Background: The double-capped 26S proteasome complex is viewed as a symmetrical particle.
Results: The ubiquitin receptor subunit Rpn13 is found in an asymmetrical ratio.
Conclusion: Rpn13 ubiquitin receptor may dictate preference in substrate recruitment and processing.
Significance: The ubiquitin receptor Rpn13 defines asymmetry and thus imposes directionality in the 26S proteasome.
Keywords: Proteasome, Protein Complexes, Protein Degradation, Proteolytic Enzymes, Ubiquitin, S5a, Ubiquitin Receptor, rpn13, Stoichiometry
Abstract
The 26S double-capped proteasome is assembled in a hierarchic event that is orchestrated by dedicated set of chaperons. To date, all stoichiometric subunits are considered to be present in equal ratios, thus providing symmetry to the double-capped complex. Here, we show that although the vast majority (if not all) of the double-capped 26S proteasomes, both 19S complexes, contain the ubiquitin receptor Rpn10/S5a, only one of these 19S particles contains the additional ubiquitin receptor Rpn13, thereby defining asymmetry in the 26S proteasome. These results were validated in yeast and mammals, utilizing biochemical and unbiased AQUA-MS methodologies. Thus, the double-capped 26S proteasomes are asymmetric in their polyubiquitin binding capacity. Our data point to a potential new role for ubiquitin receptors as directionality factors that may participate in the prevention of simultaneous substrates translocation into the 20S from both 19S caps.
Introduction
The proteasome is the main cellular recycling machine that couples ATP hydrolysis to the unfolding, translocation, and degradation of damaged, misfolded, or otherwise unwanted proteins in the cell. The proteasome consists of two subcomplexes: the 20S catalytic particle, a cylindrical particle in which proteins are digested processively to small peptides, and the 19S regulatory particle (RP),4 which recognizes protein substrates, unfolds, and translocates them into the 20S catalytic core. The proteolytic particle is a 670-kDa barrel-shaped protein complex made of four stacked heptagonal rings; two outer α-rings and two inner β-rings (α1–7β1–7β1–7α1–7). The proteolytic active sites are located on the β-rings, facing the lumen of the barrel, sequestered from the bulk solution (1, 2). The gated channel in the α-ring through which substrates enter the 20S particle is narrow (∼15 Å) (3), thus constricting entry only to unfolded polypeptides. Consequently, a globular substrate must undergo unfolding by the ATPases of the 19S complex (Rpt1–6) prior to translocation and digestion within the 20S particle.
Orchestrated transcription of proteasomal subunits has been shown to be mediated by a single transcription factor both in yeast and mammals (4–6). Yet, this mode of regulation is subject for additional regulatory steps such as transcript stability, translation, post-translational modifications, and protein stability. Accordingly, additional regulation at the protein level must ensure the accurate ratio of proteasomal subunits.
Proteasomal assembly is an orderly process requiring dedicated chaperones (7–9). This process progresses through defined intermediates as exemplified in the assembly of the hexameric ATPase ring (Rpt1–6). The process initiates by formation of three dimers of specific Rpt subunits and is guided by three chaperones (9). Hexamerization serves as a platform for 19S RP assembly and can be thus regarded as an initiating step in this process (7–9). Final steps of RP assembly entail the incorporation of the two proteasomal resident ubiquitin receptors. S5a/Rpn10 was the first ubiquitin-binding proteasomal subunit to be discovered (10, 11) and was shown to bind ubiquitin chains through an ubiquitin-interacting motif. Because inactivation of this subunit (in yeast) had almost no phenotype (11), researchers looked for additional subunits. Subsequently, another subunit of the 19S complex, Rpn13/ADRM1, which docks to Rpn2 through its N-terminal domain, was also shown to bind lysine 48-based polyubiquitin chains (12, 13). Interestingly, although both ubiquitin and proteasomes are essential, the inactivation of Rpn10 and Rpn13 in yeast does not appear to be lethal. Rather, a synthetic phenotype results when mutations that affect the ubiquitin binding sites of Rpn10 and Rpn13 are combined, suggesting a functional linkage between these two ubiquitin receptors (12). Prior studies have implicated Rpn10 and Rpn13 both in the initial binding of polyubiquitinated substrate, as well as in the commitment step (14, 15), which appear to be the crucial event in productive interaction (commitment) between polyubiquitinated substrates and the proteasome (16). Yet, it is unknown how ubiquitinated proteins associate with the 26S proteasome and whether this binding step necessarily commits the substrate to degradation. Rpn10 and Rpn13 have been proposed to be sufficiently close in proximity to interact with the same polyubiquitin chain and thus to possibly comprise one binding site (17). Recent localization studies of the 19S subunits demonstrated that Rpn10 and Rpn13 are peripheral and are located on two opposing ends of the 19S lid (18–20). These studies suggested that the distance between the two receptors allow binding of single tetra-ubiquitin chain simultaneously by both Rpn10 and Rpn13, supporting a potential synergistic mode of action of the two integral 19S ubiquitin receptors (12). In light of this synergistic effect on polyubiquitin binding, we set out to evaluate the ubiquitin receptors incorporation into double-capped (DC) proteasomes.
EXPERIMENTAL PROCEDURES
Haploid and Diploid Yeast Strain Used in This Study
See Table 1 for more details.
TABLE 1.
