Background: PFL is a glycyl radical enzyme (GRE) activated by a radical AdoMet-activating enzyme (PFL-AE).
Results: Equilibrium constants for PFL-AE binding to PFL and AdoMet are determined, and the effects of substrates on activation are quantified.
Conclusion: In vivo, PFL-AE exists largely in complex with PFL and AdoMet.
Significance: GREs play key roles in anaerobic metabolism, but their activation is poorly understood.
Keywords: Circular Dichroism (CD), Electron Paramagnetic Resonance (EPR), Radicals, S-Adenosylmethionine (AdoMet), Surface Plasmon Resonance (SPR)
Abstract
The activation of pyruvate formate-lyase (PFL) by pyruvate formate-lyase activating enzyme (PFL-AE) involves formation of a specific glycyl radical on PFL by the PFL-AE in a reaction requiring S-adenosylmethionine (AdoMet). Surface plasmon resonance experiments were performed under anaerobic conditions on the oxygen-sensitive PFL-AE to determine the kinetics and equilibrium constant for its interaction with PFL. These experiments show that the interaction is very slow and rate-limited by large conformational changes. A novel AdoMet binding assay was used to accurately determine the equilibrium constants for AdoMet binding to PFL-AE alone and in complex with PFL. The PFL-AE bound AdoMet with the same affinity (∼6 μm) regardless of the presence or absence of PFL. Activation of PFL in the presence of its substrate pyruvate or the analog oxamate resulted in stoichiometric conversion of the [4Fe-4S]1+ cluster to the glycyl radical on PFL; however, 3.7-fold less activation was achieved in the absence of these small molecules, demonstrating that pyruvate or oxamate are required for optimal activation. Finally, in vivo concentrations of the entire PFL system were calculated to estimate the amount of bound protein in the cell. PFL, PFL-AE, and AdoMet are essentially fully bound in vivo, whereas electron donor proteins are partially bound.
Introduction
Pyruvate formate-lyase (PFL)2 supplies the citric acid cycle with acetyl-CoA during anaerobic glycolysis by catalyzing the reaction of pyruvate + CoA ⇌ acetyl-CoA + formate and is a central enzyme in anaerobic metabolism of Escherichia coli and other facultative anaerobes. PFL is among the growing list of glycyl radical enzymes (1), which play key roles in anaerobic metabolism in microbes, including the reduction of ribonucleotides to deoxyribonucleotides (2), synthesis of benzylsuccinate (3), and conversion of choline to trimethylamine (4). The defining feature of a glycyl radical enzyme is the presence of a stable and catalytically essential glycyl radical in the active site. The glycyl radical is generated by an activating enzyme that belongs to the radical S-adenosylmethionine (AdoMet) superfamily; these radical AdoMet activases utilize a [4Fe-4S] cluster and AdoMet to generate the glycyl radical by direct H-atom abstraction. These glycyl radical enzymes and their activating enzymes are notoriously difficult to study due to the oxygen sensitivity of both the glycyl radical in the glycyl radical enzyme and the [4Fe-4S] cluster in the activating enzymes.
PFL is constitutively expressed in E. coli; however, its expression increases 10–12-fold under anaerobic conditions (5, 6). The enzyme is in an inactive state when produced and must be activated by an activating enzyme (PFL-AE) under anaerobic conditions before catalysis can occur (6–8). PFL exists as a dimer with one active site per subunit (6, 9, 10) and has been shown to exhibit half-site reactivity (5, 11–13). X-ray crystal structures of PFL have revealed that each active site is buried ∼8 Å from the surface of the enzyme (9, 10). These data, together with the evidence that activation requires direct H-atom abstraction from an active site glycine residue (PFL Gly-734) by a deoxyadenosyl radical generated in the PFL-AE active site (14–18), suggest that significant conformational changes of one or both proteins are required during the activation process. Recent biophysical and biochemical studies indeed support a two-state model for PFL, in which the closed state that has been structurally characterized can be converted to an open state in which the glycyl radical loop of PFL is more solvent-exposed (11). This conversion to the open state is favored in the presence of PFL-AE (11).
PFL-AE is a radical S-adenosylmethionine (AdoMet or AdoMet) enzyme that utilizes an iron-sulfur cluster and AdoMet to activate PFL via pro-S hydrogen abstraction from Gly-734 (14, 19). The [4Fe-4S] cluster of PFL-AE is coordinated by the cysteines of a conserved CX3CX2C motif, with the fourth unique iron coordinated by AdoMet (20–23, 32). PFL-AE cycles between two different oxidation states during hydrogen abstraction, [4Fe-4S]2+/1+, with the [4Fe-4S]1+ being the state capable of reductive cleavage of AdoMet to generate the glycyl radical (12). In vivo, it is believed that PFL-AE is reduced by flavodoxin, which is first reduced by either NADP+:flavodoxin oxidoreductase or pyruvate:flavodoxin oxidoreductase (7, 8, 24–26).
The activation of PFL by PFL-AE involves intriguing issues of protein-protein interactions, associated protein conformational changes, and protected generation and transfer of highly reactive carbon radical species. In this publication, we provided biophysical insight into interactions between PFL and PFL-AE using surface plasmon resonance under anaerobic conditions, and we explore the roles of AdoMet and PFL substrates on this interaction. Our own data together with some previously published work has allowed us to estimate the degree to which the PFL system components are bound in complexes in vivo and to provide a more complete understanding of the conditions under which PFL activation occurs.
