Background: Dipeptidyl-peptidases (DPPs) are key factors for amino acid metabolism and bacterial growth of asaccharolytic Porphyromonas gingivalis.
Results: DPP5, which is specific for Ala and hydrophobic residues, is expressed in the periplasmic space of P. gingivalis.
Conclusion: DPP5 was discovered in prokaryotes for the first time.
Significance: The discovery of DPP5 expands understanding of amino acid and energy metabolism in prokaryotes.
Keywords: Amino Acid, Metabolism, Peptidases, Periodontal Disease, Subcellular Fractionation
Abstract
Porphyromonas gingivalis, a Gram-negative asaccharolytic anaerobe, is a major causative organism of chronic periodontitis. Because the bacterium utilizes amino acids as energy and carbon sources and incorporates them mainly as dipeptides, a wide variety of dipeptide production processes mediated by dipeptidyl-peptidases (DPPs) should be beneficial for the organism. In the present study, we identified the fourth P. gingivalis enzyme, DPP5. In a dpp4-7-11-disrupted P. gingivalis ATCC 33277, a DPP7-like activity still remained. PGN_0756 possessed an activity indistinguishable from that of the mutant, and was identified as a bacterial orthologue of fungal DPP5, because of its substrate specificity and 28.5% amino acid sequence identity with an Aspergillus fumigatus entity. P. gingivalis DPP5 was composed of 684 amino acids with a molecular mass of 77,453, and existed as a dimer while migrating at 66 kDa on SDS-PAGE. It preferred Ala and hydrophobic residues, had no activity toward Pro at the P1 position, and no preference for hydrophobic P2 residues, showed an optimal pH of 6.7 in the presence of NaCl, demonstrated Km and kcat/Km values for Lys-Ala-MCA of 688 μm and 11.02 μm−1 s−1, respectively, and was localized in the periplasm. DPP5 elaborately complemented DPP7 in liberation of dipeptides with hydrophobic P1 residues. Examinations of DPP- and gingipain gene-disrupted mutants indicated that DPP4, DPP5, DPP7, and DPP11 together with Arg- and Lys-gingipains cooperatively liberate most dipeptides from nutrient oligopeptides. This is the first study to report that DPP5 is expressed not only in eukaryotes, but also widely distributed in bacteria and archaea.
Introduction
Porphyromonas gingivalis, a Gram-negative rod, is a bacterium associated with severe forms of periodontal disease, which leads to loss of teeth in adults (1, 2). Over 47% of American adults suffer from periodontitis, the most common infectious disease in humans (3). In addition to dental diseases, recent studies have also reported that P. gingivalis is related with systemic diseases, such as cardiovascular diseases (4), decreased kidney function (5), and rheumatoid arthritis (6). The bacterium does not ferment carbohydrates, but grows under strict anaerobic conditions using proteinaceous substrates as carbon and energy sources. Although anaerobic energy production in P. gingivalis is poorly understood, biochemical examinations (7, 8), whole genome information (9, 10), and transcriptome analysis (11) have led to drafts of the configurations of the entire metabolism and transport pathways.
Glu and Asp, the most consumed amino acids in P. gingivalis (7, 8, 12), are located in the central pathway of amino acid metabolism (9, 13). They are mutually converted in the cytoplasm by δ-1-pyrroline-5-carboxylate dehydrogenase (PGN_1401) and ornithine aminotransferase (PGN_1403), respectively. Asp undergoes deamination by aspartate ammonia-lyase (PGN_0377) and is converted to fumarate, which is thought to be a major terminal electron acceptor of the respiratory chain in the organism. In addition to energy production, amino acid metabolism is important for understanding bacterial virulence, because the end products of short-chain fatty acids, e.g. butyrate, exhibit cytotoxicity against periodontal epithelial cells (14). In fact, the concentration of butyrate is increased in subgingival fluid in periodontal patients (15), and these compounds are proposed to be involved in the initiation and progression of these diseases (16). Hence, amino acid metabolism is an important issue for both anaerobic asaccharolytic metabolism and the pathogenicity of P. gingivalis.
To incorporate amino acids, P. gingivalis possesses potent proteolytic activities toward extracellular proteinaceous nutrients. First, proteins are degraded to oligopeptides by the cysteine endopeptidases Arg-gingipain (Rgp)3 and Lys-gingipain (Kgp) (17–20). Subsequently, oligopeptides are cleaved into dipeptides from the N terminus by dipeptidyl-peptidases (DPPs). To date, 3 DPPs, i.e. DPP4, DPP7, and DPP11, have been identified, of which DPP4 is highly specific for Pro and less potently for Ala at the penultimate position from the N terminus (P1) (MEROPS classification: S9.013) (21), whereas DPP7 prefers aliphatic and aromatic residues (22). We recently reported that DPP7 also releases dipeptides with a non-hydrophobic P1 residue, if the N-terminal (P2) residue is hydrophobic (23). The third and novel enzyme DPP11 (S46.002) shares a 38.7% amino acid sequence identity with DPP7 (S46.001), and is specific for Asp and Glu (24). When Pro is located at the third position from the N terminus, DPPs do not cleave the Xaa2–Pro3 bond, although prolyl tripeptidase A (PTP-A) cleaves the Pro3–Xaa4 bond (25). These liberated di- and tripeptides are efficiently incorporated into bacterial cells (8), possibly through oligopeptide transporters (13). The importance of DPPs and PTP-A for bacterial metabolism was confirmed in a study that showed that the growth of P. gingivalis with simultaneously deleted dpp4, dpp7, and ptp-A was retarded (26). Growth retardation has also been observed in a dpp11-disrupted strain (24).
The present study was performed to fully clarify the DPP members in P. gingivalis. Examination of dipeptide production repertoires among wild-type, gingipain-null, and dpp4, dpp7, and dpp11 triple gene-disrupted strains suggested the existence of additional DPP activities responsible for cleavage of Met-Leu-, Lys-Ala-, and Gly-Phe-methylcoumaryl-7-amide (MCA). Examination of putative peptidases revealed that PGN_0756 tentatively annotated as prolyl oligopeptidase, represented the remaining activity. Based on enzymatic properties and amino acid sequence similarity, PGN_0756 was identified as DPP5 (EC = 3.4.14.-, S09.012), which was found to be specific for Ala and hydrophobic amino acids at the P1 position. To our knowledge, this is the first report of DPP5 distribution in a prokaryotic organism.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes and DNA-modifying enzymes were purchased from Takara Bio (Otsu, Japan) and New England Biolabs (Ipswich, MA), and KOD-Plus-Neo DNA polymerase from Toyobo (Osaka, Japan). Oligonucleotide primers were from Fasmac (Atsugi, Japan). Commercially unavailable dipeptidyl-MCAs (Leu-Arg-, Val-Arg-, Leu-Lys-, Ala-Arg-, Ala-Asn-, Leu-Gln-, Gly-Arg-, Lys-Lys-, Thr-Asp-, Val-Asp-, Ile-Asp-, and His-Asp-MCA) were prepared from the appropriate peptidyl-MCA through digestion with thermolysin from Bacillus thermoproteolyticus rokko, bovine trypsin (Sigma), or staphylococcal glutamyl endopeptidase GluV8, as previously described (24). Benzyloxycarbonyl (Z)-Phe-Arg-, Arg-Arg-, t-butyloxycarbonyl (boc)-Val-Leu-Lys-MCA, and MCA peptides used to produce dipeptidyl MCA were obtained from the Peptide Institute (Osaka, Japan); Met-Leu-, Arg-Arg-, Gly-Phe-, Ser-Tyr-, Thr-Ser-, and Gly-Gly-MCA were from Bachem (Dubendorf, Switzerland). Leu-Asp-, Leu-Glu-, and Z-Leu-Leu-Gln-MCA were synthesized by Thermo Fisher Scientific (Ulm, Germany) and TORAY (Tokyo, Japan); and Lys-Phe-, Lys-Leu-, and Lys-Val-MCA by Scrum (Tokyo, Japan). Alkaline phosphatase-conjugated anti-rabbit Ig and streptavidin were purchased from Promega. Isoleucine thiazolidide hemi-fumarate (P32/98) was from Focus Biomolecules (Plymouth Meeting, PA), sitagliptin from Sigma, and vildagliptin from LKT Laboratories (St. Paul, MN). SYPRO Ruby protein gel stain was from Invitrogen. Formyl cellulofine was purchased from Seikagaku Corporation (Tokyo, Japan).
Bacterial Strains and Growth Conditions
P. gingivalis ATCC 33277 was purchased from American Type Culture Collection (Manassas, VA) and other strains used are shown in Table 1. Bacterial cells were grown in anaerobic bacteria culture media (Eiken Chemical, Tokyo, Japan) in the presence of menadione (1 μg/ml, Sigma) with or without appropriate antibiotics (0.7 μg/ml of tetracycline, 10 μg/ml each of ampicillin, erythromycin, and chloramphenicol) at 35 °C under an anaerobic condition (90% N2, 5% H2, 5% CO2). P. gingivalis cells were harvested in the early stationary phase and washed twice with ice-cold phosphate-buffered saline (PBS) at pH 7.4. Escherichia coli XL-1 Blue was cultured in Luria-Bertani broth containing 75 μg/ml of ampicillin for plasmid manipulation and expression of recombinant proteins.
TABLE 1.
