Background: The function of protein 4.1B in behaviors of motile cells remains poorly understood.
Results: Deficiency of 130-kDa 4.1B resulted in impaired cell adhesion, spreading, migration, and stress fiber formation.
Conclusion: Protein 4.1B plays an important role in modulating cell adhesion and motility.
Significance: Our results identify a novel function for protein 4.1B and have implications for understanding the mechanisms of 4.1 function.
Keywords: Actin, Adhesion, Cytoskeleton, Membrane, Migration
Abstract
Protein 4.1B is a member of protein 4.1 family, adaptor proteins at the interface of membranes and the cytoskeleton. It is expressed in most mammalian tissues and is known to be required in formation of nervous and cardiac systems; it is also a tumor suppressor with a role in metastasis. Here, we explore functions of 4.1B using primary mouse embryonic fibroblasts (MEF) derived from wild type and 4.1B knock-out mice. MEF cells express two 4.1B isoforms: 130 and 60-kDa. 130-kDa 4.1B was absent from 4.1B knock-out MEF cells, but 60-kDa 4.1B remained, suggesting incomplete knock-out. Although the 130-kDa isoform was predominantly located at the plasma membrane, the 60-kDa isoform was enriched in nuclei. 130-kDa-deficient 4.1B MEF cells exhibited impaired cell adhesion, spreading, and migration; they also failed to form actin stress fibers. Impaired cell spreading and stress fiber formation were rescued by re-expression of the 130-kDa 4.1B but not the 60-kDa 4.1B. Our findings document novel, isoform-selective roles for 130-kDa 4.1B in adhesion, spreading, and migration of MEF cells by affecting actin organization, giving new insight into 4.1B functions in normal tissues as well as its role in cancer.
Introduction
Protein 4.1B (4.1B) is a member of a family of adaptors between the actin cytoskeleton and membrane proteins and lipids (for reviews, see Refs. 1–3). The protein 4.1 family has four members (4.1R, 4.1B, 4.1G, and 4.1N) that are encoded by four paralogous genes (4). One common feature of this family is that their mRNAs all undergo extensive alternative splicing, leading to generation of multiple isoforms (5). It has long been assumed that different isoforms possesses diverse functions (5), but functional evidence is limited as yet.
At the protein level, these proteins contain four highly conserved functional domains: the membrane-binding FERM domain (6–8), a FERM-adjacent (FA)4 regulatory domain (9), a spectrin-actin binding domain (SABD) (10, 11), and a C-terminal domain (CTD) unique to the 4.1 proteins (12). These conserved domains are interspersed by non-conserved regions that are distinct between 4.1 proteins; U1 is an N-terminal headpiece (HP), U2 is a non-conserved sequence between the FA domain and SABD, and U3 is the sequence separating SABD and CTD. The FERM domain represents a genetically mobile module that is shared with a variety of other animal proteins (about 50 in humans) that are typically at the interface of the cytoskeleton and membranes (1, 6, 13, 14).
Extensive molecular and functional studies have been performed on the prototypical member of the family, 4.1R. For example, it has been shown that the differences in promoter utilization can lead to usage of three translation initiation sites, ATG1 in exon 2, ATG2 in exon 4, and ATG3 in exon 8 (15–18). Functional studies demonstrated that 4.1R plays diverse roles in erythrocytes as well as other cell types (19–26). By contrast, studies on 4.1B have so far been mainly focused on two aspects: in tumor and metastasis suppression (27–30) and in the organization of peripheral myelinated axons (31–33).
4.1B was discovered both as a paralog of 4.1R (4) and as a tumor suppressor named DAL-1 (deleted in adenocarcinoma of the lung) (34). The gene encoding 4.1B (EPB41L3) is expressed in most mammalian tissues (35). It has been associated with suppression of lung, meningioma, beta cell, breast, ovarian, and prostate cancer progression or metastasis as well as the motility of cancer cell lines (for reviews, see Refs. 1 and 14). Mouse genetics point to a further role in organization of the nervous system. Transgenic mice lacking the full-length (∼130 kDa) isoform of 4.1B have impaired gait and motility associated with ultrastructural anomalies in myelinated axons (31, 32). In both aspects of function, 4.1B interacts with cell adhesion molecules, notably CADM1/TSLC1 (8, 36) in relation to tumor suppression and CASPR/paranodin/neurexin-4 (CNTP1) (37) in relation to the organization of paranodes and juxtparanodes of myelinated axons.
To further explore the physiological function of 4.1B, we generated primary mouse embryonic fibroblast (MEF) cells from wild type and 4.1B knock-out mice for functional analysis. These cells express two 4.1B isoforms: 130 and 60 kDa. Translation of the 130-kDa isoform is initiated in exon 2. The 60-kDa isoform lacks the headpiece; translation is initiated in exon 9 (a previously undescribed initiation site within the FERM domain). Taking advantage of the fact that only 130-kDa 4.1B is absent in the 4.1B knock-out MEF cells, we explored the function of this specific isoform. Lack of the 130-kDa 4.1B led to impaired cell adhesion, spreading, and directional migration. 130-kDa 4.1B-deficient MEF cells also failed to form actin stress fibers when cultured on fibronectin-coated surface. We find that 4.1B interacts with F-actin, but in contrast to 4.1R it does not interact directly with β1 integrin. Taken together, these findings suggest that 130-kDa 4.1B mediates cellular behaviors of motile cells by affecting actin skeleton organization.
EXPERIMENTAL PROCEDURES
Mice
The wild type C57BL/6 mice were purchased from The Jackson Laboratory. The 4.1B knock-out mice in a mixed background (38) were kindly provided by Dr. J. Kissil (The Wistar Institute). These mice were generated by targeting the second coding exon of the 4.1B locus, which contains sequences coding for ATG1 (38). The 4.1B knock-out mice were backcrossed onto C57BL/6 pure background for >10 generations. All the mice were housed at the animal facility of New York Blood Center under pathogen-free conditions accredited by the American Association for Laboratory Animal Care. Animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee.
Reagents for Western Blot and Immunofluorescence
All anti-4.1 antibodies were characterized and used in our previously published studies (39). Alexa Fluor 488-conjugated secondary antibody against mouse, rat, or rabbit IgG, monoclonal anti β-actin antibody, Texas Red®-X phalloidin for the staining of F-actin, TO-PRO3 for the staining of nucleus, and Alexa 488 Fluor-labeled wheat germ agglutinin were all purchased from Invitrogen. Monoclonal anti-mouse β1 integrin (clone 9EG7), which recognizes the active form of β1 integrin, and monoclonal anti mouse β1 integrin (clone MB1.2), which recognizes total β1 integrin, were purchased from BD Biosciences. Phosphatidylethanolamine-conjugated anti mouse CD49e (Integrin α5 chain) was also purchased from BD Biosciences. GelCode® Blue stain reagent was purchased from Thermo Scientific.
Preparation of MEF Cells and Cell Culture
Preparation of primary MEF cells from wild type and 4.1B knock-out mice day-13.5 embryos were performed as described before (40). Primary MEF cells were cultured in DMEM containing 10% FBS. For serum starvation experiments, MEF cells were plated in DMEM containing 0.1% FBS and then incubated at 37 °C for 18 h in humidified condition. Three independently isolated lines were used in the experiments reported here.
Western Blot Analysis
MEF cell lysates were extracted using ice-cold radioimmune precipitation assay buffer (Millipore) in the presence of proteinase inhibitor mixture (Sigma) followed by clarification of extracts by centrifugation at 14,000 rpm for 10 min at 4 °C. Protein concentrations were measured by Bradford assay (Bio-Rad). 40-μg protein extracts were subjected to 10% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated with primary antibodies overnight at 4 °C in PBST containing 5% nonfat dry milk (Bio-Rad). After washing, the membranes were incubated with the corresponding second antibody. Signals were detected by ECL system (Thermo Scientific). When indicated, densities were quantified using Image J.
Cloning of 4.1B cDNA from MEF Cells
Total RNA was isolated from wild type and 4.1B knock-out MEF cells by the RNeasy mini kit (Qiagen). 1 μg of RNA was reverse-transcribed into cDNA using random primers and M-MuLV reverse transcriptase (New England Biolabs). Reverse transcription was performed according to the manufacture's instructions (New England Biolabs). Primers used to amplify 4.1B transcripts were: 4.1BATG1FOR, 5′-ATGACGACCGAATCAGGATCAGACTCAG-3′; 4.1BATG3FOR, 5′-ATGCCAAAAAGCTCTCCATGTACGGGGT-3′; 4.1BRE, 5′-TCAATCCTCTCCGTCCTCTGGTGTGATT-3′. PCR was performed in a 50-μl reaction mixture containing 2×Hotstar Taq Plus Master Mix DNA Polymerase (Qiagen), 10 μm primer each, 200 ng of template cDNA, and double distilled H2O. Cycling conditions were 30 s at 94 °C for denaturation, 30 s at 55 °C (for 4ATG1–4.1B), and 60 °C (for ATG3–4.1B) for annealing, 4 min (for ATG1–4.1B) and 2 min 30 s (ATG3–4.1B) at 68 °C for extension, and a final extension for 5 min at 72 °C. Cycle numbers were 45 for ATG1–4.1B and 35 cycles for ATG3–4.1B.