EXPERIMENTAL PROCEDURES
Protein Preparation and Small Molecules
PFL-AE and PFL was grown and purified as published previously (12, 20, 27, 28). PFL-AE was quantified using ϵ280 nm = 39.4 mm−1 cm−1, which was in agreement with the Bradford assay (29) using a correction factor of 0.65 (19). Two batches of PFL-AE were prepared for AdoMet binding assays and PFL activation assays. Iron assays were performed on both batches, and iron content was determined to be 2.83 ± 0.03/protein for AdoMet binding assays and 3.96 ± 0.02/protein for PFL activation assays by a previously published method (30). PFL was quantified using either the Bradford assay or ϵ280 nm = 178 mm−1cm−1, with both techniques giving identical values (29). The PFL-AE and PFL extinction coefficients were obtained using the ExPASy ProtParam tool. S-adenosylmethionine was synthesized using AdoMet synthetase and purified as described previously (22). The small molecule substrates pyruvate, oxamate, and coenzyme A that were used in PFL activation assays were obtained from Sigma Aldrich and were of the highest commercially available quality and used without further purification.
PFL and PFL-AE-binding Studies
All experiments were carried out in triplicate under anaerobic conditions using a Biacore X-100 with the Plus package in a Coy chamber (Coy Laboratories, Grass Lake, MI). The ligand protein (PFL-AE) was attached to a CM5 sensor chip using standard thiol coupling procedures as recommended by the manufacturer (31). PFL-AE contains six cysteine residues (Cys-12, Cys-29, Cys-33, Cys-36, Cys-94, Cys-102) and only Cys-29, Cys-33, and Cys-36 are involved in coordinating the iron sulfur cluster (32). Mutagenesis studies show that Cys-29, Cys-33, and Cys-36 are the only cysteine residues essential for catalysis (33); therefore Cys-12, Cys-94, and Cys-102 are potential targets for thiol coupling.
Thiol coupling was performed at a flow rate of 5 μl/minute, and all injections lasted 400 s. A 1:1 (v/v) mixture of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide·HCl (EDC) and N-hydroxysuccinimide (200 mm EDC and 50 mm N-hydroxysuccinimide) was injected on the CM5 sensor chip to activate the carboxylic acid moieties using the Biacore X-100. 2-(2-Pyridinyldithio)ethaneamine hydrochloride was freshly prepared at 80 mm in 0.1 m borate buffer at pH 8.5. The ligand protein PFL-AE was injected at a concentration of 22.7 mg/ml, resulting in a baseline increase of 557 resonance units. The unreacted 2-(2-pyridinyldithio)ethaneamine hydrochloride was blocked by injecting 50 mm l-cysteine in 20 mm HEPES, 10 mm NaCl, pH 5.5, and the signal was decreased by 224 resonance units. We estimate that 360 RU of PFL-AE was coupled to the CM5 biosensor. Experiments were run at 25 °C in 20 mm HEPES, 10 mm NaCl, pH 7.4. Analytes were injected at a flow rate of 30 μl/min with a contact time of 180 s and a dissociation time of 60 s. Experimental sensograms were corrected by subtracting the response from the control flow cell. After each experiment, immobilized PFL-AE was regenerated using 20 mm HEPES, 500 mm KCl, 0.005% polysorbate 20, 200 mm imidazole, pH 7.4, with a regeneration time of 180 s, which completely removed the PFL and restored the preinjection baseline. Typical experiments included two blank cycles with buffer followed by three trials, each separated by one blank cycle. Five experimental PFL concentrations were prepared for each trial by making a 3-fold dilution with a maximum concentration of 10 μm PFL dimer. Triplicate experiments resulted in similar Rmax values indicating that PFL-AE was not damaged during regeneration. Sensograms for PFL-AE and PFL binding in single cycle kinetics mode were fit to a Langmuir 1:1 interaction model using BIAevaluation software (from GE Healthcare) available on the Biacore X-100 plus package.
AdoMet Binding Studies
CD experiments were run in triplicate under anaerobic conditions using a Jasco-710 spectropolarimeter at room temperature. Visible region measurements were collected using a 1-cm path length cuvette, and far-UV spectra were run using a 0.1-mm path length cuvette. For visible region scans, the sensitivity of the Jasco-710 was set to 100 millidegrees, with a data pitch of 0.1 nm, in continuous scan mode at a speed of 100 nm/min, with a response of 1 s, a bandwidth of 1.0 nm, and an accumulation of three scans. The parameters for far-UV scans were exactly the same as the performed in the visible region except a scan rate of 50 nm/min was used from 195 to 260 nm. The buffer used for all CD experiments was 20 mm HEPES, 250 mm NaCl, 1 mm DTT, pH 7.4, and PFL-AE concentrations were in the range of 50–120 μm in the visible region and 30 μm in the far-UV region. During AdoMet binding experiments, small volumes of concentrated AdoMet were titrated into the cuvette, and CD data were collected from 300 to 800 nm. A control experiment was also performed where buffer was titrated in place of AdoMet to show AdoMet binding was responsible for the changes in the CD spectrum and to provide data for dilution correction of the AdoMet binding experiments. The AdoMet binding results are an average of triplicate data analyzed using the change in ellipticity at 400 nm, which was divided by the maximum change in ellipticity and fit to Equation 1.
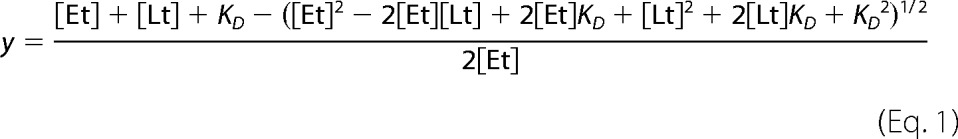 |
The total PFL-AE concentration is represented by the variable Et in the equation below. Lt represents the total AdoMet concentration titrated during the assay, and KD is the equilibrium constant.