P. gingivalis strains used in this study
| Strain | Genotype | Source or Ref. |
|---|---|---|
| ATCC 33277 | Wild-type | ATCC |
| KDP136 | kgp-2::cat rgpA2::[ermF ermAM] rgpB2::tetQ | 28 |
| NDP200 | dpp4::cepA | This study |
| NDP300 | dpp5:: [ermF ermAM] | This study |
| NDP400 | dpp7::tetQ | This study |
| NDP320 | dpp5::[ermF ermAM]dpp7::tetQ | This study |
| NDP210 | dpp4::cepA dpp7::tetQ | This study |
| NDP211 | dpp4::cepA dpp5::[ermF ermAM]dpp7::tetQ | This study |
| NDP212 | dpp4::cepA dpp5::[ermF ermAM]dpp7::tetQ dpp11::cat | This study |
| NDP511 | dpp4::cepA dpp7::tetQ dpp11:: [ermF ermAM] | This study |
Plasmids and Expression of DPPs in E. coli
DNA fragments encoding the full-length form of putative DPP3 (MER034609/PGN_1645: Val2–Glu906), putative prolyl oligopeptidase (MER034615/PGN_0756: Asn2–Lys691), and putative Ala-DPP (MER034613/PGN_1694: Lys2–Leu841) were obtained by PCR using genomic DNA. After digestion with BglII for DPP3, BamHI for prolyl oligopeptidase, and NcoI for Ala-DPP, the DNA fragment was inserted into the BamHI (DPP3 and prolyl oligopeptidase) or NcoI (Ala-DPP) site of pQE60. Expression plasmids for P. gingivalis DPP4 (MER004211/PGN_1469: Asp23–Leu723), DPP7 (MER014366/PGN_1479: Met3–Ile712), and DPP11 (MER034626/PGN_0607: Lys2–Pro720) were also used (23, 24). DPPs were expressed in E. coli as described (24). Briefly, overnight cultures of E. coli were added to 2 volumes of Luria-Bertani broth containing 75 μg/ml of ampicillin and 0.2 mm isopropyl β-d-thiogalactopyranoside, then further incubated at 30 °C for 4 h. Bacterial cells were lysed with lysis/washing buffer (20 mm Tris-HCl, pH 8.0, 0.1 m NaCl containing 10 mm imidazole) supplemented with 0.4 mg/ml of lysozyme and 10 μg/ml of leupeptin. Recombinant proteins were purified using a Talon metal affinity resin (Takara Bio, Tokyo, Japan) column according to the manufacturer's protocol. Bound proteins were eluted with 0.1 m imidazole (pH 8.0) containing 10% (v/v) glycerol and stored at −80 °C until use.
Construction of P. gingivalis Strains with Disrupted dpp Genes
To construct a dpp4 (PGN_1469) deletion mutant, 5′- and 3′-untranslated regions (UTR) of dpp4 were PCR amplified from the chromosomal DNA of P. gingivalis ATCC 33277 with sets of primers (DPP4-5R-1 and DPP4-5R-cep-comp, DPP3-5R-cep, and DPP4-cep-3R-1-comp, respectively) (Table 2). A cepA-gene cassette was amplified with a set of primers (cep-3R-1, 3R-comp) and pCR4-TOPO as a template. Nested PCR was performed with a mixture of the two UTR fragments and the cepA gene using primers (DP4-5R2, DP4-3R-com2), then the obtained 3-kb fragment was introduced into P. gingivalis by electroporation, resulting in NDP200 (dpp4::cepA). pUC119-SH-Δdpp7-tet (26) was linearized with ApaLI and introduced into NDP200, resulting in NDP210 (dpp4::cepA dpp7::tetQ). For disruption of the gene encoding PGN_0756 (annotated as prolyl oligopeptidase and identified as DPP5 in this study), both the 5′- and 3′-UTRs of the gene (PGN_0756) were amplified with sets of primers (5DPP5Up and 3DPP5Up-erm, 5ErmDPP5Up and 3Erm-DPP5Dwn, respectively), and an erythromycin-resistant gene cassette was amplified with a set of primers (5Erm-DPP5-Up, 3Erm-DPP5Dwn) and pYKP301 (26). Next, the second PCR was performed with a mixture of these fragments and primers (5DPP5Up, 3DPP5Dwn). The obtained DNA fragment was introduced into NDP210, resulting in NDP211 (dpp4::cepA dpp5::[ermF ermAM] dpp7::tetQ). To construct dpp4-5-7-11 deletion mutants, DNA fragments corresponding to the 5′-UTR of PGN_0608 and 3′-UTR of dpp11 (PGN_0607) were amplified with sets of primers (5DPP11Up-2 and 3DPP11Up-catR, 5DPP11Dwn-catR and 3DPP11Dwn-2, respectively), and the chloramphenicol-resistant gene cat was amplified from pACYC184 with primers (5CatR-DPP11Up, 3CatR-DPP11Dwn). A 3.5-kb DNA fragment composed of these fragments was amplified with a mixture of three DNA fragments and a set of primers (5DPP11Up-2, 3DPP11Dwn-2), then introduced into NDP211, resulting in NDP212 (dpp4::cepA dpp5::[ermF ermAM] dpp7::tetQ dpp11::cat). Similarly, to construct a dpp5-desrupted mutant, P. gingivalis ATCC 33277 was electroporated with a DNA fragment composed of the 5′- and 3′-UTR of dpp5 and the erythromycin gene cassette, resulting in NDP300 (dpp5::[ermF ermAM]); linearized pUC119-SH-Δdpp7-tet was introduced into NDP300, producing NDP320 (dpp5::[ermF ermAM]dpp7::tetQ). pUC119-SH-Δdpp7-tet was also introduced into both the wild-type and NDP500 (dpp11::[ermF ermAM]) (24), resulting in NDP400 (dpp7::tetQ) and NDP510 (dpp7::tetQ dpp11::[ermF ermAM]), respectively. NDP510 was further transformed with the dpp4::cepA fragment, resulting in NDP511 (dpp4::cepA dpp7::tetQ dpp11::[ermF ermAM]).
TABLE 2.
Primers used for the expression or disruption of DPP genes
Restriction sites are shown as italics. The first six primers on the list were used for expression and the others for gene disruption (see in detail in the text).
| Name | Sequence (5′-3′) |
|---|---|
| 5PgDPP3V2BglII | AACAGTAGATCTGTTTTATACGGGAGAGAAC |
| 3PgDPP3E906BglII | GAACATAGATCTCTCGTCCTTTGGGTAGAAG |
| 5PgDPP5N2BamHI | CGATCAGGATCCAACAAAAAAATCTTTTCGATGAT |
| 3PgDPP5K648BamHI | GACTCAGGATTCCTTCTTCAGCCAACGATCCAGCC |
| 5PgDPPAlaK2NcoI | TCAGACCATGGGCAAAAAATCATTACTC ATGCT |
| 3PgDPPAla L841NcoI | CTAGACCATGGAGAGATTCACAGGAGGATA GAG |
| DPP4-5R-1 | GTGGGCAACGTCAAGCTCT |
| DPP4-5R-cep-comp | TGCTTCACACCACGATCCATCTCTCGGCG |
| DPP4-5R-cep | GATGGATCGTGTGAAGCATCTTCGATGCTG |
| DPP4-cep-3R-1-comp | AGCAACTGAGCCGATAGTGATAGTGAACGG |
| cep-3R-1 | TCACTATCGGCTCAGTTGCTTGATGGGGC |
| 3R-comp | TAGCTCTCTCTCATATTTGCC |
| DP4-5R2 | GAGATCGAACTGCACAAACG |
| DP4-3R-comp2 | GCCTTATGTCATTTTTCAATCG |
| 5DPP5Up | CCGAAACAAGGGTAGGGGCTATC |
| 3DPP5Up-erm | CAATAGCGGAAGCTGATTTATTACTGTATTGCG |
| 5DPP5Dwn-erm | GATCCTCTAGAGCCCGTCACAAACGAAAGGAGG |
| 3DPP5Dwn | CGATACGAAAAGGGGGCTGTACGG |
| 5erm-DPP5Up | CAGTAATAAATCAGCTTCCGCTATTGCTTTTTTGC |
| 3erm-DPP5Dwn | CGTTTGTGACGGGCTCTAGAGGATCCCCGAAGCTG |
| 5DPP11Up-2 | GTTCCTGCCGAATGGAAATAGAAAG |
| 3DPP11Up-catR | GATTTTTTTCTCCATTATTACTTCGTTTTATTTTGT GTG |
| 5DPP11Dwn-catR | GGGCGGGGCGTAAAGCTATAAGAGGTGTGTCGAACGTC |
| 3DPP11Dwn-2 | ATGCTCAAATATACAAAATAACTCCC |
| 5catR-DPP11Up | TAAAACGAAGTAATAATGGAGAAAAAAATCACTGGATATAC |
| 3catR-DPP11Dwn | CACCTCTTATAGCTTTACGCCCCGCCCTGCCACTCATC |
Hydrolyzing Activity toward Dipeptidyl MCA
Recombinant peptidases (1–50 ng) were incubated with 20 μm dipeptidyl-MCA in 200 μl of reaction solution composed of 50 mm sodium phosphate (pH 7.0), 5 mm EDTA, and 0.1 m NaCl at 37 °C for 30 min. In several experiments, pH was varied using 50 mm sodium phosphate buffer and the NaCl concentrations were varied from 0 to 1.6 m. Fluorescence intensity was measured with excitation at 380 nm and emission at 460 nm with a Fluorescence Photometer F-4000 (Hitachi, Tokyo, Japan). To determine the enzymatic parameters, recombinant proteins (5 ng) were incubated with various concentrations of dipeptidyl-MCA (0.5–160 μm) in the presence of 0.1 m NaCl at pH 6.7. Data were analyzed using a nonlinear regression curve fitted to the Michaelis-Menten equation using the GraphPad Prism software program (San Diego, CA). Averages and S.E. were calculated from 4 independent measurements.
