Background: We recently identified CSF3R mutations in chronic neutrophilic leukemia. The most common mutation, T618I, signals without ligand through an undefined mechanism.
Results: CSF3R T618I abrogates an O-glycosylation event and increases receptor dimerization.
Conclusion: The constitutive activity of CSF3R T618I may be explained by loss of O-glycosylation.
Significance: This study illustrates the role of O-linked glycosylation in protein function and oncogenesis.
Keywords: Cytokine, Glycoprotein, Leukemia, Neutrophil, Receptor Modification, GCSFR T595I, Myeloproliferative Neoplasm
Abstract
Mutations in the CSF3 granulocyte colony-stimulating factor receptor CSF3R have recently been found in a large percentage of patients with chronic neutrophilic leukemia and, more rarely, in other types of leukemia. These CSF3R mutations fall into two distinct categories: membrane-proximal mutations and truncation mutations. Although both classes of mutation have exhibited the capacity for cellular transformation, several aspects of this transformation, including the kinetics, the requirement for ligand, and the dysregulation of downstream signaling pathways, have all been shown to be discrepant between the mutation types, suggesting distinct mechanisms of activation. CSF3R truncation mutations induce overexpression and ligand hypersensitivity of the receptor, likely because of the removal of motifs necessary for endocytosis and degradation. In contrast, little is known about the mechanism of activation of membrane-proximal mutations, which are much more commonly observed in chronic neutrophilic leukemia. In contrast with CSF3R truncation mutations, membrane-proximal mutations do not exhibit overexpression and are capable of signaling in the absence of ligand. We show that the Thr-615 and Thr-618 sites of membrane-proximal mutations are part of an O-linked glycosylation cluster. Mutation at these sites prevents O-glycosylation of CSF3R and increases receptor dimerization. This increased dimerization explains the ligand-independent activation of CSF3R membrane-proximal mutations. Cytokine receptor activation through loss of O-glycosylation represents a novel avenue of aberrant signaling. Finally, the combination of the CSF3R membrane proximal and truncation mutations, as has been reported in some patients, leads to enhanced cellular transformation when compared with either mutation alone, underscoring their distinct mechanisms of action.
Introduction
The colony-stimulating factor 3 receptor (CSF3R, also known as granulocyte colony-stimulating factor receptor) is the receptor for CSF3 (also known as granulocyte colony-stimulating factor), the growth factor that instructs myeloid hematopoietic cells to differentiate down the neutrophil lineage (1). Upon ligand binding, CSF3R signals through two tyrosine kinase pathways: the JAK (2, 3) as well as the SRC kinase pathways (4–7). Activation of these signaling pathways by CSF3R leads to both proliferation and differentiation of precursor cells into mature neutrophils. Mice lacking CSF3R or CSF3 are neutropenic (1, 8).
Activating mutations in CSF3R are emerging as important drivers of leukemia. Initial evidence for the role of CSF3R in leukemogenesis came from patients with severe congenital neutropenia who were chronically treated with CSF3 ligand to increase their neutrophil levels and subsequently acquired CSF3R truncation mutations in cells of the myeloid progenitor compartment. Severe congenital neutropenia patients who acquire these truncation mutations are at significantly elevated risk for the development of acute myeloid leukemia (9, 10). Furthermore, CSF3R mutations have been found in a small fraction (0.5–1%) of patients with de novo acute myeloid leukemia (11, 12), underscoring the potential for CSF3R to be a driver of myeloid leukemia. Finally, we have shown recently that CSF3R mutations are found in ∼60% of patients with atypical chronic myeloid leukemia (aCML)3 or chronic neutrophilic leukemia (CNL) (12). A subsequent report found the CSF3R mutation frequency in CNL patients to be as high as 83% but much lower in patients with aCML(13). These patient cohorts of CNL/aCML exhibited both truncation and membrane-proximal mutations, and the majority of patients exhibited membrane proximal mutations. The most frequent of these membrane-proximal mutations is T618I (T595I when using a numbering system that does not include the signal peptide) (12, 13). CSF3R truncation and membrane-proximal mutations can occur in combination on the same allele in patients with severe congenital neutropenia/acute myeloid leukemia (11) or CNL (12).
Functional analyses of truncation and membrane-proximal CSF3R mutations have been suggestive of distinct mechanisms of activation. Truncation mutations result in ligand hypersensitivity and preferential activation of SRC family kinases as well as sensitivity to SRC kinase inhibitors. Mechanistically, truncation mutations induce dramatic increases in cell surface expression of the receptor, which have been reported to occur because of loss of endocytic and degradation motifs (14–16). In contrast, membrane-proximal mutations exhibit ligand-independent signaling with preferential activation of JAK kinases and sensitivity to JAK kinase inhibitors (12, 17). Membrane-proximal mutations do not result in any change in receptor expression compared with the wild type, and the alternative mechanistic processes that underlie dysregulation of CSF3R activity by membrane-proximal mutations remain unknown. Here we show that the sites of these membrane-proximal mutations are part of an O-linked glycosylation cluster and that mutation of these residues leads to a loss of O-glycosylation of the receptor. This coincides with increased receptor dimerization, explaining ligand-independent signaling induced by membrane-proximal mutations.
EXPERIMENTAL PROCEDURES
Plasmid Construction
MSCV-IRES-GFP and p3XFLAG-CMV-14 (Sigma-Aldrich) were made Gateway cloning-compatible using the Gateway vector conversion system (Invitrogen). The pcDNA3.2/V5-DEST Gateway vector was purchased from Invitrogen. The CSF3R transcript variant 1 (NM_000760.2) pDONR vector was purchased from GeneCopoeia. CSF3R mutations were made using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). For the CSF3R 3X FLAG and V5 constructs, stop codons were removed by site-directed mutagenesis prior to cloning into Gateway destination vectors. Wild-type and mutated CSF3R cDNA was transferred into MSCV-IRES-GFP, p3XFLAG-CMV-14, or pcDNA3.2/V5-DEST using Gateway LR Clonease enzyme mixture (Invitrogen).