Genotypes of the diploid and haploid yeast and their parental strains
Strains were validated by PCR and the ability to grow on the indicated relevant selective media. When possible, expression of introduce epitopes was verified by immunoblot.
| Strain number | Strain | Genotype |
|---|---|---|
| 1 | Rpn11-TEV-ProA (21) | MATa; lys2-801; leu2-3, 2-112; ura3-52; his3-Δ200; trp1-1; rpn11::RPN11-TEV-ProA (HIS3) |
| 2 | Rpn11-TEV-ProA (this study) | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; rpn11::RPN11-TEV-ProA (NAT) |
| 3 | Rpn11-HA | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; rpn11::RPN11-HA (hph) |
| 4 | Rpn11-HA/Rpn11-TEV-ProA (matting of 2 and 3) | MATa/MATα; his3Δ1/his3Δ1; leu2Δ0/ leu2Δ0; met15Δ0/MET15; LYS2/lys2Δ0; ura3Δ0/ura3Δ0; rpn11::RPN11-TEV-ProA (NAT)/rpn11::RPN11-HA (hph) |
| 5 | HA-Rpn10 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; rpn10:: HA-RPN10 (NAT) |
| 6 | HA-Rpn10/Rpn11-TEV-ProA (matting of 1 and 5) | MATa/MATα; lys2-801/lys2Δ0; leu2-3, 2-112/leu2Δ0; ura3-52/ura3Δ0; his3-Δ200/his3Δ1; trp1-1/TRP1; rpn11::RPN11-TEV-ProA (HIS3)/RPN11; RPN10/rpn10::HA-RPN10 (NAT) |
| 7 | HA-Rpn13 | MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; rpn13:: HA-RPN13 (NAT) |
| 8 | HA-Rpn13/Rpn11-TEV-ProA (matting of 1 and 7) | MATa/MATα; lys2-801/lys2Δ0; leu2-3, 2-112/leu2Δ0; ura3-52/ura3Δ0; his3-Δ200/his3Δ1; trp1-1/TRP1; rpn11::RPN11-TEV-ProA (HIS3)/ RPN11; RPN13/rpn13::HA-RPN13 (NAT) |
Proteasome Purification and Activity
Yeast proteasome purifications were performed from the relevant strains as described previously (21); briefly, yeast cells were disrupted by glass beads in MS buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 2 mm MgCl2, 1 mm ATP, 10% glycerol) and absorbed to homemade epoxy conjugated IgG beads (Bio-Rad). Where applicable, proteasome release from the matrix was performed by addition of TEV protease. Mammalian proteasome purifications and velocity gradients were performed as described previously (22); briefly, cells were lysed in TNH buffer (20 mm Hepes, pH 7.5, 100 mm NaCl, 1 mm EDTA, 1.5 mm MgCl2, 1% Triton X-100, 2 mm ATP, 1 mm DTT), and cellular lysate was subjected to the indicated immunopurification or immunoblot. Where indicated, velcade (10 μm, 60 min) or cyclohexamide (100 μg/ml) were added. Expression vectors encoding GFP and GST fusions to PSMD14 were described previously (22). S5a and Rpn13 fusions were constructed by standard molecular biology procedures and expressed from pCDNA3.1 vectors, and fusion proteins were expressed as N terminus (S5a) or C terminus (Rpn13). Transient transfections into 293 cells were performed using a calcium phosphate procedure. The various PA28 isoforms were subcloned as C terminus-tagged proteins into the pCDNA3.1 plasmid and expressed transiently in 293 cells. For PA28 experiments, cells were lysed in buffer B (25 mm Tris, pH 7.5, 5 mm MgCl2, 0.2% Nonidet P-40, 2 mm ATP, and 1 mm DTT). PSMD14 and Rpn13 proteasome symmetry evaluations were performed from high molecular weight proteasomal glycerol velocity gradient fractions. Immunopurifications of proteasomes was performed by PSMA1 or by specific epitope tag antibodies as indicated.
Proteasomal and tag antibody source were as follows. Mammalian proteasomes used were: PSMA1 hybridoma (22), Rpn13 (Bethyl and rabbit polyclonal), PSMD14 (rat polyclonal), P97 loading control (rabbit polyclonal). Yeast proteasomal subunits were Rpt subunits (Biomol), Rpn10 and Rpn11 (rat polyclonal), Pre3 (Biomol), and actin-loading control (Biomol). The sources for tagged epitope antibodies were FLAG (M2 sigma), HA (12CA5), GST (rabbit polyclonal), and GFP (rat monoclonal).
Determining the Molecular Ratios of Rpn10 and Rpn13 in the DC 26S Proteasome and the Intact 19S Particle by AQUA Peptide Standards
AQUA peptides were selected based on the criteria outlined by Gerber et al. (23), tested, and found suitable for accurate MS quantification. The following peptides were synthesized by Sigma and served in the MS analysis in yeast: FATLGIDPPK(13C15N15) for Rpt1; IFALVFSSNE(R13C15N) for Rpn13; VLSTFTAEFG(K13C15N) for Rpn10. For MS analysis from 293 cells, the following peptides were used: ALDEGDIALL(K13C15N) for Rpt1; VAHLAL(K13C15N) for S5a; GDVEAFA(K13C15N) for Rpn13.
MS Quantification using AQUA Peptides
Proteasomes from HEK293 cells were affinity-purified by a PSMA1 immunopurification and released from the IgG beads through a competitive peptide elution. Affinity-purified 26S proteasome and 19S complexes from yeast were released from the IgG beads through TEV digestion and separated on 5–10% gradient native gel. Following electrophoresis and staining, the protein bands corresponding to the 26S proteasome and the intact 19S complex were excised and extensively washed with MiliQ water. The proteins in the gel were reduced (3 mm DTT, 10 mm ammonium bicarbonate, and 10% acetonitrile), modified with 12 mm iodoacetamide in the same buffer, and trypsinized (modified trypsin (Promega)) at a 1:10 enzyme-to-substrate ratio. Following initial calibration, 50 (for mammalian) or 300 (for yeast) fmol of each AQUA peptide were spiked into the mixture of the resulting tryptic peptides. The mixture was resolved by reversed phase chromatography on 0.075 X 200-mm fused silica capillaries (J&W Scientific) packed with Reprosil reversed phase material (Dr. Maisch, GmbH, Germany). Peptides were eluted with 65-min linear gradients of 5 to 45% followed by 15 min of 95% acetonitrile with 0.1% formic acid in water at flow rates of 0.25 μl/min. Mass spectrometry was performed by an ion-trap mass spectrometer (OrbitrapXP, Thermo) in a positive mode using, repetitively, full MS scans followed by multistage activation collision induces dissociation of the seven most dominant ions selected from the first MS scan. The mass spectrometry data were analyzed using the Sequest software (version 3.31, Thermo-Fisher) searching against the yeast or human section of the UniProt database. The peak area of the AQUA peptides and the corresponding sample peptides were calculated using ICIS with mass tolerance of 0.005 Da. Each of the AQUA experiments was repeated at least three times with two additional concentrations of the AQUA peptides giving rise to similar results.