Subcellular Fractionations
All procedures were carried out at 4 °C or on ice according to a previously reported method (27), with slight modifications. Briefly, a 20-ml culture of P. gingivalis KDP136, a gingipain-null mutant (28), in the log-phase was centrifuged at 6,000 × g for 15 min. The supernatant was filtrated with a 0.45-μm membrane filter, and used for measurement of enzymatic activity as the extracellular fraction. A part of the supernatant was precipitated with 10% trichloroacetic acid on ice for 30 min and collected by centrifugation at 12,500 × g for 30 min, then washed with 30 ml of ice-cold acetone and used for SDS-PAGE and immunoblotting. Bacterial cells were washed with 40 ml of PBS, resuspended in 20 ml of PBS, and an aliquot was used as whole cells. Bacterial cells collected by centrifugation were resuspended in 4 ml of 0.25 m sucrose in 5 mm Tris-HCl (pH 7.5), and left for 10 min on ice. Cells were precipitated at 12,500 × g for 15 min, resuspended in 4 ml of 5 mm Tris-HCl, and mixed gently for 10 min to disrupt the outer membrane. The supernatant was obtained by centrifugation at 6,000 × g for 15 min and collected as the periplasmic fraction containing the outer membranes. The resultant precipitate was washed with 40 ml of PBS, then resuspended in 4 ml of PBS and used as spheroplasts. Spheroplast suspensions were sonicated by pulse for 10 times for 10 s each with a 2-s interval in an ice-water bath. After removal of unbroken spheroplasts and debris by centrifugation at 6,000 × g for 20 min, the supernatant (cytosolic fraction) was obtained by ultracentrifugation at 150,000 × g for 1 h, then the precipitate was suspended in 20 ml of PBS, centrifuged at 150,000 × g for 1 h, and then resuspended in 0.1 ml of PBS (inner membrane fraction).
Separately, a total membrane fraction was prepared according to a previously reported method (29), with slight modifications. Briefly, after washing with PBS, bacterial cells from a 200-ml culture were suspended in 20 ml of 10 mm HEPES (pH 7.5) containing 0.15 m NaCl, 0.2 mm phenylmethylsulfonyl fluoride, and 0.1 mg/ml of leupeptin, then disrupted in a French press at 100 m pascal in the presence of 10 μg/ml of RNase and 10 μg/ml of DNase through 2 passes. After centrifugation at 6,000 × g for 20 min, the supernatant was further centrifuged at 100,000 × g for 1 h. After removal of supernatant, the precipitate was suspended in 3 ml of HEPES buffer, and then a 1.5-ml aliquot (total membrane fraction) was separated by sucrose density gradient (26–68%, w/v) centrifugation at 100,000 × g for 48 h in a Hitachi RPS40T. Fractions (0.5 ml) were collected from the top.
Homology Modeling
Sequences for PGN_0756 were submitted to the ESyPred3D web server (30) and SWISS-MODEL. From the homology models constructed, one in pdb format was returned by the ESyPred3D. The program performs all steps of homology modeling including comparisons of amino acid sequences (PSI-BLAST of NCBI NR data bank then scored against the PDB), multiple sequence and pairwise alignments (combination of ClustalW, Dialign2, Match-Box, Multalin, and PRRP), and model building (MODELLER).
Miscellaneous
A polyclonal antibody against P. gingivalis PGN_0756 (DPP5), both DPP5- and anti-DPP5-antibody cellulofine resins were prepared as previously described (24). Native DPP5 were purified from the periplasmic and cytosolic fractions using anti-DPP5 antibody- cellulofine resin. Protein samples were separated on SDS-PAGE at a polyacrylamide concentration of 10%, and either stained with Coomassie Brilliant Blue or SYPRO Ruby, or electrically transferred to a polyvinylidene fluoride membrane and then subjected to immunoblotting analysis or N-terminal sequencing (24).
Antiserum against the 35-kDa hemin-binding protein (HBP35) localized in the outer membrane (31) was a kind gift from Dr. M. Shoji. The presence of the 13-kDa biotin-containing protein (BCP) localized in the inner membrane was visualized by blotting using alkaline phosphatase-conjugated streptavidin, according to a method reported in a previous study (32). Protein concentrations were determined using the bicinchoninic acid method (Pierce) with BSA as the standard.
RESULTS
Dipeptide Production in P. gingivalis Wild Type and KDP136
First, we determined cell-associated DPP and endopeptidase activities in P. gingivalis wild-type and gingipain-null mutant KDP136 cells (28). DPP activities in culture supernatant samples from both strains were negligible, as previously reported (24). In addition to activities toward boc-Phe-Ser-Arg-MCA by Rgp and boc-Val-Leu-Lys-/boc-Glu-Lys-Lys-MCA by Kgp, dipeptidyl substrates, such as Arg-Arg-MCA and Leu-Lys-, and Lys-Lys-MCA, were hydrolyzed in the wild-type cells, indicating the presence of Arg- and Lys-specific DPP activities (Fig. 1A). Both endopeptidase and DPP activities toward Arg and Lys were abolished in KDP136 cells, suggesting that Arg and Lys-DPP activities were mediated by Rgp and Kgp, respectively. Activities toward Gly-Pro-MCA by DPP4, Met-Leu-MCA by DPP7, and Leu-Asp-MCA by DPP11 were demonstrated to some extent in the wild-type, and interestingly, these activities were elevated in KDP136 cells, probably to compensate for the loss of gingipain activities. This phenomenon is in accord with a previous report that noted that DPP4 and PTP-A activities were elevated in a porT mutant strain that does not express functional gingipains (32). We also observed elevated DPP activities in a porT-disrupted strain (data not shown), further indicating that achievement of DPP activities in P. gingivalis is relevant to neither gingipain activities nor the type IX (PorSS) secretion system, which mediates the transport of gingipains.
FIGURE 1.
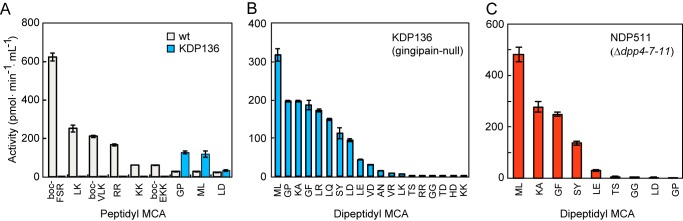
Peptidase activity of P. gingivalis. Peptidyl MCA-hydrolyzing activities of whole cells were determined. A, wild-type and KDP136; B, KDP136 (gingipain-null); C, NDP511 (Δdpp4-7-11). Values are shown as the mean ± S.D. (n = 3).
Detailed measurements of DPP activities in KDP136 cells showed that the hydrolyzing activity toward Met-Leu-MCA was predominant, followed by that for Gly-Pro-, Lys-Ala-, Gly-Phe-, Leu-Arg-, Leu-Gln-, Ser-Phe-, Leu-Asp, Leu-Glu-, and Val-Asp-MCA (Fig. 1B). This profile could be explained by actions of the 3 defined DPPs: i.e. Gly-Pro-MCA by DPP4; Lys-Ala-MCA by both DPP7 and DPP4; Met-Leu-, Gly-Phe-, Leu-Arg-, and Ser-Tyr-MCA by DPP7; and Leu-Asp-, Leu-Glu-, and Val-Asp-MCA by DPP11, whereas Leu-Gln-MCA may be cleaved by DPP7 because of its hydrophobic P2 preference (33). Several substrates such as Thr-Ser- and Gly-Gly-MCA were scarcely released by the bacterium, as suspected based on the substrate preferences of the 3 DPPs and a previous report that noted that no peptidase was found to be responsible for Thr, Ser, and Gly in P. gingivalis (34). Thr-Asp- and His-Asp-MCA were also scarcely hydrolyzed, because DPP11 prefers a hydrophobic P2 residue (24).
To address whether the 3 DPPs and gingipains are full members or an unidentified peptidase is still involved in dipeptide production, we produced mutant strain NDP511, in which dpp4, dpp7, and dpp11 were simultaneously disrupted. Although NDP511 convincingly lost activities for Gly-Pro-MCA mediated by DPP4, and for Leu-Asp- and Leu-Glu-MCA mediated by DPP11, we unexpectedly found significant hydrolyzing activities toward Met-Leu-, Lys-Ala-, Gly-Phe-, and Ser-Tyr-MCA, which had been implicated to be mediated by DPP7 (Fig. 1C). This result indicated the existence of an unidentified DPP possessing a DPP7-like substrate specificity.
Dipeptidyl Peptidase Activity of PGN_0756
We searched for candidates among the 2,090 protein-coding sequences of P. gingivalis ATCC 33277 (10), based on sequence similarity to the defined DPPs generally composed of 650–750 amino acid residues. Consequently, 3 genes for putative DPP3 (PGN_1645), putative prolyl oligopeptidase (PGN_0756), and putative Ala-DPP (PGN_1694) emerged. We expressed these in E. coli and studied the properties of the recombinant proteins. Apparent molecular weights were in accord with their calculated molecular masses (Fig. 2A). An attempt to detect the peptidase activity of putative Ala-DPP, listed as an unassigned peptidase in the S9C family (MER034613) in the MEROPS database (35), was not successful using currently available substrates, thus we did not analyze it further. The 100-kDa putative DPP3 (Mr = 102,897 in Val2–Glu906 with vector tag Mr = 1416.5) exhibited activity with a preference for Arg at the P1 position and the highest activity for Arg-Arg-MCA (Fig. 2B), in accord with the properties of eukaryotic (36) and Bacteroides thetaiotaomicron DPP3 (37). Therefore, PGN_1645 truly represents P. gingivalis DPP3. However, the observation that KDP136 did not hydrolyze Arg-Arg-MCA strongly suggested that DPP3 did not participate in extracellular dipeptide production in P. gingivalis and that it may be localized in the cytoplasm, as reported for B. thetaiotaomicron DPP3 (37).
FIGURE 2.