Plasmids Construction and Preparation of Recombinant Proteins
For mammalian cell expression, ATG1–4.1B and ATG-3 4.1B were cloned into pEGFP-C3 vector using restriction enzymes XhoI and BamHI upstream and downstream, respectively. The primers used were: ATG1–4.1BFOR, 5′-AAACTCGAGATGACGACCGAATCAGGATCAGACTCAG-3′; ATG3–4.1BFOR, 5′-AAACTCGAGATGCCAAAAAGCTCTCCATGTACGGGGT-3′; 4.1BREV, the reverse primer was 5′-AAAGGATCCCGTCAATCCTCTCCGTCCTCTGGTGTGATT-3. Construction of His-4.1R was described previously (41). 4.1B 130-kDa protein was cloned into pET-31b(+) with NsiI and XhoI upstream and downstream, respectively. The cytoplasmic domain of β1 integrin was cloned into pMAL-c2X vector using restriction enzymes SmaI and XhoI. The fidelity of all constructs was confirmed by sequencing. The expression and purification of His-tagged and MBP-tagged proteins were described previously (26).
Immunofluorescence
For confocal immunofluorescence microscopy assay, cells were grown on Lab-Tek™ Chambered Coverglass (Thermo Scientific) pre-coated with 10 μg/ml fibronectin. Cells were fixed with 1% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 in 0.25% paraformaldehyde, PBS for 15 min at room temperature followed by blocking in 10% horse serum, 0.1% Triton X-100 in PBS for 30 min to minimize nonspecific antibody binding. The cells were then incubated with primary antibodies at 4 °C overnight. After washing 3 times with PBS, the cells were incubated with the appropriate second antibody at room temperature for 30 min. For analysis of the actin cytoskeleton, cells were stained with Texas Red-phalloidin, diluted 1:40 in Dako® antibody diluent for 30 min at room temperature followed by 3 washes with PBS. TO-PRO-3 was used to stain the nucleus. Images were collected on a Zeiss LSM510 META confocal microscope using 25×/NA 0.8 objective or 63×/NA 1.4 oil immersion objective.
Adhesion Assay
Sub-confluent cells were trypsinized, resuspended in DMEM with 10% FBS medium, and then seeded at a density of 2 × 104 per well in 96-well plates precoated with 10 μg/ml fibronectin. After 1 or 3 h of incubation at 37 °C, floated cells were washed away with PBS. The adherent cells were fixed with 4% paraformaldehyde. After washing 2 times with PBS, the cells were stained with 2% crystal violet for 10 min. After two times washing with PBS, the color was liberated with 2% SDS and measured at 560 nm using a FLUOstar Omega Multi-mode microplate reader (BMG Labtech).
Spreading Assay
MEF cells were plated on Lab-Tek™ Chambered Coverglass (Thermo Scientific) precoated with 10 μg/ml fibronectin and allowed to spread for 1 or 3 h at 37 °C in complete medium. Cells were fixed in 1% paraformaldehyde for 30 min at room temperature and then labeled with Alexa Fluor 488-conjugated wheat germ agglutinin (Invitrogen) for 30 min to better visualize the cell outlines. Cells were chosen randomly, and surface areas were measured by LSM Image Browser (Version 4.2). Two-tailed Student's t tests were applied to test the statistical significance of the data.
Transwell Migration Assay
For migration assays, 8-μm-diameter pore transwell cell culture inserts (BD Biosciences) were placed in 6-well plates. The underside of the insert and the bottom of the plate surface were coated with 10 μg/ml fibronectin at 4 °C overnight. Cells suspended in serum-starved medium were seeded in the upper chamber of the insert (5 × 105/well), and the complete medium was added to the lower chamber. Then cells were incubated for 8 h to be allowed to migrate through the pores from the insert to the lower side of the membrane of the insert. At the end of the transwell migration assays, the chamber upper side was cleaned with a cotton swab, and the bottom side was stained for 1 h with crystal violet (Sigma) in 2% ethanol. Filters were then imaged with a Leica inverted microscope. Five representative images (×10 magnification) were randomly captured for each insert and used to manually count the number of cells present. Results are presented as the mean number of cells per field ± S.D.
Transient Transfection
0.1 × 106 cells were plated in 6-well plates 1 day before transfection. FuGENE® HD transfection reagent (Promega) was used. The transfection was processed after the manufacturer's instruction. 48 h after transfection, 1.7 × 104 cells were plated and allowed to grow for 2 days to show the location of GFP tagged protein in single or sub-confluent cells; 3.5 × 104 cells were plated and allowed to grow 1 more day after cells were confluent to show the location of GFP tagged protein in completely confluent cells.
Co-immunoprecipitation
MEF cells were lysed with ice-cold 1× radioimmune precipitation assay buffer (50 mm HEPES, pH 8.3, 420 mm KCl, 0.1% Nonidet P-40, 1 mm EDTA) in the presence of proteinase mixture (Sigma) for 30 min on ice. Supernatant was collected after centrifugation at 14,000 × g at 4 °C for 10 min, and the concentration of protein in the supernatant was determined by the Bradford method using BSA as standard (Bio-Rad). 500 μg of extract was incubated with either 5 μg of anti-4.1B-HP or anti-β-actin antibody in 500 μl of co-immunoprecipitation buffer (Active Motif) at 4 °C overnight with rotation. The immunoprecipitates were isolated with magnetic Protein-G beads (Millipore) and separated by 10% SDS-PAGE.
Pulldown Assay
MBP-tagged cytoplasmic domain of β1 integrin was used to pull down 4.1B or 4.1R. Amylose resin (New England Biolabs) was washed and then suspended in 50% PBS. 100 μl of MBP-tagged recombinant cytoplasmic domain of β1 integrin at the concentration of 2 μm was coupled to 5 μl of amylose resin at room temperature for 1 h. Beads were pelleted and washed 5 times with buffer containing 150 mm NaCl, 50 mm Tris·HCl, 1 mm NaN3, 1 mm EDTA, pH 7.4, 0.05% Tween 20. Then 100 μl of His-tagged 80-kDa 4.1R or His-tagged 130-kDa 4.1B at the concentration of 2 μm was added to the bead pellets. The mixture was incubated for 1 h at room temperature, pelleted, washed, and then analyzed by SDS-PAGE. The binding was detected by Western blot using anti-His antibody.
Flow Cytometry
Wild type or 4.1B knock-out MEF cells were serum-starved for 24 h. The cells were trypsinized and washed twice with 0.5% BSA in PBS solution. The cells were stained with monoclonal anti-integrin-β1 antibody (clone MB1.2, which recognizes total surface β1 integrin) or rat anti-mouse CD29 antibody (Clone 9EG7, which recognizes the active form of β1 integrin) in 0.5% BSA in PBS solution for 30 min on ice. The cells were washed twice and incubated with phosphatidylethanolamine-conjugated anti-rat secondary antibody for a further 30 min on ice. Flow cytometric analysis was performed on a FACS Canto flow cytometer (BD Biosciences), and flow data overlay plots were produced using the Cell Quest Pro software (BD Biosciences).
β1 Integrin Trafficking Assay
The experiment was performed as previously described (42). Briefly, 1.7 × 104 MEF cells were plated in Lab-Tek™ Chambered Coverglass (Thermo Scientific) precoated with 10 μg/ml fibronectin and cultured for 24 h. The cells were then starved for 2 h at 37 °C in DMEM with 0.5% bovine serum albumin (Sigma) followed by a 1-h pulse with 10 μg/ml antibody to mouse β1 integrin (9EG7) at 4 °C. To see the surface labeling of β1 integrin after a 1-h pulse, cells were fixed, and surface-labeled β1 integrin was visualized. The chase phase was performed in prewarmed complete culture medium at 37 °C for 10 min, 2 h, and 4 h. At the end of every chase, cells were quenched by an acid rinse (0.5% acetic acid, 0.5 m NaCl, pH 3.0) for 45 s. This treatment led to the removal of >90% externally bound antibody. To visualize the internalized β1 integrin, the cells were then fixed, permeabilized, and stained with Alexa Fluor® 488-conjugated anti rat antibody.
RESULTS
Two 4.1B Isoforms Are Expressed in Primary MEF Cells
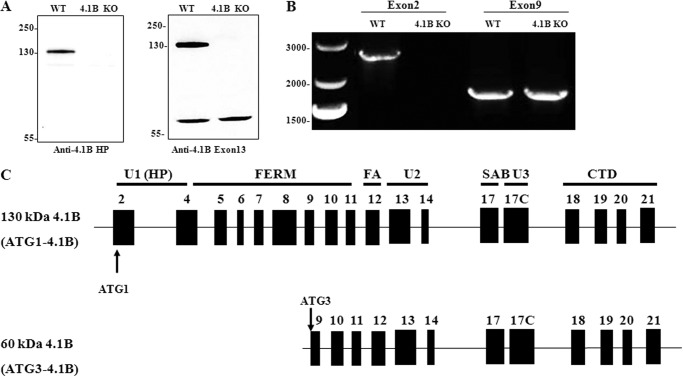
As a first step to investigate the role of 4.1B in MEF cells, we examined the expression of 4.1B by Western blot. For this, we used two antibodies, anti-HP (antibody to peptide within the N-terminal headpiece encoded by exon 2) and anti-exon 13. So far as is known, exon 13 is a constitutive exon, expressed in all 4.1B isoforms. Fig. 1A shows that anti-4.1B HP antibody recognized a 130-kDa band in wild type but not 4.1B knock-out MEF cells. However, when anti-4.1B exon 13 antibody was used in addition to the 130-kDa band, a 60-kDa band was also observed that was present in both wild type and 4.1B knock-out MEF cells.
