality.10–13 With the emergence of the NAP1/027/BI strain, there is a critical need for a highly sensitive and rapid method of detection and strain typing in health-care settings.4,14
Current methods of diagnosing C. difficile include stool culture, toxin testing, enzyme immunoassays, and PCR. Although stool culture and cytotoxicity tests provide high sensitivity and specificity, these methods are impractical in most clinical settings, because they require 2 to 3 days to complete, during which time clinicians must rely on empirical treatment of disease with antibiotics.3,15,16 Commercial enzyme immunoassay kits for C. difficile toxins A and B are highly specific and rapid, but often lack sensitivity, which can be as low as 65%.16–20 Several multiplex PCR assays have been reported to identify various genes associated with C. difficile, including two commercially available tests, one of which has the ability to differentiate between the NAP1/027/BI strain and other toxigenic strains of C. difficile.15,21–26
Here, we describe a novel PCR assay coupled with a small-volume, real-time platform allowing for simple and rapid detection of three C. difficile genes: tcdB, cdtB, and tcdC; the last is included for detection of the hypervirulent NAP1/027/BI variant. These genes code, respectively, for toxin B, binary toxin, and the TcdC protein suspected to regulate toxin production.27,28 These genes were selected as targets for our assay based on the roles these toxins and proteins play in conferring a virulent or hypervirulent phenotype. Our assay has the ability to indicate the presence of markers of both PCR ribotype 027 and 078 hypervirulent strains for future point-of-care testing. This proof-of-concept study demonstrates a simple quantitative real-time PCR (qPCR) platform as a potential tool for cost-effective and timely clinical decision making.
Materials and Methods
Primer Design and Evaluation
The pathogenicity locus in the C. difficile genome contains two genes coding the toxins A (tcdA) and B (tcdB), as well as three accessory genes (tcdC, tcdD, and tcdE). The tcdC gene is a putative negative regulator of tcdA and tcdB gene expression.27,29 Some strains also produce a third toxin, known as binary toxin, encoded by the gene cdtB. The tcdA and tcdB genes are typically used as markers for the presence of the organism, but other molecular markers are required for detection of the hypervirulent NAP1/027/BI strain or similar hypervirulent strains. Hallmarks of the NAP1/027/BI strain include increased production of toxins A and toxin B, as well as production of binary toxin. The strain also carries an 18-bp deletion in the tcdC gene, which is thought to play a role in increased toxin production.30
Three target gene segments were thus targeted for the subtyping assay, to detect not only the presence of toxigenic C. difficile but also the hypervirulent genotype. Our assay targets the tcdB gene, which codes for toxin B; this toxin is present in nearly all toxigenic strains and serves as first-line detection for the presence of the disease-causing organism.29 We also target the regulatory gene tcdC, with the ability to detect both the 18-bp deletion in the hypervirulent NAP1/027/BI strain and larger deletions in other strains that are also considered to be hypervirulent (078).31 The last target is cdtB, expressed by the NAP1/027/BI strain. The cdtB gene does not occur in all disease-causing C. difficile strain; it codes for the binary toxin and is associated with of hypervirulence. Full-genome C. difficile gene sequences from 15 strains were acquired from the National Center for Biotechnology Information and were aligned to the C. difficile 630 reference sequence using multiPipMaker,32 a multiple alignment tool designed to handle such large-scale sequences. The consensus sequences for three target genes (tcdB, cdtB, and tcdC) specific to the C. difficile genome were analyzed by visual inspection and with Primer3 Plus,33 to identify 100% conserved regions suitable for amplification by PCR. Information on the strains used for the alignment is summarized in Table 1.
Table 1.
Strain Information for Alignment of C. difficile Sequences
| Strain | Year | Country | Ribotype | NAP type | Accession no.∗ |
|---|---|---|---|---|---|
| 630† | 1982 | Switzerland | 012 | ND | AM180355.1 |
| CF5 | 1995 | Belgium | 017 | ND | FN665652 |
| M68 | 2006 | Ireland | 017 | ND | FN668375.1 |
| M120 | 2007 | UK | 078 | ND | FN665653.1 |
| BI-9 | 2001 | USA | 001 | ND | FN668944.1 |
| CD196 | 1985 | France | 027 | NAP1 | FN538970.1 |
| BI-1 | 1988 | USA | 027 | NAP1 | FN668941.1 |
| R20291 | 2006 | UK | 027 | NAP1 | FN545816.1 |
| QCD-37x79 | 2005 | Canada | ND | NAP1 | NZ_CM000658.1 |
| QCD-76w55 | 1988 | USA | ND | NAP1a/001 | NZ_CM000661.1 |
| QCD-97b34 | 2004 | Canada | ND | NAP1b/006 | NZ_CM000657.1 |
| K744 | 2004–2006 | Canada | 078 | NAP8 | HQ639679 |
| AE70 | 2004–2006 | Canada | 027 | NAP1 | HQ639675 |
| AE16 | 2004–2006 | Canada | 027 | NAP1 | HQ639671 |
| AE978 | 2004–2006 | Canada | 027 | NAP1 | HQ639678 |
| CD98 | 2004–2006 | Canada | 078 | NAP7 | HQ639674 |
The region of the tcdC gene to be amplified through PCR was designed to include the 18-bp deletion characteristic of the genome of the hypervirulent C. difficile strain NAP1/027/BI. For each primer pair, the primers were optimized to anneal between 53°C and 60°C (Table 2). The self-dimerization and heterodimerization energies of each of the primer pairs were evaluated in silico, using the online folding tool DINAMelt (http://mfold.rna.albany.edu; DINAMelt Server, RNA Institute, SUNY Albany, Albany, NY),34 to ensure limited formation of primer dimers. Several candidate primer pairs for each gene were obtained from Integrated DNA Technologies (Coralville, IA). Multiple iterations of primer design and validation of the primers in the multiplex PCR platform were completed, to identify the optimum grouping of three primer pairs to use on the platform.
Table 2.