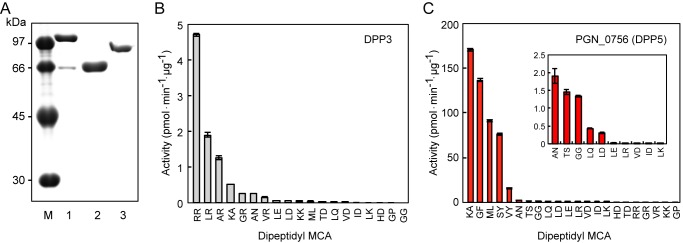
Expression and characterization of putative DPP3, PGN_0756 (DPP5), and Ala-DPP. A, recombinant proteins (0.5 μg) of putative DPP3 (lane 1), PGN_0756 (lane 2), and Ala-DPP (lane 3) were separated on SDS-10% (w/v) polyacrylamide gels. Lane M, low-molecular weight markers. The peptidase activities of DPP3 (B) and PGN_0756 (DPP5) (C), were determined with dipeptidyl MCA substrates. Values are shown as the mean ± S.D. (n = 3). Ala-DPP showed no DPP activity (see text).
In contrast to these unpromising results, the recombinant form of PGN_0756/MER034615 (Mr = 77,453 at Met1–Lys684) migrating as a 66-kDa band on SDS-PAGE demonstrated high dipeptidyl peptidase activity toward Lys-Ala-, Gly-Phe-, Met-Leu-, and Ser-Tyr-MCA, and it slowly hydrolyzed Val-Tyr-MCA (Fig. 2C). Interestingly, this repertoire substantially coincided with that of NDP511 (compare Figs. 1C and 2C).
Although the PGN_0756 gene has been annotated as prolyl oligopeptidase, because of sequence identity to the S9-family Pro-specific peptidases, it never hydrolyzed Gly-Pro-MCA (Fig. 2C) and we also found that it shares a 28.5% amino acid sequence identity with another S9 family member, Aspergillus fumigatus DPP5 (38). This homology value is even higher than that of Pro-specific P. gingivalis DPP4 (14.2%) and PTP-A (14.8%) (Fig. 3). Furthermore, in accordance with no preference for Pro, inhibitor analysis using P32/98 (39), sitagliptin (40), and vildagliptin (41), potent inhibitors of human DPP4, demonstrated that neither had an effect on Lys-Ala-MCA hydrolysis by PGN_0756, whereas these compounds completely abrogated the activity of P. gingivalis DPP4 toward Lys-Ala- and Gly-Pro-MCA (Fig. 4). These results certainly indicated that PGN_0756 is enzymatically distinct from prolyl oligopeptidase and DPP4, but it represents an orthologue of A. fumigatus DPP5. Thus, we designated PGN_0756 as DPP5, the fourth P. gingivalis DPP involved in extracellular production of dipeptides. Because DPP5 has been reported to be distributed only in eukaryotes to date (42), this is the first study to identify DPP5 in prokaryotes.
FIGURE 3.
Comparison of amino acid sequences related to S9 family peptidases. The amino acid sequences of P. gingivalis DPP5 (PgDPP5, pgn:PGN_0756), A. fumigatus DPP5 (AfDPP5, DPP5 ASPLC), P. gingivalis DPP4 (PgDPP4, pgn:PGN_1469), and P. gingivalis PTP-A (PgPTPA, pgn:PGN_1149) were aligned by COBALT. Hyphens represent gaps introduced for maximal matching. Common and conserved amino acids are marked by asterisks and dots, respectively. Underlines represent the N-terminal amino acid sequences determined with recombinant DPP5, and the yellow box is that of native DPP5. The signal peptide in A. fumigatus DPP5 is written in bold italics. Three amino acids essential for serine proteases are boxed in red. The amino acid sequence chosen for three-dimensional homology modeling is marked by an arrow (Pro33–Lys684), in which predicted β-strands and α-helixes shown in Fig. 12 are noted with red arrows and blue coils, respectively. The Glu195–Glu196 motif conserved in DPP4 (48) and a potential motif (Asp144-Glu145) in P. gingivalis DPP5 are boxed in green.
FIGURE 4.
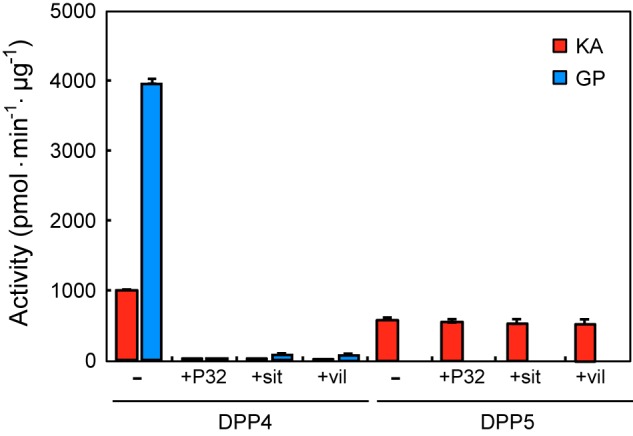
Effects of human DPP4 inhibitors on P. gingivalis DPP4 and DPP5 activities. The activities of DPP4 and DPP5 were determined with 20 μm Lys-Ala-MCA or Gly-Pro-MCA in the absence and presence of 250 μm P32/P98 (P32), sitagliptin (sit), and vildagliptin (vil).
Effects of dpp5 Gene Disruption in P. gingivalis
We constructed several types of dpp5-disrupted strains and examined their dipeptidyl peptidase activities toward Met-Leu-, Lys-Ala-, Gly-Phe-, and Ser-Tyr-MCA, which were hydrolyzed in the Δdpp4-7-11 strain NDP511. The activity for Met-Leu-MCA was moderately decreased in both the Δdpp5 (NDP300) (72.2%) and Δdpp7 (NDP400) (61.2%) strains, whereas the decrease was less significant in the Δdpp4 strain (NDP200) (83.0%) (Fig. 5). Activities toward Lys-Ala- and Gly-Phe-MCA were significantly reduced in the Δdpp5 strain to 23.3 and 14.9%, respectively, as compared with the wild-type, whereas the decrease was lowest in both Δdpp4 and Δdpp7 strains. These results suggest a major contribution of DPP5 with these hydrolyzes.
FIGURE 5.
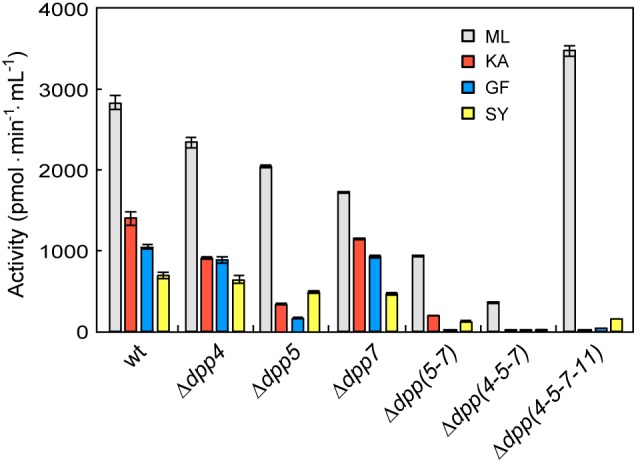
DPP activities of dpp-disrupted P. gingivalis strains. The peptidase activities of P. gingivalis ATCC 33277 wild type (wt), NDP200 (Δdpp4), NDP300 (Δdpp5), NDP400 (Δdpp7), NDP320 (Δdpp5-7), NDP211 (Δdpp4-5-7), and NDP212 (Δdpp4-5-7-11) were determined using 4 dipeptidyl-MCA substrates whose activities remained in NDP511 (Δdpp4-7-11) (see Fig. 2C). Values are shown as the mean ± S.D. (n = 3).
A Δdpp5-7-double knock-out strain (NDP320) resulted in abrogation of Gly-Phe-MCA hydrolysis, and significant decreases in Lys-Ala- and Ser-Tyr-MCA hydrolyzes. Met-Leu hydrolyzing activity was also diminished in NDP320, whereas these remaining activities were nearly abolished in the Δdpp4-5-7-triple knock-out strain (NDP211). NDP211 showed only 12.3% Met-Leu-MCA activity as compared with the wild type. It was also observed that NDP211 lost hydrolyzing abilities with Lys-Ala- and Ser-Tyr-MCA, indicating an additive contribution of DPP4 for their degradation. Taken together, P. gingivalis DPP5 truly participated in hydrolysis of the 4 substrates.
We also found that Met-Leu-MCA hydrolyzing activity was markedly (9.8-fold) increased in the Δdpp4-5-7-11 quadruple-disrupted strain (NDP212) as compared with triple-disrupted NDP211 and was even higher than that of the wild-type. Similarly, Ser-Tyr-MCA hydrolysis was increased by 6.3-fold in NDP212 over that in NDP211. The overshoot of activity for Met-Leu-MCA strongly suggested expressional induction of an unknown peptidase(s) to compensate for the four-DPP defects. We also observed an increase in activities for succinyl-Leu-Leu-Val-Tyr- and Z-Leu-Leu-Leu-MCA in NDP212 (data not shown), suggesting that the entity mediating the hydrolysis of Met-Leu-MCA was not DPP, but likely an endopeptidase with preference for hydrophobic residues.
N-terminal Sequence Analysis
The N terminus of the recombinant 66-kDa molecule expressed from Asn2 to Lys684 was Lys4 (Fig. 3), indicating cleavage at the Lys3–Lys4 bond in E. coli, and thus, a non-requirement of N-terminal processing to exert the DPP activity. Moreover, a minor species with the N terminus of Ala17 was also present. On the other hand, the N terminus of native 66-kDa DPP5 purified by anti-DPP5 antibody-cellulofine was Thr25 (Fig. 3), indicating the cleavage at the Gly24–Thr25 bond.
Size Exclusion Gel Chromatography
Recombinant DPP5 was subjected to size exclusion gel chromatography by use of Sephacryl S200HR. DPP5 was eluted as a single peak at 140 kDa, indicating its dimeric conformation under non-denaturing conditions (data not shown).
