Background: Cancer cells are characterized by elevated mitochondrial ROS. The dismutases SOD1 and SOD2 regulate ROS.
Results: SOD2 is down-regulated following oncogenic activation in breast cancers. However, SOD1 is overexpressed, and its inhibition by LCS-1 leads to mitochondrial fragmentation.
Conclusion: In the absence of SOD2, inhibition of SOD1 abolishes the integrity of the mitochondria.
Significance: Our data suggest a SOD switch during transformation.
Keywords: Cancer, Glucose Metabolism, Mitochondria, Reactive Oxygen Species (ROS), Superoxide Dismutase (SOD), ROS
Abstract
Cancer cells are characterized by elevated levels of reactive oxygen species, which are produced mainly by the mitochondria. The dismutase SOD2 localizes in the matrix and is a major antioxidant. The activity of SOD2 is regulated by the deacetylase SIRT3. Recent studies indicated that SIRT3 is decreased in 87% of breast cancers, implying that the activity of SOD2 is compromised. The resulting elevation in reactive oxygen species was shown to be essential for the metabolic reprograming toward glycolysis. Here, we show that SOD2 itself is down-regulated in breast cancer cell lines. Further, activation of oncogenes, such as Ras, promotes the rapid down-regulation of SOD2. Because in the absence of SOD2, superoxide levels are elevated in the matrix, we reasoned that mechanisms must exist to retain low levels of superoxide in other cellular compartments especially in the intermembrane space of the mitochondrial to avoid irreversible damage. The dismutase SOD1 also acts as an antioxidant, but it localizes to the cytoplasm and the intermembrane space of the mitochondria. We report here that loss of SOD2 correlates with the overexpression of SOD1. Further, we show that mitochondrial SOD1 is the main dismutase activity in breast cancer cells but not in non-transformed cells. In addition, we show that the SOD1 inhibitor LCS-1 leads to a drastic fragmentation and swelling of the matrix, suggesting that in the absence of SOD2, SOD1 is required to maintain the integrity of the organelle. We propose that by analogy to the cadherin switch during epithelial-mesenchymal transition, cancer cells also undergo a SOD switch during transformation.
Introduction
Reactive oxygen species (ROS)3 are produced as a result of the leakage of electrons from the electron transport chain and their reaction with oxygen. Superoxide is the first species to be produced, and it accumulates in both the matrix and the intermembrane space of the mitochondria. Superoxide is converted to hydrogen peroxide through the activity of the dismutases SOD1 and SOD2. SOD2 localizes to the matrix, where its activity is regulated by the deacetylase SIRT3 (1). SOD1 is, however, mainly present in the cytosol, but a fraction localizes to the intermembrane space. Cancer cells are characterized by elevated levels of ROS (2, 3). A recent study indicated that the expression of SIRT3 is abolished or decreased in 87% of breast cancers (4).
However, because excessive accumulation of superoxide in the mitochondria would result in damage to mitochondrial DNA, proteins, and lipids, ultimately leading to irreversible damage to the organelle (2), we reasoned that in the absence of SIRT3, other mechanisms must be up-regulated to limit the levels of mitochondrial ROS and maintain the integrity of the organelle. As SOD1 is the other dismutase in the mitochondria, we aimed at analyzing SOD1 in a variety of breast cancer models.
EXPERIMENTAL PROCEDURES
Immunohistochemistry
The tissues were processed using the Histostain-Plus broad spectrum (diaminobenzidine) substrate kit for peroxidase (Invitrogen). Briefly, antigen retrieval was performed by boiling in 10 mm sodium citrate. Endogenous peroxidase activity was blocked with 3% hydrogen peroxidase for 20 min followed by 30 min of blocking. Tissues were incubated overnight in the cold room with anti-SIRT3 and anti-SOD1 antibodies and detected with biotinylated anti-rabbit (DAKO) for 20 min and streptavidin peroxidase for 10 min. Slides were developed with chromogen diaminobenzidine. Sections were then counterstained in hematoxylin for 30 s and mounted with DePEX. Staining was scored based on the intensity of staining as 0, 1+.2+, or 3+. Slides with a score of 2+ or above were considered positive.