PCR Primers for Amplification of C. difficile Genes tcdB, tcdC, and cdtB
| Primer name | Primer sequence | Tm (°C) at 12 mmol/L Mg2+ | Amplicon length (bp) |
|---|---|---|---|
| tcdB Fwd | 5′-CTGGAGAATCTATATTTGTAG-3′ | 54.9 | 328 |
| tcdB Rev | 5′-GCAGTTGATACTAATTCAAC-3′ | 56.4 | |
| tcdC Fwd | 5′-CTCAAAAAACAGAAATAGAAAC-3′ | 56.7 | 300 |
| tcdC Rev | 5′-ACCTCATCACCATCTTC-3′ | 56.8 | |
| cdtB Fwd | 5′-GCAGTTAAGTGGGAAGATAG-3′ | 59.1 | 190 |
| cdtB Rev | 5′-TCCATACCTACTCCAACAAT-3′ | 59.6 |
Fwd, forward; Rev, reverse; Tm, melting temperature.
Clinical Sample Collection and Isolation of Clinical DNA
Double-stranded DNA was isolated from anonymized stool samples obtained from patients at Rhode Island Hospital, Miriam Hospital, and Newport Hospital (all in Rhode Island). Approval was obtained from the Institutional Review Board of Rhode Island Hospital/Lifespan (approval no. 211049-2), which waived the need for written or verbal informed consent from the participants; the exemption was granted on the basis of the research involving previously collected diagnostic specimens from subjects who could not be identified, either directly or indirectly. Anonymized stool samples were obtained from patients whose stool specimens had already been obtained for evaluation of suspected C. difficile during clinical care by physicians not involved with the present study. Samples were determined as positive or negative for C. difficile with a GeneOhm Cdiff PCR kit (BD Diagnostics, La Jolla, CA) on a LightCycler qPCR system (Roche Diagnostics, Indianapolis, IN). Stool samples were stored at −20°C. DNA was isolated using a QIAamp stool DNA isolation kit (Qiagen, Valencia, CA; Hilden, Germany) and quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Isolated DNA was stored at −20°C.
PCR for C. difficile Detection
Target sequences were amplified by PCR, with isolated DNA added to a final reaction volume of 10 μL. Unless otherwise specified, the reaction mix contained 1× Taq polymerase buffer (New England Biolabs, Ipswich, MA), 12.0 mmol/L MgCl2 (Life Technologies, Carlsbad, CA), 0.1 μg/μL bovine serum albumin, 0.2 mmol/L of each dNTP (New England Biolabs), 0.1 U/μL Taq polymerase (New England Biolabs), 0.267 μmol/L of each tcdC primer, 0.267 μmol/L of each cdtB primer, and 0.167 μmol/L of each tcdB primer. After an initial heating step of 5 minutes at 94°C, the samples were amplified for 40 cycles of 30 seconds at 94°C, 30 seconds at 65°C to 53°C for 30 seconds (decreasing 0.5°C per cycle and held constant thereafter), and 72°C for 1 minute; the samples were then held at 72°C for 5 minutes for a final extension step. Singleplex reactions were performed with the same conditions, except that 1.5 mmol/L MgCl2 was used and the annealing temperatures were 55°C for tcdB and tcdC and 57°C for cdtB. Target sequences were amplified either using a conventional thermal cycler (Bio-Rad Laboratories, Hercules, CA) or on our droplet sandwich platform (2 μL reaction mix). The product size was determined using Agilent DNA 1000 chips on an Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara CA).
Droplet Sandwich Platform
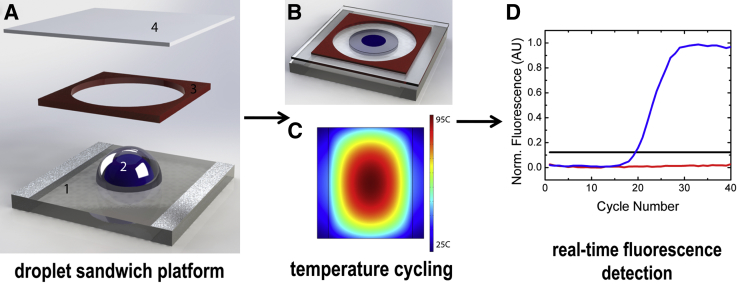
Schematics of the droplet sandwich platform and the workflow are shown in Figure 1. The platform consists of an indium tin oxide (ITO)–coated glass resistive heater, an imaging spacer, and a coverslip to prevent evaporation during thermal cycling.35,36 A custom-written proportional-integral-derivative controller was used to control the droplet temperature and was manually tuned to match the cycling conditions for the PCR protocol. The setup includes a 2-μL droplet of PCR mix surrounded by mineral oil, which produces a disk-shaped compound droplet (Figure 1, A and B). Temperature and fluorescence signals are collected simultaneously using a thermocouple (type E) and a photomultiplier tube. The ITO-coated glass surface is optically transparent, which makes fluorescence detection simple using a standard inverted fluorescence microscope and photomultiplier tube. A typical output graph of fluorescence over time is presented in Figure 1D. Removable fluorinated ethylene propylene tape was applied to the glass surface for each experiment to prevent sample carryover and contamination and to prevent adsorption of the PCR reagents to the glass surface.36 This method is also useful in that droplets act as individual reaction vessels, which also help limit sample carryover. All qPCR experiments were performed with SYBR Green I dye (Life Technologies). A heating profile for the ITO-coated glass when 15 V is applied to the resistive surface was generated by COMSOL Multiphysics 4.3b simulation software (Burlington, MA; Stockholm, Sweden) (Figure 1C). The temperature profile shows the temperature cycling achieved using the droplet platform at the center of the radial heating profile. The platform requires low voltage (approximately 15 V) and uses a microfan for cooling. The average cycle time is approximately 100 seconds; the ramp rate is approximately 3.0°C/second for heating and approximately 2.0°C/second for cooling. An important advantage of droplet-based PCR over traditional PCR methodologies is the small sample volumes, which have decreased thermal diffusion distances and thus allow for faster heat transfer.
Figure 1.
Components of the droplet sandwich platform and workflow. A: Exploded view of the platform components: ITO-coated glass (1) with a compound droplet (2) surrounded by a spacer (3) and covered with a coverslip (4). B: The fully assembled platform effectively sandwiches the compound droplet in a reaction chamber. The slide dimensions are 40 mm × 40 mm, and the compound droplet is approximately 2.8 mm in diameter. C: Heating profile generated by COMSOL Multiphysics 4.3b simulation software. The ITO-coated glass resistive surface heats radially up to 95°C when 15 V is applied to the resistive surface. D: Plot of DNA amplification with fluorescent intercalating dye shows positive samples (blue), negative samples with no change over time (red), and the calculated threshold (black). Fluorescence is collected in real time during the extension phase of PCR, using a photomultiplier tube. AU, arbitrary units; Norm., normalized.