NaCl and pH Dependence
DPP5 activity for Lys-Ala-MCA showed a bell-shaped dependence on NaCl concentration, with the maximal activity at 0.1 m (Fig. 6A). We also examined the effects of NaCl on DPP4 and DPP7 activities, because both DPPs could hydrolyze Lys-Ala-MCA. DPP4 hydrolyzed Lys-Ala-MCA at the maximal rate in the absence of NaCl and the activity was gradually decreased at higher concentrations. DPP7 showed very low, but constant activity at a wide range of NaCl concentrations.
FIGURE 6.
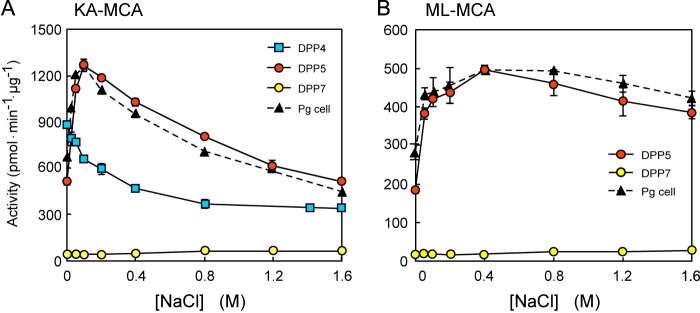
Effects of NaCl on activities of DPPs. A, the activities of DPP4 (square), DPP5 (red circle), and DPP7 (yellow circle) were determined with Lys-Ala-MCA at 0–1.6 m NaCl. The activity of P. gingivalis wild-type cells was also determined and plotted as an arbitrary unit (triangle). Values are shown as the mean ± S.D. (n = 3). B, the activities of DPP5 (red circle) and DPP7 (yellow circle) were determined with Met-Leu-MCA. The activity of P. gingivalis cells (triangle) was also determined and plotted as an arbitrary unit. Values are shown as mean ± S.D. (n = 3).
The salt dependence of DPP5 for Met-Leu-MCA (Fig. 6B), as well as Gly-Phe- and Ser-Tyr-MCA (data not shown) was similar to that of Lys-Ala-MCA, although changes in the activity became moderate and the optimal NaCl concentration was shifted from 0.1 to 0.4 m. Although DPP7 most efficiently hydrolyzed Met-Leu-MCA among the dipeptidyl MCAs tested (23), the activity was nearly constant and substantially lower than that of DPP5 at all NaCl concentrations. DPP4 and DPP11 do not hydrolyze Met-Leu-MCA (23). It should be noted that the activities in P. gingivalis for Lys-Ala- and Met-Leu-MCA exhibited nearly the same NaCl dependence as those of DPP5, strongly suggesting that these activities are mostly mediated by DPP5 in the bacterial cells.
The optimal pH for DPP5 activity toward Lys-Ala-MCA was 6.7 (Fig. 7), which is identical to that of A. fumigatus DPP5 (38). The pH optimum of DPP7 was slightly shifted to basic pH around 7 and that of DPP4 was further to pH 7.5 in consistent to previous studies (21, 22).
FIGURE 7.
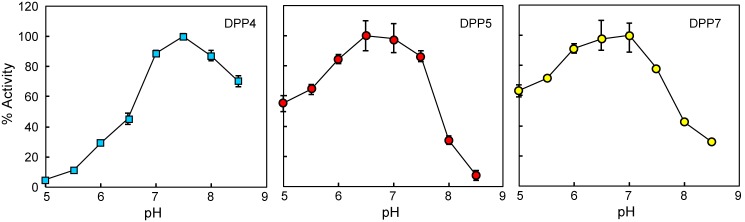
pH profile of DPP4, DPP5, and DPP7. The activities of DPP4, DPP5, and DPP7 were determined with Gly-Pro-, Lys-Ala-, and Met-Leu-MCA, respectively, at pH 5–8.5, in 50 mm phosphate buffer without (DPP4 and DPP7) or with 0.1 m NaCl (DPP5). Values are shown as the mean ± S.D. (n = 3).
Substrate Specificity of DPP5 Compensates That of DPP7
Enzymatic parameters of DPP5 were determined by fitting a nonlinear regression curve to the Michaelis-Menten equation at pH 6.7 in the presence of 0.1 m NaCl (Table 3). DPP5 possessed the smallest Km for Lys-Leu-MCA (145.7 μm), followed by Lys-Phe-MCA (151.5 μm), and possessed the largest kcat/Km value for Gly-Phe-MCA (13.01 μm−1 s−1), followed by Lys-Ala-MCA (11.02 μm−1 s−1). As previously reported (33), the substrates with hydrophobic P1 residue could also be hydrolyzed by DPP7 and the order of Thr-Ser hydrolysis activity by DPP7 was Met-Leu-, Ser-Tyr-, Gly-Phe-, and Lys-Ala-MCA, with kcat/Km values of 10.62, 3.92, 1.17, and 0.32 μm−1 s−1, respectively. Therefore, when comparing the activities between DPP5 and DPP7, the superiority of P1 position Ala should be reinforced in DPP5. To further address P1 position specificity, we synthesized 3 additional MCA substrates with Lys fixed as the P2 position residue and altered P1 position residues, i.e. Ala, Phe, Leu, and Val. DPP5 had the largest kcat/Km value for Lys-Ala-MCA, followed by Lys-Phe- and Lys-Leu-MCA, and possessed the smallest value for Lys-Val-MCA (Table 3).
TABLE 3.
Enzymatic parameters of P. gingivalis DPP5 and hydrophobic indexes of amino acids at the P1 position
Enzymatic parameters of DPP5 were determined at 37 °C at pH 6.7 in the presence of 0.1 m NaCl. Values are shown as the mean ± S.E. (n = 3).
| MCA peptide | kcat | Km | kcat/Km | Hydrophobic index at P2 position | Hydrophobic index at P1 position |
|---|---|---|---|---|---|
| s−1 | μm | μm−1 s−1 | |||
| Gly-Phe- | 5638.1 ± 403.2 | 396.1 ± 51.3 | 13.01 | 0 | 100 |
| Lys-Ala- | 7577.4 ± 637.4 | 687.6 ± 11.0 | 11.02 | −23 | 41 |
| Met-Leu- | 5561.9 ± 701.3 | 701.4 ± 103.8 | 7.93 | 74 | 97 |
| Lys-Phe- | 743.6 ± 55.2 | 151.5 ± 55.2 | 4.91 | −23 | 100 |
| Ser-Tyr- | 1329.0 ± 100.0 | 296.1 ± 30.7 | 4.49 | −5 | 63 |
| Lys-Leu- | 489.8 ± 20.3 | 145.7 ± 10.2 | 3.36 | −23 | 97 |
| Lys-Val- | 299.3 ± 17.8 | 219.8 ± 19.6 | 1.36 | −23 | 76 |
The values obtained for kcat/Km strongly suggested that Ala and hydrophobic amino acids are suitable at the P1 position for the activity of DPP5. This substrate specificity was roughly in accord with that of A. fumigatus DPP5, although it has been reported to further hydrolyze His-Ser-2-(4-methoxy)naphthylamide (38). Therefore, hydrolyzing activity especially toward His-Ser was a major obstacle for comprehensive understanding of DPP5 substrate specificity. To obtain general information, activities toward 21 dipeptidyl-MCA substrates were plotted against hydrophobic indexes (43) of P1 and P2 position residues (Fig. 8, A and B). Consequently, a strong positive correlation was observed between the log of activities and hydrophobic indexes of the P1 position residues. In contrast, no correlation was recognized with hydrophobic indexes of the P2 position residues. Taken together, we concluded that DPP5 prefers hydrophobic residues and Ala at the P1 position, and has no hydrophobic preference at the P2 position.
FIGURE 8.
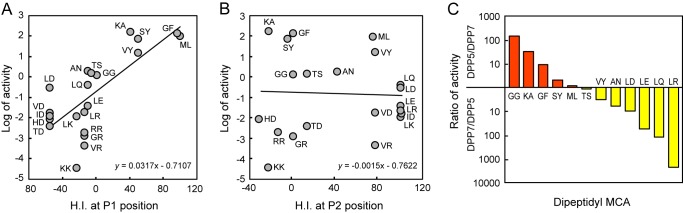
Amino acid preference at P1 and P2 positions of DPP5. Peptidase activities of DPP5 were determined in the presence of 0.1 m NaCl, then plotted against hydrophobic index values (H.I.) (43) of P1 (A) and P2 (B) position residues. Regression lines were calculated for P1 residues with R2 = 0.472 (p = 0.0004) and P2 residues with R2 = 0.0015 (p = 0.862). C, the ratio of the activity of DPP5 (Fig. 2C) per that of DPP7 (33) was plotted from the substrates ranked in the top 10 of either DPP5 or DPP7.
When the top 10 cleavable substrates for either DPP5 or DPP7 were aligned in respect to the ratios of DPP5 to DPP7 activities, Gly-Gly-, Lys-Ala-, Gly-Phe-, and Ser-Tyr-MCA were preferentially hydrolyzed by DPP5 (Fig. 8C). On the other hand, Leu-Arg-, Leu-Gln-, Leu-Glu-, Leu-Asp-, Ala-Asn-, and Val-Tyr-MCA were preferentially hydrolyzed by DPP7. This selectivity can be primarily explained by the properties of DPP5 (Fig. 2C) and those of DPP7 reported in our previous study (23), i.e. hydrophobic and Ala preference at the P1 position and no preference at P2 of DPP5, and hydrophobic preference at the P1 and P2 positions of DPP7.