FIGURE 1.
Expression of 4.1B in MEF cells. A, immunoblot analysis of 4.1B expression in WT and 4.1B KO MEF cells. 40 μg of protein lysates were probed with polyclonal rabbit antibody against 4.1B head piece or 4.1B Exon 13. B, amplification of 4.1B cDNAs from MEF cells. The ATG1 or ATG3 4.1B transcripts were amplified as described under “Experimental Procedures.” Sizes of each PCR product, expressed in base pair (bp), are indicated. C, exon maps and protein structure of MEF cell 4.1B. Translation initiation sites are indicated (ATG-1, ATG-3). Domains are shown as described in the Introduction.
To characterize the exon composition of these two 4.1B isoforms and to confirm that the 60-kDa band is indeed 4.1B, we amplified 4.1B transcripts by reverse transcript-PCR (RT-PCR). Previous studies have documented two translation initiation sites for 4.1B, ATG1 in exon 2 and ATG2 in exon 4 (43). The fact that the 130-kDa band is recognized by anti-HP antibody implies that it is translated from ATG1 located in exon 2. Known isoforms of 4.1B translated from ATG2 are 100 kDa (43). Therefore, the size of the 60-kDa band suggests that the translation of this 4.1B isoform is initiated downstream of both ATG1 and ATG2. The next in-frame ATG occurs in exon 9 (amino acid 285 in the full-length protein, Uniprot:Q9WV92), which we term ATG3. Thus, for RT-PCR, we used two forward primers starting from ATG1 and ATG3. Amplification of 4.1B coding region using forward primers starting from the ATG1 or ATG3 and a reverse primer at the end of the coding region (exon 21) resulted in amplification of a 2628-bp and a 1814-bp PCR product, respectively (Fig. 1B).
Consistent with Western blot analysis, the 2628-bp transcript was amplified from wild type but not 4.1B knock-out MEF cells, whereas the 1814-bp transcript was amplified from both wild type and 4.1B knock-out MEF cells. PCR products were cloned in plasmid vectors; 4 individual 2628-bp products were fully sequenced as were 4 1814-bp products; sequences of the replicates were identical to each other. Their sequences are given in the EMBL database with the accession number for the 60-kDa 4.1B HG738869 and the accession number for the 130-kDa HG738870.
As shown in Fig. 1C, 130-kDa 4.1B contains exons encoding full-length FERM (exons 4–11), FA (exon 12) and full-length CTD (exons 18–21). In terms of SAB domain, whereas exon 17 is present, exon 16 is missing. The U2 and U3 regions are encoded by exons 13–14 and exon 17C, respectively. The 60-kDa form is identical to the 130-kDa form but lacks polypeptide encoded by exons 2–8; therefore, it lacks the headpiece (exon 2 and the beginning of exon 4), the whole of the N-terminal lobe of the FERM domain, and about half of the central lobe (exons 4–8).
Thus we have identified two distinct 4.1B isoforms in MEF cells. The finding that the 60-kDa 4.1B is still present in 4.1B knock-out MEF cells suggests that the 4.1B knock-out mice do not represent a complete knock-out.
Distinct Localization of the Two 4.1B Isoforms in MEF Cells
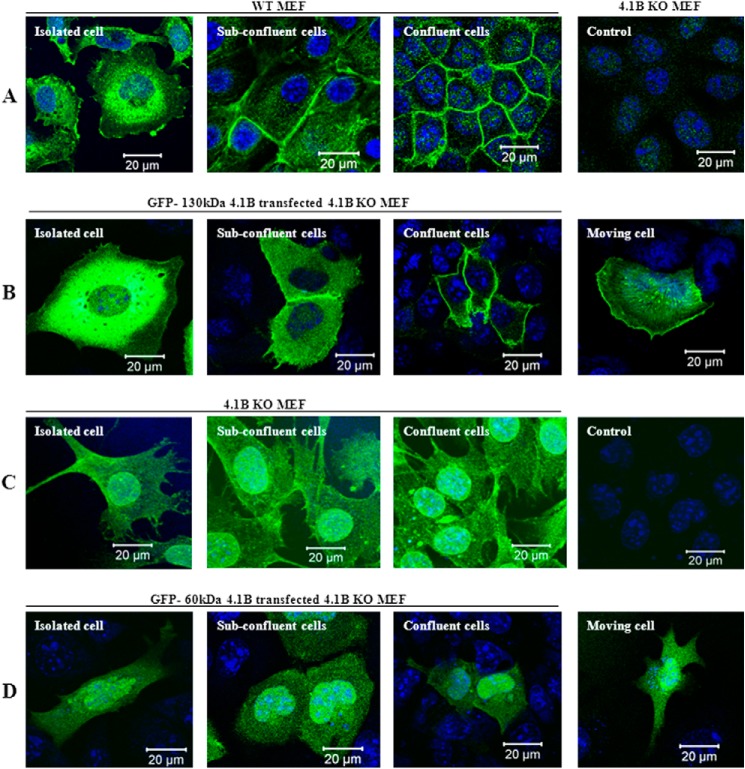
To examine the localization of endogenous 130-kDa 4.1B, we stained the MEF cells using anti-4.1B HP antibody (this only recognizes 130kDa 4.1B but not 60-kDa 4.1B; see Fig. 1A). Fig. 2A shows that in isolated MEF cells, the 130-kDa 4.1B is predominantly located at perinuclear region. It is also located in cytoplasm and plasma membrane primarily at ruffling edges (Fig. 2A, left). Under subconfluent conditions, when cells start to make contact, in addition to the cytoplasmic and perinuclear distribution, it is recruited to areas of cell-cell contact (Fig. 2A, second panel). In completely confluent cells, the 130-kDa 4.1B is very strongly enriched at cell-cell contacts (Fig. 2A, third panel). As a control, probing 4.1B knock-out MEF cells with this antibody revealed only a weak background stain (Fig. 2A, fourth panel).
FIGURE 2.
Localization of 4.1B in MEF cells. A, localization of endogenous 130-kDa 4.1B. MEF cells were cultured on fibronectin-coated surface to reach different confluent conditions as indicated. The cells were then fixed and stained using anti-4.1B HP antibody. 4.1B KO MEF cells were used as the negative control. B, localization of exogenously transfected GFP-130-kDa 4.1B. GFP-130-kDa 4.1B was transiently expressed in 4.1B KO MEF cells. The cells were cultured as described, then fixed. The images were taken using confocal microscopy. C, localization of endogenous 60-kDa 4.1B. 4.1B KO MEF cells were cultured on fibronectin-coated surface to reach different confluent conditions as indicated. The cells were then fixed and stained using anti-4.1B exon 13 antibody. Cells stained with secondary antibody only were used as a negative control. D, localization of exogenously transfected GFP-60-kDa 4.1B. GFP-60-kDa-4.1B was transiently expressed in 4.1B KO MEF cells. The cells were cultured as described, then fixed. The images were taken using confocal microscopy. All pictures were taken with oil-immersed 63× objective. Scale bar, 20 μm.
We also examined the localization of exogenously transfected GFP-130kDa 4.1B. Fig. 2B shows that localization of GFP-130kDa 4.1B is similar to that of endogenous 130kDa 4.1B (Fig. 2B, first to third panels). Furthermore, in moving cells, the GFP-130-kDa 4.1B is accumulated at the leading edge with additional evidence of stress fiber location (Fig. 2B, fourth panel).
Furthermore, we examined the localization of both endogenous 60-kDa-4.1B and exogenously expressed GFP-60-kDa-4.1B. As shown in Fig. 2, C and D, both endogenous and exogenous 60-kDa-4.1B are diffusely distributed in cytoplasm as well as in nucleus under all conditions (isolated cells, subconfluent, complete confluent, and moving cells). These data indicate that differential mRNA splicing of 4.1B controls cellular location and that polypeptide encoded by exons 2–8 is required for location at sites of cell-cell contact and in motility-related structures.
Expression and Localization of Other 4.1 Family Members in Wild Type and ATG1–4.1B-deficient MEF Cells
To examine the expression of 4.1 family members in MEF cells and to examine the effect of 130kDa 4.1B deficiency on other 4.1 family members, we compared the expression and localization of 4.1R, 4.1N, and 4.1G in wild type and ATG1–4.1B-deficient MEF cells.