PFL Activation Studies
EPR spectra were measured on a Bruker ER-200D-SRC spectrometer at 12 and 60 K for PFL-AE and PFL, respectively, with a frequency of 9.37 GHz. The EPR microwave power was set to 0.06 milliwatt (for examining the glycyl radical of PFL) and 1.59 milliwatt (for PFL-AE) with a modulation frequency of 100 kHz and a 5 gauss modulation amplitude for all samples; all spectra were the sum of four scans. PFL activation reactions were carried out under anaerobic conditions in an mBraun box with <1 ppm O2. PFL-AE was added to an EPR tube at 100 μm with a volume of 350 μl in 100 mm Tris, 250 mm NaCl 10 mm DTT, 100 μm 5-deazariboflavin, pH 7.4 and photoreduced for a time course of 0, 5, 10, 20, 30, and 60 min with a 500-watt halogen bulb. Photoreduced PFL-AE was then added to PFL to make a 1:1 ratio at a final concentration of 50 μm each of PFL-AE and PFL in the presence of 500 μm AdoMet. One PFL substrate was then added to each EPR sample (10 mm pyruvate, 10 mm oxamate, 100 μm CoA, or no substrate), and components were mixed. Samples were pipetted into a clean EPR tube and wrapped in foil before being incubated for 20 min to provide time for the reaction to go to completion. EPR samples were then flash frozen in liquid N2 and stored in a liquid N2 Dewar until the EPR spectrum could be measured. The concentration of the PFL glycyl radical was determined using a K2(SO3)2NO standard according to previously described methods (11, 12, 34). The Km values for PFL substrates have been determined previously to be 2 mm for pyruvate and 7 μm for CoA (5). Equilibrium constants have also been determined for PFL small molecule binding, yielding a KD of 2 mm for oxamate and 100 μm for pyruvate (10). Under the conditions employed during these experiments, we can therefore confidently say that the substrates are at sufficiently high concentrations to interact with PFL and should be close to fully bound.
In Vivo Concentrations of the PFL System
Protein purifications and two-dimensional gel electrophoresis studies provide information on the number of protein copies per cell for the PFL system in E. coli grown under anaerobic conditions in minimal medium and supplemented with glucose (5, 6, 24, 26, 35). A more recent study using cell microscopy determined cell volume for E. coli cells under some of the most commonly used growth conditions (36). We selected the cell volume that corresponded to growth under anaerobic conditions in minimal media and supplemented with glucose to determine the in vivo concentrations for proteins of the PFL system.
RESULTS
PFL-AE Binding Interactions with PFL
Surface plasmon resonance binding experiments were performed under anaerobic conditions to investigate the interaction between PFL and the oxygen-sensitive PFL-AE. We determined the KD for this interaction to be 1.1 ± 0.2 μm at 25 °C (Fig. 1). The association rate for complex formation was determined to be 1028 ± 34 (m−1 s−1). When compared with other biological systems, this rate is very slow and on the low end for protein-protein interactions; this indicates that the association rate is limited by large conformational changes rather than by diffusion (37). Indeed, conformational changes are evident in the crystal structure of PFL-AE upon binding of AdoMet and the 7-mer peptide analog of the PFL active site (32). Conformational changes have also been detected in PFL upon binding of PFL-AE when the active site loop on PFL must unfold to interact with the binding site in PFL-AE (11).
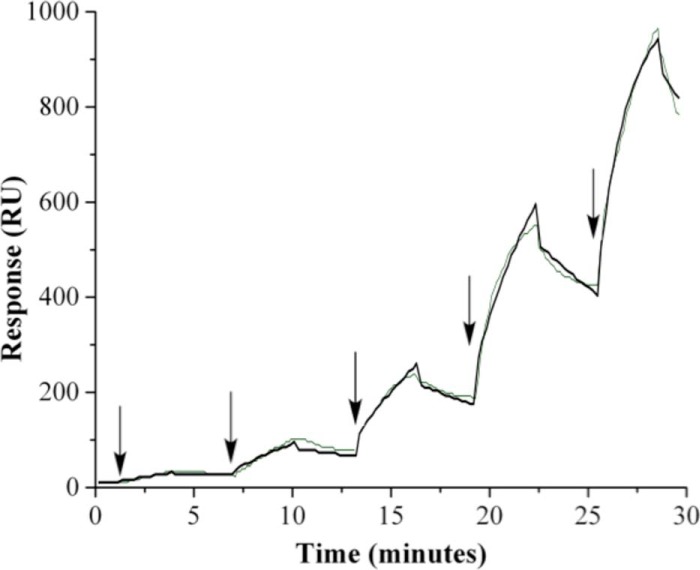
FIGURE 1.
PFL-AE and PFL binding kinetics using surface plasmon resonance under anaerobic conditions. PFL-AE was covalently attached to a CM5 biochip using thiol-coupling procedures. The analyte PFL was injected at a rate of 30 μl/minute in 20 mm HEPES, 10 mm NaCl, pH 7.4 at 25 °C for 180 s to measure association rates. Experiments were run in triplicate using five experimental injections of 0.123, 0.370, 1.11, 3.33, and 10 μm PFL dimer (arrows indicate the start of each injection) in single cycle kinetics mode. At the end of each injection of PFL, buffer was injected for 60 s to dissociate the analyte; these regions show a decrease in response. The green trace corresponds to experimental data, and the black trace corresponds to a 1:1 Langmuir interaction model fit of the experimental data. At the end of each cycle, PFL-AE was regenerated using 20 mm HEPES, 500 mm KCl, 0.005% polysorbate 20, 200 mm imidazole, pH 7.4, for 180 s, which completely removed the bound PFL. RU, resonance units.
Electrostatic interactions between proteins lead to association rates that are much faster than the rate of diffusion; given the slow rate of association for PFL-AE with PFL, it is therefore reasonable to assume that electrostatic interactions do not play a significant role in PFL-AE and PFL binding (37). These data are further corroborated by activity assay data that show ionic strength does not affect PFL activity in the range of 0.1–1.6 m KCl (38). The dissociation rate for the PFL-AE·PFL complex was determined to be 1.17 ± 0.16 × 10−3 s−1, indicating that the complex exhibits reasonable stability. When the same PFL-AE and PFL binding data were examined using affinity analysis, a KD of 3.4 ± 2.2 μm was determined for the interaction, which is with in error of the equilibrium constant based on association and dissociation rates.