Subcellular Localization of DPP5 and Growth of Four DPP Gene-disrupted P. gingivalis
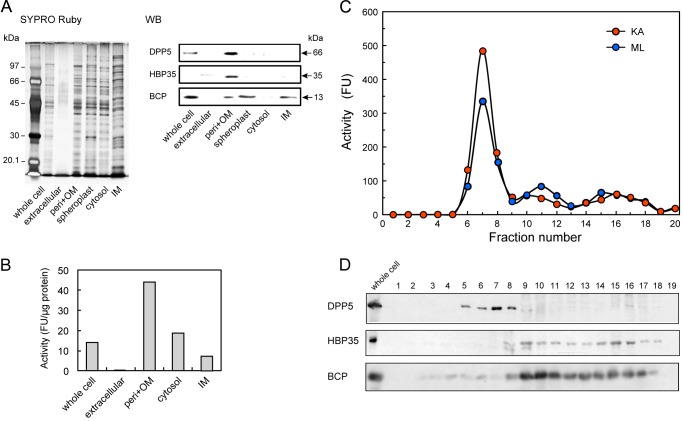
Subcellular localization of DPP5 was investigated with KDP136, to avoid the effect of massive proteolytic degradation by gingipains. Western blotting demonstrated that DPP5 and HBP35 were predominantly localized in the periplasm/outer membrane fraction (Fig. 9). The highest DPP5 activity toward Lys-Ala-MCA was shown in the periplasm/outer membrane fraction, although a part of that activity was also observed in both the cytosol and inner membrane fractions. Noticeably, Lys-Ala-MCA hydrolyzing activity was not observed in the extracellular fraction. Periplasmic localization of DPP5 was ascertained by sucrose density gradient centrifugation of unwashed total membrane fractions. The activity monitored by hydrolyzes of Lys-Ala- and Met-Leu-MCA, and the 66-kDa DPP5 immunoblot were detected in the upper fractions (Fig. 9, C and D, fractions 5–8). In contrast, HPB35 was observed predominantly in the outer membrane (fractions 14–16) and BCP was in the inner membrane fractions (fractions 9–11). These results clearly indicated predominant periplasmic localization of DPP5 in P. gingivalis.
FIGURE 9.
Subcellular localization of DPP5. A, whole cell, extracellular fraction, periplasmic fraction containing the outer membranes (peri+OM), spheroplast, cytosol, and the inner membrane fraction (IM) (1 μg of proteins) were separated on SDS-PAGE, then proteins were visualized with SYPRO Ruby. DPP5 and HBP35 were detected by immunoblotting, and a 13-kDa biotin-containing protein (BCP) by streptavidin. Lane LM, low-molecular weight marker. B, the activity toward Lys-Ala-MCA was determined with the same fractions shown in panel A without spheroplast. C, the total membrane fraction was subjected to sucrose density gradient centrifugation. The separated samples were fractionated (0.5-ml each) from the top of the tube, then an aliquot (5 μl) was used to determine the activity with Lys-Ala- and Met-Leu-MCA. D, fractions of panel C were separated on SDS-PAGE, and DPP5, HBP35, and BCP were detected.
The growth of NDP212, which lost 4 identified DPP genes, was significantly more retarded than the wild-type when the organisms were cultured in anaerobic bacteria culture medium (Fig. 10). However, growth retardation in KDP136 was more remarkable, suggesting that oligopeptidase production by gingipains is more critical under the present culture condition.
FIGURE 10.
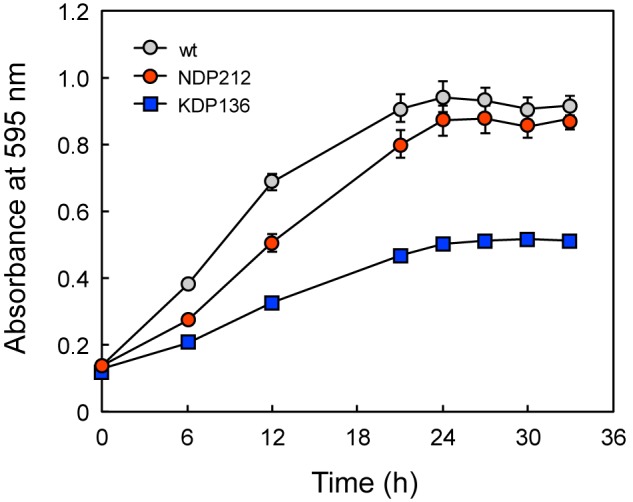
Effects of DPP and gingipain gene disruptions on cell growth. P. gingivalis ATCC 33277 wild-type (gray circle), NDP212 (Δdpp4-5-7-11, red circle), and KDP136 (gingipain-null, square) were grown at 35 °C, and absorbance at 595 nm was monitored. Values are shown as the mean ± S.D. (n = 3).
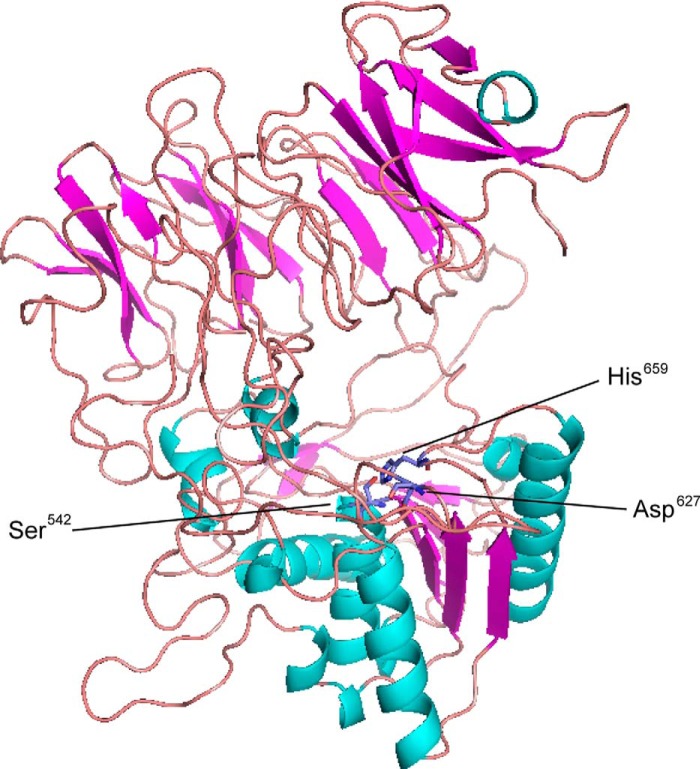
Modeling of Three-dimensional Structure of DPP5
Three-dimensional structures of DPP5 were predicted by submission of the full amino acid sequences to the fully automated protein structure homology-modeling server ESyPred3D (30). As shown in Fig. 11, a predicted structure was obtained by use of Stenotrophomas maltophilia DPP4 (Protein Data Bank code 2ecf) (44) as a template, which shares a 21.0% amino acid identity with P. gingivalis DPP5. Similar to that structure, this model contained a C-terminal peptidase domain with an α/β hydrolase-fold, and the catalytic triad (Ser542, Asp627, His659) exists as a large cavity formed between an N-terminal propeller domain-like region and the C-terminal peptidase domain.
FIGURE 11.
Modeling of three-dimensional structure of P. gingivalis DPP5. Homology modeling of the three-dimensional structure of P. gingivalis DPP5 was performed using the ESyPred3D program (30), which utilized the structure of S. maltophilia DPP4 (Protein Data Bank code 2ecf) (44) as a platform. The diagram was drawn using PDBjViewer, and β-strands and α-helixes shown are noted with red arrows and blue coils, respectively. Three amino acids of the catalytic triad are indicated.
DISCUSSION
In the present study, we identified and characterized P. gingivalis DPP5 encoded by the PGN_0756 gene, which has been annotated as prolyl oligopeptidase. This is the first report of DPP5 being demonstrated in prokaryotes, and it is the fourth DPP known to be expressed in the bacterium. P. gingivalis DPP5 is composed of 684 amino acids, exists as a dimeric form and is predominantly localized in the periplasm. The N terminus of native DPP5 was Thr25, indicating Met1–Gly24 as a signal peptide.
DPP5 is classified in the S9 serine peptidase family (EC = 3.4.14.-), which liberates N-terminal dipeptide Xaa–Yaa from a polypeptide, where Yaa is Ala and hydrophobic residues except Pro, with kcat/Km values of 1–13 μm−1 s−1, and no preference for Xaa, showing the highest activity in the presence of 0.1–0.4 m NaCl with an optimum pH of 6.5–7.0. This pH optimum is identical to A. fumigatus DPP5 (38). The substrate specificity of DPP5 partially overlaps and also compensates for that of DPP7, thus expanding the dipeptide production repertoire in P. gingivalis.