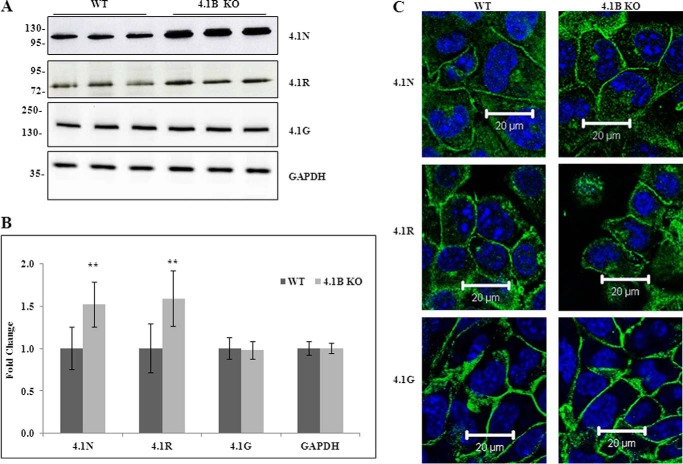
Fig. 3A shows that 4.1R, 4.1G, and 4.1N are also expressed in MEF cells. Notably, both 4.1R and 4.1N are up-regulated, whereas 4.1G is unchanged in 130-kDa 4.1B-deficient MEF cells. Quantitative analysis from three independent experiments is shown in Fig. 3B, and it reveals a 1.5-fold increase for both 4.1R and 4.1N (in both cases p < 0.01). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as control in these analyses.
FIGURE 3.
Expression and localization of 4. 1N, 4.1R, and 4.1G in WT and 4.1B KO MEF cells. A, immunoblot analysis. Total lysates (40 μg protein) were probed with antibodies against 4.1N HP, 4.1R exon13, and 4.1G HP. GAPDH was used as the loading control. B, quantitative analysis of immunoblot. The results from three independent experiments were shown. **, p < 0.01. C, immunofluorescence staining. Cells were cultured on fibronectin-coated surface. One day after the cells reached confluence, the cells were fixed and stained using antibodies as indicated. All pictures were taken with oil-immersed 63× objective. Scale bar, 20 μm.
Immunofluorescence staining (Fig. 3C) showed that 4.1R, 4.1G, and 4.1N are located at sites of cell-cell contact in plasma membrane of confluent cells in both wild type and 130-kDa 4.1B-deficient MEF cells, indicating that the localization of these 4.1 family members are not affected by the lack of 130-kDa 4.1B.
Impaired Adhesion, Spreading, and Migration of 130-kDa 4.1B-deficient MEF Cells
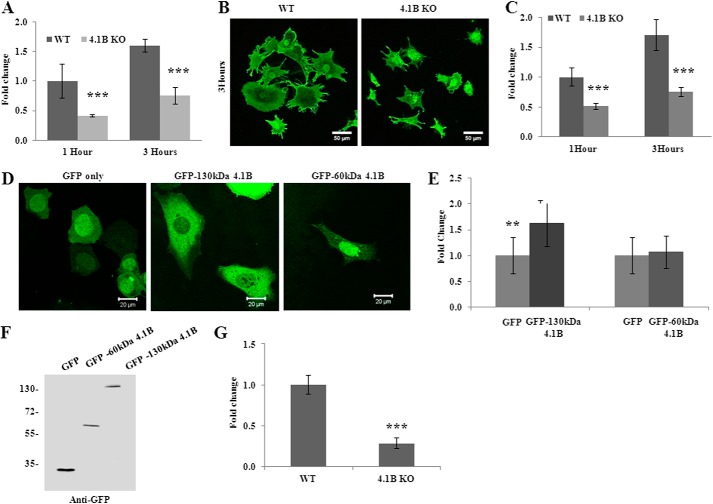
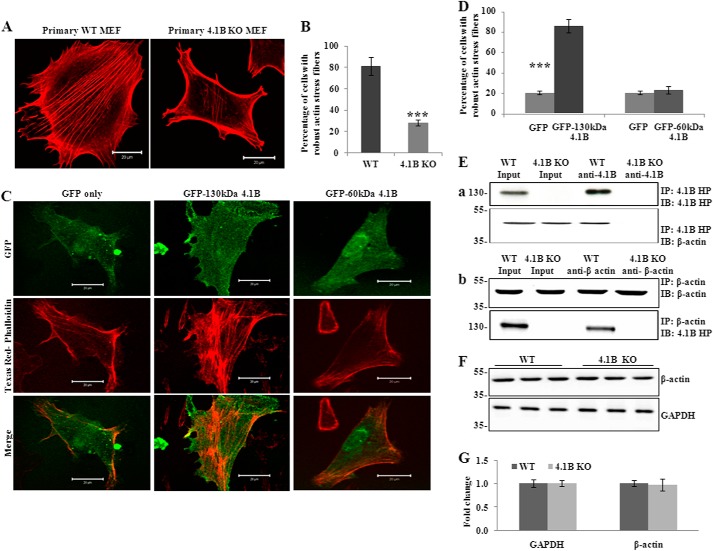
The finding that the 130-kDa 4.1B isoform is localized at the leading edge of the moving cells suggests a possible role of the 130-kDa 4.1B in cellular behaviors such as adhesion, spreading, and migration. To test this, we examined the adhesion of the cells to the fibronectin-coated surface. Fig. 4A shows that at 1 or 3 h after plating, the adhesion of 130-kDa 4.1B-deficient MEF cells to the surface was 60 and 40% less than that of wild type MEF cells, respectively. We then examined the cell spreading on fibronectin-coated surface. Representative images of cells that were allowed to spread for 3 h are shown in Fig. 4B; note the reduced spreading areas of 130-kDa 4.1B-deficient MEF cells. Quantitative analysis from 45 cells revealed a 50% reduction in the spreading area of the 4.1B KO MEF cells at 1 and 3 h (Fig. 4C). Importantly, the impaired spreading was rescued by re-expression of GFP-130-kDa 4.1B but not by GFP-60-kDa 4.1B (Fig. 4, D and E). The expression of GFP-130-kDa 4.1B and GFP-60-kDa 4.1B in 4.1B KO MEF cells was confirmed by Western blot using the anti-GFP antibody (Fig. 4F).
FIGURE 4.
Impaired adhesion, spreading, and migration of 4.1B KO MEF cells. A, cell adhesion. Cells were plated on fibronectin-coated 96-well plates and incubated for 1 or 3 h. The adherent cells were stained with crystal violet, and the staining intensity was quantified by spectrophotometry at 560 nm. The quantitative analysis from three independent experiments was shown. ***, p < 0.001. B, cell spreading. Cells were seeded and allowed to spread for 1 or 3 h. The cells were labeled with Alexa Fluor 488-conjugated wheat germ agglutinin, and the images were collected by Zeiss confocal microscope. The representative pictures of cells that were allowed to spread for 3 h were shown. Pictures were taken with a 25× objective. Scale bars, 20 μm. C, quantitative analysis of cell spreading. The mean surface area from 45 cells of each type was calculated using LSM 5 Pascal software. ***, p < 0.001. D and E, rescue of impaired cell spreading. 4.1B KO MEF cells were transiently transfected with GFP, GFP-130-kDa 4.1B, or GFP-60-kDa 4.1B. The cell spreading after 3 h was measured as described above. D, representative images. E, quantitative analysis from 30 cells of each type was shown. **, p < 0.01. Pictures were taken with an oil-immersed 63× objective. Scale bars, 20 μm. F, immunoblot analysis of transfected 4.1B. 40 μg of cell lysates were probed with polyclonal rabbit antibody against GFP. G, Transwell assay. 8-μm-diameter pore transwell cell culture inserts were placed in 6-well plates, and the bottom surface of insert was coated with fibronectin. 5 × 105 were seeded on top of the insert and incubated for 8 h. The cells migrated to the bottom of the well were fixed and stained with crystal violet, and cell numbers were counted. The averages from three experiments are shown. ***, p < 0.001.
Directional migration of wild type and 4.1B KO MEF cells was examined using a transwell migration assay. Fig. 4G shows that the proportion of 4.1B KO MEF cells that migrated toward fibronectin through the pore of the transwell cell culture inserts is ∼30% relative to wild type cells. Collectively, these data demonstrate a novel role for 130-kDa 4.1B in regulating cell adhesion, spreading, and migration.
Altered Actin Skeleton Organization of 130-kDa-4.1B-deficient MEF Cells
Protein 4.1R associates with actin and plays important roles in actin organization (20, 22, 44). Because 4.1R and other members of the family have a common domain that binds actin (the SABD) (11), protein 4.1B may likewise have roles in actin organization. To explore the mechanisms by which lack of 130-kDa 4.1B led to impaired cell adhesion, spreading, and migration, we examined the actin stress fiber formation of wild type and 130-kDa 4.1B-deficient MEF cells. For this, cells were allowed to spread for 3 h, and actin filaments were stained with Texas Red®-X phalloidin. Fig. 5A shows representative images of endogenous actin filaments in these cells. Quantitative analysis is shown in Fig. 5B, which reveals that >80% of wild type MEF cells exhibited robust actin stress fibers. In contrast, >70% of 130-kDa-4.1B-deficient MEF cells failed to form the well defined actin stress fiber. Instead, actin filaments in these cells were restricted to the cell periphery and often ran parallel to the plasma membrane.
