Background: The mechanism of marine collagen degradation is largely unknown.
Results: Myroicolsin, a subtilisin-like protease from a marine bacterium, was characterized, and its collagenolytic mechanism was studied.
Conclusion: Myroicolsin has a novel domain structure and a unique collagen degradation mechanism compared with other subtilisin-like proteases.
Significance: This study provides new insights into the mechanism of subtilisin-like proteases' collagenolysis and marine nitrogen cycling.
Keywords: Bacteria, Biodegradation, Collagen, Enzyme Mechanisms, Serine Protease, Collagenolytic Mechanism, Deep Sea Sediment, Nitrogen Cycling, Structural Characteristics, Subtilisin-like Protease
Abstract
Collagen is an insoluble protein that widely distributes in the extracellular matrix of marine animals. Collagen degradation is an important step in the marine nitrogen cycle. However, the mechanism of marine collagen degradation is still largely unknown. Here, a novel subtilisin-like collagenolytic protease, myroicolsin, which is secreted by the deep sea bacterium Myroides profundi D25, was purified and characterized, and its collagenolytic mechanism was studied. Myroicolsin displays low identity (<30%) to previously characterized subtilisin-like proteases, and it contains a novel domain structure. Protein truncation indicated that the Pro secretion system C-terminal sorting domain in the precursor protein is involved in the cleavage of the N-propeptide, and the linker is required for protein folding during myroicolsin maturation. The C-terminal β-jelly roll domain did not bind insoluble collagen fiber, suggesting that myroicolsin may degrade collagen without the assistance of a collagen-binding domain. Myroicolsin had broad specificity for various collagens, especially fish-insoluble collagen. The favored residue at the P1 site was basic arginine. Scanning electron microscopy and atomic force microscopy, together with biochemical analyses, confirmed that collagen fiber degradation by myroicolsin begins with the hydrolysis of proteoglycans and telopeptides in collagen fibers and fibrils. Myroicolsin showed strikingly different cleavage patterns between native and denatured collagens. A collagen degradation model of myroicolsin was proposed based on our results. Our study provides molecular insight into the collagen degradation mechanism and structural characterization of a subtilisin-like collagenolytic protease secreted by a deep sea bacterium, shedding light on the degradation mechanism of deep sea sedimentary organic nitrogen.
Introduction
The degradation of marine organic nitrogen is an important part of the global nitrogen cycle. High molecular weight particulate organic nitrogen (PON)3 is present as chitin and peptidoglycan at the surface of the ocean, whereas it is mostly present as biological and chemical hydrolysis-resistant amides on the deep sea floor (1). Deep sea sediments contain at least 25% of the global ocean nitrogen burial. Most PON in the sediments can be degraded by microorganisms into dissolved organic nitrogen, which can then participate in ocean nitrogen cycling (2, 3). Therefore, sedimentary PON degradation by microorganisms is an important step in ocean nitrogen cycling. Collagen is the most abundant fibrous protein in all higher organisms, including marine animals (4–6). Due to its tight, complicated structure and insolubility in water, collagen is resistant to common proteases and can only be degraded by a limited number of collagenolytic proteases. Collagen is therefore an important component of deep sea sedimentary PON, and its degradation by the extracellular collagenolytic proteases from various deep sea microorganisms is an important biological process for the release of fixed nitrogen into the global nitrogen cycle. The collagen monomer is composed of three helical polypeptide chains containing a repeating Gly-Xaa-Yaa sequence. Collagen monomers assemble into stable collagen fibrils in a parallel staggered arrangement, and various covalent cross-links within or between fibrils maintain the stability of the fibrillar structure (7–9). Collagen fibrils further aggregate to form collagen fibers and other superfibrillar structures through interdigitation with proteoglycans in the extracellular matrix (10).