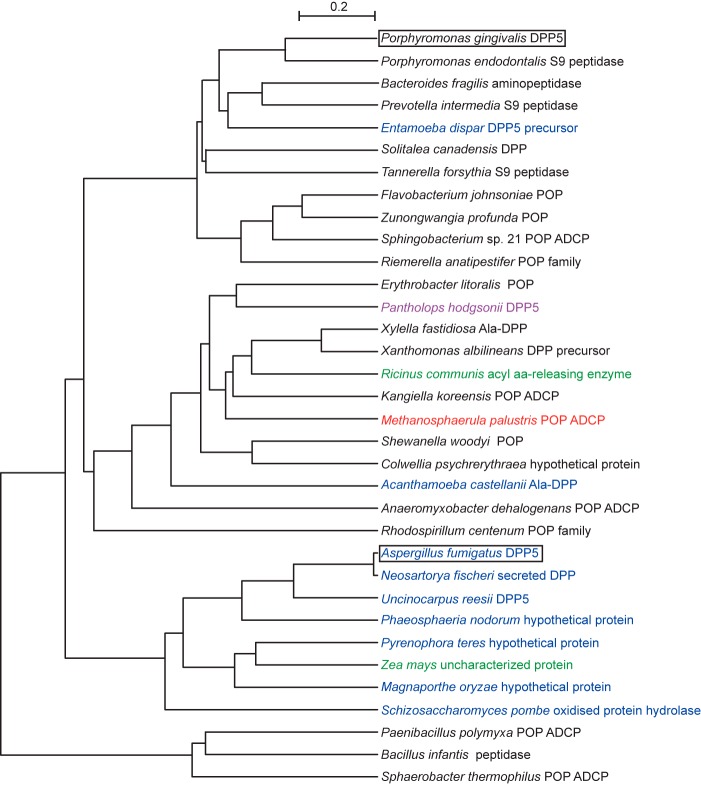
The amino acid identity between P. gingivalis and A. fumigatus DPP5 is 28%, although they exhibited similar enzymatic characteristics. Until recently, 26 proteins had been nominated as DPP5 in fungi, including Trichphyton rubrum, Ajellomyces capsulata, and Schizosscharomyces pombe. Based on the discovery of P. gingivalis DPP5, a homology search by KEGG BLASTP produced a long list of P. gingivalis DPP5-like molecules, in which the 250 proteins were obtained at the amino acid identity higher than 25% (supplemental Table S1). Those are distributed in eubacteria including the phyla Bacteroidetes, Proteobacteria, Firmicutes, Actinobacteria, Chloroflexi, Acidobacteria, Chloroflexi, archaeon (Methanosphaerula palustris), and eukaryotes including Fungi, Plantae, and Animalia. Currently, they are annotated as either prolyl oligopeptidase, Ala-DPP, unclassified DPP, or a hypothetical protein, etc., except for the cases of fungal DPP5. At least, we confirmed that an S9 peptidase from Porphyromonas endodontalis possessed the substrate specificity identical to that of P. gingivalis DPP5.4
DPP5, prolyl oligopeptidase, as well as DPP4 belong to the S9 peptidase family, which is divided into four subclasses based on the sequence characteristics around the essential Ser542: GGSXGGLL, where X is typically Asn or Ala (S9A), GWSYGGY (9SB), GGSYGG (S9C), and GGHSYGAFMT (S9D) (35). On this criterion, fungal and P. gingivalis DPP5 are categorized in the S9C. Similarly, 98% of 250 homologues of P. gingivalis DPP5 possess the sequence G(G/A)S(Y/F/W)GG. Thus, if substitution from Gly to Ala at the second position and from Tyr to Phe or Trp at the fourth position are admitted, most of the homologues are members of S9C. In other words, the S9C subfamily may be equivalent to DPP5. Relevant distance among 34 representatives is observed in a phylogenetic tree (Fig. 12), which indicates that DPP5 is ubiquitously distributed within bacteria, eukaryotic species, as well as archaea and plays a role in amino acid metabolism. In addition, bacterial DPP5 especially in pathogenic bacteria may function as a virulence factor, the same as in fungi (38, 45). It should be prudently noted that molecules from Animalia (Pantholops hodgsonii) and those from Plantae (Ricinus communis and Zea mays) will be evaluated in further studies.
FIGURE 12.
Phylogenetic tree of P. gingivalis DPP5-related proteins. A phylogenic tree of 34 representatives (supplemental Table S1) was constructed by rooted phylogenetic tree with branch length (UPGMA) with the CLUSTAL W Multiple Sequence Alignment Program. P. gingivalis and A. fumigatus DPP5 are boxed. Members were from eubacteria (black), archaea (red), animal (violet), plant (green), and other eukaryotic species (blue). Prolyl oligopeptidase ADCP, peptidase S9 prolyl oligopeptidase active site domain-containing protein.
In contrast to fungal DPP5 (38) and the P. gingivalis predominant endopeptidase gingipain (17–20, 46), P. gingivalis DPP5 is neither secreted nor included in vesicles, but rather predominantly localized in the periplasm, which seems suitable for its function to produce nutrient dipeptides. In the periplasm, diffusion of products is limited and they could be readily incorporated into the cells via transporters localized in the inner membrane. P. gingivalis DPP5 does not possess a C-terminal conserved sequence for anchoring with the outer membrane. In addition, subcellular translocation of DPP5 was independent of the type IX secretion system (PorSS) that mediates transportation of outer membrane-associated proteins including gingipains (46) and HBP35 (31), because the porT mutant did not show lateralization of the localization and activity of DPP5.5
Although the dpp5 gene PGN_0756 has been annotated as a prolyl oligopeptidase, it did not exhibit Pro specificity, but preferred Ala and hydrophobic amino acids. It has been reported that the S1 pocket of DPP4 is hydrophobic and small, which accounts for the Pro and Ala restrictions at this position (47). Hence, it could be speculated that the S1 pocket shares some similarity between DPP5 and DPP4, whereas that of DPP5 is further specialized to exclude Pro. This speculation was supported by the finding that the DPP4-specific inhibitors had no effect on the activity of DPP5. Furthermore, in contrast to their P1 preference, the similarity between DPP5 and DPP4 could be noted in the S2 pocket, i.e. both had no preference at the P2 residue, indicating similarity of the fine structure of the S2 pocket. Structurally, it has been reported that the Glu-Glu motif localized in the N-terminal propeller domain is conserved in P. gingivalis DPP4 (Glu195–Glu196, Fig. 3) and is essential for recognition of the N-terminal amine in a substrate peptide (48). However, as shown in the multiple sequence alignment, this motif is not present in P. gingivalis DPP5. Instead, Asp144–Glu145 of DPP5 might function as that motif. To understand the molecular mechanism of the substrate preference of DPP5, structural analysis using crystallography is required and a study by our group is in progress.
In conclusion, DPP5 was identified and biochemically characterized for the first time in the prokaryotic species P. gingivalis. In the bacterium, four DPPs, i.e. DPP4, DPP5, DPP7, and DPP11, are the major members together with gingipains responsible for dipeptide production. Such a combination of peptidases with altered substrate specificities certainly expands the dipeptide production repertoire, providing benefit for P. gingivalis to obtain proteinaceous energy sources from the environment.
Supplementary Material
Acknowledgments
We express our gratitude to Dr. M. Shoji (Nagasaki University) for providing the anti-HGP35 antiserum and helpful discussions. We also thank Dr. K. Sato, Dr. H. Yukitake (Nagasaki University), Dr. K. Ito (Kyushu University of Health and Welfare), and Dr. Y Nakajima (Setsunan University) for helpful discussions.
This work was supported in part by Grants-in-aid for Scientific Research from the Japan Society for the Promotion of Science (to Y. O.-N., T. K. N., T. K., and S. K.) and a grant from the Institute for Fermentation, Osaka (to T. K. N.).

This article contains supplemental Table S1.
S. M. A. Rouf, Y. Ohara-Nemoto, and T. K. Nemoto, unpublished observation.
Y. Ohara-Nemoto, T. Ono, and T. K. Nemoto, unpublished observation.
- Rgp and Kgp
- Arg- and Lys-gingipains, respectively
- BCP
- the 13-kDa biotin-containing protein
- HBP35
- the 35-kDa hemin-binding protein
- boc-
- t-butyloxycarbonyl-
- DPP
- dipeptidyl-peptidase
- MCA
- 4-methylcoumaryl-7-amide
- P32/98
- isoleucine thiazolidide hemifumarate
- PTP-A
- prolyl tripeptidase A
- Z
- benzyloxycarbonyl.