Cell Culture
MCF-10A, MCF7, MDA-MB-231, and MDA-MB157 were grown in DMEM medium supplemented with 10% fetal calf serum and antibiotics (Invitrogen). BT474 and T47D were grown in RPMI medium supplemented with 10% fetal calf serum and antibiotics (Invitrogen).
Immunoblot Analysis
For protein extractions, cells were washed three times in ice-cold PBS and lysed in 200 μl of ice-cold lysis buffer (50 mm Tris, pH 7.5, 250 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 50 mm NaF, 0.2 mm Na3VO4, 1 g/ml leupeptin, 1 g/ml pepstatin, 100 g/ml PMSF, 1 mm DTT). Lysates were clarified by centrifugation at 13,000 rpm for 20 min at 4 °C, and the protein concentration of the supernatant was assayed using Bio-Rad protein assay (Bio-Rad). Immunoprecipitations were performed as described previously (5), and for immunoblot analysis, proteins were separated by SDS-PAGE electrophoresis, transferred to nitrocellulose membrane (PerkinElmer Life Sciences), and probed with the following antibodies: SIRT3 (rabbit, Millipore), SOD1 (rabbit, Santa Cruz Biotechnology), SOD2 (rabbit, Millipore), actin (mouse, Zymed Laboratories Inc.), and prohibitin (mouse, Calbiochem).
Mitochondrial Fractionation
Subcellular fractionation was performed as described previously (17). Briefly, cells were resuspended in buffer A (300 mm sucrose, 20 mm HEPES-KOH, pH 7.5,1 mm EDTA, 10 mm KCl, 1 mm dithiothreitol (DTT), 1.5 mm MgCl2, and protease inhibitors) and homogenized by 30 strokes with a Dounce homogenizer. The mitochondrial and cytoplasmic fraction (supernatant) was separated from the nuclear fraction (pellet) by centrifugation at 800 × g for 10 min. The supernatant was further centrifuged at 10,000 × g for 20 min, and the enriched mitochondrial fraction was washed twice with buffer A and lysed in buffer B (Tris acetate, pH 8, 10% Nonidet P-40, 5 mm CaCl2, 1 mm DTT, and protease inhibitors).
Transmission Electron Microscopy
Cells were fixed at 4 °C overnight, in 2.5% glutaraldehyde, 4.0% paraformaldehyde in 0.1 m phosphate buffer, and were processed by the electron microscopy facility at Mount Sinai. Samples were examined using a Hitachi 7650 transmission electron microscope.
Dismutase Activity Measurement
Cell lines were allowed to grow in culture for 48 h. Cells were washed twice with cold PBS, detached with a rubber policeman, collected, and spun down at 1,000 × g. Cells were resuspended in 200 μl of 20 mm HEPES buffer, pH 7.2, containing 1 mm EGTA, 210 mm mannitol, and 70 mm sucrose and homogenized with an 18-gauge needle. Lysates centrifuged for 20 min at 10,000 × g. SOD activity levels were quantified with the use of the superoxide dismutase assay kit (Cayman Chemicals item number 706002).
Superoxide Measurement
Cells were treated with either DMSO or LCS-1 the next day. 48 h after treatment, cells were detached with 0.05% trypsin EDTA, collected in the appropriate growth medium, and spun down at 1,000 × g for 5 min. Cells were stained with MitoSOX Red mitochondrial superoxide indicator (Invitrogen item number M36008) as per the manufacturer's instructions for 20 min at 37 °C. Cells were spun down at 1,000 × g for 5 min and washed in ice-cold PBS. Superoxide levels were quantified by FACS with the use of a BD FACSCanto.