Collagenolytic proteases are widely distributed among many peptidase families. Among the collagenolytic proteases, several mammalian matrix metalloproteinases (MMPs, peptidase family M10), serine proteases isolated from Uca pugilator (family S1), and cathepsins K and L from animals (family C1) have been extensively studied, and their collagen degradation mechanisms have been explored (11–18, 20). Some bacterial extracellular metalloproteases in the M9 family, primarily from the Vibrio (21) and Clostridium strains (22, 23), also show strong collagenolytic activity. The structures and collagen degradation mechanisms of these collagenolytic proteases are different.
The S8 family, which is also known as the subtilisin family, is the second largest family of serine proteases (11). Subtilisin, the prototype of the S8 family, is one of the most studied bacterial serine proteases because of its industrial importance (24). Some subtilisin-like proteases from environmental and pathogenic microorganisms have been demonstrated to be collagenolytic proteases. The thermostable protease secreted by Geobacillus collagenovorans MO-1 was the first S8 collagenolytic protease to be studied (25, 26). Several subtilisin-like proteases with strong collagenolytic activity are secreted by the pathogens Stenotrophomonas maltophilia and Acanthamoeba spp. and may serve as targets for the development of therapeutic agents (27–29). Nevertheless, there are few studies about subtilisin-like collagenolytic proteases secreted by marine bacteria. Deseasin MCP-01 from the deep sea bacterium Pseudoalteromonas sp. SM9913 is a collagenolytic protease of the S8 family with a C-terminal polycystic kidney disease (PKD) domain that is responsible for collagen binding (30). AcpII from the deep sea bacterium Alkalimonas collagenimarina AC40, another S8 collagenolytic protease, contains a protease-associated domain to regulate the access of substrates to the catalytic domain (31). The collagen degradation mechanisms of subtilisin-like collagenolytic proteases are rarely studied.
Myroides profundi D25 is a protease-secreting bacterium isolated from the deep sea sediment of the southern Okinawa Trough at a water depth of 1245 m (32). Our previous study showed that the D25 strain produces a novel elastinolytic metalloprotease, myroilysin, which also has strong collagen-swelling ability and cooperates with collagenase in collagen hydrolysis (33). In this study, a novel collagenolytic protease secreted by strain D25, designated myroicolsin, was purified and characterized. Gene cloning and sequence analysis showed that this enzyme is a novel subtilisin-like protease containing a unique domain structure. The function of some domains and propeptides of myroicolsin was studied by protein truncation, and the collagenolytic mechanism of myroicolsin was revealed in detail through microscopic observation and biochemical analysis. Finally, a collagen degradation model was proposed based on our results.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Experimental Materials
M. profundi D25 isolated from the deep sea sediment of the southern Okinawa Trough was used for protease production (32). Escherichia coli DH5α (Transgen Biotech) was used as the host for gene cloning. pET-22b(+) (Novagen) was used for the construction of expression vectors, and E. coli BL21 (DE3) (Transgen Biotech) was used as the host for protein expression. Insoluble type I collagen fiber (bovine Achilles tendon) was purchased from Worthington. Type II and IV collagens were purchased from BD Biosciences. α-Casein, elastin-orcein, azo dye-impregnated collagen, and decorin from bovine articular cartilage were purchased from Sigma. Gelatin was purchased from Boston Biomedical Inc., and trypsin was from Beijing Solarbio Co. Fish collagen was extracted from Cyprinus carpio skin using the method described previously (34). Acid-soluble collagen was prepared by dissolving insoluble bovine type I collagen in acetic acid and then dialyzing against 20 mm Tris-HCl buffer (pH 8.5) at 4 °C overnight. All peptides were synthesized by China Peptides Co., Ltd.
