Abstract
Aberrant regulation of DNA methylation is characteristic of cancer cells and clearly influences phenotypes of various malignancies. Despite clear correlations between DNA methylation and patient outcome, tests that directly measure multiple-locus DNA methylation are typically expensive and technically challenging. Previous studies have demonstrated that the prognosis of patients with acute myeloid leukemia can be predicted by the DNA methylation pattern of 18 loci. We have developed a novel strategy, termed microsphere HpaII tiny fragment enrichment by ligation-mediated PCR (MELP), to simultaneously analyze the DNA methylation pattern at these loci using methylation-specific DNA digestion, fluorescently labeled microspheres, and branched DNA hybridization. The method uses techniques that are inexpensive and easily performed in a molecular laboratory. MELP accurately reflects the methylation levels at each locus analyzed and segregates patients with acute myeloid leukemia into prognostic subgroups. Our results demonstrate the usefulness of MELP as a platform for simultaneous evaluation of DNA methylation of multiple loci.
Cancer has been traditionally considered a genetic disease, involving a series of mutations that activate oncogenes and inactivate tumor suppressors.1 Although mutagenic events are critical for carcinogenesis, recent work has unequivocally demonstrated that cancer is also characterized by dysregulation of chromatin structure that involves the DNA itself (eg, CpG methylation) and its associated histones.2 Because these epigenetic modifications are at least partially responsible for influencing tumorigenic processes, it is not surprising that several studies have demonstrated that prognosis of certain tumors can be predicted from analysis of epigenetic features.3–5
An example of a tumor type that shows clear dysregulation of epigenetic modifications is acute myeloid leukemia (AML).6,7 Many of the recurrent mutations seen in AML, including those in DNMT3A, IDH1, IDH2, MLL, and EZH2, influence DNA methylation, DNA hydroxymethylation, or histone modification.8–11 Dysregulated DNA methylation at specific loci, such as CDKN2B and MGMT, has been found in many cases of AML.12 Moreover, studies of global methylation have shown DNA methylation in leukemic blasts is distinct from that seen in normal CD34+ cells, and that DNA methylation patterns alone can segregate AML samples into categories with significant clinical and biological features.3 Indeed, a DNA methylation analysis using only 18 loci was shown to distinguish prognostic subgroups of AML, and this methylation-based classifier retained significance in a multivariate analysis that included factors used clinically for determining patient prognosis.3
Despite the clear implications of epigenetics for tumor biological features and patient prognosis, studies involving multiple-locus DNA methylation of cancers have lagged behind those assessing DNA sequence variations. One reason is the lack of robust multiplex assay platforms that are amenable for high-throughput laboratory use. Most assays that probe DNA methylation are technically challenging because they use sodium bisulfite treatment of DNA, which can cause sample degradation.13,14 In addition, examination of methylation at multiple loci requires either nucleotide microarrays or high-throughput sequencing technologies, both of which require extensive investment for materials and equipment.
To circumvent the technical challenges involved in routine epigenetic analysis, we have developed a novel method to determine DNA methylation status that uses analytical techniques commonly used in molecular laboratories. As a proof of principle of the utility of this assay and to directly compare it with well-established tests for DNA methylation, we have applied our novel technique to measure DNA methylation at 18 loci previously shown to carry prognostic significance in patients with AML.3 Our method, conceptually based on the HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) assay, does not use bisulfite treatment. Rather, it uses methylation-sensitive restriction digestion, followed by oligonucleotide ligation and PCR.15 Examination of methylation levels is performed by flow cytometric analysis of fluorescent microspheres, thereby alleviating the need for microarrays or high-throughput sequencing technologies. We demonstrate that this methylation assay, designated microsphere HELP (MELP), accurately recapitulates genome-wide HELP of AML samples, both in terms of DNA methylation status at individual loci and with a global classifier relating DNA methylation to patient outcome. Thus, MELP may prove to be an appropriate technology for evaluation of DNA methylation in diseases associated with dysregulated epigenetic status.
Materials and Methods
Samples and DNA Preparation
Samples for the development of MELP and the comparison between HELP and MELP were from patients treated on study protocols of the Dutch-Belgian Hemato-Oncology Cooperative Group (HOVON) and have been previously described.3 The sets of patients used as training, test, and validation cohorts are all from this group of patients, and the samples in each cohort were drawn from those previously studied.3 Additional samples to establish initial characteristics of MELP are primary deidentified AML apheresis samples purchased from the Stem Cell and Xenograft Core of the University of Pennsylvania (Philadelphia). All samples were obtained after patient consent on a University of Pennsylvania Institutional Review Board–approved protocol and cryopreserved as viable cells in 10% dimethyl sulfoxide.
DNA Preparation and Ligation-Mediated PCR
Procedures for preparing DNA and performing ligation-mediated PCR have been previously described.3 Briefly, DNA preparation from 5 million cells was performed with the Qiagen Puregene kit (Qiagen, Valencia, CA), following the manufacturer’s protocol for DNA extraction from buffy coat samples. MspI, HpaII (NEB, Ipswich, MA), and mock digests of 1 μg of DNA were performed at 37°C overnight, followed by 16°C overnight T4 ligase-mediated ligation of pre-annealed JHpaII 12 and JHpaII 24 linkers. Subsequent PCR amplification using JHpaII 24 primers was performed as described. For most reactions, PCR was performed in 100 μL total volume for 20 cycles. For reactions in which input DNA was serially diluted, PCR with 11 cycles of amplification was performed.
