Background: Gluconeogenesis is an important physiological pathway in response to fasting and stress.
Results: DBC1 regulates gluconeogenesis and PEPCK expression by a mechanism that is at least in part explained by the Rev-erbα and the deacetylase SIRT1.
Conclusion: A new role for DBC1as a regulator of PEPCK expression is described.
Significance: We aim to understand the molecular mechanisms of glucose metabolism and response to fasting.
Keywords: Gluconeogenesis, Metabolism, Nuclear Receptors, Signal Transduction, SIRT1, Deleted In Breast Cancer 1, Rev-erbα
Abstract
Liver gluconeogenesis is essential to provide energy to glycolytic tissues during fasting periods. However, aberrant up-regulation of this metabolic pathway contributes to the progression of glucose intolerance in individuals with diabetes. Phosphoenolpyruvate carboxykinase (PEPCK) expression plays a critical role in the modulation of gluconeogenesis. Several pathways contribute to the regulation of PEPCK, including the nuclear receptor Rev-erbα and the histone deacetylase SIRT1. Deleted in breast cancer 1 (DBC1) is a nuclear protein that binds to and regulates both Rev-erbα and SIRT1 and, therefore, is a candidate to participate in the regulation of PEPCK. In this work, we provide evidence that DBC1 regulates glucose metabolism and the expression of PEPCK. We show that DBC1 levels decrease early in the fasting state. Also, DBC1 KO mice display higher gluconeogenesis in a normal and a high-fat diet. DBC1 absence leads to an increase in PEPCK mRNA and protein expression. Conversely, overexpression of DBC1 results in a decrease in PEPCK mRNA and protein levels. DBC1 regulates the levels of Rev-erbα, and manipulation of Rev-erbα activity or levels prevents the effect of DBC1 on PEPCK. In addition, Rev-erbα levels decrease in the first hours of fasting. Finally, knockdown of the deacetylase SIRT1 eliminates the effect of DBC1 knockdown on Rev-erbα levels and PEPCK expression, suggesting that the mechanism of PEPCK regulation is, at least in part, dependent on the activity of this enzyme. Our results point to DBC1 as a novel regulator of gluconeogenesis.
Introduction
Hepatic gluconeogenesis plays a key role in the maintenance of systemic glucose levels during health and disease. During fasting, circulating hormones promote the de novo synthesis of glucose (1, 2), ensuring delivery of energy to glucose-dependent tissues. However, up-regulation of glucose production in the liver may also play a role in the development of hyperglycemia in diabetes (3). Regulation of gluconeogenesis mainly targets the expression of the enzyme phosphoenolpyruvate carboxykinase (PEPCK3, also known as PCK1) because this enzyme catalyzes a committed step in the gluconeogenic pathway (1, 4). PEPCK is regulated by several signaling systems, including the glucagon-cAMP and the insulin-AKT pathways (1). In addition, like many other metabolically relevant enzymes, PEPCK displays a circadian regulation (5). Central to the metabolic circadian rhythms are the nuclear receptors Rev-erbs (6). Rev-erbα, the most studied member of the family, is a heme receptor (7, 8) that represses transcription of target genes via an interaction with the nuclear corepressor (NCoR) (6). Two studies have demonstrated that Rev-erbα is a transcriptional repressor of PEPCK in liver cells (8, 9), and a central role for Rev-erbα in the control of hepatic metabolism has been proposed (10, 11).
The relative abundance of Rev-erbα present in a cell is in part controlled by its ubiquitin-mediated proteasomal degradation (12). We have shown recently that deleted in breast cancer 1 (DBC1) binds and stabilizes Rev-erbα, preventing its degradation (13). In fact, in DBC1 KO mice, the hepatic levels of Rev-erbα are lower than in wild-type littermates. More importantly, our previous study demonstrated that DBC1 regulates circadian rhythms that are Rev-erbα-dependent (13).
DBC1 is a nuclear protein that binds and regulates the activity of several nuclear receptors (14) and enzymes involved in epigenetic processes, such as the methyltransferase SUV39H1 (15) and the deacetylases HDAC3 (16) and SIRT1 (17, 18). Not much is known about the molecular cues that modulate the interaction between DBC1 and its partners. However, we have shown previously that the interaction between DBC1 and SIRT1 is disrupted under fasting conditions (19) and that the kinases AMP-activated protein kinase (AMPK) and PKA regulate the interaction between both proteins (20). SIRT1, a NAD-dependent deacetylase, regulates glucose metabolism in the liver through deacetylation of several targets, although different models provide conflicting results (21–27). It appears that SIRT1 deacetylates PGC1α (21) and FOXO (Forkhead box) (28), increasing their transcriptional activity. We previously showed that mice knocked out for DBC1 display higher SIRT1 activity (19) and are protected against high-fat diet-induced liver steatosis (19), suggesting that DBC1 plays an important role in the regulation of liver metabolism (19).
In this study, we explored whether DBC1 is important for glucose metabolism and PEPCK regulation and the mechanisms involved in this process. Our data reveal that DBC1 is a novel regulator of PEPCK expression and gluconeogenesis by a mechanism that involves, at least in part, both Rev-erbα and SIRT1.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Unless specified otherwise, all reagents and chemicals were purchased from Sigma-Aldrich. The Rev-erbα antagonist SR8278 was from Tocris Bioscience. Anti-human SIRT1 and anti-mouse SIRT1, phospho-AKT, AKT, tubulin, and GAPDH antibodies were from Cell Signaling Technology. Anti-DBC1 antibodies were from Bethyl Laboratories. The anti-mouse PEPCK antibody was from Cayman Chemical. Anti-human PCK1, anti-Rev-erbα, and anti HA antibodies were from Abcam. Anti-actin and anti-FLAG antibodies were from Sigma.