Protease Purification
M. profundi D25 was cultured at 15 °C for 84 h in the fermentation medium containing 0.2% (w/v) yeast extract, 1% (w/v) gelatin, and artificial seawater (pH 8.0) with continuous shaking. The culture was centrifuged (10,000 rpm, 20 min), and the supernatant was precipitated by 55% saturation with solid ammonium sulfate powder. The precipitate was collected by centrifugation (10,000 rpm, 10 min) and dissolved in 50 mm Tris-HCl buffer (pH 9.5, buffer A). After dialysis against buffer A for desalination, the proteases in the solution were purified on a pre-equilibrated DEAE-Sepharose Fast Flow column (Amersham Biosciences) with a linear gradient of 0–0.8 m NaCl in buffer A. The fractions with collagenolytic activity were collected and further purified on a Sephadex G75 column with 50 mm Tris-HCl buffer (pH 8.5, buffer B). All purification procedures were conducted at 0–10 °C. Protease purity was analyzed by 12.5% SDS-PAGE. The purified protease was named myroicolsin.
Protein Measurement and Enzyme Assays
Protein concentration was measured using a BCA protein assay kit (Thermo) with bovine serum albumin (BSA) as the standard. The activities of myroicolsin toward multiple types of collagen and gelatin were assessed according to the method described by Worthington (35). Briefly, myroicolsin was incubated for 5 h with insoluble collagen (bovine type I collagen or fish collagen) or for 0.5 h with soluble collagen (type II or type IV) or gelatin. The amino acids liberated were quantified using the colorimetric ninhydrin method with l-leucine as the standard. One unit of enzyme activity was defined as the amount of enzyme that released 1 nmol of l-leucine/h from insoluble collagen fibers or the amount that released 1 nmol of l-leucine/min from soluble collagens or gelatin. The activity against azo dye-impregnated collagen was measured according to the method of Abraham and Breuil (36). The activity toward casein was determined at 40 °C using the method of He et al. (37). The elastinolytic activity was determined with the method described by Chen et al. (33). The activities against synthetic peptides were measured at 25 °C using a previous method (38).
Cloning and Sequencing of the Myroicolsin Gene
The purified myroicolsin in the SDS-polyacrylamide gel was blotted onto a polyvinylidene difluoride membrane (Sequi-Blot PVDF membrane, Bio-Rad), and its N-terminal amino acid sequence was determined by Edman degradation on a PROCISE491 sequencer (Applied Biosystems). Inhibitor assays indicated that myroicolsin was a serine protease. Based on the N-terminal sequence of myroicolsin and the conserved sequence in the catalytic domain of serine proteases (39), the myroicolsin gene containing 2040 bp was obtained by PCR and thermal asymmetric interlaced PCR (40). After verification with high fidelity Fastpfu DNA polymerase (Transgen Biotech), the nucleotide sequence of this gene was submitted to the GenBankTM database under accession number JF514144.
Expression and Purification of the Mutants of Myroicolsin
The DNA fragments encoding mutants of myroicolsin were amplified by PCR using the genomic DNA of M. profundi D25 as a template. The recombinants were expressed in E. coli BL21 (DE3) with the pET-22b(+) expression vector and induced with isopropyl-β-d- thiogalactopyranoside. The constructed recombinants included the following: Δβ/C (β-jelly roll domain and C-pro-secre-tail truncated mutant), Δβ (β-jelly roll domain truncated mutant), Δlinker/β (linker and β-jelly roll domain truncated mutant), and CT-β (expression of β-jelly roll domain). For Δβ/C, Δβ, and Δlinker/β, the cultures were incubated at 15 °C for 3–5 days with 0.05 mm isopropyl-β-d- thiogalactopyranoside. The supernatants were then collected and purified by the same method used for wild-type myroicolsin. For recombinant CT-β, the culture was induced with 0.2 mm isopropyl-β-d- thiogalactopyranoside at 20 °C for 12 h. After centrifuging the culture, the intracellular target proteins were released by sonication in an ice bath and were further purified with a His-Bind column (Novagen).