RESULTS
The Ubiquitin Receptor Rpn10 Is Present on Both Ends of Doubly Capped 26S Proteasomes
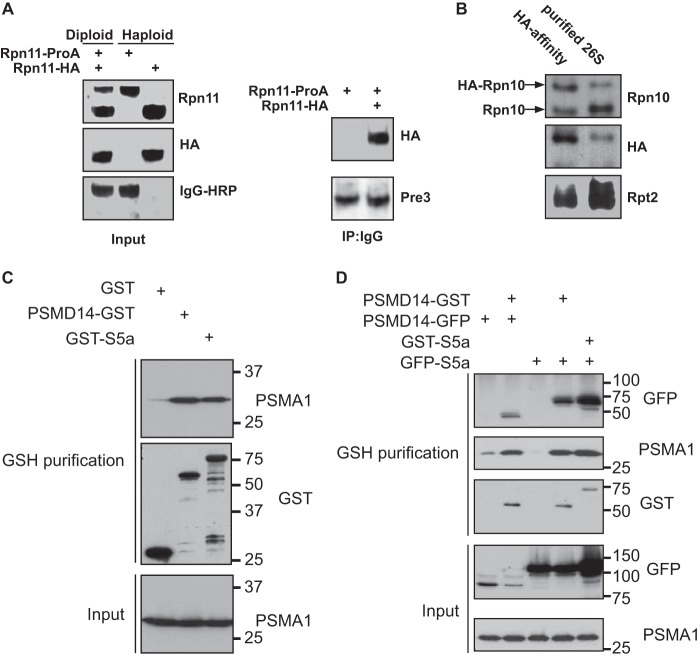
Previous studies raised the possibility that in each DC 26S complex, only one of the 19S regulatory particles has an integral Rpn10 subunit (24, 25). This is an attractive possibility, as it may provide the basis for preventing concomitant processing of substrate by two 19S particles in the DC 26S proteasome. To directly evaluate this possibility, we took advantage of the fact that yeast can be maintained in haploid or diploid life cycles. The rationale guiding this experiment was to generate a diploid yeast strain expressing two distinct versions of Rpn10 (owing to specific epitopes and differential MW). If each 26S proteasome contains only one Rpn10, a 26S complex purified through a tag unique to one of the expressed variants of Rpn10, would not contain the other version of Rpn10. In contrast, if 26S proteasomes include an Rpn10 subunit in each of the two decorating 19S regulatory particles, the purified proteasomes should contain both versions of Rpn10. As a control, we generated a diploid strain expressing two differentially tagged versions of Rpn11 (Rpn11-TEV-ProA and Rpn11-HA; Table 1). Rpn11 is an essential subunit that is presumed to be present in both 19S complexes decorating the same 20S particle. Purifying 26S complexes via the ProA tag of Rpn11 gave rise to a uniformly catalytic particle proteasome as judged by EM and native gels (data not shown). Upon ProA proteasome purification from the Rpn11 diploid strain harboring both Rpn11 tags, an HA-tagged Rpn11 was present, suggesting a 2:1 molar ratio between the tagged subunits (Rpn11) and the 26S particle, thereby demonstrating the validity of this approach (Fig. 1A). We constructed an Rpn11-TEV-ProA diploid strain harboring only one genomic copy of HA-tagged Rpn10, whereas the other Rpn10 copy was untagged (Table 1). 26S proteasomes from the above strain were affinity-purified on IgG beads, released from the matrix by TEV protease and then immunopurified with an HA antibody. As shown in Fig. 1B, comparable amounts of untagged and HA-tagged versions of Rpn10 were detected in the HA pulldown, thus demonstrating the occupancy of two Rpn10 molecules in a DC 26S proteasome.
FIGURE 1.
Rpn10 is present in the 19S complexes on both ends of a DC 26S proteasome. A, left panel: Western blot analysis of haploid strains expressing Rpn11-proA or Rpn11–3×HA, as well as diploid yeast expressing both differentially tagged versions of Rpn11. Right panel: 26S proteasome affinity purified on IgG beads were released from the matrix by TEV protease and analyzed for the present of HA-tagged Rpn11. Pulldown of complete proteasome were verified by immunodetection of a 20S subunit in the immunoprecipitate from both haploid and diploid strains. B, 26S proteasome were affinity purified using IgG-linked beads from a diploid strain expressing 3×HA-tagged and wild-type Rpn10 as well as Rpn11-ProA (Table 1). The 26S particles were released from the matrix by TEV protease and were then incubated with agarose beads decorated with HA antibodies. The immunoprecipitate was subjected to Western blot analysis using anti Rpn10, HA, and Rpt2 antibodies. Both wild-type and 3×HA-tagged Rpn10 were detected indicating that the two Rpn10 versions are present at least in some of the 26S proteasomes. Co-precipitation of other proteasomal subunits was confirmed by the detection of Rpt2. C, 293 cells were transfected with the indicated expression plasmids and subjected to glutathione affinity purification. Proteasomal purification by the GSH matrix was evaluated by immunoblots toward PSMA1. D, 293 cells were transfected with the indicated expression plasmids, and cell lysates were fractionated by velocity gradient centrifugation. Proteasome-containing fractions were pooled and subjected to glutathione affinity purification. Existence of doubly bound PSMD14 and S5a proteasome complexes was determined by a GFP immunoblot.
To test the universality of these findings, we transversed to mammalian cells and simultaneously expressed two tagged versions of S5a, the mammalian homologue of Rpn10. We expressed in 293 cells both GST-S5a and GFP-S5a, and as a control, we also expressed two differentially tagged versions of PSMD14 (an ortholog of the yeast Rpn11 subunit). As seen in Fig. 1C, GST N-terminal tagging of either PSMD14 or S5a did not perturb association with the proteasome (note the reactivity of the PSMA1 immunoblot). Co-expression of double-tagged PSMD14 or S5a, followed by glutathione affinity purification, indicated the presence of both tagged proteins in the same DC 26S particle. This is noted by the GFP-tagged version of S5a or PSMD14 being detected in the purified 26S complex (Fig. 1D, lanes 2 and 5), suggesting a 2:1 molar ratio between the tagged subunits and the 20S particle. Due to the similar MW of GFP and GST, we were unable to distinguish between both tagged forms upon co-expression in an S5a immunoblot (data not shown). These findings further support the results obtained in yeast, we thus conclude that each of the two 19S regulatory complexes decorating a 20S catalytic particle may contain a copy of Rpn10 both in yeast and in mammals.