Animal Handling and Studies
All mice used in this study were maintained at the Mayo Clinic animal facility. All experimental protocols were approved by the Institutional Animal Care and Use Committee at the Mayo Clinic (protocol A33209 and A52112), and studies were performed according to the methods approved in the protocols. The production and characterization of DBC1 KO mice has been described before (19). All studies were performed in animals that were between 12 and 36 weeks old. Mice were fed normal chow or a 60% high-fat diet as described before (19). A glucose tolerance test was performed after 16 h of fasting. Mice were injected intraperitoneally with 1.5 g of dextrose/kg. To perform the insulin sensitivity test and the pyruvate tolerance test, DBC1 WT or KO mice were fasted for 6 h. After this, mice were challenged with a single dose of either 0.5units/kg of insulin or 1.5g/kg of pyruvate intraperitoneally. All mice had access to water during the fasting period. Blood glucose was measured from the tip of the tail vein using an AlphaTRAK blood glucose monitoring system (Abbott). For the glucose tolerance test and the pyruvate tolerance test, the area under the curve was calculated as the net incremental area over the baseline. For the insulin sensitivity test, the blood glucose values were normalized to time 0, and the total area under the curve was calculated. For shorter fasting periods, food was removed 1 h before the start of the dark cycle.
Cell Culture
HepG2 and 293T cells were cultured in Dulbecco's modified Eagle's medium (5 g/liter glucose) with 10% FBS, glutamine, and penicillin/streptomycin (Invitrogen). In all experiments with HepG2 cells, the cells were switched to serum-free medium 16 h before harvesting. Transfection of HA/Myc or FLAG-Rev-erbα in 293T cells was performed in 6-well plates using Lipofectamine 2000 (Invitrogen) and 1 μg of total DNA. To generate 293T stable cell lines overexpressing FLAG-DBC1 or HA/Myc-Rev erbα, cells were cotransfected with a vector that confers resistance to puromycin and the vector with the cDNA for DBC1 or Rev-erbαα using Lipofectamine 2000. 48 h after transfection, the cells were diluted and selected with puromycin for 15 days. Individual cells were grown to generate clones, which were screened by immunofluorescence and Western blot analysis with anti-tag antibodies.
siRNA
All siRNAs were from Dharmacon (Lafayette, CO). The siRNA duplex against DBC1 was 5′-CAGUUGCAUGACUACUUUUU (sense). SMARTpool siRNAs were used to knock down SIRT1. Non-targeting siRNA 3 was used as a control (D001210-03-20). Transfections in HepG2 and 293T were performed with 100 nm siRNA on day 1. Cells were split on the following day, transfected again on the third day, and harvested 96 h after the first transfection.
Western Blot Analysis
Mouse tissues and cultured cells were lysed in NETN buffer (20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40) supplemented with 5 mm NaF, 50 mm 2-glycerophosphate, 1 mm Na3VO4, and protease inhibitor mixture (Roche). For experiments to detect acetylation, 5 μm trichostatin A and 5 mm nicotinamide were also added. Homogenates were incubated at 4 °C for 30 min under constant agitation and then centrifuged at 11,200 × g for 10 min at 4 °C. Cell and tissue lysates were analyzed by Western blot analysis with the indicated antibodies. Western blots were developed using secondary antibodies and SuperSignal West Pico chemiluminescent substrate (Pierce). Films were scanned, and bands were quantified by densitometry using ImageJ (http://rsbweb.nih.gov/ij/).
Real-time PCR
Total RNA was prepared and retrotranscribed as described before (13). TaqMan probes for DBC1, PEPCK, and GAPDH were obtained from Applied Biosystems. The relative amount of mRNA was calculated using the ΔCT method using GAPDH as an internal control.
Site-directed Mutagenesis
Point mutations were performed using a QuikChange directed mutagenesis kit (Stratagene) following the instructions of the manufacturer.
Statistics
Values are presented as mean ± S.D. of three to five experiments unless indicated otherwise. The significance of differences between means was assessed by one-way analysis of variance or two-tailed Student's t test. A p value of less than 0.05 was considered significant.
RESULTS
Lack of DBC1 Results in Elevated Gluconeogenesis
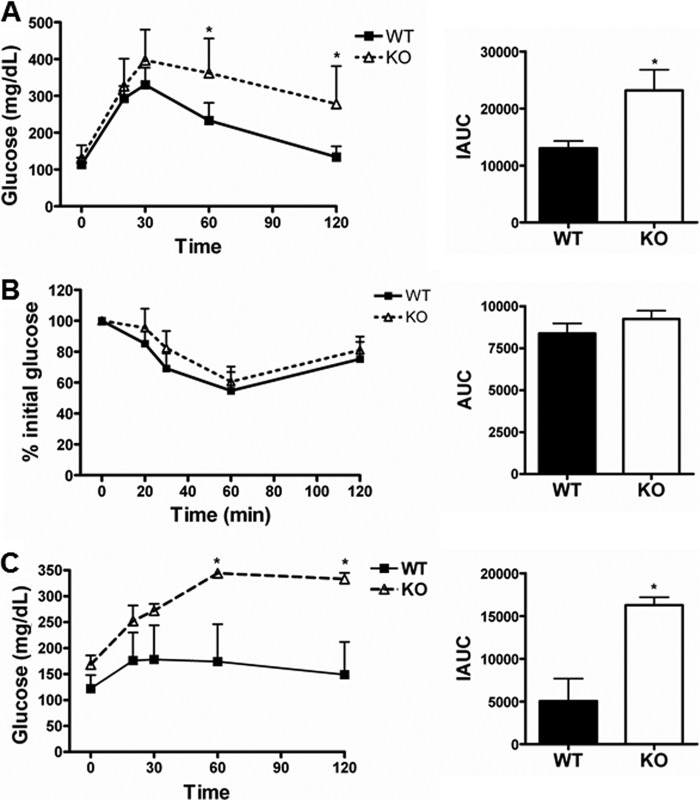
To characterize the role of DBC1 in glucose homeostasis, we first performed glucose tolerance tests in WT and DBC1 KO mice fed regular chow. We observed that DBC1 KO male mice reached higher levels of blood glucose than WT mice after an intraperitoneal challenge of glucose (Fig. 1A). We also observed that DBC1 KO mice display higher blood glucose levels in the fed state (Fig. 1B) than WT mice, although glycemia was not different in the fasted state (Fig. 1A). We then evaluated whether insulin release and insulin sensitivity were responsible for the differences in glucose tolerance between WT and DBC1 KO mice. We found that insulin release was similar between genotypes (Fig. 1C). There was no significant difference in insulin sensitivity (Fig. 1D). Because the glycemia is a balance between glucose clearance and glucose production, we sought to assess the contribution of gluconeogenesis to the observed glucose intolerance. To this end, we compared the response of wild-type and DBC1 KO mice to a pyruvate challenge, a precursor for glucose synthesis. As can be seen in Fig. 1E, KO mice reached higher levels of blood glucose than their WT littermates (Fig. 1B). These findings point to elevated gluconeogenesis in the DBC1 KO mice compared with the WT counterparts.