2-Hydroxyethidium Measurement
MDA-MB-231 cells were treated as described for superoxide measurement but were prepared and run on Beckman System Gold HPLC with UV-visible and fluorescence detectors to determine 2-hydroxyethidium concentration. Extraction of samples and HPLC analysis were performed according to a published method (6), and the λexcitation = 510 nm and λemission = 595 nm signals were used for quantitation. Protein concentration was determined after cell lysis by the CB X protein assay (GE Healthcare) for use as a normalizing factor. Linearity of the assay was established on our system by comparing peak areas with a standard curve prepared from a mito-hydroethidine standard (R2 = 0.96). Amount (nanomoles per milligram of protein) of 2-hydroxyethidium in the samples was determined by comparing the peak areas with a standard (150 pmol) of mito-hydroethidine injected after assay of the samples, chosen to match the response of the unknowns.
RESULTS AND DISCUSSION
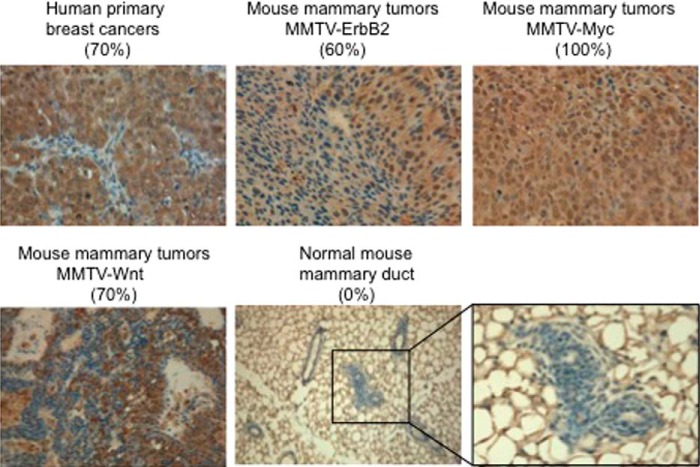
We found that SOD1 is overexpressed in 70% (21/30) of human primary breast cancers (Fig. 1) but also in three different mouse models of mammary tumors, namely the murine mammary tumor virus (MMTV)-ErbB2 (60%), MMTV-Myc (100%), and MMTV-Wnt (70%) transgenic mice (Fig. 1). However, SOD1 was not detected in normal mammary ducts (Fig. 1). We concluded that the overexpression of SOD1 is observed in 60–100% of tumor sections tested in three mammary tumor models where tumorigenesis is driven by different oncogenes. Therefore, the overexpression of SOD1 is not linked to the expression of a specific oncogene.
FIGURE 1.
SOD1 is overexpressed in breast and mammary cancers. Immunohistochemistry of SOD1 was performed on the indicated tissue sections. The percentage of samples positive for SOD1 staining is shown in each case. A representative image of the staining is shown.
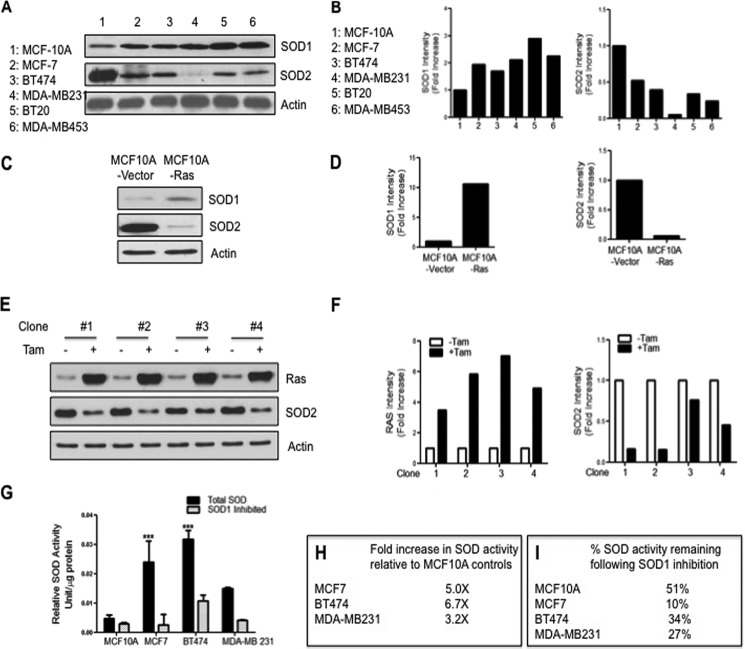
We next tested whether the overexpression of SOD1 is also observed in a panel of five breast cancer cell lines. The non-tumorigenic cell line, MCF-10A, was used as a control. We found that SOD1 level was the lowest in MCF-10A cells, whereas it was overexpressed in all five breast cancer cell lines (Fig. 2, A and B). The expression of SIRT3 was also decreased when compared with MCF-10A in all breast cancer cell lines (data not shown). Further, because SOD2 levels were never determined in this panel of cell lines, we also analyzed SOD2 by Western analysis. We found that SOD2 is reduced by 50–90% in these cell lines (Fig. 2, A and B).