REFERENCES
- 1. Lamont R. J., Jenkinson H. F. (1998) Life below the gum line. Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62, 1244–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holt S. C., Ebersole J. L. (2005) Porphyromonas gingivalis, Treponema denticola, and Tanneralla forsythia. The “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000 38, 72–122 [DOI] [PubMed] [Google Scholar]
- 3. Eke P. I., Dye B. A., Wei L., Thornton-Evans G. O., Genco R. J. (2012) Prevalence of periodontitis in adults in the United States, 2009 and 2010. J. Dent. Res. 91, 914–920 [DOI] [PubMed] [Google Scholar]
- 4. Iwai T., Inoue Y., Umeda M., Huang Y., Kurihara N., Koike M., Ishikawa I. (2005) Oral bacteria in the occluded arteries of patients with Buerger disease. J. Vasc. Surg. 42, 107–115 [DOI] [PubMed] [Google Scholar]
- 5. Kshirsagar A. V., Offenbacher S., Moss K. L., Barros S. P., Beck J. D. (2007) Antibodies to periodontal organisms are associated with decreased kidney function. The Dental atherosclerosis risk in communities study. Blood Purif. 25, 125–132 [DOI] [PubMed] [Google Scholar]
- 6. Detert J., Pischon N., Burmester G. R., Buttgereit F. (2010) The association between rheumatoid arthritis and periodontal disease. Arthritis Res. Ther. 12, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takahashi N., Sato T., Yamada T. (2000) Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by Porphyromonas gingivalis. J. Bacteriol. 182, 4704–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takahashi N., Sato T. (2001) Preferential utilization of dipeptides by Porphyromonas gingivalis. J. Dent. Res. 80, 1425–1429 [DOI] [PubMed] [Google Scholar]
- 9. Nelson K. E., Fleischmann R. D., DeBoy R. T., Paulsen I. T., Fouts D. E., Eisen J. A., Daugherty S. C., Dodson R. J., Durkin A. S., Gwinn M., Haft D. H., Kolonay J. F., Nelson W. C., Mason T., Tallon L., Gray J., Granger D., Tettelin H., Dong H., Galvin J. L., Duncan M. J., Dewhirst F. E., Fraser C. M. (2003) Complete genome sequence of the oral pathogenic Bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185, 5591–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naito M., Hirakawa H., Yamashita A., Ohara N., Shoji M., Yukitake H., Nakayama K., Toh H., Yoshimura F., Kuhara S., Hattori M., Hayashi T., Nakayama K. (2008) Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 15, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Høvik H., Yu W. H., Olsen I., Chen T. (2012) Comprehensive transcriptome analysis of the periodontopathogenic bacterium Porphyromonas gingivalis W83. J. Bacteriol. 194, 100–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mazumdar V., Snitkin E. S., Amar S., Segrè D. (2009) Metabolic network model of a human oral pathogen. J. Bacteriol. 191, 74–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meuric V., Rouillon A., Chandad F., Bonnaure-Mallet M. (2010) Putative respiratory chain of Porphyromonas gingivalis. Future Microbiol. 5, 717–734 [DOI] [PubMed] [Google Scholar]
- 14. Tsuda H., Ochiai K., Suzuki N., Otsuka K. (2010) Butyrate, a bacterial metabolite, induces apoptosis and autophagic cell death in gingival epithelial cells. J. Periodont. Res. 45, 626–634 [DOI] [PubMed] [Google Scholar]
- 15. Niederman R., Buyle-Bodin Y., Lu B. Y., Robinson P., Naleway C. (1997) Short-chain carboxylic acid concentration in human gingival crevicular fluid. J. Dent. Res. 76, 575–579 [DOI] [PubMed] [Google Scholar]
- 16. Pöllänen M. T., Salonen J. I. (2000) Effect of short chain fatty acids on human gingival epithelial cell keratins in vitro. Eur. J. Oral Sci. 108, 523–529 [DOI] [PubMed] [Google Scholar]
- 17. Chen Z., Potempa J., Polanowski A., Wikstrom M., Travis J. (1992) Purification and characterization of a 50-kDa cysteine proteinase (gingipain) from Porphyromonas gingivalis. J. Biol. Chem. 267, 18896–18901 [PubMed] [Google Scholar]
- 18. Fujimura S., Shibata Y., Nakamura T. (1992) Comparative studies of three proteases of Porphyromonas gingivalis. Oral Microbiol. Immunol. 7, 212–217 [DOI] [PubMed] [Google Scholar]
- 19. Pike R., McGraw W., Potempa J., Travis J. (1994) Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J. Biol. Chem. 269, 406–411 [PubMed] [Google Scholar]
- 20. Kadowaki T., Yoneda M., Okamoto K., Maeda K., Yamamoto K. (1994) Purification and characterization of a novel arginine-specific cysteine proteinase (argingipain) involved in the pathogenesis of periodontal disease from the culture supernatant of Porphyromonas gingivalis. J. Biol. Chem. 269, 21371–21378 [PubMed] [Google Scholar]
- 21. Banbula A., Bugno M., Goldstein J., Yen J., Nelson D., Travis J., Potempa J. (2000) Emerging family of proline-specific peptidases of Porphyromonas gingivalis. Purification and characterization of serine dipeptidyl peptidase, a structural and functional homologue of mammalian prolyl dipeptidyl peptidase IV. Infect. Immun. 68, 1176–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banbula A., Yen J., Oleksy A., Mak P., Bugno M., Travis J., Potempa J. (2001) Porphyromonas gingivalis DPP-7 represents a novel type of dipeptidylpeptidase. J. Biol. Chem. 276, 6299–6305 [DOI] [PubMed] [Google Scholar]
- 23. Rouf S. M., Ohara-Nemoto Y., Hoshino T., Fujiwara T., Ono T., Nemoto T. K. (2013) Discrimination based on Gly and Arg/Ser at position 673 between dipeptidyl-peptidase (DPP) 7 and DPP11, widely distributed DPPs in pathogenic and environmental Gram-negative bacteria. Biochimie 95, 824–832 [DOI] [PubMed] [Google Scholar]
- 24. Ohara-Nemoto Y., Shimoyama Y., Kimura S., Kon A., Haraga H., Ono T., Nemoto T. K. (2011) Asp- and Glu-specific novel dipeptidyl peptidase 11 of Porphyromonas gingivalis ensures utilization of proteinaceous energy sources. J. Biol. Chem. 286, 38115–38127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banbula A., Mak P., Bugno M., Silberring J., Dubin A., Nelson D., Travis J., Potempa J. (1999) A novel enzyme with possible pathological implications for the development of periodontitis. J. Biol. Chem. 274, 9246–9252 [DOI] [PubMed] [Google Scholar]
- 26. Oda H., Saiki K., Tonosaki M., Yajima A., Konishi K. (2009) Participation of the secreted dipeptidyl and tripeptidyl aminopeptidases in asaccharolytic growth of Porphyromonas gingivalis. J. Periodontal Res. 44, 362–367 [DOI] [PubMed] [Google Scholar]
- 27. Nguyen K. A., Travis J., Potempa J. (2007) Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J. Bacteriol. 189, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi Y., Ratnayake D. B., Okamoto K., Abe N., Yamamoto K., Nakayama K. (1999) Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274, 17955–17960 [DOI] [PubMed] [Google Scholar]
- 29. Murakami Y., Imai M., Nakamura H., Yoshimura F. (2002) Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur. J. Oral Sci. 110, 157–162 [DOI] [PubMed] [Google Scholar]
- 30. Lambert C., Léonard N., De Bolle X., Depiereux E. (2002) ESyPred3D. Prediction of proteins 3D structures. Bioinformatics 18, 1250–1256 [DOI] [PubMed] [Google Scholar]
- 31. Shoji M., Shibata Y., Shiroza T., Yukitake H., Peng B., Chen Y. Y., Sato K., Naito M., Abiko Y., Reynolds E. C., Nakayama K. (2010) Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis. Its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 10, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen K. A., Zylicz J., Szczesny P., Sroka A., Hunter N., Potempa J. (2009) Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology 155, 328–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rouf S. M., Ohara-Nemoto Y., Ono T., Shimoyama Y., Kimura S., Nemoto T. K. (2013) Phenylalanine 664 of dipeptidyl peptidase (DPP) 7 and phenylalanine 671 of DPP11 mediate preference for P2-position hydrophobic residues of a substrate. FEBS Open Bio. 3, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suido H., Nakamura M., Mashimo P. A., Zambon J. J., Genco R. J. (1986) Arylaminopeptidase activities of oral bacteria. J. Dent. Res. 65, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 35. Rawlings N. D., Barrett A. J., Bateman A. (2010) MEROPS. The peptidase database. Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ellis S., Nuenke J. M. (1967) Dipeptidyl arylamidase III of the pituitary. Purification and characterization. J. Biol. Chem. 242, 4623–4629 [PubMed] [Google Scholar]
- 37. Vukelić B., Salopek-Sondi B., Špoljarić J., Sabljić I., Meštrović N., Agić D., Abramić M. (2012) Reactive cysteine in the active-site motif of Bacteroides thetaiotaomicron dipeptidyl peptidase III is a regulatory residue for enzyme activity. Biol. Chem. 393, 37–46 [DOI] [PubMed] [Google Scholar]
- 38. Beauvais A., Monod M., Debeaupuis J. P., Diaquin M., Kobayashi H., Latgé J. P. (1997) Biochemical and antigenic characterization of a new dipeptidyl-peptidase isolated from Aspergillus fumigatus. J. Biol. Chem. 272, 6238–6244 [DOI] [PubMed] [Google Scholar]
- 39. Pederson R. A., White H. A., Schlenzig D., Pauly R. P., McIntosh C. H., Demuth H. U. (1998) Improved glucose tolerance in Zucker fatty rats by oral administration of the dipeptidyl peptidase IV inhibitor isoleucine thiazolidide. Diabetes 47, 1253–1258 [DOI] [PubMed] [Google Scholar]
- 40. Herman G. A., Bergman A., Liu F., Stevens C., Wang A. Q., Zeng W., Chen L., Snyder K., Hilliard D., Tanen M., Tanaka W., Meehan A. G., Lasseter K., Dilzer S., Blum R., Wagner J. A. (2006) Pharmacokinetics and pharmacodynamic effects of the oral DPP-4 inhibitor sitagliptin in middle-aged obese subjects. J. Clin. Pharmacol. 46, 876–886 [DOI] [PubMed] [Google Scholar]
- 41. Ahrén B., Landin-Olsson M., Jansson P. A., Svensson M., Holmes D., Schweizer A. (2004) Inhibition of dipeptidyl peptidase-4 reduces glycemia, sustains insulin levels, and reduces glucagon levels in type 2 diabetes. J. Clin. Endocrinol. Metab. 89, 2078–2084 [DOI] [PubMed] [Google Scholar]
- 42. Monod M., Beauvais A. (2013) Dipeptidyl-peptidases IV and V of Aspergillus. In Handbook of Proteolytic Enzymes, 3rd Ed., pp. 3392–3394, Elsevier, London, UK [Google Scholar]
- 43. Monera O. D., Sereda T. J., Zhou N. E., Kay C. M., Hodges R. S. (1995) Relationship of side chain hydrophobicity and α-helical propensity on the stability of the single-stranded amphipathic α-helix. J. Pept. Sci. 1, 319–329 [DOI] [PubMed] [Google Scholar]
- 44. Nakajima Y., Ito K., Toshima T., Egawa T., Zheng H., Oyama H., Wu Y. F., Takahashi E., Kyono K., Yoshimoto T. (2008) Dipeptidyl aminopeptidase IV from Stenotrophomonas maltophilia exhibits activity against a substrate containing a 4-hydroxyproline residue. J. Bacteriol. 190, 7819–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vermout S., Baldo A., Tabart J., Losson B., Mignon B. (2008) Secreted dipeptidyl peptidases as potential virulence factors for Microsporum canis. FEMS Immunol. Med. Microbiol. 54, 299–308 [DOI] [PubMed] [Google Scholar]
- 46. Sato K., Naito M., Yukitake H., Hirakawa H., Shoji M., McBride M. J., Rhodes R. G., Nakayama K. (2010) A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rosenblum J. S., Kozarich J. W. (2003) Prolyl peptidases. A serine protease subfamily with high potential for drug discovery. Curr. Opin. Chem. Biol. 7, 496–504 [DOI] [PubMed] [Google Scholar]
- 48. Rasmussen H. B., Branner S., Wiberg F. C., Wagtmann N. (2003) Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat. Struct. Biol. 10, 19–25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.