FIGURE 1.
DBC1 KO mice are more gluconeogenic than WT littermates. A, glucose tolerance test and net incremental area under the curve (IAUC) in WT and DBC1 KO mice fed regular chow. B, blood glucose in WT and DBC1 KO mouse males fed ad libitum. C, insulin release after a glucose challenge. D, insulin sensitivity in WT and DBC1 KO mice. E, pyruvate tolerance test and net incremental area under the curve in WT and DBC1 KO mice fed regular chow. *, p < 0.05; n = 4–8.
Increased gluconeogenesis is a hallmark of type II diabetes and is usually accompanied by obesity. To study glucose metabolism in obese mice, we fed DBC1 WT and KO mice a hypercaloric high-fat diet. We performed a glucose tolerance test after 16 weeks of high-fat diet and found that both genotypes became glucose-intolerant, although the glucose tolerance was more altered in DBC1 KO mice (Fig. 2A). Akin to what we observed when mice were fed normal chow, feed glycemia was higher in the DBC1 knockout mice (180 mg/dl) compared with wild-type mice (140 mg/dl), although insulin sensitivity was similar between genotypes (Fig. 2B). However, when we performed a pyruvate tolerance test, the response of DBC1 KO mice on the high-fat diet was exaggerated in comparison with WT mice (Fig. 2C). Therefore, the effect of the high-fat diet on gluconeogenesis was more pronounced in mice lacking DBC1. Taken together, our results demonstrate that the absence of DBC1 leads to glucose intolerance driven mainly by elevated gluconeogenesis.
FIGURE 2.
Effect of a high-fat diet on glucose metabolism of DBC1 KO mice. A, glucose tolerance test and net incremental area under the curve (IAUC) in WT and DBC1 KO mice fed a high-fat diet. B, insulin sensitivity and net area under the curve (AUC) in WT and DBC1 KO mice fed a high-fat diet. C, pyruvate tolerance test and net incremental area under the curve in WT and DBC1 KO mice fed a high-fat diet. *, p < 0.05.
DBC1 Regulates PEPCK Expression
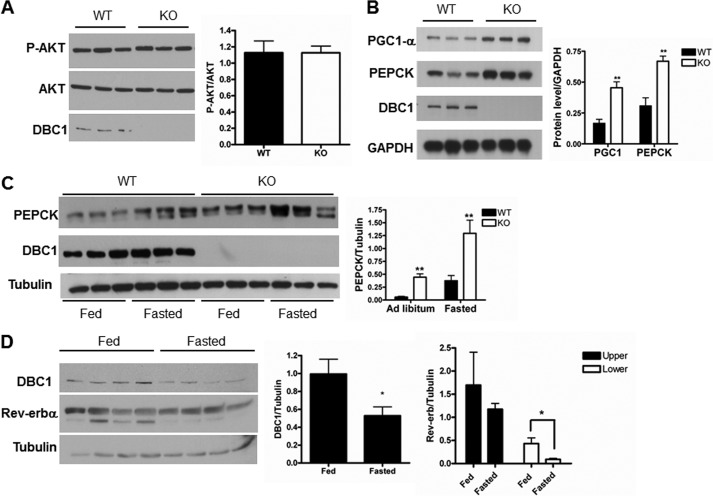
To confirm that the phenotype we observed was driven by elevated gluconeogenesis and not insulin sensitivity, we evaluated the expression level of AKT and PEPCK in the livers of WT and KO mice. The levels of phospho-AKT and AKT were similar in ad libitum conditions in both genotypes (Fig. 3A). In contrast, PEPCK levels were higher in KO mice than in WT mice under basal conditions and after 6 and 24 h of fasting (Fig. 3, B and C). These results are in agreement with glucose synthesis as the main cause of the glucose intolerance presented by the DBC1 KO mice. In addition to elevated levels of PEPCK, we also observed that the protein levels of PGC1-α, a coactivator that potentiates PEPCK transcription, were also increased in DBC1 KO livers (Fig. 3B). The fact that the expression of both PGC1-α and PEPCK is higher in KO mice is not surprising, considering that several signaling pathways regulate both genes. In fact, we hypothesized that DBC1 is upstream of a factor that is common to the regulation of PEPCK and PGC1-α. Among the transcription factors involved in the regulation of PEPCK, the heme receptor Rev-erbα has recently gained attention. We showed previously that DBC1 regulates the stability and the function of Rev-erbα (13), a repressor of PEPCK and PGC1-α (8). In fact, we showed that DBC1 stabilizes Rev-erbα and that the livers of DBC1 KO mice express lower levels of this nuclear receptor (13). So far, a possible function for DBC1 and Rev-erbα during fasting and gluconeogenesis has not been established. We speculated that if alterations in these proteins were involved in the onset of PEPCK transcription, these should occur early during fasting. To test this hypothesis, mice were fasted for a short period (3 h), and the levels of DBC1 and Rev-erbα were assessed by Western blot analysis. We found that DBC1 levels decreased about 40% in this condition (Fig. 3D). We also detected a similar decrease in Rev-erbα, specifically in the band that migrates at ∼80 kDa (Fig. 3D). It is important to note that the decrease in DBC1 levels is transient. After 24 h of fasting, its levels are similar to the fed state (Fig. 3C).