Real-Time PCR
PCR products from eight primary AML samples from the HOVON cohort were diluted 1:20 in PCR-grade water. Quantitative PCRs with 1 μL of the diluted products in 20 μL total volume were performed using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) with the same PCR conditions as those used in the initial PCR. Real-time PCR primers (final concentration, 200 nmol/L) were as follows: B2M forward, 5′-TTTCTGGCCTGGAGGCTATC-3′; B2M reverse, 5′-ACGGAGCGAGAGAGCACAG-3′; E2F1 forward, 5′-CAGCCATCAGCCACCTCTTC-3′; E2F1 reverse, 5′-TTCCAGGCACCGCTCTTC-3′; chromosome X locus forward, 5′-CCAGAAGGCTGGCACACA-3′; and chromosome X locus reverse, 5′-AAGTGCAGCGTCAGCAAGAG-3′.
Quantigene 2.0 Hybridization
Prognostic loci used for the Quantigene 2.0 (Affymetrix, Santa Clara, CA) hybridization panel have been described previously3 and are listed in Supplemental Table S1. The three loci used for normalization are as follows (human genome assembly GRCh37/hg19): normalization A) chr6: 34856156-34857019; normalization B) chr13: 53028642-53029495; and normalization C) chr19: 37958559-37955860. Sequential hybridization reactions for complexing amplicons onto fluorescent microspheres and for branched DNA signal amplification were performed with the Quantigene 2.0 assay, following the manufacturer’s protocol for RNA hybridization (Affymetrix). Specifically, 8 μL PCRs were incubated at room temperature using 5 μL of 2.5 mol/L NaOH, 5 μL of the locus-specific probe mixture, and 5 μL of lysis mixture (the latter two products provided in the Quantigene 2.0 assay) in 68 μL total volume. The reaction was neutralized by addition of 36 μL of 2 mol/L HEPES buffer. This amplicon/probe mixture was added to a 20 μL reaction mix consisting of 0.2 μL of proteinase K, 15 μL of lysis mixture, 2 μL of blocking reagent, and 1 μL of locus-specific fluorescent microspheres (all products provided in the Quantigene 2.0 assay). These hybridizations were incubated with shaking at 55°C overnight. Reactions were placed on a magnet and washed three times with wash buffer (provided in the Quantigene 2.0 assay). The reactions were then sequentially hybridized to pre-amplifier, amplifier, and biotinylated label-probe DNA in 100 μL of the appropriate buffer (provided in the Quantigene 2.0 assay). All hybridizations were performed for 1 hour at 50°C with shaking. Each hybridization was preceded by magnetic bead capture and three washes. After hybridizations, the reaction was incubated at room temperature with 4 μg/mL streptavidin-phycoerythrin in the appropriate buffer (supplied by the manufacturer). After three washes, the fluorescent microspheres were analyzed by flow cytometry on a FLEXMAP three-dimensional instrument running xPONENT 4.0 software (Luminex Corporation, Austin, TX). The entire procedure was performed separately for products derived from MspI-digested, HpaII-digested, or mock-digested DNA. Amount of bound product was determined by phosphatidylethanolamine signal, whereas locus identity was determined by fluorescence signal of each microsphere. Relative methylation was determined by the ratio of phosphatidylethanolamine median fluorescence intensity of each locus in MspI-digested and HpaII-digested samples normalized to the same ratio of known hypomethylated loci.
Normalization
To identify unmethylated control loci for normalization, we selected a list of 26 candidates for which measurements were available in at least 340 samples and that met criteria based on the width of the distribution across samples and the absence of methylated outliers (density maximum, >0.55; minimum value, >1 across all samples; mean value, <4). From this list, three loci (MSPI0406S00318682, MSPI0406S00653944, and MSPI0406S00890278) were chosen to represent an unmethylated baseline within each sample. HELP or MELP methylation values were normalized by obtaining the ratio with the average methylation score from the three unmethylated control loci. In the case of HELP, the original data had previously undergone global normalization; thus, (−) normalization reflects this original normalization, whereas (+) normalization reflects the further transformation of the HELP data using the ratio to control loci previously described.
Data Sources and Statistical Analysis
All statistical analyses were performed using the R statistical package (http://www.r-project.org). R scripts and tumor sublists used to generate the results for this article are available as Supplemental Scripts S1, S2, S3, S4, and S5. MELP expression values and HOVON tumor data are available as Supplemental Tables S2 and S3. Raw MELP data files were preprocessed using an in-house Perl script. HELP data have been previously published3 (Gene Expression Omnibus; http://www.ncbi.nlm.nih.gov/geo; accession number GSE18700) and represent values that had been processed by global normalization. MassArray correlation files and subject survival data were provided by the authors.3 Comparisons between measurement modalities were performed using the Pearson correlation coefficient. To convert from HELP to MELP values, Deming regression was performed on training data using the MethComp package in R (http://www.r-project.org).