The Presence of a Single Copy of Rpn13 Enforces an a Priori Asymmetry to the 26S Complex
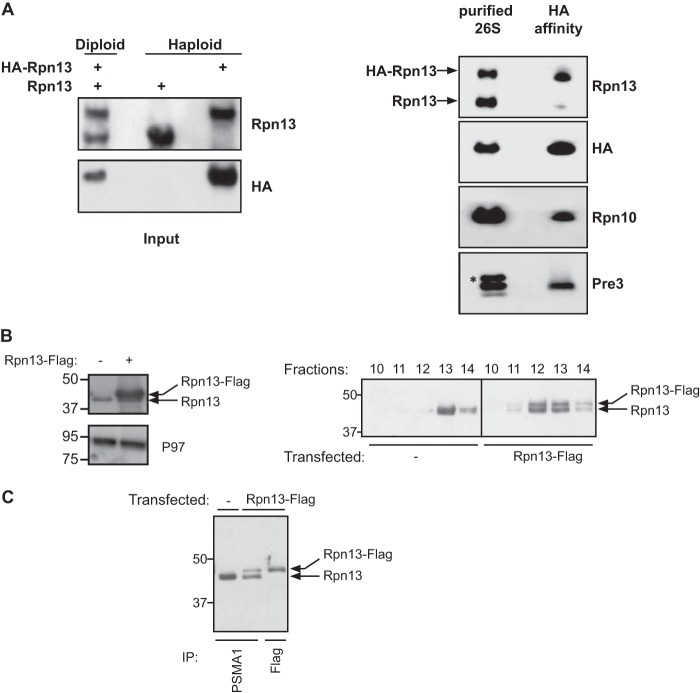
Our findings with Rpn10 support a model in which the 26S proteasome is symmetric (at least prior to substrate binding) with no preferred side for engaging polyubiquitinated substrates or release of the degradation products. Yet, the 19S contains an additional subunit (Rpn13) that was implicated in polyubiquitin binding (12, 13). To directly assess the occupancy of Rpn13 in the two 19S regulatory complexes of a single 26S proteasome, we utilized the same experimental scheme used to evaluate ratios of Rpn10/S5a and Rpn11/PSMD14 (Fig. 2). To this end, we constructed a diploid strain harboring one genomic copy of 3×HA-tagged Rpn13, as well as a genomic copy of the untagged Rpn13. In addition, one of the genomic copies of Rpn11 was fused to a protein A domain (Rpn11-TEV-ProA) (Fig. 2A and Table 1). Cell extracts from this diploid strain were used to affinity purify 26S proteasomes on IgG beads. The 26S particles were released from the matrix by TEV protease and then subjected to HA affinity purifications. As shown in Fig. 2A, only the HA-tagged version of Rpn13 was detected at the end of the double affinity purification procedure. This Rpn13 subunit was integrated in the 26S proteasome as evident from the Rpn10 and Pre3 immunoblots (Fig. 2A). Furthermore, as seen in Fig. 2A, Western blot analysis of IgG purified proteasomes demonstrate the presence of HA-tagged and untagged versions of Rpn13 in a 1:1.2 ratio, thereby eliminating the possibility of a bias preference of the HA-Rpn13 to incorporate into proteasomes.
FIGURE 2.
A single Rpn13 subunit is present in DC 26S proteasome. A, left panel: Western blot analysis of total cell lysate and HA immunoprecipitate prepared from haploid yeast strains expressing a genomic version of wild type and 3×HA N-terminally tagged Rpn13, as well as a diploid strain expressing both versions of Rpn13. Right panel: 26S proteasome were affinity-purified using IgG-linked beads from a diploid strain expressing 3×HA-tagged and wild-type Rpn13 as well as Rpn11-ProA (Table 1). The 26S particles were released from the matrix by TEV protease and were then subjected to a HA affinity purification. Affinity-purified content was revealed using anti Rpn13, HA, Rpn10, and Pre3 antibodies. Only the 3×HA-tagged Rpn13 was detected, indicating presence of a single copy of Rpn13 in a 26S proteasomes. Co-precipitation of other proteasomal subunits was confirmed by the detection of Rpn10 and Pre3. An asterisk indicates a nonspecific band. B, cell lysates from the indicated cells directly evaluated toward the total Rpn13 and P97 content (left panel). The indicated lysates were fractionated on a glycerol gradient and split into 14 equal fractions and Rpn13 content in the HMW fractions was evaluated by an Rpn13 immunoblot detecting both the endogenous and exogenous Rpn13 forms (right panel). C, proteasomal fractions (fractions 10–14) from B were subjected to a PSMA1 or a FLAG affinity purification as indicated. Note that although the PSMA1 purification from the transfected cells had a higher endogenous Rpn13 content, this isoform was completely absent from the FLAG purification.
To test the universality of these findings, we evaluated the situation in mammalian cells and expressed a FLAG-tagged Rpn13 that could be discriminated based on its higher MW from the endogenous Rpn13. Total cell lysates indicate the higher level of Rpn13 expression upon overexpression of Rpn13-Flag (Fig. 2B, left), thus excluding the possibility of Rpn13 levels being a limiting factor in particle integration. Cell lysates were fractionated on a glycerol gradient and high molecular weight proteasome containing fractions (Fig. 2B, fractions 10–14) were evaluated toward their Rpn13 content. As shown in Fig. 2B (right panel), equal expression and distribution are seen between the endogenous and exogenous expressed Rpn13 in the HMW fractions. We then performed a proteasome purification (PSMA1 IP) from both transfected and untransfected cells and revealed that upon transfection, both Rpn13 versions could be detected (with a bias toward the untagged Rpn13; Fig. 2C, lane 2). Despite this bias, when we performed a FLAG affinity purification from the transfected cells, we did not detect any endogenous untagged Rpn13 present in our purified material, implying that 26S proteasomes contain only one copy of Rpn13 unlike S5a or PSMD14. Notably, Rpn13 genomic tagging (yeast) or ectopic expression (mammals) did not alter the amount of double-tagged proteasomes as evaluated by native gel (data not shown). Taken together, our findings suggest that only a single copy of Rpn13 is present in a 26S complex, conferring an inherent asymmetry on the majority of the 26S proteasome population, a feature conserved from yeast to mammals.