FIGURE 5.
130kDa-4.1B is required for actin stress fiber formation in MEF cells. A and B, endogenous actin filament staining. Cells were cultured on the fibronectin-coated surface and stained with Texas Red-phalloidin. The quantitative analysis from 50 cells of each cell type is shown in B. ***, p < 0.001. All pictures were taken with oil-immersed 63× objective. Scale bars, 20 μm. C and D, rescue of actin stress fiber formation by GFP-130-kDa 4.1B. 4.1B KO MEF cells were transfected with GFP, GFP-130-kDa 4.1B, or GFP-60-kDa 4.1B. Cells were stained with Texas Red-phalloidin. The quantitative analysis from 30 cells of each cell type is shown in D. ***, p < 0.001. All pictures were taken were taken with oil-immersed 63× objective. Scale bars, 20 μm. E, immunoprecipitation (IP). a, 130-kDa 4.1B was immunoprecipitated from MEF cells using anti-4.1B HP antibody. 130-kDa 4.1B or actin in the immunoprecipitate was detected using anti-4.1B HP antibody or anti-actin antibody. b, actin was immunoprecipitated from MEF cells using anti-actin antibody. Actin or 130-kDa 4.1B in the immunoprecipitate was detected using the indicated antibodies. IB, immunoblot. F and G, immunoblot analysis of actin. 20 μg of cell lysates were probed with anti-actin antibody. A GAPDH immunoblot is shown as a loading control (F). Quantitative analysis from three independent experiments is shown in G.
To further analyze the role of 130-kDa 4.1B in regulating the organization of actin stress fiber, we transfected 130-kDa 4.1B-deficient MEF cells with GFP-130-kDa 4.1B or GFP-60-kDa 4.1B and examined the actin stress fiber formation. Fig. 5C shows that the impaired actin stress fiber formation was rescued by re-expression of GFP-130-kDa 4.1B but not by GFP-60-kDa 4.1B. Quantitative analysis is shown in Fig. 5D which demonstrates that the robust actin stress fiber was seen in ∼85% GFP-130-kDa 4.1B-transfected cells, whereas it was only seen in ∼20% GFP-transfected cells or GFP-60-kDa 4.1B-transfected cells.
To test if 130-kDa 4.1B can interact with actin in MEF cells, we performed a co-immunoprecipitation assay to examine the binding of 4.1B with β-actin in situ. As shown in Fig. 5E, β-actin was pulled down together with 4.1B by anti-4.1B HP antibody. Reciprocal co-immunoprecipitation assay using anti-β-actin antibody shows that 4.1B was pulled down together with β-actin. 130-kDa 4.1B-deficient MEF cells were used as a negative control. Together with the previous finding that 4.1B directly binds to actin in vitro (11), these findings suggest that 4.1B directly binds to actin and modulates its organization in MEF cells.
It has been shown that lack of 4.1R in erythrocytes resulted in reduced levels of actin and disorganized actin skeleton (20). To examine whether the failure of actin stress fiber formation observed in 130-kDa 4.1B-deficient MEF cells could also be due to decreased expression of actin, we compared the expression of actin in these cells. No differences were observed (Fig. 5, F and G).
Decreased Surface Expression of β1 and α5 Integrin in 130-kDa 4.1B-deficient MEF Cells
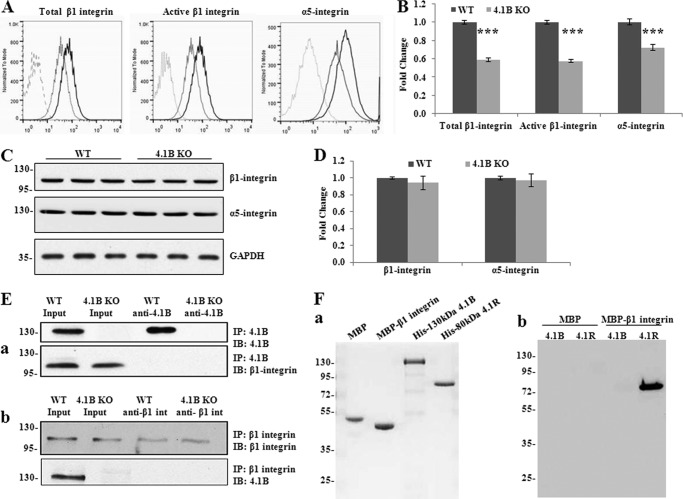
Because the 130-kDa 4.1B-deficient MEF cells showed defects in spreading and motility on fibronectin, we examined the β1 integrin in these cells. β1 integrin is a fibronectin receptor. Previous studies revealed that 4.1R regulates cell adhesion, spreading, migration, and motility of mouse keratinocytes by modulating surface expression of β1 integrin (22). Cell surface expression of β1 integrin was analyzed by flow cytometry. Both the surface expression of total β1 integrin and active form β1 integrin were decreased by 40% in 130-kDa 4.1B-deficient MEF cells (Fig. 6, A and B). As the extent of decreased surface expression and activity of β1 integrin was similar, the decreased activity was probably due to the decreased surface expression.
FIGURE 6.
Decreased surface expression and activity of β1 integrin accompanied with the loss of surface α5 integrin in 4.1B KO MEF cells. A and B, surface expression of β1 and α5 integrin. The representative profiles and quantitative analysis from three independent experiments are shown in A and B, respectively. Black line, WT; light gray, 4.1B KO; broken line, unstained. ***, p < 0.001. C and D, immunoblot analysis of β1 and α5 integrin. 40 μg of cell lysates were probed with anti-β1 integrin antibody and α5 integrin, respectively. GAPDH was used as the loading control (C). Quantitative analysis from three independent experiments was shown in D. E, immunoprecipitation (IP). a, 130-kDa 4.1B was immunoprecipitated from MEF cells using anti-4.1B HP antibody. 130-kDa 4.1B or β1 integrin in the immunoprecipitate was detected using anti-4.1B HP antibody or anti-β1 integrin antibody. IB, immunoblot. b, β1 integrin was immunoprecipitated from MEF cells using anti-β1 integrin antibody. β1 integrin or 130-kDa 4.1B in the immunoprecipitate was detected using the indicated antibodies. F, in vitro pulldown assay for β1 integrin-4.1B interaction. a, proteins used in the binding assays. 2 μg of each affinity-purified recombinant protein was separated by 10% SDS-PAGE and stained with Gel Blue; from the left, as indicated: MBP, MBP-β1 integrin cytoplasmic domain fusion protein, His-tagged 130-kDa 4.1B, His-tagged 80 kDa 4.1R. b, analysis of binding. His-tagged constructs of 4.1B or 4.1R, as in a, were mixed with either MBP or MBP-β1 integrin cytoplasmic domain. The mixtures were incubated with amylose beads and centrifuged to recover bound complexes as described under “Experimental Procedures.” Bound complexes were removed from the beads by washing with SDS, and they were analyzed by SDS-PAGE and immunoblotting with anti-His tag antibody. Note that MPB-β1 integrin cytoplasmic domain binds to 4.1R but not 4.1B and that neither 4.1 protein binds MBP alone.
It is well known that β1 integrin pairs with α5 to form the α5β1 integrin heterodimer, the main fibronectin receptor. We then examined the surface expression of α5 by flow cytometry. Fig. 6, A and B, shows that the surface expression of α5 integrin was also declined by ∼30% in 130-kDa 4.1B-deficient MEF cells. Western blot analysis revealed that the expression levels of both total β1 integrin and α5 integrin in 130-kDa 4.1B-deficient MEF cells were similar to wild type MEF cells (Fig. 6, C and D).
To examine whether 4.1B binds to β1 integrin in MEF cells, we performed a co-immunoprecipitation assay using either anti-4.1B or anti-β1 integrin antibody. Fig. 6E shows that β1 integrin was not pulled down by anti-4.1B HP antibody and that 4.1B was not pulled down by anti-β1 integrin antibody.
In vitro analysis was also consistent with lack of direct interaction between 4.1B and β1 integrin. The cytoplasmic domain of β1 integrin was expressed in Escherichia coli as a fusion with MBP as well as MBP alone; 130-kDa 4.1B and 80 kDa 4.1R were expressed as His-tagged proteins in E. coli (Fig. 6Fa) (note that for reasons that are unclear, MBP-β1 migrates faster on SDS-PAGE than MBP alone). MBP-tagged cytoplasmic domain of β1 integrin, but not MBP alone, was able to pull down 80-kDa 4.1R (as shown previously (22)) but not 130-kDa 4.1B (Fig 6Fb). It should be noted that the 4.1B we used for this in vitro binding was able to bind to cytoplasmic domain of EGF receptor (data not shown), indicating that lack of interaction with β1 integrin was not due to simple inactivity of the construct. Our data indicate that 4.1R and 4.1B control the cellular properties of β1 integrin though distinct mechanisms.