Multiplex Methylation Scores for AML
The level of methylation at a single locus is defined by the normalized ratio of the signal from the HpaII- and the MspI-digested sample. To assign a given sample to a survival group, a predictor model was trained on the 18 loci previously reported by Figueroa et al.3 Training, testing, and validation of this model were performed using the respective HOVON sample sets previously specified.3 Briefly, we obtained HELP values for the 18 predictive loci from the training group (n = 200, the original HOVON HELP training data3). Of these samples, 84 were rerun using MELP, and Deming regression was used to convert HELP-derived values for the entire 200 training samples (including the 84 on which MELP was directly performed) to MELP-scale values. Methylation values of the test set (n = 84) and validation set (n = 48) were obtained directly by MELP. The training set was used to build the classification model, and model coefficients were refined using the training and test sets, as previously described.3 A methylation outcome score (MS) is given as follows:
| (1) |
where L# is the methylation level (ie, normalized HpaII/MspI ratio) at each locus, and the associated constant (a, b, c) is the weighting factor, as determined by our training algorithm (SuperPC; http://statweb.stanford.edu/∼tibs/superpc). Each tumor in the training set receives a methylation outcome score, and cutoffs are determined by using scores that segregate the training set into thirds (tertiles). Tumors in the test and validation subsets were segregated according to these cutoffs, and survival was determined.
Results
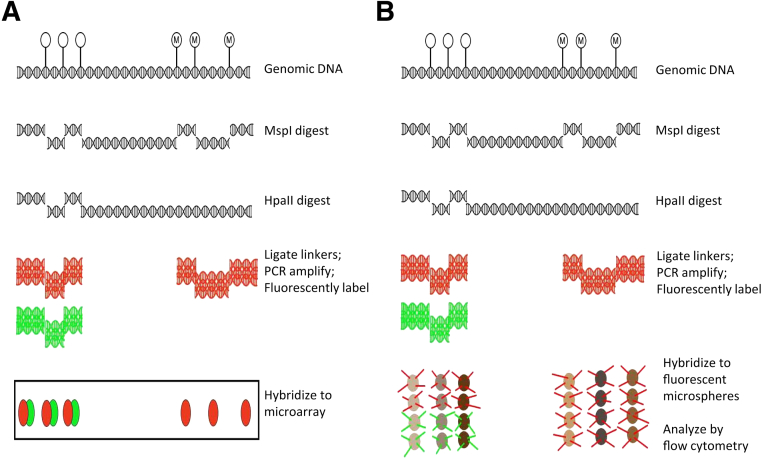
Because the HELP assay avoids sodium bisulfite treatment and was previously used to establish a DNA methylation-based classifier for AML prognosis,3 we modified this assay to make it more feasible for routine use. The HELP assay (Figure 1A) involves DNA digestion by the methylation-insensitive restriction endonuclease MspI or its methylation-sensitive isoschizomer, HpaII. After digestion, oligonucleotides are ligated onto fragment ends and linker-mediated PCR is performed with Taq polymerase.15 The conditions of polymerization favored the amplification of smaller fragments, which are fluorescently labeled and hybridized to custom-made oligonucleotide microarrays. Regions with relative hypomethylation should display similar levels of hybridization from both MspI- and HpaII-derived products, whereas those with relative hypermethylation should have a predominance of MspI-derived products. The MELP modification (Figure 1B) that we introduced replaces the oligonucleotide microarray with fluorescently labeled microspheres covalently coupled to oligonucleotides specific for the loci in the HELP-derived AML classifier. PCR products are hybridized onto these microspheres and are detected by using a flow cytometer, rather than by microarray scanning. Fluorescence properties of the microspheres identify the specific locus, and the relative signal intensity of the hybridized PCR products reflects level of methylation. These modifications make the assay highly feasible for routine use, because detection of fluorescent microspheres by flow cytometry has been adapted in several other assays.16,17
Figure 1.
A: Schematic of the HELP assay. Genomic DNA is digested with either MspI (methylation insensitive) or HpaII (methylation sensitive). The resulting fragments are ligated to linkers and PCR amplified with linker-specific primers. Amplicons are fluorescently labeled (as shown, red for amplicons from MspI-digested genomic DNA and green for HpaII-digested genomic DNA) and hybridized to oligonucleotide microarrays. Only relatively short fragments are amplified by Taq polymerase. Regions of relative hypomethylation will show signal from both MspI- and HpaII-digested DNA, whereas regions of relative hypermethylation will show a predominance of MspI signal. B: MELP modification of HELP assay. Rather than hybridizing to oligonucleotide arrays, amplicons (with red and green denoting the initial MspI and HpaII digests, respectively) are hybridized to oligonucleotides covalently linked to fluorescent microspheres with distinct fluorescent properties (depicted as shades of brown). The microspheres are subjected to flow cytometry and analyzed for locus/microsphere identity (determined by brown intensity) and for amplicon amount (determined by red/green intensity).
Initial tests using oligonucleotide-coupled microspheres for product detection demonstrated that we could specifically hybridize amplicons from the linker-mediated PCR onto fluorescent microspheres; however, the fluorescence intensity from the labeled PCR products was lower than the detection limit of the instrument. Increasing the number of PCR cycles from 20 (standard for HELP) to 55 did not lead to a subsequent increase in signal intensity, suggesting that the PCR saturates (data not shown). Thus, a method to linearly amplify signal from a fixed number of amplicons was required. Because the use of branched DNA technology has been shown to specifically detect femtogram amounts of nucleic acids, we altered our detection method to incorporate a series of branched DNA hybridization reactions (QuatigenePlex 2.0 technology; Affymetrix) before analysis. This technology has been used for RNA expression but is less well characterized for DNA and has never been used to analyze DNA methylation.