AQUA-MS Analysis of Rpn10 and Rpn13 Molar Ratio in the Intact 26S and 19S Complexes
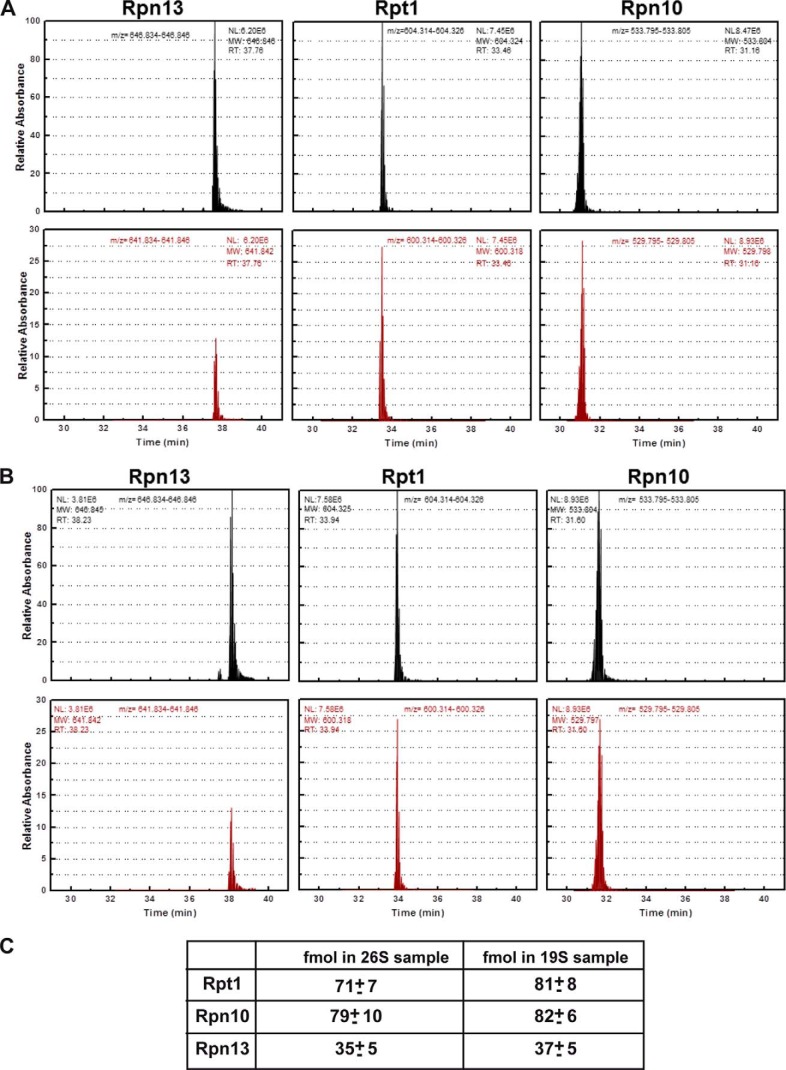
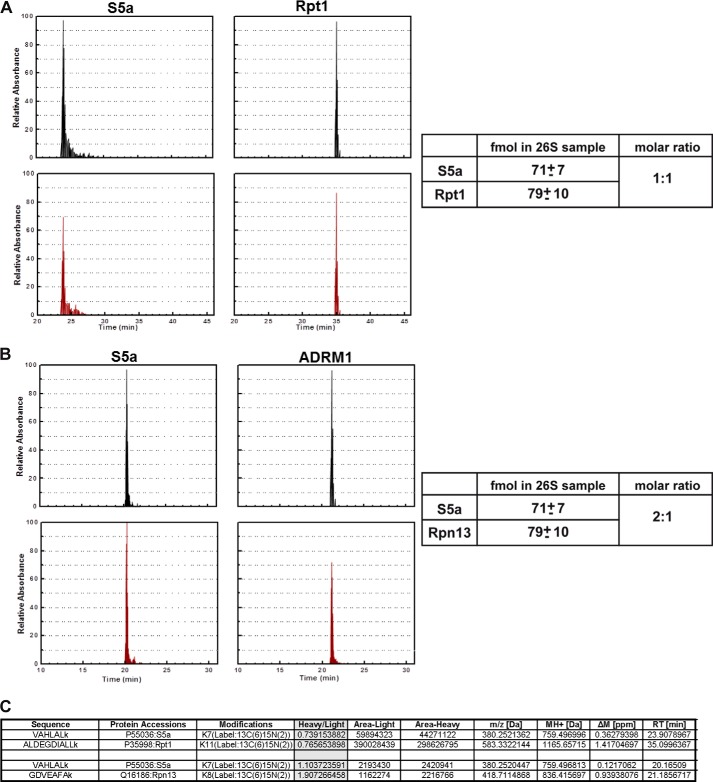
The analysis of Rpn10 and Rpn13 occupancy in a DC 26S proteasome presented above has two drawbacks. First, as the two tagged experiments are only qualitative, we cannot deduce what portion of the double capped 26S proteasome contains two Rpn10 subunits. Moreover, even semi-quantitative estimates relaying on the intensity of immunoblot is not possible as several of our experiment relay on different antibodies (Figs. 1 and 2). Thus, our findings merely demonstrate that a population of proteasomes decorated with two copies of the ubiquitin receptor Rpn10/S5a, exist. Yet, these findings could not exclude the possibility that a major portion of the 26S proteasomes are decorated with only one Rpn10 subunit and some might be missing Rpn10 altogether. Similarly, the biochemical analysis of the occupancy Rpn13 molecules in 26S proteasome can only suggest that when present, only one copy of Rpn13 could be found in a 26S complex. Again, no quantitative information on the amount of 26S complexes that do not contain Rpn13 (if existent) can be obtained. An additional possible limitation of our analysis has to do with the introduction of tags (HA, FLAG, GST, or GFP) to the subunits of interest (Rpn10/S5a, Rpn11/PSMD14, and Rpn13). Although we demonstrate that these tags have no detectable effect on the integrity, assembly, and preferential integration toward tagged or untagged versions of Rpn10/S5a and Rpn13 (Figs. 1 and 2), we could not exclude possible effects of the tags that may have escaped our analysis. To overcome these limitations and obtain direct quantitative measurements of the diversity in occupancy of Rpn10/S5a and Rpn13 in the 26S proteasome, we utilized AQUA peptide MS analysis that allows direct quantification of proteins in a given sample (23, 26). We thus set to determine the actual occupancy of Rpn10/S5a and Rpn13 in the 26S and 19S purified samples (Figs. 1 and 2) utilizing AQUA peptides corresponding to Rpn10/S5a, Rpn13, and Rpt1. Rpt1 (one of the 19S ATPases) must be present in a molecular ratio of 2:1 in the double-capped 26S proteasome (and 1:1 in the intact 19S complex) as an essential 19S subunit and thus served as a standard for 2:1 molar ratio occupancy (two subunits for one DC 26 complex) in this assay. As seen in Fig. 3, the amount of Rpn10 and Rpt1 was equal, whereas that of Rpn13 was half that of Rpn10, suggesting that indeed most DC 26S proteasomes contain only a single copy of Rpn13. Similarly, only half of the intact 19S particles contained this subunit. Notably, AQUA MS evaluation of the same subunits in the mammalian 26S proteasome revealed similar ratios between the various subunits (Fig. 4). Our ability to purify homogenous 26S proteasome suggests that our AQUA-MS analysis truly represents the content of these complexes. Taken together with our biochemical analysis, we conclude that each of the two 19S particles decorating a DC 26S proteasome contains an Rpn10 ubiquitin receptor, and only one of these 19S regulatory particles contains the Rpn13 ubiquitin receptor, enforcing asymmetry on the DC 26 proteasome at least in its capacity to bind polyubiquitinated targets.
FIGURE 3.