Results
PCR Can Selectively and Sensitively Amplify Target Genes
Each primer pair was tested individually and in conjunction with other primer pairs. The primers were tested with a template concentration of 5.0 ng/μL for two positive-control samples. In both situations, all primer pairs effectively amplified their respective targets. An electrophoresis gel plot of DNA amplified in a PCR assay is presented in Figure 2. DNA isolated from cultured C. difficile reference strain 630 (C. difficile ATCC BAA-1382) was used, or DNA isolated from the C. difficile strain VA17 (provided by Dr. Curtis Donskey). The VA17 strain is designated by pulsed-field gel electrophoresis as the North American Profile 1 (NAP1), by PCR as ribotype 027, and by restriction endonuclease analysis as type BI and is thus known as the NAP1/027/BI strain.
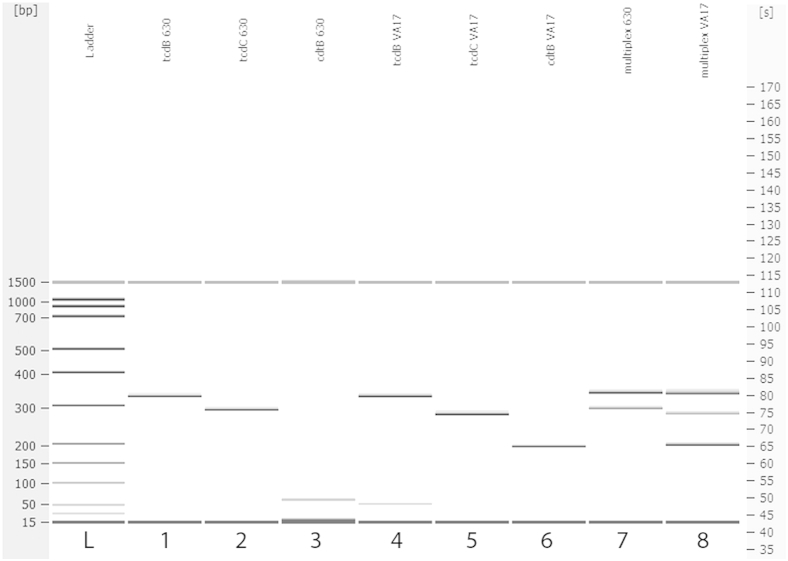
Figure 2.
Electrophoresis gel plot of single-gene and multiplex PCR using C. difficile reference strain 630 and the NAP1/027/BI cultured isolate VA17. The template is indicated at the top of each lane. Lanes 1 and 4, tcdB primer set; lanes 2 and 5, tcdC primer set; lanes 3 and 6, cdtB primer set; lanes 7 and 8, multiplex results for the reference strain 630 and the hypervirulent VA17 isolate, respectively.
In lanes 1, 2, 3, and 7 (Figure 2), reference strain 630 was used as a template and in lanes 4, 5, 6, and 8 the NAP1/027/BI strain was used. Lanes 1 and 4 show amplification of a region within the tcdB gene, which is identical for both templates, at 328 bp. In lanes 2 and 5, cdtB primer pairs were used that show amplification of the binary toxin gene for the NAP1/027/BI strain, at 190 bp, but not for the reference strain 630 (because this binary toxin gene is not present in the reference genome and similar strains). Lanes 3 and 6 show amplification of the tcdC gene for both strains; however, the 18-bp deletion in the tcdC gene for the NAP1/027/BI strain is apparent in the downward shift of the band. The sizing results from the Agilent Bioanalyzer system indicate an amplicon size of 300 bp for the C. difficile reference strain 630 and of 282 bp for the VA17 strain, a difference of 18 bp. These findings corroborate that our primer set can detect the mutation in the tcdC gene associated with virulence in NAP1/027/BI C. difficile strains.27,28,37 We also obtained multiplex results for the reference genome 630 and the NAP1/027/BI strain (Figure 2).
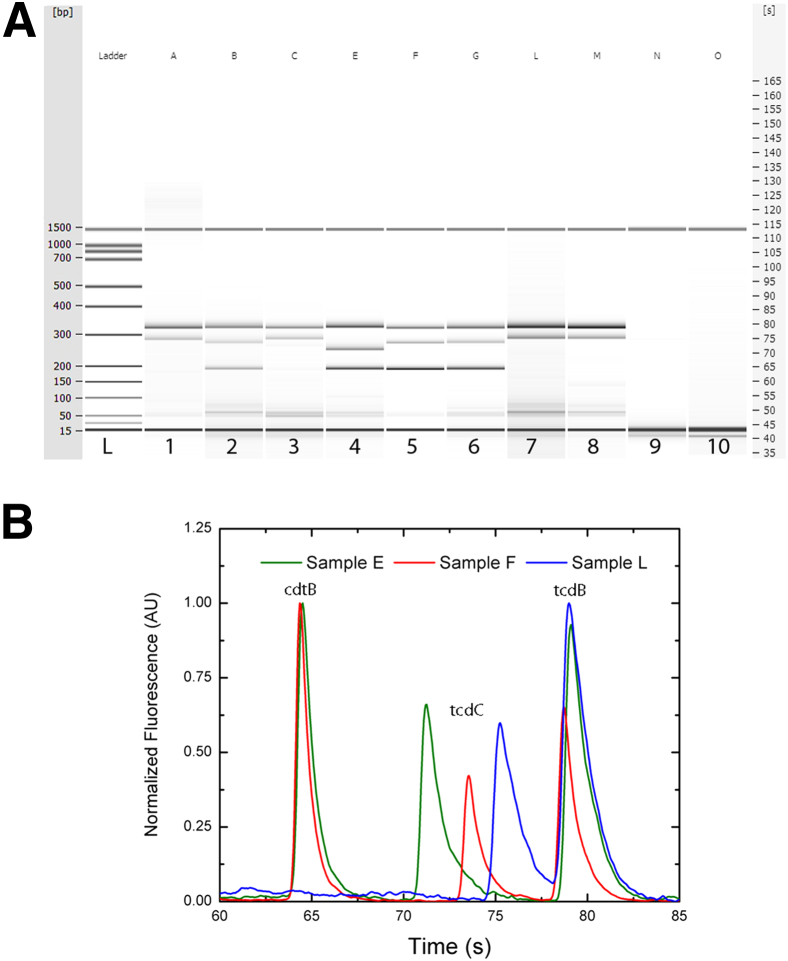
PCR was also performed on DNA isolated from 20 PCR-positive patient stool samples and 12 PCR-negative samples obtained from the Rhode Island Hospital Clinical Microbiology Laboratory. An electrophoresis gel plot shows amplified products from the multiplex PCR assay using DNA isolates from eight positive and two negative representative clinical stool samples (Figure 3A). Samples B, E, F, and G show all three amplicons, the largest corresponding to tcdB, the intermediate size corresponding to tcdC, and the smallest corresponding to cdtB. In samples A, C, L, and M, amplification of tcdB and tcdC is shown. Virulence genes were detected in all of the positive patient samples and in none of the negative samples (Table 3) and (Figure 3A).