AdoMet Binding Studies with PFL-AE
The CD spectrum of PFL-AE exhibits λmax values of 305 and 430 nm with shoulders at 345 and 630 nm and λmin values of 380 and 550 nm. When AdoMet is titrated into a solution of PFL-AE, there are dramatic changes in the CD spectrum with multiple isosbestic points (Fig. 2A). The CD spectrum of PFL-AE with AdoMet bound has λmax values of 305, 410, 495, and 690 nm with shoulders at 365 and 630 nm and λmin values of 345 and 560 nm. The spectral changes upon titration with AdoMet have allowed us to determine that PFL-AE in the as isolated form binds AdoMet with a KD of 7.6 ± 1.9 μm (Fig. 2B). Results from the Knappe lab using reconstituted PFL-AE with similar iron content shows that only holo-PFL-AE binds AdoMet with an equilibrium constant of 3 μm, in close agreement with our data (33). The Km for AdoMet has been determined previously as 2.8–7 μm (25, 38).
FIGURE 2.
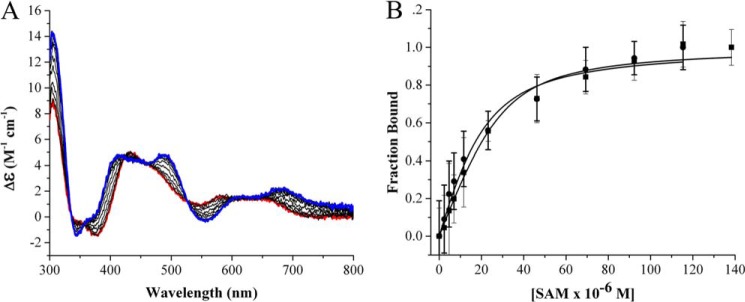
A, visible region circular dichroism of PFL-AE showing the [4Fe-4S] cluster is perturbed upon AdoMet binding under anaerobic conditions. Red, as isolated PFL-AE; blue: PFL-AE with AdoMet (SAM) fully bound. The PFL-AE concentration was 50 μm, and AdoMet concentrations were in the range of 0–140 μm; the assay was performed in 20 mm HEPES, 250 mm NaCl, 1 mm DTT, pH 7.4 at 25 °C. B, AdoMet binding data for PFL-AE (●) and AdoMet binding to the PFL-AE/PFL complex (■). PFL-AE and PFL (when present) concentrations were 50 μm each; other buffer conditions were as indicated for A. The data were analyzed using changes in ellipticity at 400 nm divided by total change in ellipticity, which was then plotted as function of AdoMet concentration and fit to the quadratic binding equation. CD parameters were set to a sensitivity of 100 millidegrees, with a data pitch of 0.1 nm, in continuous scan mode with a scan rate of 100 nm/minute, a scan range of 300–800 nm, response of 1 s, bandwidth of 1.0 nm, and an accumulation of three scans; all measurements were performed using a 1-cm path length anaerobic cuvette.
By using our experimentally determined affinity of PFL-AE for PFL, we were able to set up binding experiments in which PFL-AE was essentially fully bound to PFL prior to titrating in AdoMet; in this way we were able to monitor binding of AdoMet to the PFL-AE·PFL complex (Fig. 2). The PFL-AE·PFL complex exhibited essentially the same affinity for AdoMet as PFL-AE alone, with a KD of 5.7 μm ± 1.7.
We used far-UV circular dichroism studies to see if changes in secondary structure occur during AdoMet binding. Interestingly, there was no difference in PFL-AE secondary structure in the presence or absence of AdoMet (Fig. 3). In either case the protein appears to be well folded. The aggregate data suggests AdoMet binding alters the environment of the iron-sulfur cluster without inducing changes in secondary structure.
FIGURE 3.
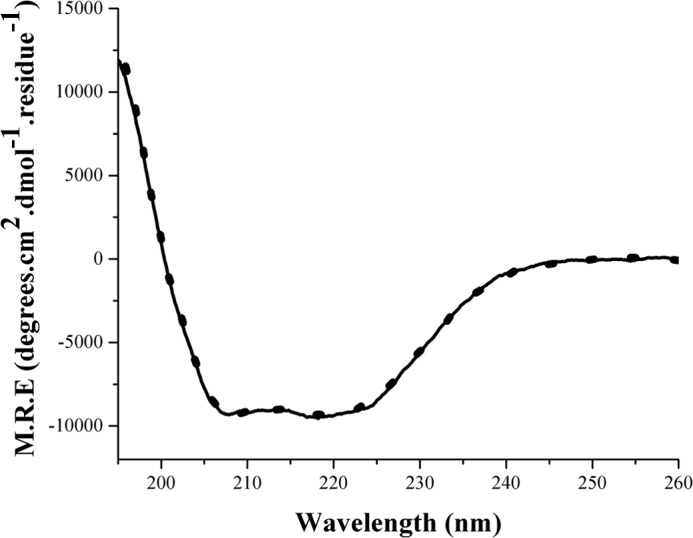
Far-UV CD of PFL-AE in the absence (dotted line) and presence of AdoMet (solid line) under anaerobic conditions in 20 mm HEPES, 250 mm NaCl, pH 7. 4 at 25 °C. CD parameters were as follows: sensitivity was 100 millidegrees, data pitch was 0.1 nm, scan range was 195–260 nm, in continuous scan mode at a rate of 50 nm/minute, response of 4 s, bandwidth of 0.1 nm, and an accumulation of three scans; all measurements were performed using a 0.1-mm path length cuvette. M.R.E., mean residue ellipticity.