AQUA MS determination of molar ratios of Rpn10 and Rpn13 in yeast 26S and 19S particles. Top plots represent AQUA peptide elution profiles corresponding to 300 fmol peptide of the indicated proteasome subunits. Lower rows represent elution profiles of endogenous proteolytic peptides corresponding to the AQUA peptides detected in the 26S (A) and 19S (B) samples. The 26S and 19S particles were excised from native gels, trypsinized, spiked with 300 fmol of the indicated AQUA peptides, separated on reversed phase chromatography, and analyzed by tandem mass spectrometry. C, amounts in fmol of AQUA peptide corresponding to Rpt1, Rpn10, and Rpn13 illustrating ratios of 2:2:1, respectively. The known concentration of each AQUA peptide was spiked into the mixture of the proteasome tryptic peptides. The peak area of the AQUA peptide and the corresponding sample peptide were calculated using ICIS with mass tolerance of 0.005 Da and the ratio between them was determined. The unknown concentration of the sample protein was calculated using this ratio and the known concentration of the AQUA peptide. The AQUA experiment was repeated least three times with two additional concentrations of the AQUA peptides giving rise to similar results. Note the 1:2:2 ratio of Rpn13/Rpt1/Rpn10 in both the DC 26S proteasomes and the intact 19S complexes.
FIGURE 4.
AQUA MS determination of molar ratios of Rpn10 and Rpn13 in mammalian 26S proteasome. Top plots represent AQUA peptide elution profiles corresponding to 50 fmol peptide of the indicated proteasome subunits. Lower rows represent elution profiles of endogenous proteolytic peptides. Analysis and ratio calculations were performed on mammalian 26S proteasomes as in Fig. 3. AQUA peptides detected in 26S samples comparing Rpt1 versus S5a (A) or S5a versus Rpn13 (B) are presented with the calculated ratios. Raw data of the indicated peptides are presented (C).
Rpn13 Presence in a Designated Ingress 19S Cap
Asymmetry of a DC proteasome may imply that an incoming substrate is preferably designated toward one side of the proteasome rather than the other. Other than the 19S, one of the most intensively studied activators of the proteasome is PA28αβ ((27); also known as REG or the 11S regulator), a heteroheptameric complex of 28-kDa subunits. PA28 binds to the 20S proteasome, but in contrast to the 19S, PA28 lacks ATPase activity and the ability to bind ubiquitin conjugates. It has been proposed that PA28 could facilitate the exit of peptide products from chimeric 19S-20S-PA28 proteasomes by providing an exit channel, thus providing directionality (28, 29). In fact the ability of PA28 to dilate the 20S entry port was demonstrated through structural studies of its homoheptameric homolog from trypanosomes, PA26 (29). To determine whether the Rpn13 containing cap functions as ingress versus egress cap, we took advantage of the fact that PA28 forms hybrid proteasomes (PA28–20S-19S) in vivo (30), reasoning that such a hybrid proteasome could only support ingress of a polyubiquitinated substrate from the ATPase containing cap, as the PA28 cap could not support unfolding of an incoming globular substrate (31). We ectopically expressed in mammalian cells PA28 α and β subunits and monitored the presence of Rpn13 in the in vivo assembled PA28–20S-19S hybrid proteasome. As noted in Fig. 5, upon purification of PA28α, both 20S components (PSMA1) and 19S components (PSMD14 and Rpn13) could be detected, indicating that the Rpn13 ubiquitin receptor resides within an ingress designated cap. Although this conclusion may seem obvious, it is noteworthy that in a DC 26S proteasome, a designated egress 19S cap also contains the additional ubiquitin receptor (Rpn10/S5a). As recently reported (32), this result may imply importance of structural non-ubiquitin binding related features of Rpn10/S5a in the 19S complex.