Impaired β1 Integrin Trafficking in 130-kDa 4.1B-deficient MEF Cells
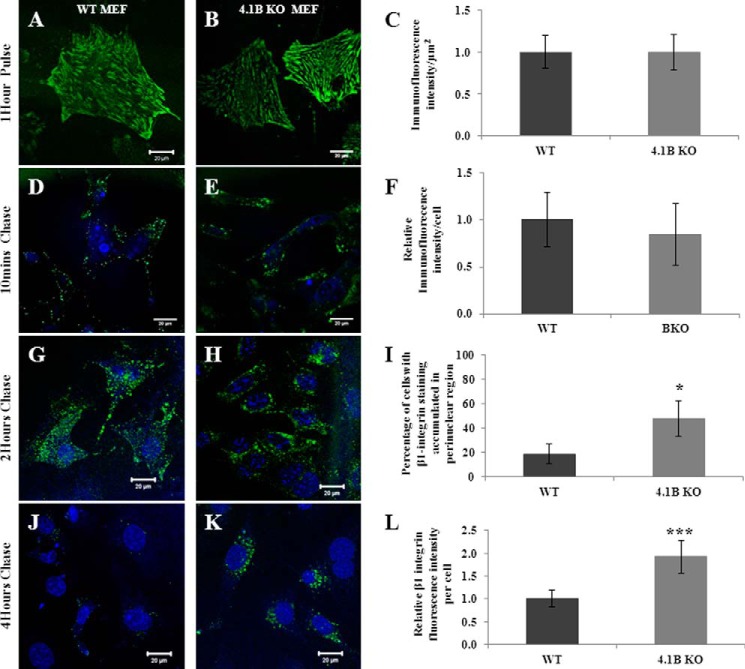
Integrins are continuously internalized and recycled to the cell membrane (45). To explore the mechanisms by which a lack of 130-kDa 4.1B leads to the decreased surface expression of β1 integrin, we performed fluorescence pulse-chase studies using 9EG7 anti-mouse β1 integrin monoclonal antibody to monitor the distribution of internalized β1 integrin over time. Fig. 7, A and B, shows the representative images of surface labeling of β1 integrin on WT and 4.1B-deficient MEF cells after a 1-h pulse at 4 °C. Quantitative analysis from 50 cells of each type is shown in Fig. 7C, which demonstrates that the relative β1 integrin immunofluorescence intensity per μm2 in 130-kDa 4.1B-deficient MEF cells is similar to that of WT MEF cells. Fig. 7, D–F, shows that the internalized β1 integrin after a 10-min chase was similar between WT and 4.1B-deficient MEF cells, implying that the initial antibody uptake is comparable between the two cell types. After 2 h of chase, internalized β1 integrin in WT cells were mainly dispersed in peripheral vesicles (Fig. 7G); in contrast, in 4.1B-deficient cells, the internalized β1 integrin vesicles were accumulated around the perinuclear region (Fig. 7H). Quantitative analysis reveals that although the perinuclear accumulation of β1 integrin was seen in <18% WT cells, it was seen in >64% of 4.1B-deficient cells (Fig. 7I). Upon 4 h of chase, the WT cells were mostly devoid of fluorescence (Fig. 7J, indicating that β1 integrin recycling is almost complete); however, at the same time point, β1 integrin was still observed at the perinuclear region within 4.1B-deficient cells (Fig. 7K). Quantitative analysis reveals that after 4 h of chase, around 2 times β1 integrin were retained in 4.1B-deficient cells than that of WT cells (Fig. 7L). These findings together imply that β1 integrin trafficking was impaired in the absence of 130-kDa 4.1B.
FIGURE 7.
Impaired β1 integrin trafficking in 130-kDa 4.1B-deficient MEF cells. A–C, pulse phase. MEF cells were pulse-labeled with 9EG7 monoclonal antibody for 1 h at 4 °C. Quantitative analysis was from 50 cells of each type. D–F, the internalized β1 integrin after 10 min chase. After a 10-min chase, the cells were quenched, and internalized β1 integrin were visualized. Quantitative analysis was from 50 cells of each type. G–I, the internalized β1 integrin after 2 h chase. After a 2-h chase, the cells were quenched, and internalized β1 integrin were visualized. Quantitative analysis was from 50 cells of each type. *, p < 0.05. J–L, the internalized β1 integrin after 4 h chase. Cells were treated with same procedures after a 4-h chase as described above. The fluorescence intensity of the cells (50 cells of each type) were measured and quantified using Image J. ***, p < 0.001. All pictures were taken with a 25× objective. Scale bars, 20 μm. Note that a single exposure setting was used for all fluorescence quantifications.
DISCUSSION
Because the discovery that 4.1B/DAL-1 is associated with lung cancer (34), much attention has focused on the role of 4.1B as a tumor suppressor (for reviews, see Refs. 14 and 46). However, most such studies have been conducted using cancer cell lines. The physiological function of 4.1B in primary cells (i.e. non-tumor derived) remains largely unexplored. In the present study using primary MEF cells derived from wild type and 4.1B knock-out mice, we found that 130-kDa 4.1B, but not 60-kDa 4.1B, is required for normal adhesion, spreading, and migration of MEF cells. Furthermore, 130-kDa 4.1B interacts with actin and is required for formation of stress fibers; it also controls cell surface expression of β1 integrin by mediating β1 integrin trafficking.
It has been well documented that by utilizing alternative first exons as well as alternative pre-mRNA splicing of numerous internal exons, the 4.1 genes can encode a remarkable array of tissue-specific and cell- and development-specific protein isoforms (5). One aspect of this is that the headpiece of 4.1R controls the activity of the FERM domain (47); differential expression of the headpiece during erythrocyte development is taken as reflecting functional differentiation of 4.1R isoforms in this pathway (48). In the case of 4.1B, a high molecular mass isoform, 200 kDa, has a selective role in the Golgi trafficking of several transmembrane proteins (39). Our data here bring new focus on the specific cellular functions of 4.1B isoforms in that only one of two 4.1B isoforms is required for the normal adhesion, spreading, and migration phenotypes of a primary cell type.
The 130-kDa 4.1B-deficient cells also appear to have a mechanism that attempts to compensate for loss of 130-kDa 4.1B. 4.1R and 4.1N are up-regulated in these cells (Fig. 3). Similar patterns of compensation are seen in other systems; in the 4.1R knock-out mouse (which has ATG1 and ATG2 knocked out), 4.1G is up-regulated in CD4+ cells, keratinocytes, and cardiomyocytes (22, 23, 49); 4.1N also is up-regulated in CD4+ cells (23). Nevertheless, despite their comparatively close sequence similarity as well as a range of common binding activities, such compensation is inadequate physiologically.
Data in Fig. 1 identify a novel translation initiation codon in exon 9 that leads to the generation of a 60-kDa protein. Interestingly, this 60-kDa isoform is still present in MEF cells and in kidney (39) as well as many other tissues (data not shown) of 4.1B knock-out mice. The 4.1B transgenic mice used here were made by targeting exon 3 (38). Initiation of translation in exon 9 should not be affected by this transgenic construct; hence, these mice represent an incomplete knock-out.
Although a large body of evidence from clinical samples and tumor cell lines strongly suggest the role of 4.1B as a tumor suppressor and metastasis suppressor (for reviews, see Refs. 14 and 46), the lack of spontaneous tumor formation in 4.1B knock-out mice prompted Yi et al. (38) to conclude that 4.1B does not function as a tumor suppressor. Our data suggest that this conclusion may be compromised, because this mouse strain is an incomplete knock-out.
Data reported here on primary cells stand in interesting contrast to some of the data reported for 4.1B/DAL-1 in cancer lines. For example, in Schwannoma cells, it has been reported that 4.1B impairs cell motility and disrupts the actin cytoskeleton during cell spreading and that it does not bind actin (50). However, these analyses were done by overexpression of a truncated form of 4.1B initiated at ATG2 and lacking the CTD using cell lines that already expressed native levels of 4.1B, so the experiments are not directly comparable to ours.
In sarcoma cells, loss of 4.1B has been reported to be associated with metastasis (27). Paralleling our observations on MEF cells, in sarcoma cells 4.1B is required for stress fiber formation. But sarcoma cells in which 4.1B was knocked down with siRNA displayed increased motility (27). Although the behavior of sarcoma cells reported in Cavanna et al. (27) stands in contrast to the present report, there are significant methodological differences between the two accounts. The cell types used were different. Moreover, in our transwell assays we examined the behavior of cells 8 h after seeding, whereas Cavanna et al. (27) analyzed cells that had been grown for 24 h or more on substrates.
There is increasing evidence for a role for interactions between the 4.1 proteins and integrins (22, 51–54). Data in Fig. 6 indicate that 4.1B does not bind β1 integrin directly, unlike 4.1R (22), although both control the level of cell surface expression and activity (albeit in different cell systems). 4.1B has been shown to bind β8 integrin via the CTD (53), and the two cooperate together in heart morphogenesis (51). 4.1B and 4.1G together regulate spreading of primary astrocytes (52). Conservation of the CTD is high between the four 4.1 proteins, and so all 4.1 proteins bind β8 integrin (53). 4.1R, therefore, has the prospect of cross-linking different integrin complexes via the FERM domain (β1 integrin) and CTD (β8 integrin). Whether analogous processes operate with 4.1B is unclear; in the future it will be important to understand whether or not the 4.1B FERM domain also binds another integrin.