Cell Culture and Transfection
293T17 cells (ATCC) were maintained in DMEM (Invitrogen) with 10% FBS (Atlanta Biologicals), l-glutamine, penicillin/streptomycin (Invitrogen), and amphotericin B (HyClone). Ba/F3 cells were maintained in RPMI 1640 medium with 10% FBS, 15% WEHI-conditioned medium (a source of IL3), l-glutamine, penicillin/streptomycin, and amphotericin B. 293T17 cells were transfected using FuGENE 6 (Promega). Murine retrovirus was produced by cotransfecting the CSF3R-MSCV-IRES-GFP construct and the EcoPac helper plasmid (a gift from Dr. Rick Van Etten, Tufts University, Boston, MA) into 293T17 cells and harvesting supernatants 48 h later.
Immunoblotting
Transfected cells were lysed using cell lysis buffer (Cell Signaling Technologies) containing complete mini protease inhibitor mixture tablets (Roche), spun at 12,000 rpm for 10 min to pellet cell debris, and then mixed with sample buffer (75 mm Tris (pH 6.8), 3% sodium dodecyl sulfate, 15% glycerol, 8% β-mercaptoethanol, and 0.1% bromphenol blue) and heated at 95 °C for 5 min. Lysates were run on Criterion 4–15% Tris-HCl gradient gels (Bio Rad), transferred to a PVDF membrane, and blocked in Tris-buffered saline with Tween (TBST) with 5% BSA. Blots were probed with anti-GCSF receptor (CSF3R) antibody (Abcam, catalog no. ab126167), anti-actin (Ab-1) mouse mAb (JLA20) (Calbiochem, catalog no. CP01), Rb anti-pSTAT3 Tyr-705 (Cell Signaling Technology, catalog no. 9131), Rb anti-STAT3 (Cell Signaling Technology, catalog no. 9132), Ms anti-V5 (Invitrogen, catalog no. R960-25), or Ms anti-FLAG M2 (Invitrogen, catalog no. F3165). Anti-rabbit or anti-mouse IgG HRP conjugate secondary antibodies (Promega) were used, and blots were developed using SuperSignal West Pico chemiluminescent substrate (Pierce). Blots were imaged on a Lummi imager (Roche Applied Science) and quantified using ImageJ.
Endoglycosidase Digestion
N-linked or O-linked oligosaccharides were removed using peptide-N-glycosidase F or O-glycosidase and neuraminidase, respectively, according to the protocol of the manufacturer (New England Biolabs).
O-Glycosylation Labeling
CSF3R mutants were transfected into 293T17 cells. At the time of transfection, cells were incubated with 50 μm N-azidoacetylgalactosamine (GalNAz) (Thermo Scientific) for 48 h. Cells were washed and then incubated in 1 ml of PBS with 1% FBS. Dibenzylcyclooctyne-sulfo-link-biotin conjugate (Click Chemistry Tools) was added to the cells at 30 μm, incubated at room temperature in the dark for 30 min, and then lysed in cell lysis buffer (Cell Signaling Technologies) containing complete mini protease inhibitor mixture tablets (Roche). Biotinylated proteins were precipitated using streptavidin-agarose (Thermo Scientific) and then subjected to immunoblot analysis for CSF3R.
Coimmunoprecipitation
FLAG-tagged constructs were immunoprecipitated from cell lysates by incubation with anti-FLAG M2 affinity gel (Sigma-Aldrich) for 1 h at 4 °C on a rotator. Beads were washed by spinning through cell lysis buffer containing 15% sucrose. Proteins were disassociated from beads using SDS loading buffer for 5 min at room temperature and then subjected to immunoblotting analysis.
Mouse Bone Marrow Colony Assays
All mouse work was performed according to a protocol approved by the Oregon Health and Science University Institutional Animal Care and Use Committee. Colony assays were performed as described previously (12). Bone marrow was harvested from 6- to10-week-old female BALB/c mice (The Jackson Laboratory), stimulated with IL3, IL6, and stem cell factor overnight, and then spinoculated with murine retrovirus expressing CSF3R constructs on two subsequent days. 2.5 × 104 cells were then plated in 1 ml of MethoCult® M3234 methylcellulose medium for mouse cells without cytokines (Stemcell Technologies) in triplicate. Mouse bone marrow colonies larger than 50 cells were counted between days 5 and 7.
Ba/F3 Cell Cytokine-independent Growth Assays
Ba/F3 cells were infected with murine retroviral supernatants by addition of Polybrene and spinoculation at 30 °C for 90 min at 2500 rpm. GFP-positive Ba/F3 cells infected with murine retroviruses were sorted by FACSaria (BD Biosciences). Cells were expanded after being washed three times and then plated at a density of 5 × 105 cells/ml in RPMI 1640 medium with 10% FBS, penicillin/streptomycin, l-glutamine, and amphotericin B. Viable cell counts were obtained using propidium iodide exclusion on a Guava personal cell analysis system (Millipore).