FIGURE 2.
SOD1 is overexpressed in breast cancer cell lines and contributes to the majority of the dismutase activity. A, Western blots of SOD1 and SOD2 in a panel of breast epithelial cell lines. MCF-10A cells are transformed but non-malignant, although all others are breast cancer cell lines. Tubulin was used as a loading control. B, graphs of the quantification of SOD1 and SOD2 levels from Western blots shown in A. C, Western blots of SOD1 and SOD2 in MCF-10A and MCF-10A stable clones expressing the oncogene Ras constitutively. D, graphs of the quantification of SOD1 and SOD2 levels from Western blots shown in C. E, Western blots of SOD1 and SOD2 in four different clones of MCF-10A stable clones expressing Ras upon treatment with tamoxifen (Tam). F, graphs of the quantification of Ras and SOD2 levels from Western blots shown in E. G, total dismutase activity was determined in all indicated cell lines in the presence or absence of pretreatment with the SOD1 inhibitor potassium cyanide at a concentration of 2 mm. Error bars indicate mean ± S.D. ***, p < 0.001. H, table of -fold increase in total SOD activity in breast cancer cell lines relative to MCF-10A control cells. I, table of the percentage of total SOD activity remaining following SOD1 inhibition.
We further tested the relative expression levels of SOD1 and SOD2 in MCF-10A cells expressing the oncogene Ras constitutively. We found that although SOD1 is elevated by 10-fold in Ras-expressing cells when compared with the MCF-10A control cells, SOD2 was reduced by 95% (Fig. 2, C and D). The same result was observed in MCF-10A cells expressing the oncogene Myc (data not shown). In addition, the levels of SOD1 and SOD2 were determined in four independent MCF-10A clones where the expression of Ras is inducible by treatment with tamoxifen. We found that in all four clones, induction of Ras for only 48 h was sufficient to lead to the reduction of SOD2 by 30–90% (Fig. 2, E and F). No effect was observed on SOD1 during this time frame (data not shown). This result indicates that the expression of SOD2 is rapidly down-regulated upon activation of oncogenes. However, although the up-regulation of SOD1 is only observed later, it nevertheless is uniformly observed in all cell lines tested. Because the overexpression of SOD1 is found in all five cell lines tested as well as in 70% of human breast cancers and 60–100% of all mouse mammary tumors tested, collectively these results indicate that it is a frequent event.
The combined decrease in SOD2 and increase in SOD1 suggests that SOD1 may be the main dismutase activity in breast cancer. To test this possibility, we next measured total dismutase activity in a panel of breast cancer cells. We found that MCF7, BT474, and MDA-MB-231 breast cancer cells showed a 5-, 6.7-, and 3.2-fold increase, respectively, in dismutase activity when compared with MCF-10A cells (Fig. 2, G and H). To determine the relative contribution of SOD1 or SOD2 to the total dismutase activity in these cells, we first incubated cells in potassium cyanide to inhibit the activity of SOD1 and measured the dismutase activity. We found that inhibition of SOD1 drastically reduced their dismutase activity to 10% in MCF7, 34% in BT474, and 27% in MDA-MB-231 cells (Fig. 2, G and I).