Our findings leave a conundrum in relation to the activities of 130-kDa 4.1B. Clearly, the 60-kDa cannot compensate for the absence of the 130-kDa, and yet the 60-kDa form contains all the best characterized functional sites. The 60-kDa 4.1B sequence retains the binding site for CADM1/TSLC1 (8), a cell adhesion molecule that binds to the C-terminal lobe of the FERM domain, the SABD (which binds actin) (11). and the CTD (which binds β8 integrin) (53). It also retains exons equivalent to the “core” of 4.1R (55), which bind IQGAP1 and recruit it to the leading edge of motile cells (44). Exons 2–8 must, therefore, encode the critical activities that distinguish the 130- from the 60-kDa protein. In relation to the known structure for 4.1 proteins, the major interactive sites lost in the MEF 60-kDa 4.1B would be the N-terminal lobe and the fully folded structure of the central lobe of the FERM domain. In 4.1R these bind multiple transmembrane proteins (20) as well as being required for interaction with phosphatidylinositol-4,5-biphosphate (56). One possibility is that lack of exons 2–8 means that 60-kDa 4.1B cannot be recruited to the plasma membrane via FERM domain interactions, and so activities of the SABD and/or CTD that require membrane localization cannot function. We also note that both 130-and 60-kDa 4.1B proteins lack exon 16, which is associated with high affinity spectrin-actin binding (11); presumably the critical activities of 130-kDa 4.1B do not include high affinity spectrin-actin binding.
Our findings on 4.1B reported here give new insight into isoform-specific functions of this protein, which is required both in normal physiological processes and in tumor suppression and metastasis. In relation to motility-related processes, they have implications in both normal physiology and cancer. A major challenge for the future will be to define the activities encoded in exons 2–8, which encode isoform-specific functions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK 26263 and DK 32094. This work was also supported by Natural Science Foundation of China Grants 81171905 and 81272187 and by an American Society of Hematology (ASH) bridge grant (to X. A.).
- FA
- FERM-adjacent
- SABD
- spectrin-actin binding domain
- CTD
- C-terminal domain
- HP
- headpiece
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Baines A. J., Lu H.-C., Bennett P. M. (2014) The Protein 4.1 family. Hub proteins in animals for organizing membrane proteins. Biochim. Biophys. Acta 1838, 605–619 [DOI] [PubMed] [Google Scholar]
- 2. Baines A. J. (2010) The spectrin-ankyrin-4.1-adducin membrane skeleton. Adapting eukaryotic cells to the demands of animal life. Protoplasma 244, 99–131 [DOI] [PubMed] [Google Scholar]
- 3. Mohandas N., Gallagher P. G. (2008) Red cell membrane. Past, present, and future. Blood 112, 3939–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peters L. L., Weier H. U., Walensky L. D., Snyder S. H., Parra M., Mohandas N., Conboy J. G. (1998) Four paralogous protein 4.1 genes map to distinct chromosomes in mouse and human. Genomics 54, 348–350 [DOI] [PubMed] [Google Scholar]
- 5. Parra M., Gee S., Chan N., Ryaboy D., Dubchak I., Mohandas N., Gascard P. D., Conboy J. G. (2004) Differential domain evolution and complex RNA processing in a family of paralogous EPB41 (protein 4.1) genes facilitate expression of diverse tissue-specific isoforms. Genomics 84, 637–646 [DOI] [PubMed] [Google Scholar]
- 6. Chishti A. H., Kim A. C., Marfatia S. M., Lutchman M., Hanspal M., Jindal H., Liu S. C., Low P. S., Rouleau G. A., Mohandas N., Chasis J. A., Conboy J. G., Gascard P., Takakuwa Y., Huang S. C., Benz E. J., Jr., Bretscher A., Fehon R. G., Gusella J. F., Ramesh V., Solomon F., Marchesi V. T., Tsukita S., Tsukita S., Hoover K. B. (1998) The FERM domain. A unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci. 23, 281–282 [DOI] [PubMed] [Google Scholar]
- 7. Han B. G., Nunomura W., Takakuwa Y., Mohandas N., Jap B. K. (2000) Protein 4.1R core domain structure and insights into regulation of cytoskeletal organization. Nat. Struct. Biol. 7, 871–875 [DOI] [PubMed] [Google Scholar]
- 8. Busam R. D., Thorsell A.-G., Flores A., Hammarström M., Persson C., Öbrink B., Hallberg B. M. (2011) Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B). J. Biol. Chem. 286, 4511–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baines A. J. (2006) A FERM-adjacent (FA) region defines a subset of the 4.1 superfamily and is a potential regulator of FERM domain function. BMC Genomics 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Correas I., Speicher D. W., Marchesi V. T. (1986) Structure of the spectrin-actin binding site of erythrocyte protein 4.1. J. Biol. Chem. 261, 13362–13366 [PubMed] [Google Scholar]
- 11. Gimm J. A., An X., Nunomura W., Mohandas N. (2002) Functional characterization of spectrin-actin-binding domains in 4.1 family of proteins. Biochemistry 41, 7275–7282 [DOI] [PubMed] [Google Scholar]
- 12. Scott C., Phillips G. W., Baines A. J. (2001) Properties of the C-terminal domain of 4.1 proteins. Eur. J. Biochem. 268, 3709–3717 [DOI] [PubMed] [Google Scholar]
- 13. Moleirinho S., Tilston-Lunel A., Angus L., Gunn-Moore F., Reynolds P. A. (2013) The expanding family of FERM proteins. Biochem. J. 452, 183–193 [DOI] [PubMed] [Google Scholar]
- 14. Yu H., Zhang Y., Ye L., Jiang W. G. (2011) The FERM family proteins in cancer invasion and metastasis. Front. Biosci. 16, 1536–1550 [DOI] [PubMed] [Google Scholar]
- 15. Parra M. K., Tan J. S., Mohandas N., Conboy J. G. (2008) Intrasplicing coordinates alternative first exons with alternative splicing in the protein 4.1R gene. EMBO J. 27, 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gascard P., Nunomura W., Lee G., Walensky L. D., Krauss S. W., Takakuwa Y., Chasis J. A., Mohandas N., Conboy J. G. (1999) Deciphering the nuclear import pathway for the cytoskeletal red cell protein 4.1R. Mol. Biol. Cell 10, 1783–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conboy J. (1999) The role of alternative pre-mRNA splicing in regulating the structure and function of skeletal protein 4.1. Proc. Soc. Exp. Biol. Med. 220, 73–78 [DOI] [PubMed] [Google Scholar]
- 18. Luque C. M., Lallena M. J., Alonso M. A., Correas I. (1998) An alternative domain determines nuclear localization in multifunctional protein 4.1. J. Biol. Chem. 273, 11643–11649 [DOI] [PubMed] [Google Scholar]
- 19. Salomao M., Chen K., Villalobos J., Mohandas N., An X., Chasis J. A. (2010) Hereditary spherocytosis and hereditary elliptocytosis. Aberrant protein sorting during erythroblast enucleation. Blood 116, 267–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salomao M., Zhang X., Yang Y., Lee S., Hartwig J. H., Chasis J. A., Mohandas N., An X. (2008) Protein 4.1R-dependent multiprotein complex. New insights into the structural organization of the red blood cell membrane. Proc. Natl. Acad. Sci. U.S.A. 105, 8026–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tchernia G., Mohandas N., Shohet S. B. (1981) Deficiency of skeletal membrane protein band 4.1 in homozygous hereditary elliptocytosis. Implications for erythrocyte membrane stability. J. Clin. Invest. 68, 454–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L., Hughes R. A., Baines A. J., Conboy J., Mohandas N., An X. (2011) Protein 4.1R regulates cell adhesion, spreading, migration, and motility of mouse keratinocytes by modulating surface expression of β1 integrin. J. Cell Sci. 124, 2478–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kang Q., Yu Y., Pei X., Hughes R., Heck S., Zhang X., Guo X., Halverson G., Mohandas N., An X. (2009) Cytoskeletal protein 4.1R negatively regulates T-cell activation by inhibiting the phosphorylation of LAT. Blood 113, 6128–6137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu C., Weng H., Chen L., Yang S., Wang H., Debnath G., Guo X., Wu L., Mohandas N., An X. (2013) Impaired intestinal calcium absorption in protein 4.1R-deficient mice due to altered expression of plasma membrane calcium ATPase 1b (PMCA1b). J. Biol. Chem. 288, 11407–11415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nunomura W., Denker S. P., Barber D. L., Takakuwa Y., Gascard P. (2012) Characterization of cytoskeletal protein 4.1R interaction with NHE1 (Na+/H+ exchanger isoform 1). Biochem. J. 446, 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang S., Guo X., Debnath G., Mohandas N., An X. (2009) Protein 4.1R links E-cadherin/β-catenin complex to the cytoskeleton through its direct interaction with β-catenin and modulates adherens junction integrity. Biochim. Biophys. Acta 1788, 1458–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cavanna T., Pokorná E., Veselý P., Gray C., Zicha D. (2007) Evidence for protein 4.1B acting as a metastasis suppressor. J. Cell Sci. 120, 606–616 [DOI] [PubMed] [Google Scholar]