RESULTS
CSF3R Membrane-proximal and Truncation Mutations Have Distinct Mechanisms of Activation
We recently identified two distinct types of CSF3R mutations in patients with CNL. Membrane-proximal mutations are the most common mutations found in CNL, with truncation mutations seen less frequently. The membrane-proximal mutations (T618I and T615A) are point mutations in the extracellular domain of CSF3R and are located in proximity to the membrane, whereas the truncation mutations are nonsense or frameshift mutations that result in premature truncation of the cytoplasmic domain of the receptor (Fig. 1A). We have shown previously that membrane-proximal mutations confer ligand-independent CSF3R signaling, leading to rapid cellular transformation, whereas truncation mutations confer ligand hypersensitivity and delayed cellular transformation (12). To validate this difference in requirement for ligand from earlier findings, we introduced CSF3R T618I, CSF3R Q741*, and the CSF3R wild type into the murine pro-B cell line Ba/F3. Ba/F3 cells normally depend on IL3 for proliferation and survival. However, certain oncogenes (including mutant CSF3R) or alternative cytokines (including CSF3) can act as a surrogate for IL3 and promote cell growth in the absence of IL3. After starving cells of IL3, we tested the growth of the CSF3R wild type or mutant-expressing Ba/F3 cells over concentration gradients of CSF3 and found that CSF3R wild type-expressing cells exhibited a dose-dependent requirement for ligand to sustain cell growth. Cells expressing truncation mutant CSF3R were also dependent on ligand, although they exhibited enhanced growth compared with the wild type at low and intermediate ligand concentrations. In contrast, cells expressing the membrane-proximal mutant CSF3R T618I exhibited high rates of growth in the absence or presence of ligand without any change over the concentration gradient (Fig. 1B).
FIGURE 1.
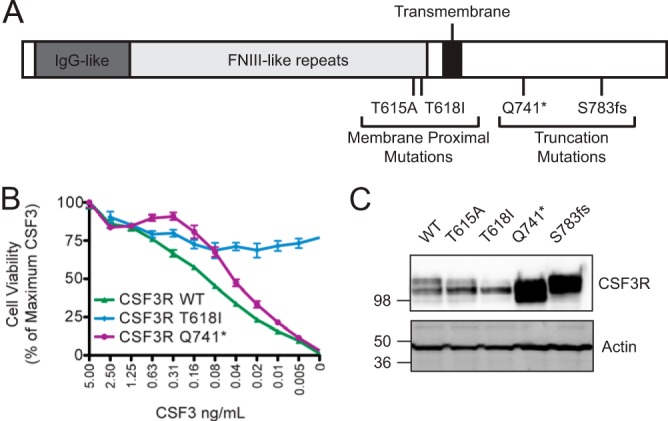
CSF3R membrane-proximal mutations have a mechanism of action distinct from CSF3R truncation mutations. A, schematic of the location of CSF3R membrane-proximal and truncation mutations. IgG-like and FNIII-like repeat regions are shown. The transmembrane domain is shown in black. Membrane-proximal mutations (T615A and T618I) and truncation mutations (Q741*, D771fs, and S783fs) are marked. B, Ba/F3 cells expressing the wild type, the membrane-proximal mutant, or truncation mutant CSF3R were starved of IL3 and plated over a dose gradient of CSF3 ligand for 3 days. Cell viability was assessed using CellTiter 99® AQueous One Solution Cell Proliferation Assay (Promega) and plotted as a percentage of viability at the highest CSF3 concentration (5 ng/ml) for each cell line. Values represent mean ± S.E., n = 3. C, the wild type, the membrane-proximal mutant, or truncation mutant CSF3R were transiently transfected into 293T17 cells, and whole cell lysates were subjected to immunoblot analysis using antibodies specific for CSF3R or actin. These experiments were performed at least twice with consistent results.
To better understand the mechanistic processes that could account for these functional differences between truncation and membrane-proximal CSF3R mutations, we began by transiently expressing the CSF3R wild type, two CSF3R truncation mutants (Q741* and S783fs)m and two CSF3R membrane-proximal mutants (T615A and T618I) in 293T17 cells. The truncation mutations were highly overexpressed compared with wild-type CSF3R (Fig. 1C). This overexpression is consistent with prior reports that truncation mutations result in the loss of endocytic and degradation motifs (14–16, 18). Unlike the truncation mutants, the CSF3R membrane-proximal mutants (T615A and T618I) did not appear overexpressed relative to wild-type CSF3R (Fig. 1C), further supporting the notion that they have a distinct mechanism of activation from the truncation mutations. Interestingly, the membrane-proximal mutants did have an altered banding pattern relative to the wild type. Although wild-type CSF3R and the truncation mutants generally exhibit two bands, the T615A mutant and especially the T618I mutant consisted predominantly of just the lower of the two bands.