Multiplex MELP Linearity and Specificity
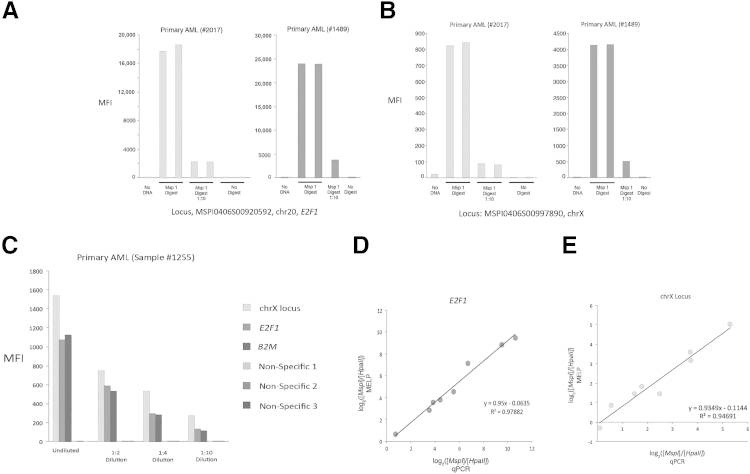
Given the novelty of the assay technology used, we first determined if the assay is quantitative, and if it could be used to accurately determine the relative amounts of PCR products from the MspI and HpaII reactions. To this end, we digested genomic DNA from primary AML samples with MspI, performed linker-mediated PCR, and detected fluorescent signal from specific PCR products after both hybridization to microspheres and branched DNA signal amplification. As shown in Figure 2, A and B, reproducible levels of fluorescence are seen for each locus, and a 10-fold dilution of PCR product results in a similar decrease in signal intensity. As expected, the assay requires restriction enzyme digest, because use of either undigested DNA or no DNA yields minimal signal. In addition, absolute median fluorescent intensities range from <100 to >20,000. Thus, the linearity of the assay appears to extend over at least a 2-log range. Because the negative controls typically display a median fluorescent intensity of <10, the linearity is likely to be close to a 3-log range. This 3-log range of linearity is consistent with results using the QuantigenePlex 2.0 assay to quantitate RNA expression.18 Overall, these controls demonstrate linear amplification and detection of MspI-digested PCR products.
Figure 2.
The MELP assay is both quantitative and specific. A and B: Linker-mediated amplification of MspI-digested DNA was performed. Amplicons were hybridized to fluorescent microspheres and detected by branched DNA hybridization. A 1:10 dilution of amplicons is also assayed. Results are shown for two independent primary AML samples at E2F1 (A) and an unnamed locus on chromosome X (B), both of which are in the HELP-defined methylation classifier for AML. C: A similar assay to A and B was performed, but input DNA was diluted 1:2, 1:4, and 1:10, rather than amplicon dilution, as in A and B. Results are shown for the two loci in A and B and for B2M. In addition, fluorescent microspheres coupled to nonspecific oligonucleotides were included to determine background signal. D and E: Relative amplicon quantitation by MELP compared with quantitation by PCR. MELP was performed on primary AML samples, and signal ratio from MspI and HpaII digests of E2F1 (D) and an unnamed locus on chromosome X (E) were normalized to ratios of the hypomethylated locus, B2M. Amplicon quantitation was also determined by locus-specific qPCR. MFI, mean fluorescent intensity.
We further sought to determine whether the assay accurately reflects the number of digested genomic fragments from the original sample, rather than simply the number of amplicons in the final PCR product. We, therefore, performed a similar assay as previously described, but rather than diluting the final product, we performed 2-, 4-, and 10-fold dilutions on the MspI-digested sample before PCR. As shown in Figure 2C, the fold decrease in signal intensity closely approximates the fold dilution of the starting material. In addition, virtually no signal was obtained with fluorescent microspheres that are covalently coupled to non-specific oligonucleotides, thus confirming the specificity of the assay.
As a final verification of the quantitative accuracy and the locus specificity of our detection method, we compared detection and quantitation by our hybridization method with quantitative PCR (qPCR). The entire MELP assay (including branched DNA hybridization and flow cytometry) was performed on DNA from eight primary AML samples and analyzed MspI- and HpaII-derived products at two loci that are part of the HELP-determined methylation classifier. In parallel, we performed the same assay but used qPCR for amplicon quantitation at these two loci. Furthermore, a known hypomethylated locus, B2M (β2 microglobulin), was analyzed in both assays for signal normalization.3 As shown in Figure 2D, the normalized ratios of MspI and HpaII products are virtually identical between the two assays at both loci, indicating that our method of detection is as specific and as quantitative as qPCR.