PFL Activation Studies
Although nearly all reports of in vitro PFL activation include the PFL substrate pyruvate or its analog oxamate in the activation mixtures, the roles of these molecules in activation have remained unclear. Previous work has shown that photoreduction of PFL-AE results in time-dependent conversion to the EPR-active [4Fe-4S]1+ cluster state, and in the presence of PFL, AdoMet, and oxamate, there is stoichiometric generation of the glycyl radical on PFL concomitant with cluster oxidation (12). We reproduced these assays to examine the roles, if any, of PFL substrates on the activation process. Activation assays were carried out by photoreducing PFL-AE for set amounts of time by using deazariboflavin and exposing the PFL-AE mixture to an intense halogen lamp. The PFL-AE reduction was quantified by using EPR spectroscopy, as the amount of the catalytically active [4Fe-4S]1+ state can be determined by comparison with a CuII(EDTA) standard; all activation samples described below and illustrated in Fig. 4 for a given time point had the same starting amount of [4Fe-4S]1+ cluster. PFL, with or without added PFL substrates, was added to the reduced PFL-AE, and the amount of glycyl radical generated was quantified by EPR spectroscopy. In these assays, samples containing either pyruvate or oxamate exhibited a stoichiometric conversion of [4Fe-4S]1+ cluster of PFL-AE to the glycyl radical on PFL (Fig. 4). After PFL-AE was photoreduced for 60 min and mixed with PFL in the presence of pyruvate, the concentration of the glycyl radical was determined to be 44 ± 5 μm and in the presence of oxamate, the glycyl radical was determined to be 46 ± 5 μm. A similar activation of PFL in the presence of the PFL substrate CoA resulted in only 12 ± 3 μm glycyl radical, despite the same starting amount of PFL-AE [4Fe-4S]1+ cluster as the samples used for PFL activation in the presence of pyruvate or oxamate. Activation of PFL in the absence of PFL substrates yielded 13 ± 3 μm glycyl radical.
FIGURE 4.
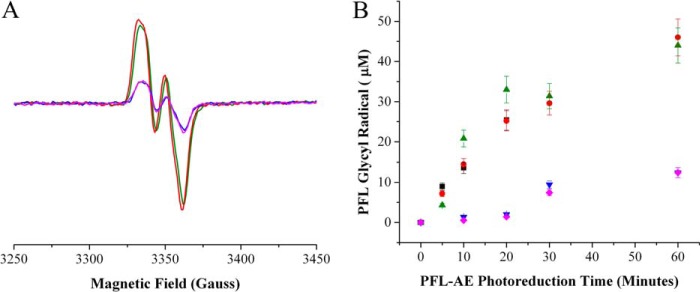
PFL activation by photoreduced PFL-AE in the presence of AdoMet and different PFL substrates. A, EPR data of the PFL glycyl radical in the presence of different PFL substrates after being mixed with PFL-AE photoreduced for 60 min: red, 10 mm oxamate; green, 10 mm pyruvate; blue, 100 μm CoA; and magenta, no substrate. B, the graph shows the quantity of glycyl radical formed on PFL after mixing with PFL-AE that had been photoreduced for 0–60 min; after mixing, the samples were analyzed using EPR. Colors are the same as in A with the addition of a PFL-AE standard in black, which was spin quantified for [4Fe-4S]1+ using a CuII(EDTA) standard. PFL was spin quantified using a K2(SO3)2NO standard. Samples containing 50 μm PFL-AE and 50 μm 5-deazariboflavin in 20 mm HEPES, 250 NaCl, pH 7.4, were photoreduced for 0, 10, 20, 30, and 60 min. 50 μm PFL dimer with either pyruvate, oxamate, CoA, or no substrate was added, and the samples were incubated in the dark for an additional 20 min before being frozen in liquid N2 and analyzed by EPR. Glycyl radical signals were measured at 60 K and the [4Fe-4S]1+ cluster signals were measured at 12 K to ensure that there would be no overlapping signals. EPR parameters were as follows: microwave frequency, 9.37 GHz; power, 19 milliwatt; modulation amplitude, 5 G.
In Vivo Concentrations of Proteins and Small Molecules Involved in the PFL System
In vivo concentrations of the proteins and small molecules involved in the PFL system were calculated for this study to provide a context for equilibrium constants and estimate the fraction of bound proteins and small molecules in vivo. Calculations were performed using data published by Knappe et al. (5) where the amount of protein per cell was quantified for the PFL system. Advances in cellular microscopy have allowed for the accurate determination of cytosolic volumes of E. coli cells under similar conditions (36). When combined this data has allowed us to calculate the in vivo concentrations of the proteins involved in the PFL system: 20 μm PFL, 1.1 μm PFL-AE, 2.9 μm flavodoxin, 2.3 μm NADP+:flavodoxin oxidoreductase, and 648 nm pyruvate:flavodoxin oxidoreductase. Unfortunately error calculations were not available for the polypeptide or percent soluble proteins measurements in the PFL system, so we are unable to calculate errors associated with in vivo concentrations. The in vivo concentration of AdoMet has been estimated to be in the range of 50 - 400 μm (39–41). In vivo concentrations have been calculated for pyruvate of 7.5 ± 0.5 mm (42). Under these conditions and assuming a AdoMet concentration of 50 μm, PFL, PFL-AE, AdoMet, and pyruvate would be essentially fully bound. Only a small fraction of these complexes would have the electron donor flavodoxin bound, however (Table 1).
TABLE 1.