Collagen Binding Assay
The collagen-binding ability of CT-β was analyzed by using a previous method with minor modifications (26). Five milligrams of collagen fibers were incubated with buffer B at 0 °C for 30 min, the buffer was discarded, and collagen fibers were mixed with 20 μg of CT-β in 200 μl of buffer B at 0 or 25 °C. Samples were collected at 0, 2, 5, and 8 h and subjected to SDS-PAGE analysis. Collagen fibers incubated with BSA, CT-β incubated in buffer B, and BSA incubated in buffer B were used as controls.
Scanning Electron Microscopy
Scanning electron microscopy (SEM) was used to observe the collagen fiber before and after enzymatic treatment. Five milligrams of collagen fibers were incubated with 5 μg of myroicolsin in 100 μl of buffer B at 30 °C with continuous mixing, and samples were collected at different reaction times. Collagen fibers treated with buffer B served as the negative control. The separated collagen samples were rinsed with distilled water and lyophilized. Then the samples were deposited on aluminum foil and coated with 5 nm of platinum. Imaging was performed on a Hitachi FE-S4800 scanning electron microscope operated at 2.0–5.0 kV.
Atomic Force Microscopy
The nanostructures of collagen fibrils and monomers were observed by atomic force microscopy (AFM) after collagen fibers (5 mg) were incubated with 5 μg of myroicolsin in 100 μl of buffer B at 30 °C for 48 h with continuous mixing. The samples were rinsed twice with distilled water, spread onto freshly cleaved mica, and dried in air. Imaging was performed in air in ScanAsyst mode using a Multimode Nanoscope VIII AFM (Bruker AXS) with a J-type scanner. Probe NSC11 (MikroMasch) with a cantilever length of 90 μm and a nominal spring constant of 48 newtons/m was used. The cross-section analysis was performed with the associated AFM software Nanoscope Analysis.
Enzymatic Digestion of Myroicolsin to Collagen Fibers
Two milligrams of collagen fibers were treated with 1 μm myroicolsin at 30 °C for 24 h, and the dry weight of collagen fibers before and after treatment was analyzed using an analytical balance. The amino acids released from 10 mg of collagen fibers by 1 μm myroicolsin at 30 °C were quantified using a colorimetric ninhydrin method with l-leucine as the standard. To test whether myroicolsin could release glycosaminoglycans (GAGs) from the proteoglycan within collagen fibrils simultaneously, the GAG concentration in the supernatant of the digested mixture was quantified by the dimethylmethylene blue assay with chondroitin 4-sulfate as the standard (41). Collagen fibers incubated with 5 μm trypsin or buffer B were used as the controls. To analyze the degradation of decorin, the archetypal proteoglycan of the vertebrate extracellular matrix, 0.5 mg of decorin was incubated with 0.1 μm myroicolsin in buffer B at 30 °C for 0, 5, 15, or 40 min. The hydrolyzed product was analyzed by SDS-PAGE. Decorin incubated with 0.5 μm trypsin at 30 °C was used as the control. A fluorescence spectroscopy assay was used to detect the release of pyridinolines and deoxypyridinolines from the telopeptides of collagen monomers by myroicolsin. Collagen fibers (20 mg) were incubated with 0.1 μm myroicolsin at 30 °C for 48 h. The supernatant of the digested mixture was detected on an FP-6500 spectrometer (Jasco) at an excitation wavelength of 295 nm. Collagen fibers incubated with 0.1 μm trypsin or in buffer B were used as the controls. Circular dichroism (CD) spectroscopy was performed to analyze the effect of myroicolsin on the secondary structure of collagen monomers according to a method described previously (42).
Analysis and Verification of the Cleavage Pattern of Collagen Fiber by Myroicolsin
Collagen fibers were incubated with 1 μm myroicolsin in 10 mm Tris-HCl buffer (pH 8.5) at 25 °C and 50 °C. Collagen fibers incubated with buffer were used as the control. The reactions were stopped by the addition of 1% methanoic acid, and the hydrolytic products were separated by HPLC (Shimaduz) on a C18 column (Venusil MP C18). The molecular masses of the hydrolytic products were then determined by liquid chromatography-mass spectrometry (LC/MS) at the Beijing Protein Institute Co., Ltd. The sequences of these released peptides were identified by MASCOT MS/MS Ion Research tools and ExPASy tools.