Normalization of DNA Methylation Measurements
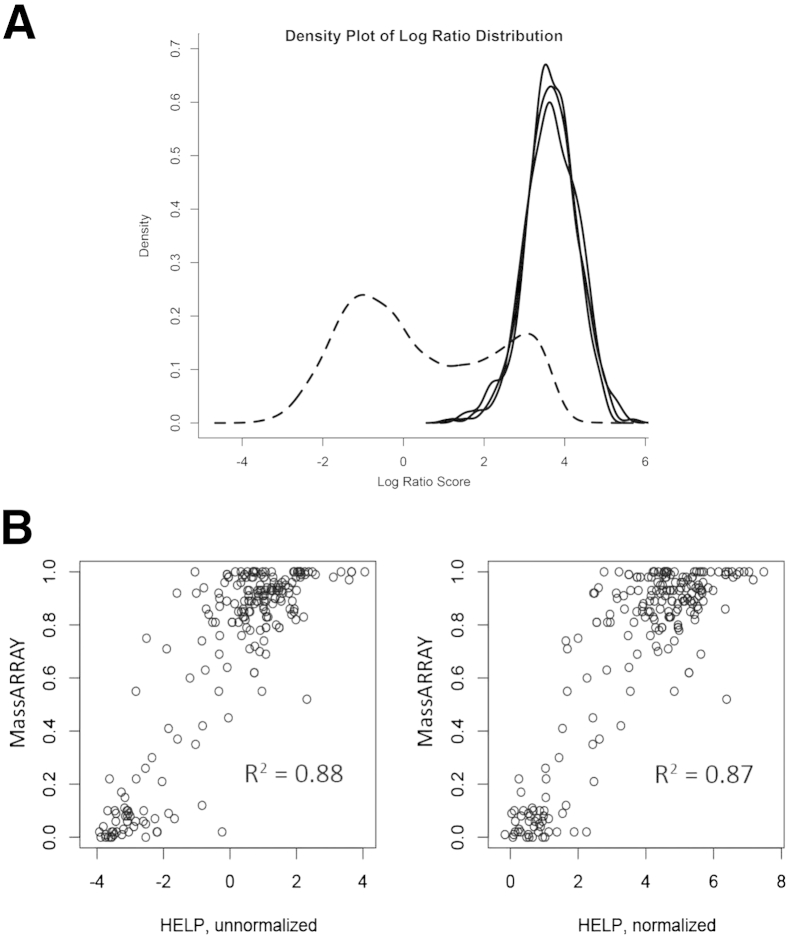
Given the robust performance of the MELP assay with a limited number of loci, we expanded it to encompass the entire 18-locus classifier used in the HELP assay to predict AML outcomes. However, one advantage of highly multiplexed HELP results that is not shared by results obtained by MELP is that expression values can be normalized using global array properties. In the absence of global normalization, it is necessary to compare measured values with those found in a known unmethylated locus. Although mitochondrial DNA is known to be unmethylated, we reasoned that the high copy number relative to autosomal DNA would not be appropriate for a PCR-based assay that could be subject to saturation. To identify autosomal, constitutively unmethylated regions from existing HELP data, we identified loci with measured values in at least 340 samples that met stringent criteria for narrow distribution and high HpaII/MspI log ratio (indicating lack of methylation). Three control loci were identified, and their consistent nonmethylated state was confirmed by plotting the distribution of their methylation scores across all HELP-analyzed samples3 and comparing it with the distribution of methylation scores for all loci in a control, CD34+ cell population (Figure 3A). Normalization was performed for HELP and MELP by calculating the average of the log HpaII/MspI ratio for these three control loci and subtracting from the unnormalized locus of interest. To demonstrate that this normalization does not significantly affect the results from HELP data that have been previously subjected to global normalization, we compared the correlation of HELP results with MassArray assessment of methylation using HELP data subjected to global normalization alone (unnormalized) or to subsequent normalization using our selected control loci (normalized) (Figure 3B). Correlation values for these comparisons were essentially indistinguishable (0.877 unnormalized, 0.873 normalized). Thus, our normalization to three hypomethylated loci provides an internal control for the assay and allows for a robust quantitative analysis of DNA methylation at the selected loci.
Figure 3.
Determination of normalizing loci. A: Smoothed density estimates reflecting the number of loci found at each methylation level are shown. Dashed line shows the bimodal distribution of relative methylation determined by HELP across all loci in normal CD34+ cells. Solid lines show the distribution of methylation determined by HELP of the three loci chosen for signal normalization. All three loci are hypomethylated in most AML samples and should, therefore, have similar fragment amounts in MspI- and HpaII-digested samples. B: Comparison of MassArray Epityper to HELP using unnormalized HELP ratios or HELP ratios normalized to ratios of the three loci shown in A.