FIGURE 3.
DBC1 KO mice display similar levels of P-AKT and elevated expression of PEPCK than WT mice. A, liver extracts of DBC1 WT and KO mice fed ad libitum were immunoblotted for P-AKT and AKT. The densitometry is shown in the right panel. B, DBC1 WT and KO mice were fasted for 6 h, and the liver extracts were immunoblotted with specific antibodies against PGC1α, PEPCK, DBC1, and GAPDH. The graph in the right panel shows the quantification of the bands by densitometry. **, p < 0.005. C, DBC1 WT and KO mice were fasted for 24 h or fed ad libitum, and the expression of the indicated proteins was determined by Western blot analysis. The graph in the right panel shows the quantification of the bands by densitometry. D, liver extracts of WT mice fasted for 3 h were immunoblotted with specific antibodies against DBC1, Rev-erbα, and tubulin. The graph in the right panel shows the quantification of the bands by densitometry. *, p < 0.05.
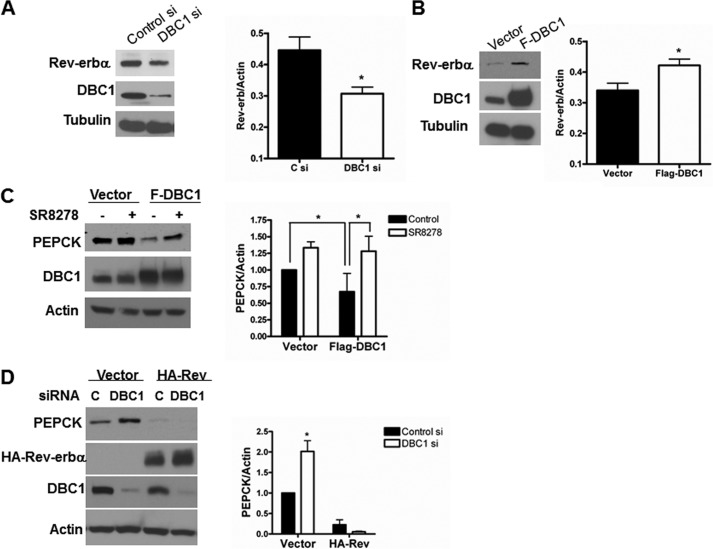
To confirm that DBC1 regulates the expression of PEPCK, we manipulated the levels of DBC1 and assessed whether there were changes in PEPCK levels. First, we knocked down DBC1 and, akin to what was observed in mice lacking DBC1, the expression of PEPCK was increased (Fig. 4A). This was accompanied by an increase in the amount of mRNA for PEPCK (Fig. 4B). Then we overexpressed DBC1 and observed a decrease in the protein and mRNA levels of PEPCK (Fig. 4, C and D), supporting our initial hypothesis that DBC1 is involved in the regulation of PEPCK.
FIGURE 4.
DBC1 regulates the expression of PEPCK in cellular models. A, HepG2 cells were knocked down with a control (C si) and DBC1 siRNA (DBC1 si), and the levels of PEPCK, DBC1, and actin were assessed by Western blot analysis. The graph in the right panel is the quantification by densitometry of PEPCK expression. *, p < 0.05; n = 5. B, the relative amount of PEPCK mRNA was determined by real-time PCR in control- (Csi) and DBC1 siRNA-transfected HepG2 cells. *, p < 0.05, **, p < 0.005; n = 3. C, FLAG-DBC1 was stably overexpressed in 293T cells. The levels of DBC1 and PEPCK were evaluated by Western blot analysis. The graph in the right panel is the quantification by densitometry of PEPCK expression. *, p < 0.05; n = 5. D, the relative level of PEPCK mRNA was determined by real-time PCR in the same F-DBC1 clone. ***, p < 0.05; n = 3.
DBC1 Regulates PEPCK Expression, at Least in Part, through Rev-erbα
To investigate whether Rev-erbα is involved in the regulation of PEPCK by DBC1, we first expanded our previous observations that manipulation of DBC1 levels affects the levels of Rev-erbα (13). Indeed, in agreement with our previous report (13) in 3T3 cells, we observed that knock down of DBC1 in HepG2 cells also resulted in a decrease in endogenous Rev-erbα (Fig. 5A) and that overexpression of DBC1 in 293T cells was accompanied by an increase in the endogenous levels of this nuclear receptor (Fig. 5B). The changes in the levels of Rev-erbα were not dramatic, and, therefore, we decided to evaluate whether they were enough to participate in the regulation of PEPCK by DBC1. With this in mind, we tested whether a Rev-erbα antagonist would rescue the decrease in PEPCK induced by overexpression of DBC1. We treated stable cell lines expressing either an empty vector (control) or overexpressing FLAG-DBC1 with the Rev-erbα antagonist SR8278 (29) for 24 h, and then the levels of PEPCK were evaluated. In accordance with our hypothesis, the treatment of the FLAG-DBC1 stable cell line with SR8278 increased PEPCK expression to a level comparable with the untreated control cells (Fig. 5C). In addition, we tested whether the small decrease in Rev-erbα induced by knockdown of DBC1 had a functional effect. For this, we generated a stable cell line that overexpresses a HA/Myc-Rev-erbα in a vector that contains multiple nuclear localization motives. Our idea was that, by forcing Rev-erbα to remain in the nucleus, there would be more stable functional Rev-erbα, even in the absence of DBC1. Certainly, when we treated this cell line with a DBC1 siRNA, the levels of Myc/HA-Rev-erbα did not decrease (Fig. 5D). In this scenario, the up-regulation of PEPCK by DBC1 was lost. However, in the empty vector-transfected cell line, a similar decrease in DBC1 led to an increase in PEPCK. Taken together, our results support, at least in part, a role for Rev-erbα in the DBC1-mediated regulation of PEPCK.