- 28. Gutmann D. H., Donahoe J., Perry A., Lemke N., Gorse K., Kittiniyom K., Rempel S. A., Gutierrez J. A., Newsham I. F. (2000) Loss of DAL-1, a protein 4.1-related tumor suppressor, is an important early event in the pathogenesis of meningiomas. Hum. Mol. Genet. 9, 1495–1500 [DOI] [PubMed] [Google Scholar]
- 29. Yu T., Robb V. A., Singh V., Gutmann D. H., Newsham I. F. (2002) The 4.1/ezrin/radixin/moesin domain of the DAL-1/protein 4.1B tumour suppressor interacts with 14-3-3 proteins. Biochem. J. 365, 783–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y., Xu R., Li G., Xie X., Long J., Wang H. (2012) Loss of expression of the differentially expressed in adenocarcinoma of the lung (DAL-1) protein is associated with metastasis of non-small cell lung carcinoma cells. Tumour Biol. 33, 1915–1925 [DOI] [PubMed] [Google Scholar]
- 31. Einheber S., Meng X., Rubin M., Lam I., Mohandas N., An X., Shrager P., Kissil J., Maurel P., Salzer J. L. (2013) The 4.1B cytoskeletal protein regulates the domain organization and sheath thickness of myelinated axons. Glia 61, 240–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cifuentes-Diaz C., Chareyre F., Garcia M., Devaux J., Carnaud M., Levasseur G., Niwa-Kawakita M., Harroch S., Girault J. A., Giovannini M., Goutebroze L. (2011) Protein 4.1B contributes to the organization of peripheral myelinated axons. PLoS ONE 6, e25043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buttermore E. D., Dupree J. L., Cheng J., An X., Tessarollo L., Bhat M. A. (2011) The cytoskeletal adaptor protein band 4.1B is required for the maintenance of paranodal axoglial septate junctions in myelinated axons. J. Neurosci. 31, 8013–8024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tran Y. K., Bögler O., Gorse K. M., Wieland I., Green M. R., Newsham I. F. (1999) A novel member of the NF2/ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res. 59, 35–43 [PubMed] [Google Scholar]
- 35. Parra M., Gascard P., Walensky L. D., Gimm J. A., Blackshaw S., Chan N., Takakuwa Y., Berger T., Lee G., Chasis J. A., Snyder S. H., Mohandas N., Conboy J. G. (2000) Molecular and functional characterization of protein 4.1B, a novel member of the protein 4.1 family with high level, focal expression in brain. J. Biol. Chem. 275, 3247–3255 [DOI] [PubMed] [Google Scholar]
- 36. Yageta M., Kuramochi M., Masuda M., Fukami T., Fukuhara H., Maruyama T., Shibuya M., Murakami Y. (2002) Direct association of TSLC1 and DAL-1, two distinct tumor suppressor proteins in lung cancer. Cancer Res. 62, 5129–5133 [PubMed] [Google Scholar]
- 37. Denisenko-Nehrbass N., Oguievetskaia K., Goutebroze L., Galvez T., Yamakawa H., Ohara O., Carnaud M., Girault J. A. (2003) Protein 4.1B associates with both Caspr/paranodin and Caspr2 at paranodes and juxtaparanodes of myelinated fibres. Eur. J. Neurosci. 17, 411–416 [DOI] [PubMed] [Google Scholar]
- 38. Yi C., McCarty J. H., Troutman S. A., Eckman M. S., Bronson R. T., Kissil J. L. (2005) Loss of the putative tumor suppressor band 4.1B/Dal1 gene is dispensable for normal development and does not predispose to cancer. Mol. Cell. Biol. 25, 10052–10059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kang Q., Wang T., Zhang H., Mohandas N., An X. (2009) A Golgi-associated protein 4.1B variant is required for assimilation of proteins in the membrane. J. Cell Sci. 122, 1091–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu J. (2005) Preparation, Culture, and Immortalization of mouse embryonic fibroblasts. Curr. Protoc. Mol. Biol. 10.1002/0471142727.mb2801s70 [DOI] [PubMed] [Google Scholar]
- 41. An X. L., Takakuwa Y., Manno S., Han B. G., Gascard P., Mohandas N. (2001) Structural and functional characterization of protein 4.1R- phosphatidylserine interaction. Potential role in 4.1R sorting within cells. J. Biol. Chem. 276, 35778–35785 [DOI] [PubMed] [Google Scholar]
- 42. Powelka A. M., Sun J., Li J., Gao M., Shaw L. M., Sonnenberg A., Hsu V. W. (2004) Stimulation-dependent recycling of integrin β1 regulated by ARF6 and Rab11. Traffic 5, 20–36 [DOI] [PubMed] [Google Scholar]
- 43. Gascard P., Parra M. K., Zhao Z., Calinisan V. R., Nunomura W., Rivkees S. A., Mohandas N., Conboy J. G. (2004) Putative tumor suppressor protein 4.1B is differentially expressed in kidney and brain via alternative promoters and 5′ alternative splicing. Biochim. Biophys. Acta 1680, 71–82 [DOI] [PubMed] [Google Scholar]
- 44. Ruiz-Sáenz A., Kremer L., Alonso M. A., Millán J., Correas I. (2011) Protein 4.1R regulates cell migration and IQGAP1 recruitment to the leading edge. J. Cell Sci. 124, 2529–2538 [DOI] [PubMed] [Google Scholar]
- 45. Bridgewater R. E., Norman J. C., Caswell P. T. (2012) Integrin trafficking at a glance. J. Cell Sci. 125, 3695–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun C. X., Robb V. A., Gutmann D. H. (2002) Protein 4.1 tumor suppressors. Getting a FERM grip on growth regulation. J. Cell Sci. 115, 3991–4000 [DOI] [PubMed] [Google Scholar]
- 47. Nunomura W., Parra M., Hebiguchi M., Sawada K., Mohandas N., Takakuwa Y. (2009) Marked difference in membrane protein binding properties of the two isoforms of protein 4.1R expressed at early and late stages of erythroid differentiation. Biochem. J. 417, 141–148 [DOI] [PubMed] [Google Scholar]
- 48. Nunomura W., Gascard P., Takakuwa Y. (2011) Insights into the function of the unstructured N-terminal domain of proteins 4.1R and 4.1G in erythropoiesis. Int. J. Cell Biol. 2011, 943272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stagg M. A., Carter E., Sohrabi N., Siedlecka U., Soppa G. K., Mead F., Mohandas N., Taylor-Harris P., Baines A., Bennett P., Yacoub M. H., Pinder J. C., Terracciano C. M. (2008) Cytoskeletal protein 4.1R affects repolarization and regulates calcium handling in the heart. Circ. Res. 103, 855–863 [DOI] [PubMed] [Google Scholar]
- 50. Gutmann D. H., Hirbe A. C., Huang Z. Y., Haipek C. A. (2001) The protein 4.1 tumor suppressor, DAL-1, impairs cell motility but regulates proliferation in a cell type-specific fashion. Neurobiol. Dis. 8, 266–278 [DOI] [PubMed] [Google Scholar]
- 51. Jung Y., Kissil J. L., McCarty J. H. (2011) β8 integrin and band 4.1B cooperatively regulate morphogenesis of the embryonic heart. Dev. Dyn. 240, 271–277 [DOI] [PubMed] [Google Scholar]
- 52. Jung Y., McCarty J. H. (2012) Band 4.1 proteins regulate integrin-dependent cell spreading. Biochem. Biophys. Res. Commun. 426, 578–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McCarty J. H., Cook A. A., Hynes R. O. (2005) An interaction between αvβ8 integrin and Band 4.1B via a highly conserved region of the band 4.1 C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 102, 13479–13483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ralston K. J., Hird S. L., Zhang X., Scott J. L., Jin B., Thorne R. F., Berndt M. C., Boyd A. W., Burns G. F. (2004) The LFA-1-associated molecule PTA-1 (CD226) on T cells forms a dynamic molecular complex with protein 4.1G and human discs large. J. Biol. Chem. 279, 33816–33828 [DOI] [PubMed] [Google Scholar]
- 55. Luque C. M., Correas I. (2000) A constitutive region is responsible for nuclear targeting of 4.1R. Modulation by alternative sequences results in differential intracellular localization. J. Cell Sci. 113, 2485–2495 [DOI] [PubMed] [Google Scholar]
- 56. An X., Zhang X., Debnath G., Baines A. J., Mohandas N. (2006) Phosphatidylinositol-4,5-biphosphate (PIP2) differentially regulates the interaction of human erythrocyte protein 4.1 (4.1R) with membrane proteins. Biochemistry 45, 5725–5732 [DOI] [PubMed] [Google Scholar]