Comparison of MELP and HELP at Individual Loci
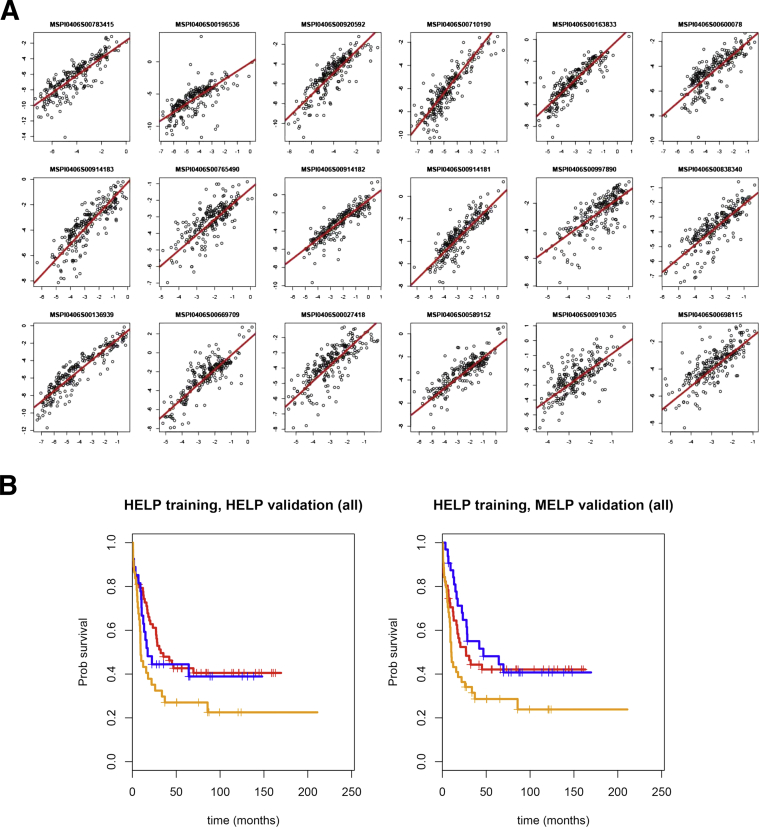
Next, we performed the MELP assay on 216 primary AML samples and determined normalized ratios between MspI- and HpaII-derived PCR products for those loci whose methylation status is prognostic for AML. The 216 samples were chosen from the HOVON data set on which HELP was previously performed (84 from the training cohort, 84 from the test cohort, and 48 from the validation cohort3). The MELP-derived methylation ratios were then compared with normalized ratios, as determined by the original HELP assay. As shown in Figure 4A, highly significant correlations between HELP and MELP are seen at all loci, indicating that MELP is virtually equivalent to HELP in determining methylation status for the 18 prognostic loci.
Figure 4.
The MELP assay accurately reflects HELP-derived data. A: Comparison of HELP-derived HpaII/MspI ratios (x axis) to MELP-derived ratios (y axis) at the 18 loci used in the methylation classifier for AML (r = 0.63 to 0.92, P < 10−12 for all loci). B: Overall survival curves of AML patients classified by the 18-locus methylation score (blue, highest score group; red, middle score group; orange, lowest score group) determined by HELP or MELP. Test and validation cohorts are shown.
Comparison of MELP and HELP in a Global Methylation Classifier of AML Patient Prognosis
The strong correlation at each individual locus between the MspI and HpaII ratios, determined by HELP and MELP, suggested that methylation status of the 18 prognostic loci, as determined by MELP, could predict outcome for patients with AML. To test this hypothesis, we first determined the linear relationship between normalized HELP and normalized MELP scores using Deming regression on training samples that had been measured using both platforms (n = 84). The regression curve was then used to convert the normalized HELP data into a normalized MELP scale, and the full training set of transformed HELP data (n = 200) previously used by Figueroa et al3 was used to train an 18-locus classifier. A predictor was generated using the SuperPC package19 (http://cran.r-project.org/web/packages/superpc/index.html) in conjunction with training and test sets. Because this classifier was trained using data that were converted to the normalized MELP scale, we then were able to use the test and validation samples to determine its performance.
By using tertile survival cut points obtained from the training data set alone, we determined the survival of methylation-classified subgroups. As shown in Figure 4B, transformed HELP data using this predictor are able to distinguish prognostic classes, and these results are consistent with the previous results of Figueroa et al,3 demonstrating that methylation (measured by HELP) can predict prognosis within this disease. To determine whether our MELP assay can also predict outcome, we performed the same analysis using samples from the test and validation samples (n = 84 and n = 48, respectively). These results (Figure 4B) demonstrate that analysis of methylation status using MELP can predict AML survival (P = 0.048), thus showing the suitability of this novel technique for disease prognosis.
Discussion
Although cancer has been traditionally thought of as a genetic disease driven by the somatic acquisition of multiple oncogenic mutations, recent studies have clearly shown that it is also an epigenetic disease and that dysregulation of chromatin structure plays a central role in tumorigenesis.2 Despite these associations, routine multilocus assessment of epigenetic phenomena is not common, due in part to the difficulty and expense involved in establishing and performing epigenetic assays. We report herein the development of a novel assay, MELP, that measures DNA methylation through the use of fluorescently labeled microspheres and branched DNA hybridization for detection of relevant amplicons. We have shown that detection of these amplicons by MELP is highly quantitative and is virtually identical to evaluation by qPCR. Analysis of DNA methylation at multiple loci by MELP is tightly correlated with similar evaluation by the HELP assay.3 Furthermore, methylation analysis at 18 loci previously shown by HELP to be prognostically relevant in patients with AML also predicts survival when measured by MELP.