FIGURE 5.
DBC1 regulates the steady levels of Rev-erbα. A, DBC1 was knocked down with specific siRNA in HepG2 cells. Expression of Rev-erbα and DBC1 was analyzed by Western blot analysis with specific antibodies. *, p < 0.05; n = 6. C si, control siRNA; DBC1 si, DBC1 siRNA. B, a stable cell line overexpressing FLAG-DBC1 was generated in 293T cells, and the levels of Rev-erbα and DBC1 were analyzed by Western blot analysis with specific antibodies. *, p < 0.05; n = 5. C, the stable FLAG-DBC1 clone or the empty vector stable clone were treated with the Rev-erbα antagonist SR8278 (10 μm) for 24 h. The expression of PEPCK was evaluated by Western blot analysis. The graph in the right panel shows the quantification of the bands by densitometry. *, p < 0.05; n = 5. D, a stable cell line overexpressing HA/Myc Rev-erbα was generated in 293T cells. Cells were knocked down with control (black bar) and DBC1-specific siRNA (white bar), and the levels of PEPCK and transfected Rev-erbα were determined by Western blot analysis. The graph in the right panel shows the quantification of the bands by densitometry. *, p < 0.05.
SIRT1 Is Important for the Regulation of PEPCK and Rev-eraα by DBC1
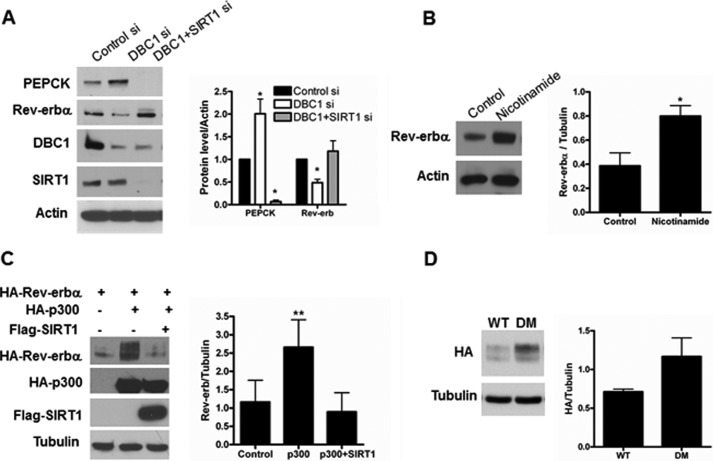
DBC1 is an endogenous inhibitor of several nuclear enzymes. Among these, SIRT1 is a NAD-dependent deacetylase involved in the regulation of gluconeogenesis. To further understand the mechanism of PEPCK regulation by DBC1, we evaluated a possible contribution of SIRT1. We compared the levels of PEPCK expression in HepG2 cells treated with a control siRNA, a DBC1 siRNA, and a combination of DBC1 and SIRT1 siRNA. As shown in Fig. 6A, in the absence of SIRT1, DBC1 is no longer able to up-regulate PEPCK. Moreover, the absence of SIRT1 led to a clear decrease in the levels of PEPCK. We also analyzed whether SIRT1 is required for the regulation of Rev-erbα. Again, we observed that, in the absence of SIRT1, a decrease in DBC1 does not result in a decrease in Rev-erbα (Fig. 6A). In addition, the treatment of cells overexpressing Rev-erbα with the sirtuin inhibitor nicotinamide resulted in increased levels of this nuclear receptor (Fig. 6B). To further explore the role of acetylation in the regulation of Rev-erbα, we transfected cells with Rev-erbα and the acetyltransferase p300 in the presence or absence of SIRT1. In agreement with our previous observations, expression of p300 increased the levels of Rev-erbα, and this effect was ablated by cotransfection with SIRT1 (Fig. 6C). p300 appears to increase levels of Rev-erbα by stabilization of the protein (supplemental Fig. 1). To assess the acetylation of Rev-erbα, we cotransfected cells with Rev-erbα and p300 and immunoprecipitated Rev-erbα. We could not detect acetylation under this condition (not shown). In addition, we could not detect acetylation of Rev-erbα immunoprecipitated from liver tissue (not shown). To dismiss the possibility that the negative results were due to technical issues, we incubated 293T cells transfected with Rev-erbα with deacetylase inhibitors and analyzed Rev-erbα posttranslational modifications by mass spectrometry. We found that lysines 400 and 591 were indeed acetylated. However, single mutants of these sites are still up-regulated by p300 (supplemental Fig. 2). Moreover, the protein levels of the single mutant (supplemental Fig. 2) and double mutant (Fig. 6D) are not lower than the WT, as one would expect if acetylation of Rev-erbα promoted its stabilization. Therefore, it is likely that p300 and SIRT1 regulate Rev-erbα levels by an indirect mechanism that involves acetylation/deacetylation events upstream of Rev-erbα. A schematic of the proposed mechanism by which DBC1 regulates transcription of PEPCK is shown in Fig. 7. Taken together, our results unveil a novel role for DBC1 in the regulation of PEPCK expression and gluconeogenesis through a mechanism that, at least in part, involves Rev-erbα and, possibly, SIRT1.