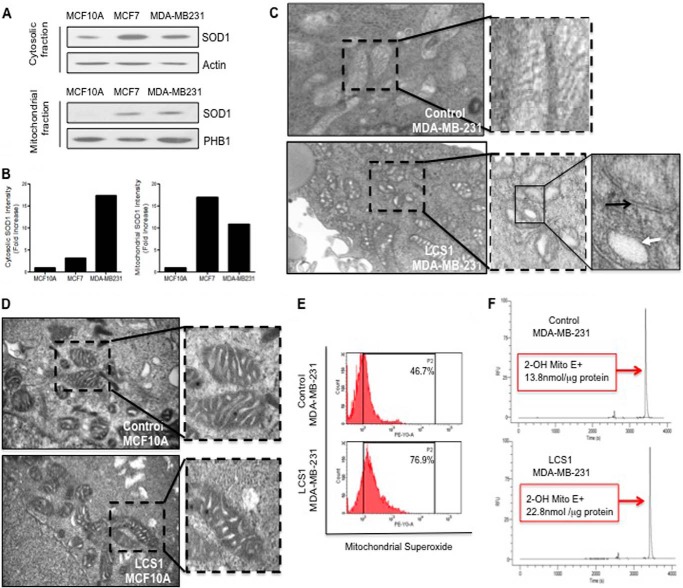
To further characterize the contribution of SOD1 in the regulation of oxidative stress in the mitochondria, we then compared the levels of SOD1 in the cytosolic and mitochondrial fractions in MCF-10A, MCF7, and MDA-MB-231 cells. We found that although SOD1 in the mitochondrial fraction of MCF-10A was undetectable, SOD1 was found in the mitochondrial fraction of MCF7 and MDA-MB-231 cells (Fig. 3, A and B). This observation is consistent with the hypothesis that mitochondrial SOD1 may be required to maintain the integrity of the organelle when the expression of SOD2 is compromised. To test this possibility, we treated MDA-MB-231 breast cancer cells with the SOD1 inhibitor, LCS-1 (7), and determined the effect of SOD1 inhibition on the morphology of the mitochondria using electron microscopy. We found that in contrast to the control-treated cells (Fig. 3C) (DMSO only), mitochondria of MDA-MB-231 cells treated with LCS-1 showed increased fragmentation and dilated cristae (Fig. 3C). In contrast, treatment of non-tumorigenic cells MCF-10A with LCS1 did not alter the morphology of their mitochondria (Fig. 3D). Further, we found that treatment with LCS-1 leads to a 1.6-fold increase (46.7–76.9%) in the level of mitochondrial superoxide as measured using MitoSOX Red assay (Fig. 3E). Replicates of this experiment revealed a statistically significant difference between LCS-1-treated versus control-treated cells (p < 0.05). However, because MitoSOX Red was reported to be nonspecific (8), we further evaluated the increase in superoxide using measurement of the superoxide-dependent product 2-hydroxyethidium by HPLC according to the previous protocol (6). We found a 1.6-fold (13.8 nmol/μg of proteins to 22.8 nmol/μg of proteins) increase in 2-hydroxyethidium concentration in cells treated with LCS1 when compared with control-treated cells (Fig. 3F). Replicates of this experiment confirmed a statistically significant difference between the LCS-1- and control-treated cells (p < 0.05). These results suggest that inhibition of SOD1 leads to an excessive level of superoxide in the mitochondria, which ultimately leads to the collapse of the integrity of the organelle.
FIGURE 3.