Figure 3.
A: Electrophoresis gel plot of multiplex PCR. A different clinical stool DNA isolate was used as template for each amplification, from eight positive samples and two negative controls. A sample identifier for the template is indicated at the top of each lane. Each positive sample (lanes 1 to 8; samples A–C, E–G, L, and M) shows the 328-bp amplicon for tcdB, followed by either a 300-bp amplicon for tcdC (lanes 1, 3, 7, and 8; samples A, C, L, and M) or a 282-bp amplicon for tcdC with the NAP1/027/BI deletion (lanes 2, 5, and 6; samples B, F, and G). Sample E (lane 4) shows a greater deletion in tcdC at 261 bp. Samples with any deletion in tcdC were also positive for binary toxin, indicated by the amplicon at 190 bp for cdtB (B, E, F, and G). Lanes 9 and 10 (samples N and O) show representative samples of clinical negative controls. B: Electropherogram for samples E, F, and L. The electropherogram image shows the peaks of the three amplicons cdtB, tcdC, and tcdB for samples E, F, and L, respectively. The base-pair shifts in the tcdC amplicon are easily visible between 70.0 seconds and 77.5 seconds; sample L shows the longest amplicon at 75.25 seconds, followed by sample F (18-bp deletion) at 73.55 seconds and then sample E (39-bp deletion) at 71.25 seconds. The peak for tcdB appears in all three samples at 79.0 seconds and cdtB in samples E and F at 64.4 seconds.
Table 3.
Summary of Virulence Genes Detected in Clinical Isolates of C. difficile
| Samples | tcdB (BD PCR)∗ | tcdB | tcdC | tcdC 18-bp deletion | tcdC 39-bp deletion | cdtB |
|---|---|---|---|---|---|---|
| Positive (n = 20) | 20 | 20 | 11 | 7 | 2 | 9 |
| Negative (n = 12) | 0 | 0 | 0 | 0 | 0 | 0 |
GeneOhm Cdiff PCR kit (BD Diagnostics).
Of the set of samples tested, we successfully amplified tcdB and tcdC in all samples, and cdtB in 9 out of 20 (Table 3). The nine samples with the amplicon for binary toxin also exhibited a deletion in the tcdC gene, as demonstrated by the amplicon shift visible on the gel electropherogram (Figure 3B). Seven of these samples display an 18-bp deletion that is typically associated with the NAP1/027/BI strain. Additionally, two samples (one of which, sample E, is shown in Figure 3) showed a larger shift in the tcdC gene (a 39-bp deletion). This larger deletion is often associated with another hypervirulent strain of C. difficile, referred to as ribotype 078. Like NAP1/027/BI strain, the 078 strain is positive for binary toxin and shows both the 18-bp deletion in tcdC that confers down-regulation of toxin management and an additional 21-bp deletion in the same gene.11,31 The base-pair shifts in the tcdC amplicon are easily visible in the electropherogram as amplification peaks (Figure 3B). The peak for tcdB in all three samples was at 79.0 seconds, and that for cdtB in two of the samples (E and F) was at 64.4 seconds (Figure 3B). TcdB was detected by our assay in all positive clinical samples and none of the negative clinical samples. Presence of tcdB in our assay also served as a standard positive control. As expected, samples negative for tcdB were additionally negative for the other two genes of interest, tcdC and cdtB. Current diagnostic methods cannot detect the presence of tcdC or cdtB, nor the deletions in the tcdC gene that are markers indicative of the NAP1/027/BI strain.
Droplet Sandwich qPCR Selectively and Sensitively Detects Target Genes
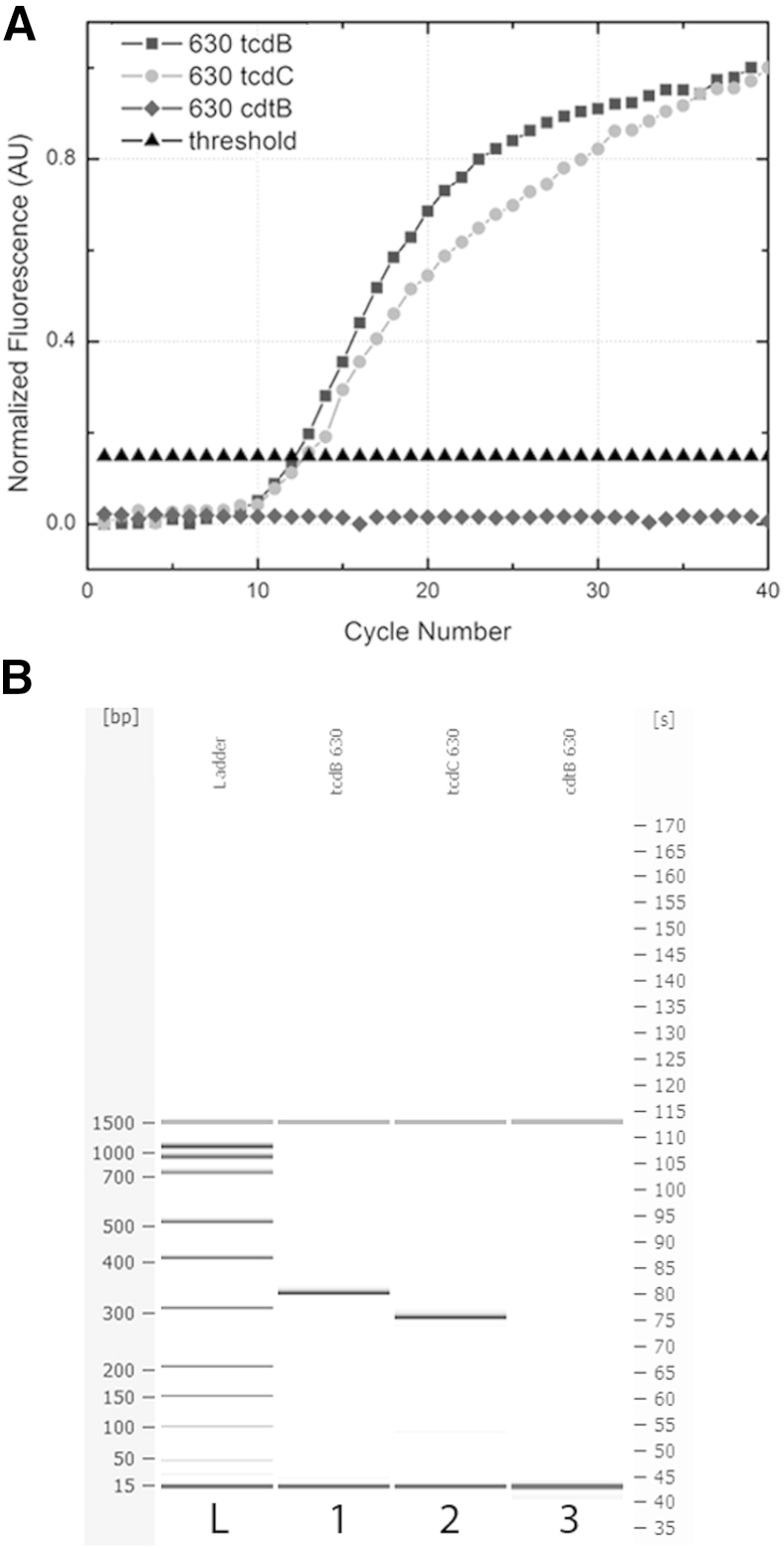
The singleplex qPCR results using the droplet platform for each amplicon of interest (tcdB, tcdC, and cdtB) for the C. difficile 630 genome are overlaid in Figure 4A. The CT values (12.2 for tcdB and 12.7 for tcdC) were calculated as 10 times the SD of the background signal, as a means of evaluating the efficiency and reproducibility of the amplification. The subsequent size analysis shows the appropriate amplicon lengths (328 bp for tcdB and 300 bp for tcdC), as well as the absence of cdtB (Figure 4B). The C. difficile 630 strain does not contain the cdtB gene, and the assay was appropriately negative for this amplicon.