FIGURE 6.
SIRT1 is involved in the regulation of PEPCK and Rev-erbα. A, DBC1 was knocked down alone or in combination with SIRT1 siRNA in HEPG2 cells, and the expression of PEPCK and Rev-erbα was evaluated by Western blot analysis. The graph in the right panel shows the quantification of the PEPCK and Rev-erbα levels normalized to actin. *, p < 0.05; n = 4. B, 293T cells were transfected with FLAG-Rev-erbα and treated with 5 mm nicotinamide for 16 h. The levels of Rev-erbα were assessed by Western blot analysis, and the quantification of the bands is shown in the graph in the right panel. *, p < 0.05; n = 3. C, 293T cells were transfected as indicated. 48 h later, the protein levels of HA-Rev-erbα, HA-p300, and SIRT1 were evaluated by Western blot analysis. The graph in the right panel shows the quantification of the bands by densitometry. **, p < 0.01; n = 3. D, 293T cells were transfected with Rev-erbα WT or the Rev-erbα K400A/K591A double mutant (DM), and the protein levels were assessed by Western blot analysis with anti-HA antibody.
FIGURE 7.

Putative mechanism of DBC1regulation of DBC1. Left, we propose that, in the fed state, DBC1 is bound to SIRT1, inhibiting it (light red). Rev-erbα (Rev) represses the transcription of PEPCK. Center, in the fasted state, DBC1 levels decrease, and it dissociates from SIRT1, which leads to an increase in its activity (dark red) and a decrease in Rev-erbα levels. These events result in derepression of PEPCK. The mechanism by which SIRT1 regulates Rev-erbα is likely indirect and not completely understood. Right, in the DBC1 KO model, SIRT1 is active, and the levels of Rev-erbα are low, which results in PEPCK expression. The model does not exclude the possibility that other targets of DBC1, such as HDAC3, SIRT1, PGC1, and FOXO1 may also play a role in the mechanism of regulation of PEPCK by DBC1.
DISCUSSION
Hepatic gluconeogenesis is an essential component of the homeostatic mechanisms that ensure proper supply of glucose to glucolytic tissues during times of food deprivation (2). However, deregulation of this process can lead to increased blood glucose levels and is an important contributor to the pathogenesis of diabetes (3). Therefore, understanding the molecular mechanisms involved in the regulation of gluconeogenesis will provide us with potential new targets to treat diabetes. A variety of hormones and dietary signals that control glucose synthesis target the expression of PEPCK, given that this enzyme catalyzes the first committed step of gluconeogenesis (4). In mouse models, overexpression of PEPCK is enough to induce glucose intolerance (30), and even a modest reduction in PEPCK expression is able to ameliorate fasting glucose levels and glucose tolerance (31).
In this report, we demonstrate a new role for the protein DBC1 in the regulation of gluconeogenesis. DBC1 levels decrease in the early hours of the fasting state, suggesting that such a decrease is one of the first events that lead to the onset of the gluconeogenic program. Mice that lack DBC1 mimic a fasted state and, therefore, display an increased production of glucose. Hence, one can infer that the natural role of DBC1 is to act as a suppressor of gluconeogenesis. Interestingly, Qiang et al. (32) showed that DBC1 knockout mice are glucose-intolerant in a normal and a high-fat diet, a phenotype that is in agreement with our findings. Ours is the first report that presents evidence that DBC1 levels are regulated in different metabolic states. Whether this regulation is transcriptional or posttranslational and which are the signaling pathways involved are a completely new avenue of research that warrants further investigation.
We have shown previously that DBC1 is important for the stability of Rev-erbα. In fact, DBC1 KO mice present lower levels of Rev-erbα in the liver and fat than wild-type littermates (13). Moreover, we demonstrated that DBC1 modulates Rev-erbα transcriptional repressor activity and that it is required for proper oscillations of clock genes, such as bmal (13). In this work, we unveil further implications of the DBC1-Rev-erbα axis, particularly in the regulation of PEPCK.
Rev-erbα has recently gained attention in the field of metabolism regulation because it connects circadian oscillations to metabolic adaptations (13). A recent report also implicates Rev-erbα in the secretion of glucagon (33). Although Rev-erbα has been linked mainly to the regulation of hepatic lipid metabolism (10, 34), previous articles show that this nuclear receptor regulates PEPCK at the mRNA level (8, 9). We show that Rev-erbα levels decrease during the first hours of starvation, an event that can contribute to derepression of the PEPCK gene. Our findings support and expand the role of Rev-erbα in the regulation of PEPCK, stressing the importance of a DBC1-Rev-erbα axis in the control of metabolic processes. Solt et al. (35) recently reported the development of Rev-erbα agonists along with its effects on circadian behavior and some aspects of metabolism. The authors find that administration of the ligand SR9009 improve lipid metabolism. However, data on glucose metabolism are not provided. Noteworthy is that Cho et al. (10) showed that, in the absence of Reverb-α and β, fasting glucose is elevated. In light of these findings and the ones presented here, it would be advisable to study the effect of Rev-erbα ligands on glucose metabolism, especially in the context of obesity.
Glucose synthesis is regulated in a circadian fashion by the repressor cryptochrome (36), and the activator Bmal (37). Hepatic Rev-erbα levels also oscillate in a circadian way (34), and we have shown oscillations of DBC1 in cells (13). It would be interesting to study whether DBC1 expression varies in vivo during the circadian cycle and if this is, in turn, connected to the oscillations in Rev-erbα and glucose synthesis.