Inhibition of SOD1 results in altered mitochondrial morphology and elevated mitochondrial superoxide. A, Western blots of SOD1 in the cytosolic or mitochondrial fractions of MCF-10A, MCF7, and MDA-MD-231 cells. B, graphs of the quantification of SOD1 and SOD2 levels in the Western blots shown in A. C, electron microscopy of the mitochondria in MDA-MB-231cells treated with vehicle only (DMSO) or the SOD1 inhibitor LCS-1 for 24 h at a dose of 1.0 μm. D, electron microscopy of the mitochondria in MCF-10A treated with vehicle only (DMSO) or the SOD1 inhibitor LCS-1 for 24 h at a dose of 1.0 μm. E, mitochondrial superoxide levels in MDA-MB-231 cells treated with vehicle only (DMSO) or 1.0 μm LCS1 for 48 h. A representative experiment out of three replicates is shown. F, MDA-MB-231 cells were treated as in D, and samples were used for measurement of 2-hydroxyethidium (2-OH Mito E+) levels by HPLC. A representative experiment out of three replicates is shown. Statistical significance was determined using Student's t test.
SOD1 has been mainly studied in the context of amyotrophic lateral sclerosis, where it is mutated in the familial form of this devastating neurological disease (9). Although SOD1 is mainly localized to the cytoplasm, a fraction of SOD1 is reported to localize to the mitochondria (10–13). The importance of this small fraction of SOD1 in the mitochondria is best illustrated by the fact that the expression of a construct of SOD1 targeted to the mitochondria is sufficient to rescue the phenotype of the SOD1 knock-out mice (14). Therefore, although mitochondrial SOD1 represents a small fraction of total cellular SOD1, it is nevertheless of major physiological importance.
In terms of cancer biology, the deregulation of SOD2 by the deacetylase SIRT3 (1, 15) has been implicated in their metabolic reprogramming (4). Our data further add to these findings by showing that SOD2 itself is reduced in all breast cancer cell lines tested and further that this reduction occurs rapidly following oncogenic activation by Ras (Fig. 2, C–F).
Our data also support the notion that SOD1 may be an important target for cancer therapy. The identification of SOD1 as a potential anticancer target by the Varmus group arose from an unbiased screen of a library of compounds and the discovery of LCS-1 (7). Although we do not rule out that LCS1 inhibits the cytoplasmic fraction of SOD1, our data suggest that the mitochondrial fraction of SOD1 may be of critical importance for the maintenance of the integrity of the organelle. We propose that the overexpression of SOD1 in cancer cells may act as an adaptation mechanism to maintain total ROS levels in the mitochondria below a critical threshold so that the integrity of the organelle is maintained.
Therefore, our results suggest that although decreased activity of SOD2 and elevated ROS have been reported to be required for the metabolic reprogramming of cancer cells (4), increased SOD1 may assist this reprogramming by avoiding excessive ROS. We propose that by analogy to the “cadherin switch” during epithelial-mesenchymal transition (16), cancer cells may also use a “SOD switch” to orchestrate the requirement for the metabolic reprogramming, while avoiding irreversible damage to the mitochondria.
Acknowledgments
Tumor sections from the MMTV mouse models were obtained from the laboratory of Dr. Rafael Mira-Lopez. Electron microscopy was performed by the shared facility at Mount Sinai, which is supported by National Institutes of Health shared resources Grant 5R24 CA095823-04, National Science Foundation Major Research Instrumentation Grant DBI-9724504, and National Institutes of Health shared instrumentation Grant 1 S10RRO 9145-01. HPLC analysis of 2-hydroxyethidium was performed at the Donald Danforth Plant Science Center, Proteomics and Mass Spectrometry Core Facility, St. Louis, MO under the supervision of Dr. Bradley S. Evans through Science Exchange.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA109482 (to D. G.). This work was also supported by the Samuel Waxman Cancer Research Foundation.
- ROS
- reactive oxygen species
- SOD
- superoxide dismutase
- DMSO
- dimethyl sulfoxide
- MMTV
- murine mammary tumor virus.