Unlike most assays that assess DNA methylation at multiple loci, numerous features of the MELP assay make it a platform that is well suited to rapidly analyze multilocus DNA methylation. MELP does not require bisulfite treatment of DNA, but rather relies on enzymatic reactions (restriction digestion, ligation, and PCR) that are routinely performed in most molecular laboratories. Evaluation of methylation is performed by measurement of fluorescent microspheres and does not require custom-made, solid-phase oligonucleotide microarrays or high-throughput sequencing technologies. Indeed, similar analysis of DNA-coupled fluorescent microspheres is used in a variety of assays, including evaluation of recurrent translocations found in AML.17,20 More important, methylation of multiple loci is evaluated simultaneously using the MELP assay. In our experiments, two reaction tubes (one each for MspI- and HpaII-digested DNA) were sufficient for the evaluation of 18 prognostic and three normalization loci. In its current format, the assay can be expanded to evaluate 80 loci without a concomitant increase in reactions. This simultaneous assessment has clear advantages over independent parallel reactions (eg, qPCR) in terms of both work flow and possibility for laboratory errors. In addition, simultaneous locus evaluation allows for reactions to be internally controlled for variations in the enzymatic and hybridization reactions. Finally, the entire assay can be performed in a relatively short time frame. DNA extraction, digestion, and linker ligation are performed on the first day, PCR and initial hybridization to the microspheres are performed on the second day, and final branched DNA hybridization and data collection and analysis are performed on the third day. Thus, results are typically obtained 2 to 4 days after sample acquisition.
Although this initial analysis of MELP has been restricted to patients with AML, the assay platform can easily be extended to other pathological conditions for which regulation of DNA methylation has been shown to have diagnostic, prognostic, or therapeutic implications. For instance, Shaknovich et al21 have shown that a DNA methylation signature can classify diffuse large B-cell lymphomas (DLBCLs) into activated B-cell or germinal center subtypes, the former of which display a more aggressive phenotype. In addition, the methylation profile of DLBCL significantly predicts outcome in patients with activated B-cell DLBCL.22 As such, determination of DNA methylation status by MELP could aid in subclassification of these tumors.
We and others have examined the role of DNA methylation as an independent prognostic indicator in AML.3,23 Clearly, survival is influenced by multiple determinants, including tumor-specific events (eg, mutations) and patient-specific factors (eg, age, ethnicity, and comorbidities). All of these factors should be assessed to determine an optimal algorithm for tumor subclassification. The establishment of MELP allows for DNA methylation analysis to be routinely incorporated in such studies.
Note Added in Proof
Subsequent to the acceptance of this manuscript for publication, we identified a correction to the analysis code that affected a subset of the samples in our original analysis. The overall results are not affected, and median-cut survival curves show a significant difference (P = 0.015). This error, which only affected the survival analysis (Figure 4B), utilized incorrect values for 17 loci in 17 samples (out of a total of 216 samples). Corrected analysis code is provided in the Supplemental Script S6.
Footnotes
This work was supported, in part, by the JP McCarthy Foundation (G.B.W.W. and A.B.), Veterans Affairs Administration grant 1I01BX000918-01 (M.C.), and NIH grant 1R01CA149566-01A1 (M.C.).
Disclosures: None declared.
Supplemental Data
R script for generating figures in the article.
Supplemental R script provides a readable version of ancillary data in Gene Expression Omnibus (http://www.ncbi.nlm.gov/geo; accession number GSE18700). Data from Figueroa et al.3 Should be renamed load_sampdata.R to run the provided R code.
List of tumors used for training, testing, and validation in both Figueroa et al3 and in the present article; should be renamed ktrain.txt to load using the accompanying R scripts.
List of tumors used for training, testing, and validation in both Figueroa et al3 and in the present article; should be renamed ktest.txt to load using the accompanying R scripts.
List of tumors used for training, testing, and validation in both Figueroa et al3 and in the present article; should be renamed kvalid.txt to load using the accompanying R scripts.