Finally, we observed that SIRT1 regulates Rev-erbα protein levels, an observation that, to our knowledge, has not been reported before. In agreement with our observations, Vieria et al. (33) suggested that, in a pancreatic cell line, SIRT1 is involved in the regulation of Rev-erbα mRNA levels. On the other hand, Rev-erbα regulates the expression of SIRT1 (38), which points to a loop of cross-regulation between these two proteins. We provide evidence that acetylation/deacetylation processes regulate Rev-erbα protein levels. However, whether SIRT1 regulates Rev-erbα by directly deacetylating it, or through an indirect mechanism, requires further research. One possibility is that activation of SIRT1 results in repression of p300, as has been shown by Bouras et al. (39), and that this, in turn, results in destabilization of Rev-erbα. The precise mechanism by which SIRT1 regulates Rev-erbα, and the physiologic consequences of this regulation, warrant further investigation.
We want to stress that the DBC1-Rev-erbα-SIRT1 pathway described in this paper may not be the only one mediating the regulation of PEPCK by DBC1. In the fasted state, and in the DBC1 KO mice, increased SIRT1 activity could result in deacetylation of PGC1α (21) and FOXO (28), which would also result in higher glucose production. Moreover, it is possible that DBC1 also regulates gluconeogenesis through histone deacetylase 3 (HDAC3), which is inhibited by DBC1 (16) and has been shown to promote glucose production (40, 41). In addition, it could also be that changes in the levels of DBC1, or changes to its binding partners, result in alterations in the epigenetic landscape of the PEPCK promoter. Finally, one can envision that DBC1 is a coordinator of all these different pathways that converge on PEPCK.
In conclusion, we describe, for the first time, a key role for the nuclear protein DBC1 as a regulator of gluconeogenesis. DBC1 may play a major role in the development of glucose intolerance and may also have a physiological role in plasma glucose maintenance during starvation. We believe that our studies reveal a new pathway involved in the regulation of gluconeogenesis and may provide a better understanding of the mechanisms that maintain body glucose homeostasis during normal physiology and disease states.
Supplementary Material
This work was supported, in whole or in part, by NIDDK, National Institutes of Health Grant DK-084055. This work was also supported by grants from the American Federation of Aging Research and from the Mayo Foundation, by the Strickland Career Development Award, by Mayo-UOFM Decade of Discovery Grant 63-01, and by Minnesota Obesity Council grant DK-50456-15.

This article contains supplemental Figs. 1 and 2.
- PEPCK
- phosphoenol pyruvatecarboxikinase
- DBC
- deleted in breast cancer.
REFERENCES
- 1. Desvergne B., Michalik L., Wahli W. (2006) Transcriptional regulation of metabolism. Physiol. Rev. 86, 465–514 [DOI] [PubMed] [Google Scholar]
- 2. Hellerstein M. K., Neese R. A., Linfoot P., Christiansen M., Turner S., Letscher A. (1997) Hepatic gluconeogenic fluxes and glycogen turnover during fasting in humans. A stable isotope study. J. Clin. Invest. 100, 1305–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gastaldelli A., Baldi S., Pettiti M., Toschi E., Camastra S., Natali A., Landau B. R., Ferrannini E. (2000) Influence of obesity and type 2 diabetes on gluconeogenesis and glucose output in humans. A quantitative study. Diabetes 49, 1367–1373 [DOI] [PubMed] [Google Scholar]
- 4. Hanson R. W., Reshef L. (1997) Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu. Rev. Biochem. 66, 581–611 [DOI] [PubMed] [Google Scholar]
- 5. Guillaumond F., Gréchez-Cassiau A., Subramaniam M., Brangolo S., Peteri-Brünback B., Staels B., Fiévet C., Spelsberg T. C., Delaunay F., Teboul M. (2010) Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol. Cell Biol. 30, 3059–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duez H., Staels B. (2009) Rev-erb-α. An integrator of circadian rhythms and metabolism. J. Appl. Physiol. 107, 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raghuram S., Stayrook K. R., Huang P., Rogers P. M., Nosie A. K., McClure D. B., Burris L. L., Khorasanizadeh S., Burris T. P., Rastinejad F. (2007) Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat. Struct. Mol. Biol. 14, 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 9. Grant D., Yin L., Collins J. L., Parks D. J., Orband-Miller L. A., Wisely G. B., Joshi S., Lazar M. A., Willson T. M., Zuercher W. J. (2010) GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbα. ACS Chem. Biol. 5, 925–932 [DOI] [PubMed] [Google Scholar]
- 10. Cho H., Zhao X., Hatori M., Yu R. T., Barish G. D., Lam M. T., Chong L. W., DiTacchio L., Atkins A. R., Glass C. K., Liddle C., Auwerx J., Downes M., Panda S., Evans R. M. (2012) Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bugge A., Feng D., Everett L. J., Briggs E. R., Mullican S. E., Wang F., Jager J., Lazar M. A. (2012) Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yin L., Joshi S., Wu N., Tong X., Lazar M. A. (2010) E3 ligases Arf-bp1 and Pam mediate lithium-stimulated degradation of the circadian heme receptor Rev-erb α. Proc. Natl. Acad. Sci. U.S.A. 107, 11614–11619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chini C. C., Escande C., Nin V., Chini E. N. (2013) DBC1 (deleted in breast cancer 1) modulates the stability and function of the nuclear receptor Rev-erbα. Biochem. J. 451, 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chini E. N., Chini C. C., Nin V., Escande C. (2013) Deleted in breast cancer-1 (DBC-1) in the interface between metabolism, aging and cancer. Biosci. Rep. 33, e00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z., Chen L., Kabra N., Wang C., Fang J., Chen J. (2009) Inhibition of SUV39H1 methyltransferase activity by DBC1. J. Biol. Chem. 284, 10361–10366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chini C. C., Escande C., Nin V., Chini E. N. (2010) HDAC3 is negatively regulated by the nuclear protein DBC1. J. Biol. Chem. 285, 40830–40837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao W., Kruse J. P., Tang Y., Jung S. Y., Qin J., Gu W. (2008) Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451, 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim J. E., Chen J., Lou Z. (2008) DBC1 is a negative regulator of SIRT1. Nature 451, 583–586 [DOI] [PubMed] [Google Scholar]