CSF3R Is O-glycosylated, and CSF3R Membrane-proximal Mutations Abrogate O-glycosylation
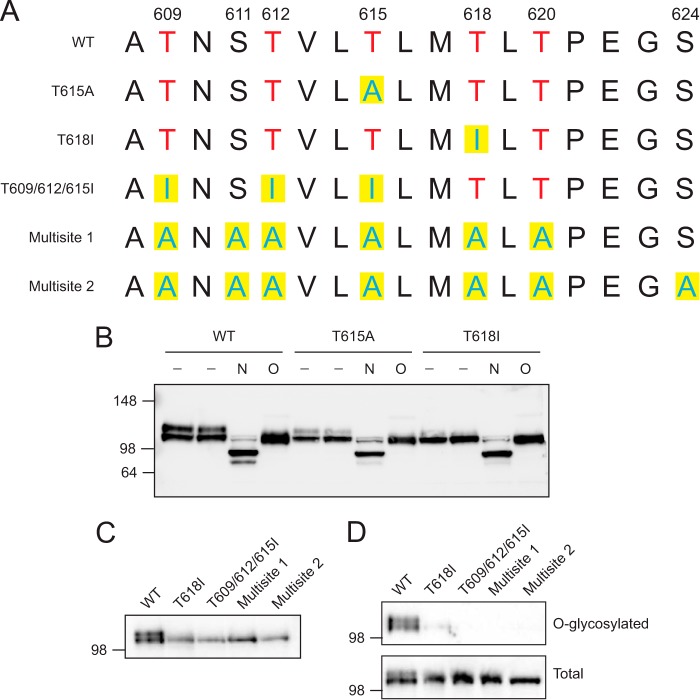
Because the mechanism of activation of CSF3R membrane-proximal mutations appeared to be distinct from to the overexpression induced by CSF3R truncation mutations, we focused on understanding how the membrane-proximal mutations cause activation of the receptor. We first looked at the primary protein sequence in the region of CSF3R membrane-proximal mutations (Fig. 2A). These mutated residues are part of a cluster of threonine residues. Although no consensus motif exists for O-glycosylation, extracellular threonine residues can be glycosylated. This prompted us to determine whether CSF3R is O-glycosylated. O-linked glycosylation was removed from wild-type CSF3R by treatment with a combination of O-glycosidase and neuraminidase (O) and then analyzed by immunoblot analysis for any shifts in size resulting from the loss of O-glycosylation (Fig. 2B). This removal of O-glycosylation did result in the elimination of the upper of the two wild-type CSF3R bands, but retention of the lower band (presumably consolidation of both bands into only the lower band form), indicating that the upper species is O-glycosylated. The shift in the molecular mass of the upper band was calculated to be ∼12 kDa. Similar enzymatic treatment of the CSF3R T615A mutant, which exhibits an upper band that is less prominent than that of the wild type under untreated conditions, also resulted in elimination of the upper band upon removal of O-glycosylation. Finally, similar enzymatic treatment of the CSF3R T618I mutation, which exhibits a nearly undetectable upper band under untreated conditions, resulted in no change of banding pattern, indicating that the baseline alteration of banding patterns for the membrane proximal mutants compared with the wild type is due to a lack of O-glycosylation. These data indicate that wild-type CSF3R is O-glycosylated and that CSF3R membrane-proximal mutation at these residues abrogates O-glycosylation of the receptor. Both the wild type and mutant banding patterns shifted in a similar fashion when treated with PNGase F (N), which removes N-linked glycosylation, indicating that there is not a general glycosylation deficiency of these mutants.
FIGURE 2.
CSF3R is O-glycosylated, and the CSF3R membrane-proximal mutations lead to a reduction in O-glycosylation. A, schematic of the CSF3R membrane-proximal mutants and CSF3R multisite mutations. The amino acid sequence surrounding the membrane-proximal point mutations is diagrammed. Threonines are shown in red, and altered amino acids are shown with yellow boxes. The positions of the serines and threonines in the O-glycosylation cluster are numbered. Additional mutant constructs with Thr-609, Thr-612, and Thr-615 mutated to isoleucine (T609I/T612I/T615I) and constructs with six (Multisite 1) or seven (Multisite 2) threonines or serines mutated to alanine are also diagrammed. B, wild-type CSF3R or membrane-proximal mutants (T615A and T618I) were transfected into 293T17 cells. Whole cell lysates were either untreated (−) or subjected to removal of N-glycosylation (N) with PNGase F or O-glycosylation (O) with O-glycosidase and neuraminidase. Lysates were then subjected to immunoblot analysis using antibody specific for CSF3R. C, immunoblot analysis of CSF3R mutant constructs shows that the Thr-609/Thr-612/Thr-615 and multisite mutants are made up of entirely the lower band in 293T17 cells. D, direct glycosylation of CSF3R mutants. O-glycosylation sites were metabolically labeled using an azide-modified galactosamine (GalNaz), which was then labeled with a DBCO-tagged biotin molecule. O-glycosylated (biotinylated) proteins were immunoprecipitated, and CSF3R pulldown was measured by immunoblot analysis. Molecular markers (kilodalton) are shown. Experiments were performed at least three times with consistent results.
To confirm that CSF3R is O-glycosylated, we performed a direct glycosylation assay. Cells were metabolically labeled using an N3-modified galactosamine residue (GalNaz) that incorporates into mucin-type O-glycosylation sites. The N3 group allows for site-specific labeling with a cyclooctene-linked biotin group that can then be used to pull down O-glycosylated proteins using streptavidin-agarose. Labeling of cells with GalNaz and biotin revealed that WT CSF3R pulled down and was, therefore, O-glycosylated. The CSF3R T618I mutant had a very slight pulldown, indicating that it greatly reduces O-glycosylation of CSF3R but does not eliminate it.
The remaining O-glycosylation of the CSF3R T618I band raises two possibilities: first, that there is an O-glycosylation site in another portion of CSF3R, and second, that there is residual glycosylation of the cluster in which Thr-618 resides. To test this possibility, we created mutations in the threonine cluster that mutated threonines Thr-609, Thr-612, and Thr-615 but left Thr-618 intact or multisite mutations in which six or seven threonines or serines in the cluster were mutated to alanines (Fig. 2A). The multisite mutations consisted primarily of the lower band (Fig. 2C) and completely abolished O-glycosylation pulldown of CSF3R (Fig. 2D), indicating that there may not be any O-glycosylation sites outside of the cluster. Interestingly, despite leaving Thr-618 intact, the T609I/T612I/T615I mutant also eliminated O-glycosylation of CSF3R, indicating that multiple threonines may be required for efficient glycosylation of the receptor.
The CSF3R T618I Point Mutation Causes Increased Receptor Dimerization
One mechanism by which loss of O-glycosylation could lead to ligand-independent signaling is by allowing for increased receptor dimerization. To test this hypothesis, wild-type or T618I CSF3R dimerization was measured by coimmunoprecipitation. For both the wild type and T618I CSF3R, plasmids encoding C-terminal FLAG or V5 epitope tags were constructed (Fig. 3A). Each pair of plasmids was transfected into 293T17 cells, which allows for robust and reproducible cotransfection of plasmids. The FLAG-tagged construct was immunoprecipitated from whole cell lysates, and dimerization was analyzed by detecting coimmunoprecipitation of the V5 construct. To control for the effects of overexpression on dimerization and ensure that there was equal expression of the WT and T618I constructs, the amount of plasmid transfected was optimized (Fig. 3, B and C). The ratio of CSF3R-V5 coimmunoprecipitate relative to the amount of direct CSF3R-FLAG immunoprecipitate was quantified, confirming the increase in dimerization of T618I over the wild type (Fig. 3D).