FIGURE 5.
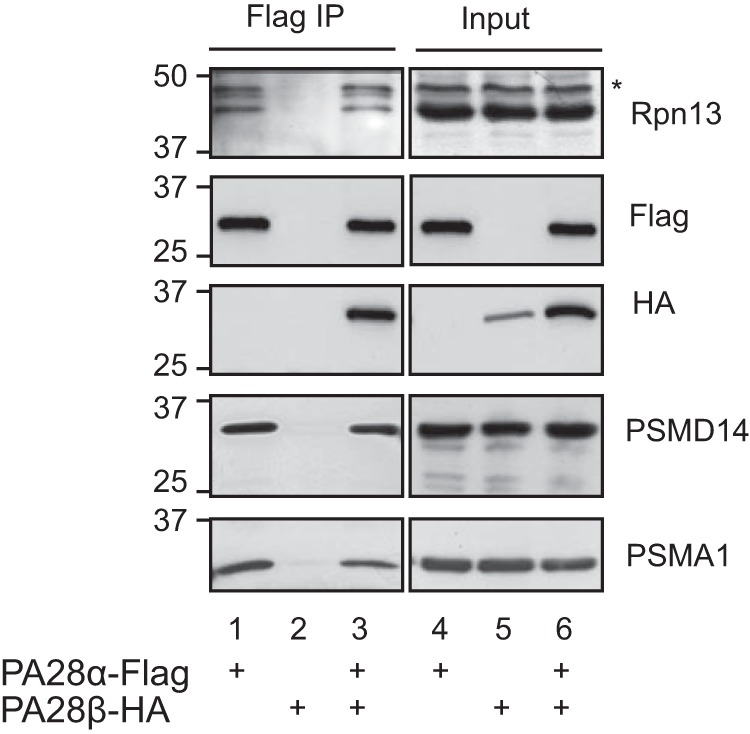
Rpn13 co-immunoprecipitates with hybrid proteasome containing the PA28 complex. 293 cells were transiently transfected with the indicated expression plasmids. Total cellular content was revealed directly (lanes 4–6; input) or were subjected to a FLAG affinity purification (lanes 1–3). PA28α-FLAG served as a positive control for the ability to purify hybrid proteasomes as indicated by the presence of 20S (PSMA1) and 19S (PSMD14) subunits. PA28β-HA served as a negative control for the FLAG immunoprecipitation when expressed without PA28α. An asterisk indicates a nonspecific band.
DISCUSSION
Our homogeneous proteasome purification enabled us to obtain quantitative data on double-capped 26S proteasomes. Although recent 26S EM structures have revealed the relative positioning of the various proteasome subunits (18–20), they did not address the quantitative aspects that may play a critical role in determination of substrate processing by the 26S proteasome. In fact, careful examination of previous data presenting proteasomal subunit staining from yeast (see Ref. 20 and supplemental Fig. S2 therein), indicate a sub-stoichiometric occupancy of Rpn13. Our purification of double-capped proteasomes from yeast and mammals indicate the existence of two Rpn11/PSMD14 as well as two Rpn10/S5a subunits in a DC proteasome (Fig. 1). These results further confirm a recent conclusion regarding S5a stoichiometry (33), whereas data raised the possibility of a non-stoichiometric amount of S5a in the proteasome (24, 25). Using the same methodology, we were unable to detect the presence of two Rpn13 copies in a DC proteasome, both in yeast and in mammals (Fig. 2). Furthermore, the mammalian integration of the FLAG-Rpn13 was biased against its incorporation compared with the endogenous isoform (Fig. 2C); yet no endogenous Rpn13 could be detected in our proteasomes purified by FLAG-Rpn13 (Fig. 2C). Although our biochemical data indicate the qualitative aspects regarding the ability of Rpn13 to bind proteasomes in a non-symmetric form, our AQUA peptide MS data confirm these results in a quantitative manner (Figs. 3 and 4). Thus, not only is Rpn13 found only in one side of a DC proteasome, it is found in this manner in the majority of the proteasome population.
Interestingly, a recent report using a high resolution EM analysis detected polyubiquitinated substrates on both ends of an in vitro-assembled DC proteasome (34); however, this result was obtained with proteasomes that lack the asymmetrical Rpn13 subunit. In several cases, proteasomal engagement has been demonstrated not to be the rate-limiting factor in substrate turnover (14, 35); therefore, there is no kinetic disadvantage in acquiring only one high affinity polyubiquitin binding 19S RP in a DC proteasome. Directional processing by the DC 26S proteasome would be advantageous if the generated peptides require 20S ring dilation to exit the 20S chamber. In this manner, directionality in a DC proteasome would ensure efficient peptide release and ensure processivity in the turnover of substrates.
Recent findings demonstrated that Rpn10 and Rpn13 both contribute to the high-affinity binding and processing of the polyubiquitin chain (Ka was reduced 2-fold upon inactivation of either receptor (14)). Taken together, these findings support the notion that synergy of both ubiquitin receptors on the same 19S cap, will favor this side of the double-capped proteasome, as the ingress cap. It is tempting to hypothesize that once substrate engage with the “ingress cap” a cooperative structural transition is transduced to the opposing cap to further diminish its capacity to recruit an additional substrate, thereby preventing simultaneous processing of substrate by both 19S capes. The a priori lack of symmetry found in the double-capped proteasome (Fig. 2) is not due to quantitative limitations, as overexpression of Rpn13-FLAG increased by several folds the total protein level without reducing endogenous Rpn13 levels (Fig. 2B) and enabled integration of both Rpn13 isoforms to proteasomal 19S RP (Fig. 2C). The existence of a sensing mechanism is an attractive possibility as it provides an additional layer of regulation in preventing simultaneous engagement of both caps with substrates. The presence of a sensing mechanism in addition to the asymmetric distribution of Rpn13 may also enable cooperative sensing between the proteasome active sites and the 20S-19S interface as reported previously (36, 37) and may also facilitate correct matching of RP to specific catalytic particle (data not shown). The molecular details of such regulatory mechanisms remain to be resolved.