- 19. Escande C., Chini C. C., Nin V., Dykhouse K. M., Novak C. M., Levine J., van Deursen J., Gores G. J., Chen J., Lou Z., Chini E. N. (2010) Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J. Clin. Invest. 120, 545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nin V., Escande C., Chini C. C., Giri S., Camacho-Pereira J., Matalonga J., Lou Z., Chini E. N. (2012) Role of deleted in breast cancer 1 (DBC1) protein in SIRT1 deacetylase activation induced by protein kinase A and AMP-activated protein kinase. J. Biol. Chem. 287, 23489–23501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 22. Wei D., Tao R., Zhang Y., White M. F., Dong X. C. (2011) Feedback regulation of hepatic gluconeogenesis through modulation of SHP/Nr0b2 gene expression by Sirt1 and FoxO1. Am. J. Physiol. Endocrinol. Metab. 300, E312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodgers J. T., Puigserver P. (2007) Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. U.S.A. 104, 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Erion D. M., Yonemitsu S., Nie Y., Nagai Y., Gillum M. P., Hsiao J. J., Iwasaki T., Stark R., Weismann D., Yu X. X., Murray S. F., Bhanot S., Monia B. P., Horvath T. L., Gao Q., Samuel V. T., Shulman G. I. (2009) SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc. Natl. Acad. Sci. U.S.A. 106, 11288–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nie Y., Erion D. M., Yuan Z., Dietrich M., Shulman G. I., Horvath T. L., Gao Q. (2009) STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat. Cell Biol. 11, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang R. H., Kim H. S., Xiao C., Xu X., Gavrilova O., Deng C. X. (2011) Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J. Clin. Invest. 121, 4477–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Banks A. S., Kon N., Knight C., Matsumoto M., Gutiérrez-Juárez R., Rossetti L., Gu W., Accili D. (2008) SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 8, 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frescas D., Valenti L., Accili D. (2005) Nuclear trapping of the forkhead transcription factor FoxO1 via SIRT-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 280, 20589–20595 [DOI] [PubMed] [Google Scholar]
- 29. Kojetin D., Wang Y., Kamenecka T. M., Burris T. P. (2011) Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem. Biol. 6, 131–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valera A., Pujol A., Pelegrin M., Bosch F. (1994) Transgenic mice overexpressing phosphoenolpyruvate carboxykinase develop non-insulin-dependent diabetes mellitus. Proc. Natl. Acad. Sci. U.S.A. 91, 9151–9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Linares A., Perales S., Palomino-Morales R. J., Castillo M., Alejandre M. J. (2006) Nutritional control, gene regulation, and transformation of vascular smooth muscle cells in atherosclerosis. Cardiovasc. Hematol. Disord. Drug Targets 6, 151–168 [DOI] [PubMed] [Google Scholar]
- 32. Qiang L., Wang L., Kon N., Zhao W., Lee S., Zhang Y., Rosenbaum M., Zhao Y., Gu W., Farmer S. R., Accili D. (2012) Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of PPARγ. Cell 150, 620–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vieira E., Marroquí L., Figueroa A. L., Merino B., Fernandez-Ruiz R., Nadal A., Burris T. P., Gomis R., Quesada I. (2013) Involvement of the clock gene Rev-erb α in the regulation of glucagon secretion in pancreatic α-cells. PLoS ONE 8, e69939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng D., Liu T., Sun Z., Bugge A., Mullican S. E., Alenghat T., Liu X. S., Lazar M. A. (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Solt L. A., Wang Y., Banerjee S., Hughes T., Kojetin D. J., Lundasen T., Shin Y., Liu J., Cameron M. D., Noel R., Yoo S. H., Takahashi J. S., Butler A. A., Kamenecka T. M., Burris T. P. (2012) Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang E. E., Liu Y., Dentin R., Pongsawakul P. Y., Liu A. C., Hirota T., Nusinow D. A., Sun X., Landais S., Kodama Y., Brenner D. A., Montminy M., Kay S. A. (2010) Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 16, 1152–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rudic R. D., McNamara P., Curtis A. M., Boston R. C., Panda S., Hogenesch J. B., Fitzgerald G. A. (2004) BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Woldt E., Sebti Y., Solt L. A., Duhem C., Lancel S., Eeckhoute J., Hesselink M. K., Paquet C., Delhaye S., Shin Y., Kamenecka T. M., Schaart G., Lefebvre P., Nevière R., Burris T. P., Schrauwen P., Staels B., Duez H. (2013) Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 19, 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bouras T., Fu M., Sauve A. A., Wang F., Quong A. A., Perkins N. D., Hay R. T., Gu W., Pestell R. G. (2005) SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem. 280, 10264–10276 [DOI] [PubMed] [Google Scholar]
- 40. Sun Z., Miller R. A., Patel R. T., Chen J., Dhir R., Wang H., Zhang D., Graham M. J., Unterman T. G., Shulman G. I., Sztalryd C., Bennett M. J., Ahima R. S., Birnbaum M. J., Lazar M. A. (2012) Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 18, 934–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mihaylova M. M., Vasquez D. S., Ravnskjaer K., Denechaud P. D., Yu R. T., Alvarez J. G., Downes M., Evans R. M., Montminy M., Shaw R. J. (2011) Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.