FIGURE 3.
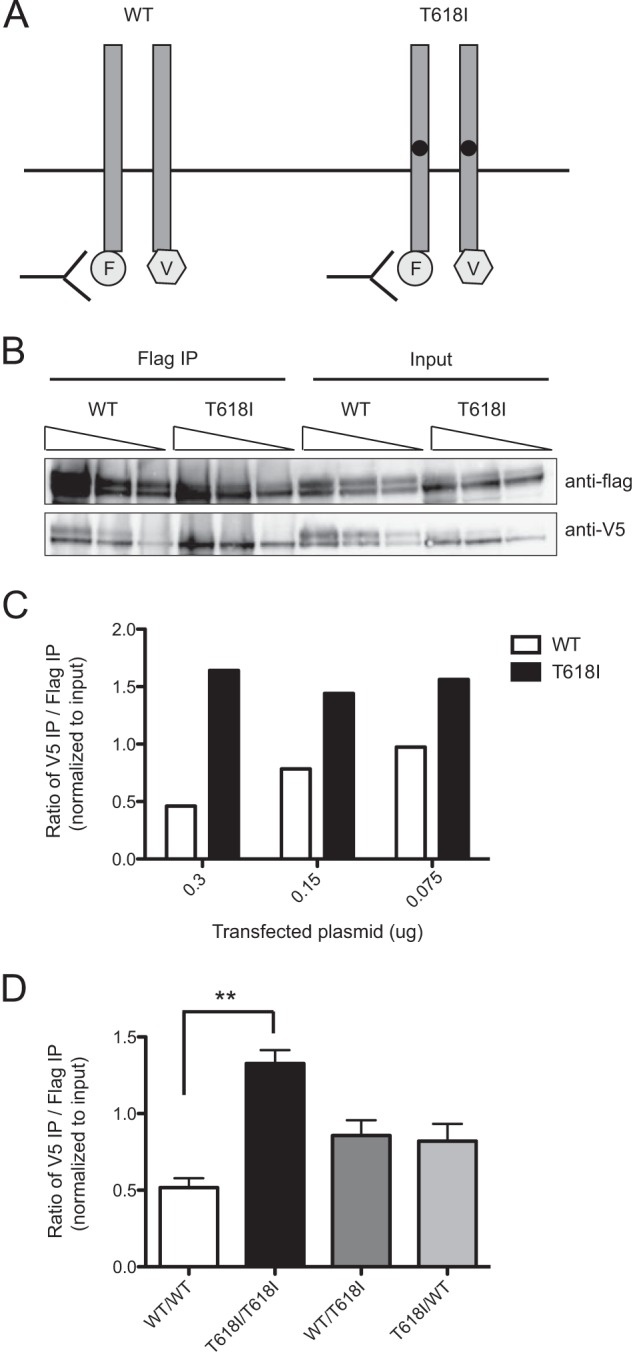
The CSF3R T618I mutation caused increased receptor dimerization. A, schematic of CSF3R dimerization studies. The CSF3R wild type and T618I constructs (0.15 μg) with either FLAG (F) or V5 (V) tags were cotransfected into 293T17 cells, and an anti-FLAG antibody was used to immunoprecipitate FLAG-tagged CSF3R. Dimerization was then assessed by immunoblot analysis with a V5-specific antibody B, FLAG- and V5-tagged CSF3R wild type or T618I constructs were cotransfected into 293T17 cells. The amounts of receptor expression from each pair of constructs were comparable between the wild type and T618I as assessed by immunoblot analysis. For each pair of constructs, 0.3, 0.15, or 0.075 μg of plasmid was transfected. Anti-FLAG affinity resin was used to immunoprecipitate (IP) FLAG-tagged CSF3R from whole cell lysates. Immunoblot analysis using an anti-FLAG antibody was used to detect the levels of FLAG pulldown. Immunoblot analysis using an anti-V5 antibody was used to detect coimmunoprecipitating CSF3R, indicative of CSF3R dimerization. C, quantification of the immunoblot analysis in B. The levels of direct FLAG immunoprecipitation and V5 coimmunoprecipitation were quantified using ImageJ for each plasmid concentration. The Ratio of V5 to FLAG is shown normalized to the respective input levels. Increased dimerization of the CSF3R T618I construct relative to the CSF3R wild type was seen at each plasmid concentration. D, FLAG- and V5-tagged CSF3R wild type or T618I constructs were cotransfected into 293T17 cells as either WT/WT, T618I/T618I, WT/T618I, or T618I/WT pairs. The ratio of V5 to FLAG is shown normalized to the respective input levels. Three replicates were performed for each condition, and the mean ± S.E. are shown. Increased dimerization of the CSF3R T618I construct relative to the CSF3R wild type was found to be statistically significant, although there was a trend of increased dimerization of WT/T618I and T618I/WT over WT/WT, but this was not statistically significant. Values represent mean ± S.E., n = 3. Statistical significance was assessed using one-way analysis of variance followed by Bonferroni's multiple comparison test. **, p < 0.01. This experiment was performed three times with consistent results.