Equilibrium constants and in vivo concentrations for the PFL system
In vivo concentrations for the PFL family were calculated using previously determined polypeptide measurements and combined with cell volume measurements of 2.9 ± 1.2 fl for E. coli cells grown in minimal media supplemented with glucose under anaerobic conditions (5, 36). In vivo concentrations under anaerobic conditions were as follows: [PFL-AE] = 1.1 μm, [PFL] = 20 μm, [Fld] = 2.9 μm, [FNR] = 2.3 μm, [PFOR] = 648 nm. Equilibrium constants were taken from previously published data (47). Error calculations could not be performed because error information was not available for polypeptide measurements (5, 48).
| Sample | KD | [Bound in vivo] | % Bound in vivo |
|---|---|---|---|
| μm | nm | ||
| FNR: Fld | 3.7 ± 1.6 μm | 826 | 36 |
| PFL-AE:PFL | 1.1 ± 0.2 μm | 1070 | 97 |
| PFL-AE:Fld | 23.3 ± 3.8 μm | 117 | 11 |
| PFL-AE:AdoMet | 7.6 ± 1.9 μm | 952 | 86 |
| PFL-AE·PFL:AdoMet | 5.7 ± 1.7 μm | 985 | 90 |
DISCUSSION
The activation of PFL was studied in this work, providing significant new information on the interactions between PFL and its activase, PFL-AE. Surface plasmon resonance binding experiments were carried out under anaerobic conditions, and the data were fit to a 1:1 interaction model with good fits. The KD value of 1.1 ± 0.2 μm, calculated for PFL and PFL-AE, is nearly identical to the Km value previously reported of 1.4 μm and agrees well with previous estimates of the KD (11, 14). The association rate between PFL-AE and PFL is on the low end for biological interactions indicating that the rate of binding is limited by large conformational changes (37).
The [4Fe-4S] cluster in PFL-AE undergoes dramatic changes to its CD spectrum as a direct consequence of AdoMet binding (Fig. 2A). However no changes in secondary structure occurred upon AdoMet binding based on far-UV CD measurements (Fig. 3). Therefore, it is assumed that the changes in the visible region CD spectrum are caused by the direct coordination of AdoMet to the unique iron of the [4Fe-4S] cluster (22, 23). These changes in the visible region CD of PFL-AE can be used to accurately determine equilibrium constants for AdoMet binding in the presence and absence of PFL. PFL-AE binds AdoMet with identical affinity within error regardless of whether PFL is bound to PFL-AE, indicating that PFL binding to PFL-AE does not affect AdoMet binding affinity. These data suggest that in vivo, the order of interaction for AdoMet binding to PFL-AE or the PFL-AE/PFL complex does not matter.
The PFL substrate pyruvate and its analog oxamate have been suggested to act as allosteric effectors required for PFL activation (8, 12, 14, 33). We used EPR spectroscopy to monitor PFL activation in the presence and absence of pyruvate, oxamate, and CoA, to determine whether they are required for PFL activation and if they have any direct affect on the amount of active enzyme produced. Our data shows that although PFL substrates are not absolutely required for activation, their presence results in significantly higher glycyl radical concentrations. When pyruvate or oxamate are incubated with PFL and reduced PFL-AE, there is a stoichiometric conversion of the [4Fe-4S]1+ cluster from PFL-AE to the glycyl radical of PFL. PFL activated in the presence of CoA or no substrate results in 3.7-fold less glycyl radical than in the presence of pyruvate or oxamate. The signal for the [4Fe-4S]1+ cluster in PFL-AE is absent in all experiments after the addition of PFL, indicating that in all cases the PFL-AE is being oxidized in the presence of PFL. The lower quantities of glycyl radical observed in the absence of pyruvate or oxamate therefore suggests that solvent quenches a portion of the PFL glycyl radical. Given that pyruvate and oxamate are known to bind in the active site of PFL (9, 10), we propose that these molecules aid in reinsertion and stabilization of the glycyl radical loop in the closed, catalytically active state of PFL (11).
In vivo concentrations for PFL-AE, PFL, flavodoxin, pyruvate:flavodoxin oxidoreductase, and NADP+:flavodoxin oxidoreductase were calculated in this work and compared with KD values to estimate the amount of bound protein in vivo. Under these conditions, PFL-AE is almost completely bound to PFL (Table 1). In vivo concentrations of AdoMet have been determined to be in the 50 - 400 μm range, (39–41) however there may be less available AdoMet for PFL-AE given the widespread use of AdoMet in many enzymatic reactions in E. coli (24, 43–46). AdoMet binds to both PFL-AE and the PFL-AE/PFL complex with the same affinity of ∼ 6 μm, so assuming an in vivo AdoMet concentration of 50 μm, PFL-AE would be essentially completely bound with AdoMet in vivo regardless of whether PFL is bound. Only 11% of cellular PFL-AE is estimated to be bound to its electron transfer partner flavodoxin at any given time in vivo, consistent with the idea that flavodoxin needs to bind only transiently to deliver an electron to the [4Fe-4S] cluster of PFL-AE.
Taken together, our data provide important new insights into the process by which a glycyl radical activating enzyme (PFL-AE) activates its substrate glycyl radical enzyme (PFL). The process involves slow binding associated with large conformational changes, likely involving movement of the glycyl radical domain of PFL and a conserved loop of PFL-AE implicated in substrate binding (11, 32). AdoMet can bind to this complex either before or after association, and binding gives rise to changes in the visible region CD spectrum of the [4Fe-4S] cluster of PFL-AE. These changes in visible CD features can be used to monitor AdoMet binding and indicate that the affinity of AdoMet for the PFL-AE·PFL complex is comparable with that for PFL-AE alone. Calculations indicate that in vivo, PFL-AE is nearly completely in the PFL-AE/AdoMet·PFL·pyruvate complex, awaiting reduction from flavodoxin to initiate catalysis.
Acknowledgments
We are grateful to GE Biacore for the Biacore X-100 Inspiration Contest and for the use of the Biacore X-100 plus model and materials for anaerobic surface plasmon resonance experiments. We thank the Dooley lab at Montana State University for the use of the CD instrument.
This work was supported, in whole or in part, by National Institutes of Health Grant 2R01GM054608-14.
- PFL
- pyruvate formate-lyase
- AdoMet
- S-adenosylmethionine
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide·HCl.