REFERENCES
- 1. Tao R., Coleman M. C., Pennington J. D., Ozden O., Park S. H., Jiang H., Kim H. S., Flynn C. R., Hill S., Hayes McDonald W., Olivier A. K., Spitz D. R., Gius D. (2010) Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell 40, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu W., Ogasawara M. A., Huang P. (2007) Models of reactive oxygen species in cancer. Drug Discov. Today Dis. Models 4, 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bellot G. L., Liu D., Pervaiz S. (2013) ROS, autophagy, mitochondria and cancer: Ras, the hidden master? Mitochondrion 13, 155–162 [DOI] [PubMed] [Google Scholar]
- 4. Finley L. W., Carracedo A., Lee J., Souza A., Egia A., Zhang J., Teruya-Feldstein J., Moreira P. I., Cardoso S. M., Clish C. B., Pandolfi P. P., Haigis M. C. (2011) SIRT3 opposes reprogramming of cancer cell metabolism through HIF1a destabilization. Cancer Cell 19, 416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Radke S., Pirkmaier A., Germain D. (2005) Differential expression of the F-box proteins Skp2 and Skp2B in breast cancer. Oncogene 24, 3448–3458 [DOI] [PubMed] [Google Scholar]
- 6. Zielonka J., Hardy M., Kalyanaraman B. (2009) HPLC study of oxidation products of hydroethidine in chemical and biological systems: ramifications in superoxide measurements. Free Radic. Biol. Med. 46, 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Somwar R., Erdjument-Bromage H., Larsson E., Shum D., Lockwood W. W., Yang G., Sander C., Ouerfelli O., Tempst P. J., Djaballah H., Varmus H. E. (2011) Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines. Proc. Natl. Acad. Sci. U.S.A. 108, 16375–16380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zielonka J., Kalyanaraman B. (2010) Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic. Biol. Med. 48, 983–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X., et al. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 [DOI] [PubMed] [Google Scholar]
- 10. Higgins C. M., Jung C., Ding H., Xu Z. (2002) Mutant Cu, Zn superoxide dismutase that causes motoneuron degeneration is present in mitochondria in the CNS. J. Neurosci. 22, RC215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaarsma D., Rognoni F., van Duijn W., Verspaget H. W., Haasdijk E. D., Holstege J. C. (2001) CuZn superoxide dismutase (SOD1) accumulates in vacuolated mitochondria in transgenic mice expressing amyotrophic lateral sclerosis-linked SOD1 mutations. Acta Neuropathol. 102, 293–305 [DOI] [PubMed] [Google Scholar]
- 12. Mattiazzi M., D'Aurelio M., Gajewski C. D., Martushova K., Kiaei M., Beal M. F., Manfredi G. (2002) Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J. Biol. Chem. 277, 29626–29633 [DOI] [PubMed] [Google Scholar]
- 13. Okado-Matsumoto A., Fridovich I. (2001) Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu, Zn-SOD in mitochondria. J. Biol. Chem. 276, 38388–38393 [DOI] [PubMed] [Google Scholar]
- 14. Fischer L. R., Igoudjil A., Magrané J., Li Y., Hansen J. M., Manfredi G., Glass J. D. (2011) SOD1 targeted to the mitochondrial intermembrane space prevents motor neuropathy in the Sod1 knockout mouse. Brain 134, 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim H. S., Patel K., Muldoon-Jacobs K., Bisht K. S., Aykin-Burns N., Pennington J. D., van der Meer R., Nguyen P., Savage J., Owens K. M., Vassilopoulos A., Ozden O., Park S. H., Singh K. K., Abdulkadir S. A., Spitz D. R., Deng C. X., Gius D. (2010) SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hazan R. B., Qiao R., Keren R., Badano I., Suyama K. (2004) Cadherin switch in tumor progression. Ann. N.Y. Acad. Sci. 1014, 155–163 [DOI] [PubMed] [Google Scholar]
- 17. Papa L., Germain D. (2014) Sirt3 regulates the mitochondrial unfolded protein response. Mol. Cell. Biol. 34, 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]