CSF3R Membrane-proximal Mutations Have Increased Oncogenic Potential When Combined with CSF3R Truncation Mutations
We showed previously that CSF3R membrane-proximal mutations are more rapidly transforming than CSF3R truncation mutations and that a fraction of CNL/aCML patients exhibit both CSF3R truncation mutations and CSF3R membrane-proximal mutations on the same allele (12). Because we now show that these mutation types result in CSF3R activation through distinct mechanisms, we next wanted to determine the functional consequences of having compound mutations. Hence, we created constructs harboring both a membrane-proximal and a truncation mutation and expressed them in 293T17 cells (compound mutations were chosen on the basis of those observed in patient samples). Expression of these constructs was measured by immunoblot analysis, and the compound mutants had similar levels of overexpression as CSF3R truncation mutations (Fig. 4A). In addition, the compound mutations exhibited a shift in banding pattern similar to the shifts observed with the membrane-proximal mutations (Fig. 4A). The compound mutations also had similar levels of activation of a marker of JAK pathway activation, pSTAT3, compared with the individual CSF3R membrane-proximal mutations (Fig. 4A). Cumulatively, these data indicate that CSF3R compound mutations exhibit biochemical and signaling features of both the membrane-proximal and truncation mutation types. To test whether the compound mutants have increased transforming potential, both the single and compound mutations were introduced into Ba/F3 cells, and cell viability was measured after IL3 withdrawal (Fig. 4B). The CSF3R compound mutations increased cellular proliferation more rapidly than the membrane-proximal or truncation mutations alone. Finally, a mouse bone marrow colony assay confirmed that a CSF3R compound mutant (T618I + D771fs) formed a greater number of colonies than either the membrane-proximal (T618I) or truncation (D771fs) mutations alone (Fig. 4C).
FIGURE 4.
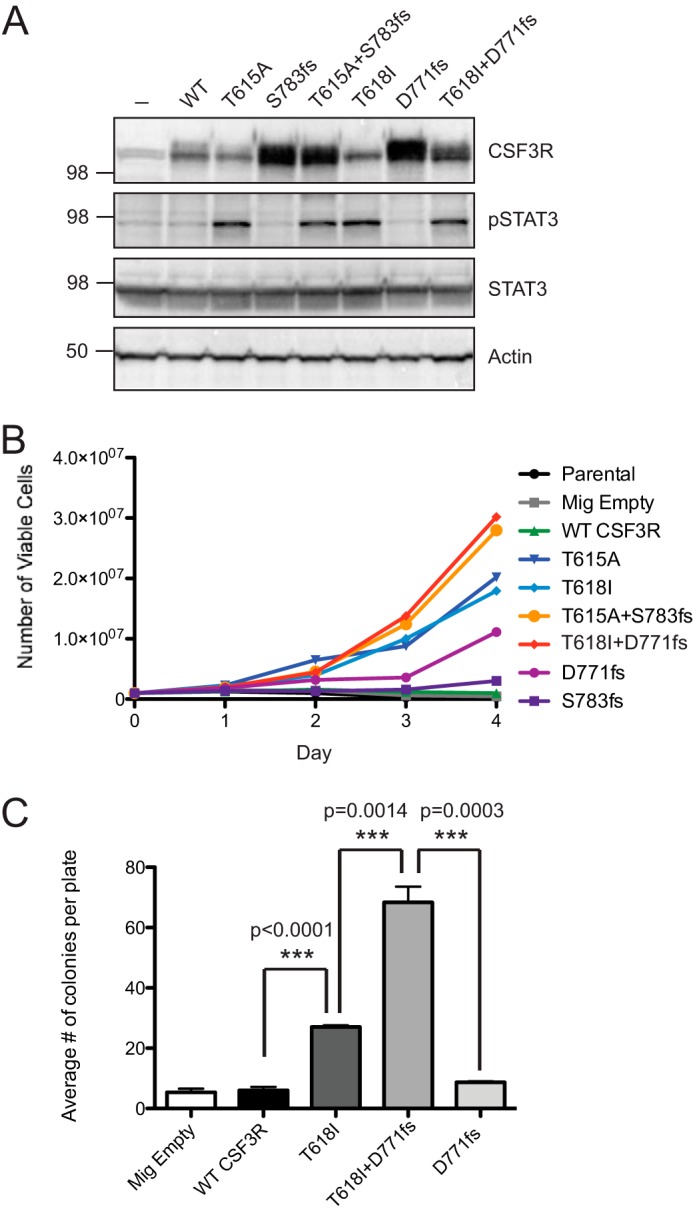
CSF3R membrane-proximal mutations have increased oncogenic potential when combined with CSF3R truncation mutations. A, CSF3R compound mutations (T615A+S783fs and T618I + D771fs) as well as the wild type and each single mutation were transfected into 293T17 cells, and whole cell lysates were subjected to immunoblot analysis using antibodies specific for CSF3R, phospho-STAT3, total STAT3, and actin. B, Ba/F3 cells expressing CSF3R individual or compound mutations were grown in medium lacking IL-3. The total number of viable cells is shown. Parental cells and cells infected with an empty vector (Mig Empty) are shown as controls. C, mouse bone marrow was infected with virus containing empty vector, wild-type CSF3R, CSF3R T618I, CSF3R D771fs, or CSF3R T618I + D771fs, and cells were plated in methylcellulose lacking CSF3 ligand. After 14 days, colonies were manually enumerated by light microscopy. Values represent mean ± S.E., n = 3. Statistical significance was assessed using one-way analysis of variance followed by Bonferroni's multiple comparison test. These experiments were performed twice with consistent results.
DISCUSSION
Mutations in CSF3R have recently emerged as a defining feature of CNL (12, 13). The two classes of CSF3R mutations have differential signaling downstream through the SRC or JAK pathways (12). There is also a substantial difference in the transforming capacity of the mutation types, with the CSF3R membrane-proximal mutations transforming cells more rapidly than truncation mutations (12). In this study, we investigated the underlying mechanisms behind these differences in mutation potency. When expressed in 293T17 cells, the CSF3R truncation mutations were highly overexpressed, consistent with prior reports that these mutations remove critical endocytic/degradation motifs (16). In contrast, the more prevalent CSF3R membrane-proximal mutations T618I and T615A were not overexpressed. This is consistent with a previous report that found no difference in the rates of endocytosis between CSF3R WT and T618I (19).