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Baylin S.B., Jones P.A. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Figueroa M.E., Lugthart S., Li Y., Erpelinck-Verschueren C., Deng X., Christos P.J., Schifano E., Booth J., van Putten W., Skrabanek L., Campagne F., Mazumdar M., Greally J.M., Valk P.J., Lowenberg B., Delwel R., Melnick A. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noushmehr H., Weisenberger D.J., Diefes K., Phillips H.S., Pujara K., Berman B.P., Pan F., Pelloski C.E., Sulman E.P., Bhat K.P., Verhaak R.G., Hoadley K.A., Hayes D.N., Perou C.M., Schmidt H.K., Ding L., Wilson R.K., Van Den Berg D., Shen H., Bengtsson H., Neuvial P., Cope L.M., Buckley J., Herman J.G., Baylin S.B., Laird P.W., Aldape K., Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Portela A., Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 6.Swerdlow S., Campo E., Harris N., Jaffe E., Pileri S., Stein H., Thielel J., Vardiman J. International Agency for Research on Cancer (IARC); Lyon: 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; pp. 110–147. [Google Scholar]
- 7.Wertheim G.B., Hexner E., Bagg A. Molecular-based classification of acute myeloid leukemia and its role in directing rational therapy: personalized medicine for profoundly promiscuous proliferations. Mol Diagn Ther. 2012;16:357–369. doi: 10.1007/s40291-012-0009-0. [DOI] [PubMed] [Google Scholar]
- 8.Ley T.J., Ding L., Walter M.J., McLellan M.D., Lamprecht T., Larson D.E. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel J.P., Gonen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O., Huberman K., Cheng J., Viale A., Socci N.D., Heguy A., Cherry A., Vance G., Higgins R.R., Ketterling R.P., Gallagher R.E., Litzow M., van den Brink M.R., Lazarus H.M., Rowe J.M., Luger S., Ferrando A., Paietta E., Tallman M.S., Melnick A., Abdel-Wahab O., Levine R.L. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rocquain J., Carbuccia N., Trouplin V., Raynaud S., Murati A., Nezri M., Tadrist Z., Olschwang S., Vey N., Birnbaum D., Gelsi-Boyer V., Mozziconacci M.J. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer. 2010;10:401. doi: 10.1186/1471-2407-10-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shih A.H., Abdel-Wahab O., Patel J.P., Levine R.L. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 12.Kraguljac Kurtović N., Krajnović M., Bogdanović A., Suvajdzić N., Jovanović J., Dimitrijević B., Colović M., Krtolica K. Concomitant aberrant methylation of p15 and MGMT genes in acute myeloid leukemia: association with a particular immunophenotype of blast cells. Med Oncol. 2012;29:3547–3556. doi: 10.1007/s12032-012-0289-6. [DOI] [PubMed] [Google Scholar]
- 13.Mill J., Yazdanpanah S., Guckel E., Ziegler S., Kaminsky Z., Petronis A. Whole genome amplification of sodium bisulfite-treated DNA allows the accurate estimate of methylated cytosine density in limited DNA resources. Biotechniques. 2006;41:603–607. doi: 10.2144/000112266. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen I.S., Krarup H.B., Thorlacius-Ussing O., Madsen P.H. High recovery of cell-free methylated DNA based on a rapid bisulfite-treatment protocol. BMC Mol Biol. 2012;13:12. doi: 10.1186/1471-2199-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueroa M.E., Melnick A., Greally J.M. Genome-wide determination of DNA methylation by Hpa II tiny fragment enrichment by ligation-mediated PCR (HELP) for the study of acute leukemias. Methods Mol Biol. 2009;538:395–407. doi: 10.1007/978-1-59745-418-6_20. [DOI] [PubMed] [Google Scholar]
- 16.Dunbar S.A., Jacobson J.W. Rapid screening for 31 mutations and polymorphisms in the cystic fibrosis transmembrane conductance regulator gene by Lminex xMAP suspension array. Methods Mol Med. 2005;114:147–171. doi: 10.1385/1-59259-923-0:147. [DOI] [PubMed] [Google Scholar]
- 17.King R.L., Naghashpour M., Watt C.D., Morrissette J.J., Bagg A. A comparative analysis of molecular genetic and conventional cytogenetic detection of diagnostically important translocations in more than 400 cases of acute leukemia, highlighting the frequency of false-negative conventional cytogenetics. Am J Clin Pathol. 2011;135:921–928. doi: 10.1309/AJCPJCW6BY0CNIHD. [DOI] [PubMed] [Google Scholar]
- 18.Cha W., Ma Y., Saif Y.M., Lee C.W. Development of microsphere-based multiplex branched DNA assay for detection and differentiation of avian influenza virus strains. J Clin Microbiol. 2010;48:2575–2577. doi: 10.1128/JCM.01979-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bair E., Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hafez M., Ye F., Jackson K., Yang Z., Karp J.E., Labourier E., Gocke C.D. Performance and clinical evaluation of a sensitive multiplex assay for the rapid detection of common NPM1 mutations. J Mol Diagn. 2010;12:629–635. doi: 10.2353/jmoldx.2010.090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaknovich R., Geng H., Johnson N.A., Tsikitas L., Cerchietti L., Greally J.M., Gascoyne R.D., Elemento O., Melnick A. DNA methylation signatures define molecular subtypes of diffuse large B-cell lymphoma. Blood. 2010;116:e81–e89. doi: 10.1182/blood-2010-05-285320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De S., Shaknovich R., Riester M., Elemento O., Geng H., Kormaksson M., Jiang Y., Woolcock B., Johnson N., Polo J.M., Cerchietti L., Gascoyne R.D., Melnick A., Michor F. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet. 2013;9:e1003137. doi: 10.1371/journal.pgen.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bullinger L., Ehrich M., Dohner K., Schlenk R.F., Dohner H., Nelson M.R., van den Boom D. Quantitative DNA methylation predicts survival in adult acute myeloid leukemia. Blood. 2010;115:636–642. doi: 10.1182/blood-2009-03-211003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
R script for generating figures in the article.
Supplemental R script provides a readable version of ancillary data in Gene Expression Omnibus (http://www.ncbi.nlm.gov/geo; accession number GSE18700). Data from Figueroa et al.3 Should be renamed load_sampdata.R to run the provided R code.
List of tumors used for training, testing, and validation in both Figueroa et al3 and in the present article; should be renamed ktrain.txt to load using the accompanying R scripts.
List of tumors used for training, testing, and validation in both Figueroa et al3 and in the present article; should be renamed ktest.txt to load using the accompanying R scripts.
List of tumors used for training, testing, and validation in both Figueroa et al3 and in the present article; should be renamed kvalid.txt to load using the accompanying R scripts.