Although lack of Rpn13 symmetry affects ubiquitin affinity to the 19S cap, it also directly implies differences in deubiquitinating enzyme capacity as UCH37 binding to the 19S cap is Rpn13-dependent (38, 39). Moreover, structural data suggest possible role for Rpn13 in Ubp6 function in the 19S cap (20) (data not shown),5 raising the possibility for a more complex scenario, in which the occupancy of Rpn13/UCH37 and Ubp6/Usp14 is coordinated. Thus, lack of Rpn13 has wide implications regarding polyubiquitin processing by the proteasome (binding and de-ubiquitination) and may be subject for further regulation.
D. Berko, O. Herkon, I. Braunstein, E. Isakov, Y. David, T. Ziv, A. Navon, and A. Stanhill, manuscript in preparation.
- RP
- regulatory particle
- DC
- double-capped
- TEV
- tobacco etch virus.
REFERENCES
- 1. Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 2. Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution [see comments]. Science 268, 533–539 [DOI] [PubMed] [Google Scholar]
- 3. Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 4. Mannhaupt G., Schnall R., Karpov V., Vetter I., Feldmann H. (1999) Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450, 27–34 [DOI] [PubMed] [Google Scholar]
- 5. Radhakrishnan S. K., Lee C. S., Young P., Beskow A., Chan J. Y., Deshaies R. J. (2010) Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steffen J., Seeger M., Koch A., Krüger E. (2010) Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 40, 147–158 [DOI] [PubMed] [Google Scholar]
- 7. Murata S., Yashiroda H., Tanaka K. (2009) Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 8. Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tomko R. J., Jr., Funakoshi M., Schneider K., Wang J., Hochstrasser M. (2010) Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol. Cell 38, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. (1994) A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269, 7059–7061 [PubMed] [Google Scholar]
- 11. van Nocker S., Sadis S., Rubin D. M., Glickman M., Fu H., Coux O., Wefes I., Finley D., Vierstra R. D. (1996) The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 16, 6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., Dikic I. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., Elsasser S., Finley D., Dikic I., Walters K. J., Groll M. (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peth A., Uchiki T., Goldberg A. L. (2010) ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol. Cell 40, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang F., Hu M., Tian G., Zhang P., Finley D., Jeffrey P. D., Shi Y. (2009) Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang N., Wang Q., Ehlinger A., Randles L., Lary J. W., Kang Y., Haririnia A., Storaska A. J., Cole J. L., Fushman D., Walters K. J. (2009) Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol. Cell 35, 280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beck F., Unverdorben P., Bohn S., Schweitzer A., Pfeifer G., Sakata E., Nickell S., Plitzko J. M., Villa E., Baumeister W., Förster F. (2012) Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 109, 14870–14875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. da Fonseca P. C., He J., Morris E. P. (2012) Molecular model of the human 26S proteasome. Mol. Cell 46, 54–66 [DOI] [PubMed] [Google Scholar]
- 20. Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., Martin A. (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 22. Stanhill A., Haynes C. M., Zhang Y., Min G., Steele M. C., Kalinina J., Martinez E., Pickart C. M., Kong X. P., Ron D. (2006) An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell 23, 875–885 [DOI] [PubMed] [Google Scholar]
- 23. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nickell S., Beck F., Scheres S. H., Korinek A., Förster F., Lasker K., Mihalache O., Sun N., Nagy I., Sali A., Plitzko J. M., Carazo J. M., Mann M., Baumeister W. (2009) Insights into the molecular architecture of the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 106, 11943–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bohn S., Beck F., Sakata E., Walzthoeni T., Beck M., Aebersold R., Förster F., Baumeister W., Nickell S. (2010) Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 20992–20997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirkpatrick D. S., Gerber S. A., Gygi S. P. (2005) The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods 35, 265–273 [DOI] [PubMed] [Google Scholar]
- 27. Ahn J. Y., Tanahashi N., Akiyama K., Hisamatsu H., Noda C., Tanaka K., Chung C. H., Shibmara N., Willy P. J., Mott J. D. (1995) Primary structures of two homologous subunits of PA28, a γ-interferon-inducible protein activator of the 20S proteasome. FEBS Lett. 366, 37–42 [DOI] [PubMed] [Google Scholar]
- 28. Köhler A., Cascio P., Leggett D. S., Woo K. M., Goldberg A. L., Finley D. (2001) The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell 7, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 29. Whitby F. G., Masters E. I., Kramer L., Knowlton J. R., Yao Y., Wang C. C., Hill C. P. (2000) Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408, 115–120 [DOI] [PubMed] [Google Scholar]
- 30. Tanahashi N., Murakami Y., Minami Y., Shimbara N., Hendil K. B., Tanaka K. (2000) Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J. Biol. Chem. 275, 14336–14345 [DOI] [PubMed] [Google Scholar]
- 31. Cascio P., Call M., Petre B. M., Walz T., Goldberg A. L. (2002) Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 21, 2636–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin Y. L., Sung S. C., Tsai H. L., Yu T. T., Radjacommare R., Usharani R., Fatimababy A. S., Lin H. Y., Wang Y. Y., Fu H. (2011) The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. Plant Cell 23, 2754–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakata E., Bohn S., Mihalache O., Kiss P., Beck F., Nagy I., Nickell S., Tanaka K., Saeki Y., Förster F., Baumeister W. (2012) Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 1479–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matyskiela M. E., Lander G. C., Martin A. (2013) Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prakash S., Inobe T., Hatch A. J., Matouschek A. (2009) Substrate selection by the proteasome during degradation of protein complexes. Nat. Chem. Biol. 5, 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kusmierczyk A. R., Kunjappu M. J., Kim R. Y., Hochstrasser M. (2011) A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat. Struct. Mol. Biol. 18, 622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kleijnen M. F., Roelofs J., Park S., Hathaway N. A., Glickman M., King R. W., Finley D. (2007) Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 14, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 38. Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., Murata S. (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25, 4524–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yao T., Song L., Xu W., DeMartino G. N., Florens L., Swanson S. K., Washburn M. P., Conaway R. C., Conaway J. W., Cohen R. E. (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 8, 994–1002 [DOI] [PubMed] [Google Scholar]