Figure 4.
A: Singleplex qPCR results for tcdB, tcdC, and cdtB amplification in C. difficile 630. The tcdB and tcdC amplicons have CT values of 12.2 and 12.7, respectively, for a starting concentration of 1.0 ng/μL. The cdtB gene is not present in the C. difficile 630 genome, and amplification did not occur. B: The corresponding electrophoresis gel plot for PCR performed on the droplet sandwich platform shows appropriate-size amplicons for tcdB at 328 bp and tcdC at 300 bp. Amplification of cdtB shows no product, and the assay is negative for that gene.
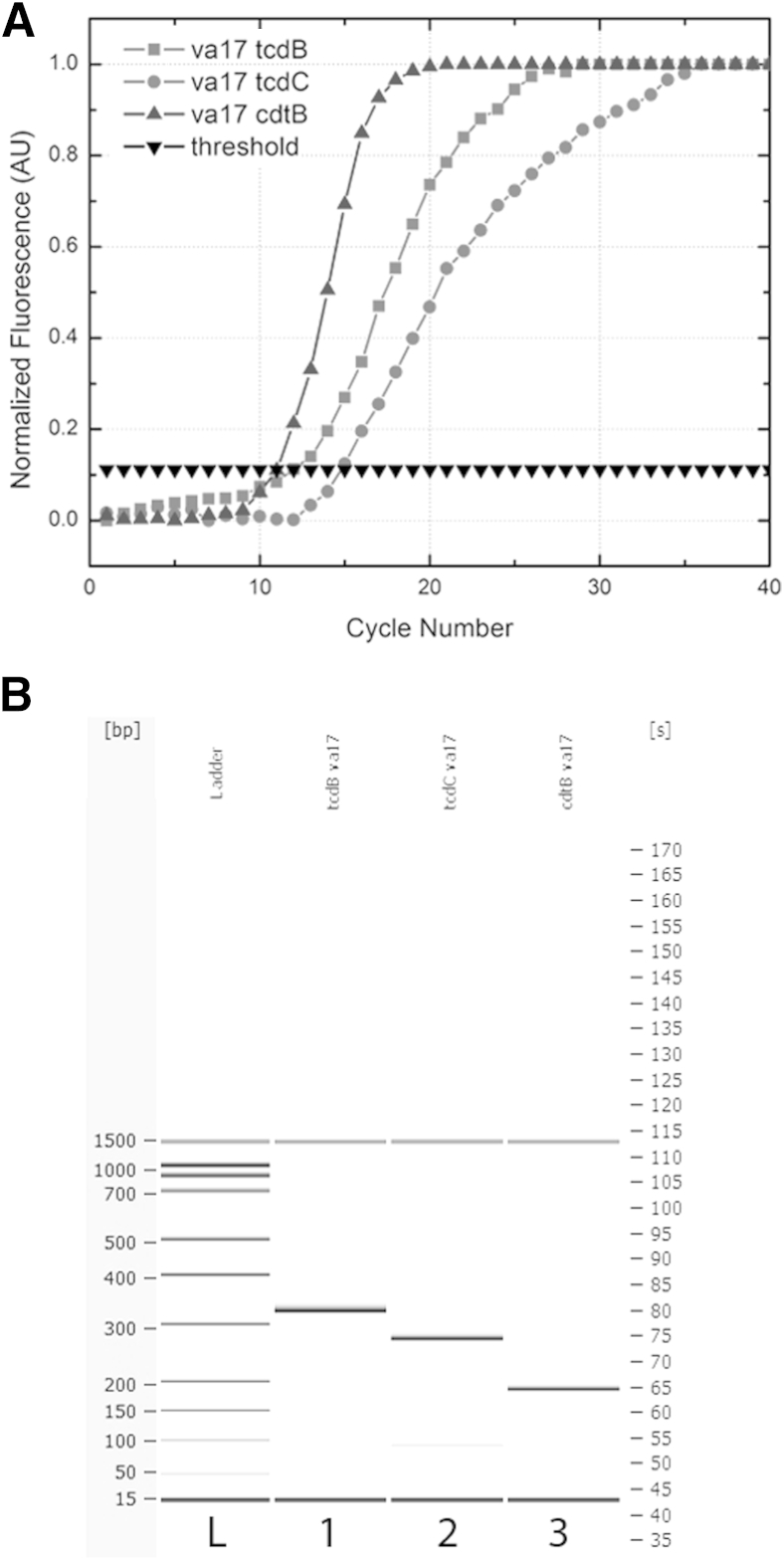
The amplification curves for each amplicon for the VA17 strain, which is of the NAP1/027/BI type, are overlaid in Figure 5A. The CT values for tcdB, tcdC, and cdtB were 11.8, 15.2, and 11.3, respectively. The amplification of the cdtB gene fragment was the most efficient, with the lowest CT value and the most exponential curve, likely because of the shorter amplicon size (190 bp). Smaller amplicons tend to amplify more efficiently for qPCR, because of better disassociation and primer annealing during temperature cycling. Additionally, both cdtB and tcdB amplicons appeared to amplify more efficiently than tcdC for this template. The proper amplicon lengths are presented in Figure 5B, including the 18-bp deletion in the tcdC gene indicative of the NAP1/027/BI strain. Overall, the standard deviation between amplification of different gene fragments at 1.0 ng/μL for this template was ±2.1 cycles (n = 3 for each template). In comparing efficiency between the 630 genome and VA17 templates for tcdB and tcdC, the standard deviation was ±0.3 and ±1.8 cycles (n = 3 for each gene fragment) respectively. Overall, this variation is low, indicating that the amplicons function with appropriate efficacy at the same template concentrations, across two different strain types, which represent the majority of clinical samples typed. The larger error in amplification of the tcdC gene could be due to differences in the templates, because one contains the 18-bp deletion and the other does not. For all genes, the no-template controls showed no change in fluorescence over time and thus graphed as flat lines. SYBR Green I was used because it is a common intercalating dye to validate the assay in real time. Melting-curve analysis could be performed with a dye such as SYBR Green I or EvaGreen (Biotium, Hayward, CA) to detect the 18-bp deletion in tcdC, as well as the other gene sequences.
Figure 5.
A: Singleplex qPCR results for tcdB, tcdC, and cdtB amplification in the VA17 NAP1/027/BI hypervirulent strain of C. difficile. The tcdB, tcdC, and cdtB amplicons have CT values of 11.8, 15.2, and 11.3, respectively, for a starting concentration of 1.0 ng/μL. B: The corresponding electrophoresis gel plot for PCR performed on the droplet sandwich platform shows, the amplicons are of the appropriate size: 328 bp for tcdB, 282 bp for tcdC, and 190 bp for cdtB. The amplicon size of tcdC indicates the presence of the 18-bp deletion associated with NAP1/027/BI.
Discussion
C. difficile colitis is a growing cause of morbidity and mortality. The hypervirulent NAP1/027/BI strain has been associated with increased severity of disease, mortality, and more frequent relapses.38,39 A number of PCR and qPCR assays have been developed to detect C. difficile.40 However, several of these assays require dedicated equipment and multiple fluorescence wavelength excitation and detection capabilities to identify the genes of interest. One commercial example is the Xpert C. difficile/epi assay (Cepheid, Sunnyvale, CA), which uses primers and probes for multiplexed detection of tcdB, tcdC, and cdtB; it is marketed to identify the NAP1/027/BI strain. Our assay using the droplet sandwich platform can eliminate the need for such expensive dedicated equipment.