Investigation of the primary protein sequence revealed a threonine cluster characteristic of O-glycosylation sites (20). O-glycosylation most often occurs on threonine or serine residues, although threonine is preferred (20). Although N-linked glycosylation of CSF3R has been reported previously (21), O-linked glycosylation of this receptor is new.
The T618I threonine-to-isoleucine substitution eliminates the majority of O-glycosylation of CSF3R. In contrast, the T615A mutation only partially prevents O-linked glycosylation, possibly because it plays a supporting role in O-glycosylation of CSF3R but is not strictly required. Interestingly, the CSF3R T618I mutation is far more common than the CSF3R T615A mutation in CNL (12), which further underscores the importance of Thr-618 in the function of CSF3R. The mutation of multiple sites in the threonine cluster completely eliminated CSF3R O-linked glycosylation, suggesting that multiple threonine residues may direct O-glycosylation.
Insight into consequences of this loss of glycosylation came from analyzing the CSF3 ligand dependence of CSF3R. Wild-type CSF3R and CSF3R truncation mutations required ligand to promote the growth of the murine pro-B Ba/F3 cell line, whereas the membrane-proximal mutations exhibited ligand-independent growth. This is consistent with previous reports showing that the T618I mutation causes mouse bone marrow colony formation in the absence of CSF3 ligand (11, 12, 19). This ligand independence of the T618I mutant may explain its relative potency over the truncation mutations, which are overexpressed but still rely on the presence of ligand for signaling.
O-linked glycosylation can affect protein tertiary structure, modulate protein aggregation, and alter protein stability (20). It can also can either increase or decrease the potency of signaling molecules. For example, O-glycosylation decreases the activity of IL-5 (22) but increases the molecular stability and activity of CSF3 (23). Because O-linked glycosylation is a bulky charged group, it may sterically inhibit CSF3R receptor dimerization or otherwise alter the conformation of the receptor to reduce its dimerization. Indeed, we found increased dimerization of the T618I mutation relative to wild-type CSF3R, consistent with the ligand-independent signaling of the mutant. Altered dimerization of CSF3R has been reported previously for the CSF3R T640N mutation (reported as the T617I mutation using the historical numbering system), which is within the transmembrane domain of CSF3R. The T640N mutation was identified in a family with congenital neutrophilia (24) and in patients with acute myeloid leukemia (25). It also causes ligand-independent activation of the receptor and has been predicted by molecular modeling to enhance receptor dimerization through stabilization of the transmembrane helices (24).
There are several patients with CNL/aCML who have both CSF3R membrane-proximal and truncation mutations. These mutations occur on the same allele, and either the truncation or point mutation can occur first (12). We found that the compound mutations exhibited both the overexpression phenotype of the truncation mutations and the shifted banding pattern of the membrane-proximal mutations, indicating that the compound mutations had features of each of the individual mutations. In both Ba/F3 cells and colony assays, the compound mutations showed increased cellular proliferative capacity. They may, therefore, be more substantially oncogenic than either mutation alone. We do not yet know whether the compound mutations confer a worse prognosis, but these molecular findings indicate that further investigation of the clinical and therapeutic relevance of these mutations is warranted.
The CSF3R mutations represent a promising therapeutic target for patients with CNL and aCML. This study identifies the molecular underpinnings of the most common mutation found in this patient population, CSF3R T618I. Loss of O-linked glycosylation sites represents a novel and exciting mechanism of cytokine receptor activation.
Acknowledgments
We thank Dorian La Tocha from the Oregon Health and Science University Flow Cytometry Core for assistance with sorting of Ba/F3 cells and Carolyn Bertozzi and Caroline Enns for advice regarding the glycosylation studies.
This work was supported by the Howard Hughes Medical Institute (to B. J. D.).
- aCML
- atypical chronic myeloid leukemia
- CNL
- chronic neutrophilic leukemia
- CSF3
- colony-stimulating factor 3
- CSF3R
- colony-stimulating factor 3 receptor
- PNGase F
- peptide-N-glycosidase F
- GalNAz
- N-azidoacetylgalactosamine.