REFERENCES
- 1. Buckel W., Golding B. T. (2006) Radical enzymes in anaerobes. Annu. Rev. Microbiol. 60, 27–49 [DOI] [PubMed] [Google Scholar]
- 2. Nordlund P., Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 [DOI] [PubMed] [Google Scholar]
- 3. Leuthner B., Leutwein C., Schulz H., Hörth P., Haehnel W., Schiltz E., Schägger H., Heider J. (1998) Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol. Microbiol. 28, 615–628 [DOI] [PubMed] [Google Scholar]
- 4. Craciun S., Balskus E. P. (2012) Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. U.S.A. 109, 21307–21312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knappe J., Sawers G. (1990) A radical-chemical route to acetyl-CoA: the anaerobically induced pyruvate formate-lyase system of Escherichia coli. FEMS Microbiol. Lett. 75, 383–398 [DOI] [PubMed] [Google Scholar]
- 6. Conradt H., Hohmann-Berger M., Hohmann H. P., Blaschkowski H. P., Knappe J. (1984) Pyruvate formate-lyase (inactive form) and pyruvate formate-lyase activating enzyme of Escherichia coli: isolation and structural properties. Arch. Biochem. Biophys. 228, 133–142 [DOI] [PubMed] [Google Scholar]
- 7. Knappe J., Schacht J., Möckel W., Höpner T., Vetter H., Jr., Edenharder R. (1969) Pyruvate formate-lyase reaction in Escherichia coli. The enzymatic system converting an inactive form of the lyase into the catalytically active enzyme. Eur. J. Biochem. 11, 316–327 [DOI] [PubMed] [Google Scholar]
- 8. Knappe J., Blaschkowski H. P., Gröbner P., Schmitt T. (1974) Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate. Eur. J. Biochem. 50, 253–263 [DOI] [PubMed] [Google Scholar]
- 9. Becker A., Kabsch W. (2002) X-ray structure of pyruvate formate-lyase in complex with pyruvate and CoA. J. Biol. Chem. 277, 40036–40042 [DOI] [PubMed] [Google Scholar]
- 10. Becker A., Fritz-Wolf K., Kabsch W., Knappe J., Schultz S., Volker Wagner A. (1999) Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. Nat. Struct. Biol. 6, 969–975 [DOI] [PubMed] [Google Scholar]
- 11. Peng Y., Veneziano S. E., Gillispie G. D., Broderick J. B. (2010) Pyruvate formate-lyase, evidence for an open conformation favored in the presence of its activating enzyme. J. Biol. Chem. 285, 27224–27231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Henshaw T. F., Cheek J., Broderick J. B. (2000) The [4Fe-4S]1+ cluster of pyruvate formate-lyase activating enzyme generates the glycyl radical on pyruvate formate-lyase: EPR-detected single turnover. J. Am. Chem. Soc. 122, 8331–8332 [Google Scholar]
- 13. Plaga W., Frank R., Knappe J. (1988) Catalytic-site mapping of pyruvate formate-lyase. Eur. J. Biochem. 178, 445–450 [DOI] [PubMed] [Google Scholar]
- 14. Frey M., Rothe M., Wagner A. F., Knappe J. (1994) Adenosylmethionine dependent synthesis of the glycyl radical in pyruvate formate-lyase by abstraction of the glycine C-2 pro-S hydrogen atom. J. Biol. Chem. 269, 12432–12437 [PubMed] [Google Scholar]
- 15. Knappe J., Elbert S., Frey M., Wagner A. F. (1993) Pyruvate formate-lyase mechanism involving the protein-based glycyl radical. Biochem. Soc. Trans. 21, 731–734 [DOI] [PubMed] [Google Scholar]
- 16. Knappe J., Neugebauer F. A., Blaschkowski H. P., Gänzler M. (1984) Post-translational activation introduces a free radical into pyruvate formate-lyase. Proc. Natl. Acad. Sci. U.S.A. 81, 1332–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Unkrig V., Neugebauer F. A., Knappe J. (1989) The free radical of pyruvate formate-lyase. Characterization by EPR spectroscopy and involvement in catalysis as studied with the substrate-analogue hypophosphite. Eur. J. Biochem. 184, 723–728 [DOI] [PubMed] [Google Scholar]
- 18. Wagner A. F., Frey M., Neugebauer F. A., Schäfer W., Knappe J. (1992) The free radical in pyruvate formate-lyase is located on glycine-734. Proc. Natl. Acad. Sci. U.S.A. 89, 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Broderick J. B., Duderstadt R. E., Fernandez D. C., Wojtuszewski K., Henshaw T. F., Johnson M. K. (1997) Pyruvate formate-lyase activating enzyme is an iron-sulfur protein. J. Am. Chem. Soc. 119, 7396–7397 [Google Scholar]
- 20. Krebs C., Broderick W. E., Henshaw T. F., Broderick J. B., Huynh B. H. (2002) Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase activating enzyme: A Mössbauer spectroscopic study. J. Am. Chem. Soc. 124, 912–913 [DOI] [PubMed] [Google Scholar]
- 21. Walsby C. J., Hong W., Broderick W. E., Cheek J., Ortillo D., Broderick J. B., Hoffman B. M. (2002) Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe-4S]+ cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 124, 3143–3151 [DOI] [PubMed] [Google Scholar]
- 22. Walsby C. J., Ortillo D., Broderick W. E., Broderick J. B., Hoffman B. M. (2002) An anchoring role for FeS Clusters: Chelation of the amino acid moiety of S-adenosylmethionine to the unique iron site of the [4Fe-4S] cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 124, 11270–11271 [DOI] [PubMed] [Google Scholar]
- 23. Walsby C. J., Ortillo D., Yang J., Nnyepi M. R., Broderick W. E., Hoffman B. M., Broderick J. B. (2005) Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “Radical AdoMet” protein superfamily. Inorg. Chem. 44, 727–741 [DOI] [PubMed] [Google Scholar]
- 24. Blaschkowski H. P., Neuer G., Ludwig-Festl M., Knappe J. (1982) Routes of flavodoxin and ferredoxin reduction in Escherichia coli. CoA-acylating pyruvate: flavodoxin and NADPH: flavodoxin oxidoreductases participating in the activation of pyruvate formate-lyase. Eur. J. Biochem. 123, 563–569 [PubMed] [Google Scholar]