We have identified the presence of C. difficile in clinical stool specimens through a series of three steps: isolation of double-stranded DNA, amplification of segments of DNA specific to C. difficile in genes of interest that may produce proteins conferring hypervirulence, and detection of those PCR products through the use of qPCR or capillary electrophoresis. Three sets of PCR primers were designed to amplify three specific regions of DNA, each located within a gene with a potential role in coding for the production of proteins involved in the severity of illness associated with C. difficile infection.
Our assay targeted segments within the tcdB gene coding for production of toxin B, segments within the cdtB gene coding for production of binary toxin, and segments within the tcdC gene with a potential role in down-regulation of production of toxins A and B.13,41 Although the 18-bp deletion in the tcdC gene is correlated with the NAP1/027/BI strain, this tcdC mutation in the NAP1/027/BI strain does not, in and of itself, lead to increased toxin production.42 The thyX gene and other insertions in the genomes of the 027 and 078 ribotypes have been linked to hypervirulence due to increased production of toxins A and B.43,44 Although our results are consistent with several studies linking hypervirulence to deletions in the tcdC gene, the genetic origin of increased toxin production in these strains has yet to be fully elucidated.27,30 Emerging information on the genetic origin of hypervirulence will provide additional targets for typing C. difficile strains and may prove useful in typing the 027 and 078 ribotypes and other strains of interest.43,44
Although most current laboratory detection methods seek to identify the tcdB gene, we have demonstrated multiplexed amplification and detection of tcdB, tcdC, and cdtB. Our technique can thus be used to detect the presence of the toxin B gene (tcdB), the binary toxin gene (cdtB), and the 18-bp deletion in the tcdC gene that may be associated with overproduction of toxins A and B in hypervirulent C. difficile strains.13,27,28 The rapid detection of these three important markers of infection not only indicates the presence of C. difficile, but also provides physicians with additional clinically relevant data regarding infection with a hypervirulent strain. This may help physicians to devise specific treatment regimens and may assist in contact isolation strategies necessary to address the challenge of C. difficile–associated diarrhea in health-care facilities and community settings.
Our droplet-based sandwich platform maintains the sensitivity and specificity required for accurate detection of genetic markers of C. difficile infection and markers associated with the hypervirulent 027 and 078 ribotypes. A major contribution of our assay is that it detects the three genes associated with hypervirulence, via several methodologies. The assay can be performed as a standard PCR with detection using electrophoresis or capillary electrophoresis or can be performed as a real-time assay. Real-time multiplexing can be achieved by using melting-curve analysis with an intercalating fluorescent dye such as SYBR Green I or EvaGreen or, alternatively, with sequence-specific probes such as molecular beacons or TaqMan probes as desired by the user.
Our results demonstrate a clinically relevant method for amplifying and detecting DNA from clinical samples, thus significantly aiding the diagnosis of C. difficile infection. Our technique of multiplex gene amplification provides a unique method that is both sensitive and specific for rapidly detecting C. difficile in patient stool samples. This method can be adapted to point-of-care testing and thus can assist physicians in developing and implementing better treatment regimens for the care of patients with C. difficile infections, particularly those with the NAP1/027/BI strain.