REFERENCES
- 1. Lieschke G. J., Grail D., Hodgson G., Metcalf D., Stanley E., Cheers C., Fowler K. J., Basu S., Zhan Y. F., Dunn A. R. (1994) Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 84, 1737–1746 [PubMed] [Google Scholar]
- 2. Nicholson S. E., Oates A. C., Harpur A. G., Ziemiecki A., Wilks A. F., Layton J. E. (1994) Tyrosine kinase JAK1 is associated with the granulocyte-colony-stimulating factor receptor and both become tyrosine-phosphorylated after receptor activation. Proc. Natl. Acad. Sci. U.S.A. 91, 2985–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tian S. S., Lamb P., Seidel H. M., Stein R. B., Rosen J. (1994) Rapid activation of the STAT3 transcription factor by granulocyte colony-stimulating factor. Blood 84, 1760–1764 [PubMed] [Google Scholar]
- 4. Zhu Q. S., Robinson L. J., Roginskaya V., Corey S. J. (2004) G-CSF-induced tyrosine phosphorylation of Gab2 is Lyn kinase dependent and associated with enhanced Akt and differentiative, not proliferative, responses. Blood 103, 3305–3312 [DOI] [PubMed] [Google Scholar]
- 5. Futami M., Zhu Q. S., Whichard Z. L., Xia L., Ke Y., Neel B. G., Feng G. S., Corey S. J. (2011) G-CSF receptor activation of the Src kinase Lyn is mediated by Gab2 recruitment of the Shp2 phosphatase. Blood 118, 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corey S. J., Burkhardt A. L., Bolen J. B., Geahlen R. L., Tkatch L. S., Tweardy D. J. (1994) Granulocyte colony-stimulating factor receptor signaling involves the formation of a three-component complex with Lyn and Syk protein-tyrosine kinases. Proc. Natl. Acad. Sci. U.S.A. 91, 4683–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corey S. J., Dombrosky-Ferlan P. M., Zuo S., Krohn E., Donnenberg A. D., Zorich P., Romero G., Takata M., Kurosaki T. (1998) Requirement of Src kinase Lyn for induction of DNA synthesis by granulocyte colony-stimulating factor. J. Biol. Chem. 273, 3230–3235 [DOI] [PubMed] [Google Scholar]
- 8. Liu F., Wu H. Y., Wesselschmidt R., Kornaga T., Link D. C. (1996) Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity. 5, 491–501 [DOI] [PubMed] [Google Scholar]
- 9. Dong F., Brynes R. K., Tidow N., Welte K., Löwenberg B., Touw I. P. (1995) Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N. Engl. J. Med. 333, 487–493 [DOI] [PubMed] [Google Scholar]
- 10. Germeshausen M., Ballmaier M., Welte K. (2007) Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis. Results of a long-term survey. Blood 109, 93–99 [DOI] [PubMed] [Google Scholar]
- 11. Beekman R., Valkhof M. G., Sanders M. A., van Strien P. M., Haanstra J. R., Broeders L., Geertsma-Kleinekoort W. M., Veerman A. J., Valk P. J., Verhaak R. G., Löwenberg B., Touw I. P. (2012) Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood 119, 5071–5077 [DOI] [PubMed] [Google Scholar]
- 12. Maxson J. E., Gotlib J., Pollyea D. A., Fleischman A. G., Agarwal A., Eide C. A., Bottomly D., Wilmot B., McWeeney S. K., Tognon C. E., Pond J. B., Collins R. H., Goueli B., Oh S. T., Deininger M. W., Chang B. H., Loriaux M. M., Druker B. J., Tyner J. W. (2013) Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N. Engl. J. Med. 368, 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pardanani A., Lasho T. L., Laborde R. R., et al. (2013) CSF3R T618I is a highly prevalent and specific mutation in chronic neutrophilic leukemia. Leukemia [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kimura A., Kinjyo I., Matsumura Y., Mori H., Mashima R., Harada M., Chien K. R., Yasukawa H., Yoshimura A. (2004) SOCS3 is a physiological negative regulator for granulopoiesis and granulocyte colony-stimulating factor receptor signaling. J. Biol. Chem. 279, 6905–6910 [DOI] [PubMed] [Google Scholar]
- 15. van de Geijn G. J., Gits J., Aarts L. H., Heijmans-Antonissen C., Touw I. P. (2004) G-CSF receptor truncations found in SCN/AML relieve SOCS3-controlled inhibition of STAT5 but leave suppression of STAT3 intact. Blood 104, 667–674 [DOI] [PubMed] [Google Scholar]
- 16. Ward A. C., van Aesch Y. M., Schelen A. M., Touw I. P. (1999) Defective internalization and sustained activation of truncated granulocyte colony-stimulating factor receptor found in severe congenital neutropenia/acute myeloid leukemia. Blood 93, 447–458 [PubMed] [Google Scholar]
- 17. Beekman R., Valkhof M., van Strien P., Valk P. J., Touw I. P. (2013) Prevalence of a new auto-activating colony stimulating factor 3 receptor mutation (CSF3R-T595I) in acute myeloid leukemia and severe congenital neutropenia. Haematologica 98, e62–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aarts L. H., Roovers O., Ward A. C., Touw I. P. (2004) Receptor activation and 2 distinct COOH-terminal motifs control G-CSF receptor distribution and internalization kinetics. Blood 103, 571–579 [DOI] [PubMed] [Google Scholar]
- 19. Mehta H. M., Glaubach T., Long A., et al. (2013) Granulocyte colony-stimulating factor receptor T595I (T618I) mutation confers ligand independence and enhanced signaling. Leukemia [DOI] [PubMed] [Google Scholar]
- 20. Van den Steen P., Rudd P. M., Dwek R. A., Opdenakker G. (1998) Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 33, 151–208 [DOI] [PubMed] [Google Scholar]
- 21. Haniu M., Horan T., Arakawa T., Le J., Katta V., Hara S., Rohde M. F. (1996) Disulfide structure and N-glycosylation sites of an extracellular domain of granulocyte-colony stimulating factor receptor. Biochemistry 35, 13040–13046 [DOI] [PubMed] [Google Scholar]
- 22. Kodama S., Tsujimoto M., Tsuruoka N., Sugo T., Endo T., Kobata A. (1993) Role of sugar chains in the in vitro activity of recombinant human interleukin 5. Eur. J. Biochem. 211, 903–908 [DOI] [PubMed] [Google Scholar]
- 23. Nissen C. (1994) Glycosylation of recombinant human granulocyte colony stimulating factor. Implications for stability and potency. Eur. J. Cancer 30, S12–14 [PubMed] [Google Scholar]
- 24. Plo I., Zhang Y., Le Couédic J. P., Nakatake M., Boulet J. M., Itaya M., Smith S. O., Debili N., Constantinescu S. N., Vainchenker W., Louache F., de Botton S. (2009) An activating mutation in the CSF3R gene induces a hereditary chronic neutrophilia. J. Exp. Med. 206, 1701–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Forbes L. V., Gale R. E., Pizzey A., Pouwels K., Nathwani A., Linch D. C. (2002) An activating mutation in the transmembrane domain of the granulocyte colony-stimulating factor receptor in patients with acute myeloid leukemia. Oncogene 21, 5981–5989 [DOI] [PubMed] [Google Scholar]