- 25. Knappe J., Blaschkowski H. P. (1975) Pyruvate formate-lyase from Escherichia coli and its activation system. Methods Enzymol. 41, 508–518 [DOI] [PubMed] [Google Scholar]
- 26. Vetter H., Jr., Knappe J. (1971) Flavodoxin and ferredoxin of Escherichia coli. Hoppe Seylers Z Physiol. Chem. 352, 433–446 [DOI] [PubMed] [Google Scholar]
- 27. Nnyepi M. R., Peng Y., Broderick J. B. (2007) Inactivation of E. coli pyruvate formate-lyase: role of AdhE and small molecules. Arch. Biochem. Biophys. 459, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Broderick J. B., Henshaw T. F., Cheek J., Wojtuszewski K., Smith S. R., Trojan M. R., McGhan R. M., Kopf A., Kibbey M., Broderick W. E. (2000) Pyruvate Formate-Lyase-Actiavting Enzyme: Strictly Anaerobic Isolation Yields Active Enzyme Containing a [3Fe-4S]+ Cluster. Biochem. Biophys. Res. Commun. 269, 451–456 [DOI] [PubMed] [Google Scholar]
- 29. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 30. Beinert H. (1978) Micro methods for quantitative determination of iron and copper in biological material. Methods Enzymol. 54, 435–445 [DOI] [PubMed] [Google Scholar]
- 31. Löfås S., Johnsson B., Edström Å., Hansson A., Lindquist G., Hillgren M., Stigh L. (1995) Methods for site controlled coupling to carboxymethyldextran surfaces in surface plasmon resonance sensors. Biosens. Bioelectron. 10, 813–822 [Google Scholar]
- 32. Vey J. L., Yang J., Li M., Broderick W. E., Broderick J. B., Drennan C. L. (2008) Structural basis for the glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc. Natl. Acad. Sci. U.S.A. 105, 16137–16141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Külzer R., Pils T., Kappl R., Hüttermann J., Knappe J. (1998) Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. Molecular properties of the holoenzyme form. J. Biol. Chem. 273, 4897–4903 [DOI] [PubMed] [Google Scholar]
- 34. Aasa R., Vänngård T. (1975) EPR signal intensity and powder shapes: A reexamination. J. Magn. Reson. 19, 308–315 [Google Scholar]
- 35. Rödel W., Plaga W., Frank R., Knappe J. (1988) Primary structures of Escherichia coli pyruvate formate-lyase and pyruvate formate-lyase activating enzyme deduced from the DNA nucleotide sequences. Eur. J. Biochem. 177, 153–158 [DOI] [PubMed] [Google Scholar]
- 36. Volkmer B., Heinemann M. (2011) Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS One 6, e23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schreiber G., Haran G., Zhou H. X. (2009) Fundamental aspects of protein-protein association kinetics. Chem. Rev. 109, 839–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wong K. K., Murray B. W., Lewisch S. A., Baxter M. K., Ridky T. W., Ulissi-DeMario L., Kozarich J. W. (1993) Molecular properties of pyruvate formate-lyase activating enzyme. Biochemistry 32, 14102–14110 [DOI] [PubMed] [Google Scholar]
- 39. Val D. L., Cronan J. E., Jr. (1998) In vivo evidence that S-adenosylmethionine and fatty acid synthesis intermediates are the substrates for the LuxI family of autoinducer sythases. J. Bacteriol. 180, 2644–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Halliday N. M., Hardie K. R., Williams P., Winzer K., Barrett D. A. (2010) Quantitative liquid chromatography-tandem mass spectrometry profiling of activated methyl cycle metabolites involved in LuxS-dependent quorum sensing in Escherichia coli. Anal. Biochem. 403, 20–29 [DOI] [PubMed] [Google Scholar]
- 41. Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., Rabinowitz J. D. (2009) Absolute metabolic concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Y. T., Bennett G. N., San K. Y. (2001) The effects of feed and intracellular pyruvate levels on the redistribution of metabolic fluxes in Escherichia coli. Metab. Eng. 3, 115–123 [DOI] [PubMed] [Google Scholar]
- 43. Bianchi V., Eliasson R., Fontecave M., Mulliez E., Hoover D. M., Matthews R. G., Reichard P. (1993) Flavodoxin is required for the activation of the anaerobic ribonucleotide reductase. Biochem. Biophys. Res. Commun. 197, 792–797 [DOI] [PubMed] [Google Scholar]
- 44. Ifuku O., Koga N., Haze S., Kishimoto J., Wachi Y. (1994) Flavodoxin is required for conversion of dethiobiotin to biotin in Escherichia coli. Eur. J. Biochem. 224, 173–178 [DOI] [PubMed] [Google Scholar]
- 45. Hall D. A., Vander Kooi C. W., Stasik C. N., Stevens S. Y., Zuiderweg E. R., Matthews R. G. (2001) Mapping the interactions between flavodoxin and its physiological partners flavodoxin reductase and cobalamin-dependent methionine synthase. Proc. Natl. Acad. Sci. U.S.A. 98, 9521–9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sekowska A., Kung H. F., Danchin A. (2000) Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2, 145–177 [PubMed] [Google Scholar]
- 47. Crain A. V., Broderick J. B. (2013) Flavodoxin cofactor binding induces structural changes that are required for protein-protein interactions with NADP+ oxidoreductase and pyruvate formate-lyase activating enzyme. Biochim. Biophys. Acta 1834, 2512–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pecher A., Blaschkowski H. P., Knappe K., Böck A. (1982) Expression of pyruvate formate-lyase of Escherichia coli from the cloned structural gene. Arch. Microbiol. 132, 365–371 [DOI] [PubMed] [Google Scholar]