Acknowledgments
We thank Joshua Leung, Zintis Inde, Christina Beck, Elizabeth Mermel, and Dr. Reese Isaacson (Brown University) for contributions to the project; Dr. Kimberly Chapin and Roberta Dickenson (Microbiology Laboratory, Rhode Island Hospital) for assistance with clinical samples; and Dr. Curtis J. Donskey (Cleveland Veterans Affairs Medical Center) for kindly furnishing cultured samples and providing strain information.
Footnotes
Supported by the National Science Foundation (CBET 0653835 to A.T.), NIH grant 1R21 A1073808-01 A1 (A.T.), and the Rhode Island Space Grant NASA Graduate Fellowship (S.L.A.).
Disclosures: None declared.
Current affiliation of A.A.S, Harvard Medical School, Harvard University, Boston, MA; of A.N., New York Presbyterian Hospital, Cornell Medical Center, New York, NY; of L.S., Nabsys, Inc., Providence, RI; of J.H.F., Yale School of Medicine, New Haven, CT.
References
- 1.Fletcher K.R., Cinalli M. Identification, optimal management, and infection control measures for Clostridium difficile-associated disease in long-term care. Geriatr Nurs. 2007;28:171–181. doi: 10.1016/j.gerinurse.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett J.G., Gerding D.N. Clinical recognition and diagnosis of Clostridium difficile infection. Clin Infect Dis. 2008;46(Suppl 1):S12–S18. doi: 10.1086/521863. [DOI] [PubMed] [Google Scholar]
- 3.Poutanen S.M., Simor A.E. Clostridium difficile-associated diarrhea in adults. CMAJ. 2004;171:51–58. doi: 10.1503/cmaj.1031189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kyne L., Hamel M.B., Polavaram R., Kelly C.P. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin Infect Dis. 2002;34:346–353. doi: 10.1086/338260. [DOI] [PubMed] [Google Scholar]
- 5.Redelings M.D., Sorvillo F., Mascola L. Increase in Clostridium difficile–related mortality rates, United States, 1999–2004. Emerg Infect Dis. 2007;13:7–9. doi: 10.3201/eid1309.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lessa F.C., Gould C.V., McDonald L.C. Current status of Clostridium difficile infection epidemiology. Clin Infect Dis. 2012;55(Suppl 2):S65–S70. doi: 10.1093/cid/cis319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He M., Miyajima F., Roberts P., Ellison L., Pickard D.J., Martin M.J., Connor T.R., Harris S.R., Fairley D., Bamford K.B., D'Arc S., Brazier J., Brown D., Coia J.E., Douce G., Gerding D., Kim H.J., Koh T.H., Kato H., Senoh M., Louie T., Michell S., Butt E., Peacock S.J., Brown N.M., Riley T., Songer G., Wilcox M., Pirmohamed M., Kuijper E., Hawkey P., Wren B.W., Dougan G., Parkhill J., Lawley T.D. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubberke E.R., Olsen M.A. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis. 2012;55(Suppl 2):S88–S92. doi: 10.1093/cid/cis335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention (CDC) Vital signs: preventing Clostridium difficile infections. MMWR Morb Mortal Wkly Rep. 2012;61:157–162. [PubMed] [Google Scholar]
- 10.Vonberg R.P., Kuijper E.J., Wilcox M.H., Barbut F., Tüll P., Gastmeier P., European C difficile-Infection Control Group. European Centre for Disease Prevention and Control (ECDC) van den Broek P.J., Colville A., Coignard B., Daha T., Debast S., Duerden B.I., van den Hof S., van der Kooi T., Maarleveld H.J.H., Nagy E., Notermans D.W., O'Driscoll J., Patel B., Stone S., Wiuff C. Infection control measures to limit the spread of Clostridium difficile. Clin Microbiol Infect. 2008;14(Suppl 5):2–20. doi: 10.1111/j.1469-0691.2008.01992.x. [DOI] [PubMed] [Google Scholar]
- 11.Carter G.P., Lyras D., Allen D.L., Mackin K.E., Howarth P.M., O'Connor J.R., Rood J.I. Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulator. J Bacteriol. 2007;189:7290–7301. doi: 10.1128/JB.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald L.C., Killgore G.E., Thompson A., Owens R.C., Jr., Kazakova S.V., Sambol S.P., Johnson S., Gerding D.N. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 13.Warny M., Pepin J., Fang A., Killgore G., Thompson A., Brazier J., Frost E., McDonald L.C. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 14.Tenover F.C., Baron E.J., Peterson L.R., Persing D.H. Laboratory diagnosis of Clostridium difficile infection. Can molecular amplification methods move us out of uncertainty? J Mol Diagn. 2011;13:573–582. doi: 10.1016/j.jmoldx.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson L.R., Manson R.U., Paule S.M., Hacek D.M., Robicsek A., Thomson R.B., Jr., Kaul K.L. Detection of toxigenic Clostridium difficile in stool samples by real-time polymerase chain reaction for the diagnosis of C. difficile-associated diarrhea. Clin Infect Dis. 2007;45:1152–1160. doi: 10.1086/522185. [DOI] [PubMed] [Google Scholar]
- 16.Chapin K.C., Dickenson R.A., Wu F., Andrea S.B. Comparison of five assays for detection of Clostridium difficile toxin. J Mol Diagn. 2011;13:395–400. doi: 10.1016/j.jmoldx.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rüssmann H., Panthel K., Bader R.C., Schmitt C., Schaumann R. Evaluation of three rapid assays for detection of Clostridium difficile toxin A and toxin B in stool specimens. Eur J Clin Microbiol Infect Dis. 2007;26:115–119. doi: 10.1007/s10096-006-0251-7. [DOI] [PubMed] [Google Scholar]
- 18.Kvach E.J., Ferguson D., Riska P.F., Landry M.L. Comparison of BD GeneOhm Cdiff real-time PCR assay with a two-step algorithm and a toxin A/B enzyme-linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection. J Clin Microbiol. 2010;48:109–114. doi: 10.1128/JCM.01630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Planche T., Aghaizu A., Holliman R., Riley P., Poloniecki J., Breathnach A., Krishna S. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect Dis. 2008;8:777–784. doi: 10.1016/S1473-3099(08)70233-0. [DOI] [PubMed] [Google Scholar]
- 20.Tenover F.C., Novak-Weekley S., Woods C.W., Peterson L.R., Davis T., Schreckenberger P., Fang F.C., Dascal A., Gerding D.N., Nomura J.H., Goering R.V., Akerlund T., Weissfeld A.S., Baron E.J., Wong E., Marlowe E.M., Whitmore J., Persing D.H. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol. 2010;48:3719–3724. doi: 10.1128/JCM.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persson S., Torpdahl M., Olsen K.E.P. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection [Erratum appeared in Clin Microbiol Infect 2009, 15:296] Clin Microbiol Infect. 2008;14:1057–1064. doi: 10.1111/j.1469-0691.2008.02092.x. [DOI] [PubMed] [Google Scholar]
- 22.Lemee L., Dhalluin A., Testelin S., Mattrat M.A., Maillard K., Lemeland J.F., Pons J.L. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004;42:5710–5714. doi: 10.1128/JCM.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Berg R.J., Kuijper E.J., van Coppenraet L.E., Claas E.C. Rapid diagnosis of toxinogenic Clostridium difficile in faecal samples with internally controlled real-time PCR. Clin Microbiol Infect. 2006;12:184–186. doi: 10.1111/j.1469-0691.2005.01301.x. [DOI] [PubMed] [Google Scholar]
- 24.Antikainen J., Pasanen T., Mero S., Tarkka E., Kirveskari J., Kotila S., Mentula S., Könönen E., Virolainen-Julkunen A.-R., Vaara M., Tissari P. Detection of virulence genes of Clostridium difficile by multiplex PCR. APMIS. 2009;117:607–613. doi: 10.1111/j.1600-0463.2009.02509.x. [DOI] [PubMed] [Google Scholar]
- 25.Barbut F., Monot M., Rousseau A., Cavelot S., Simon T., Burghoffer B., Lalande V., Tankovic J., Petit J.C., Dupuy B., Eckert C. Rapid diagnosis of Clostridium difficile infection by multiplex real-time PCR. Eur J Clin Microbiol Infect Dis. 2011;30:1279–1285. doi: 10.1007/s10096-011-1224-z. [DOI] [PubMed] [Google Scholar]
- 26.Bélanger S.D., Boissinot M., Clairoux N., Picard F.J., Bergeron M.G. Rapid detection of Clostridium difficile in feces by real-time PCR. J Clin Microbiol. 2003;41:730–734. doi: 10.1128/JCM.41.2.730-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curry S.R., Marsh J.W., Muto C.A., O'Leary M.M., Pasculle A.W., Harrison L.H. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile [Erratum appeared in J Clin Microbiol 2007, 45:2103] J Clin Microbiol. 2007;45:215–221. doi: 10.1128/JCM.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hundsberger T., Braun V., Weidmann M., Leukel P., Sauerborn M., von EichelStreiber C. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur J Biochem. 1997;244:735–742. doi: 10.1111/j.1432-1033.1997.t01-1-00735.x. [DOI] [PubMed] [Google Scholar]
- 29.Lyras D., O'Connor J.R., Howarth P.M., Sambol S.P., Carter G.P., Phumoonna T., Poon R., Adams V., Vedantam G., Johnson S., Gerding D.N., Rood J.I. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Connor J.R., Johnson S., Gerding D.N. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain [Erratum appeared in Gastroenterology 2009, 137:743] Gastroenterology. 2009;136:1913–1924. doi: 10.1053/j.gastro.2009.02.073. [DOI] [PubMed] [Google Scholar]
- 31.Goorhuis A., Bakker D., Corver J., Debast S.B., Harmanus C., Notermans D.W., Bergwerff A.A., Dekker F.W., Kuijper E.J. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. 2008;47:1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz S., Zhang Z., Frazer K.A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W. PipMaker–a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markham N.R., Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angione S.L., Inde Z., Beck C.M., Artenstein A.W., Opal S.M., Tripathi A. Microdroplet sandwich real-time RT-PCR for detection of pandemic and seasonal influenza subtypes. PLoS One. 2013;8:e73497. doi: 10.1371/journal.pone.0073497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angione S.L., Chauhan A., Tripathi A. Real-time droplet DNA amplification with a new tablet platform. Anal Chem. 2012;84:2654–2661. doi: 10.1021/ac202532a. [DOI] [PubMed] [Google Scholar]
- 37.Loo V.G., Bourgault A.M., Poirier L., Lamothe F., Michaud S., Turgeon N., Toye B., Beaudoin A., Frost E.H., Gilca R., Brassard P., Dendukuri N., Béliveau C., Oughton M., Brukner I., Dascal A. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med. 2011;365:1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 38.Petrella L.A., Sambol S.P., Cheknis A., Nagaro K., Kean Y., Sears P.S., Babakhani F., Johnson S., Gerding D.N. Decreased cure and increased recurrence rates for Clostridium difficile infection caused by the epidemic C. difficile BI strain. Clin Infect Dis. 2012;55:351–357. doi: 10.1093/cid/cis430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blossom D.B., McDonald L.C. The challenges posed by reemerging Clostridium difficile infection. Clin Infect Dis. 2007;45:222–227. doi: 10.1086/518874. [DOI] [PubMed] [Google Scholar]
- 40.O'Horo J.C., Jones A., Sternke M., Harper C., Safdar N. Molecular techniques for diagnosis of Clostridium difficile infection: systematic review and meta-analysis. Mayo Clin Proc. 2012;87:643–651. doi: 10.1016/j.mayocp.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merrigan M., Venugopal A., Mallozzi M., Roxas B., Viswanathan V.K., Johnson S., Gerding D.N., Vedantam G. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol. 2010;192:4904–4911. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cartman S.T., Kelly M.L., Heeg D., Heap J.T., Minton N.P. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol. 2012;78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakker D., Smits W.K., Kuijper E.J., Corver J. tcdC does not significantly repress toxin expression in Clostridium difficile 630ΔErm. PLoS One. 2012;7:e43247. doi: 10.1371/journal.pone.0043247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knetsch C.W., Hensgens M.P.M., Harmanus C., van der Bijl M.W., Savelkoul P.H.M., Kuijper E.J., Corver J., van Leeuwen H.C. Genetic markers for Clostridium difficile lineages linked to hypervirulence. Microbiology. 2011;157:3113–3123. doi: 10.1099/mic.0.051953-0. [DOI] [PubMed] [Google Scholar]