To confirm the P1 and P1′ residue preferences of myroicolsin, several peptides were synthesized by China Peptides Co., Ltd., based on the sequences of the peptides released from collagen polypeptide chains. Each of these peptides (250 μg) was mixed with 5 μg of myroicolsin in 100 μl of 10 mm Tris-HCl buffer (pH 8.5). After incubation at 25 °C for 1 h, the reaction was stopped by the addition of 1% methanoic acid. The molecular masses and sequences of all released peptides were analyzed as described above.
RESULTS
Purification and Characterization of the Collagenolytic Protease Secreted by M. profundi D25
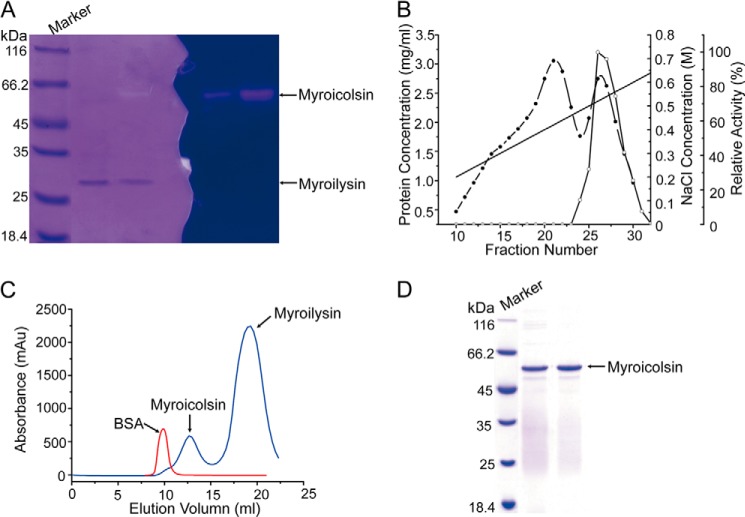
In addition to myroilysin (33), M. profundi D25 secretes another protease with high molecular mass and gelatinolytic activity (Fig. 1A). This protease was purified from strain D25 cultured in a gelatin-containing medium (Fig. 1, B–D). Approximately 7-fold purification was achieved, with a final recovery of 11.3% of the original activity. The purified protease, a monomeric enzyme (Fig. 1C) with a molecular mass of ∼56 kDa (Table 1), was named myroicolsin. Myroicolsin showed strong activity toward insoluble collagen fibers. With insoluble bovine type I collagen fiber as the substrate, the optimal temperature of myroicolsin was 60 °C, and 10% of the activity was retained at 0 °C (Table 1). Because collagen is more vulnerable to denaturation to gelatin at temperatures ≥50 °C, the high activity of myroicolsin at 60 °C may result from its gelatinolytic activity. The half-time of myroicolsin activity at 60 °C was only 10 min, indicating that myroicolsin is thermally labile without substrate protection (Table 1). Myroicolsin displayed the greatest collagenolytic activity at pH 8.5 (Table 1), which rapidly decreased at pH below 6.0 or above 10.0; greater than 60% activity was retained between pH 7.0 and 9.5. The activity of myroicolsin toward collagen was detected in the presence of 0–4 m NaCl. Myroicolsin showed the highest activity in 0.5 m NaCl (114.69 ± 3.24%), and more than 50% of the highest activity was retained in 4 m NaCl. The collagenolytic activity of myroicolsin was inhibited by most of the metal ions tested. However, 4 mm Ca2+ markedly increased the activity by 92.49% (Table 2). The activity of myroicolsin was almost completely abolished by the serine protease inhibitor p-amidinophenylmethylsulfonyl fluoride, suggesting that myroicolsin is a serine protease. The chelators EDTA and EGTA also inhibited the collagenolytic activity of myroicolsin (Table 2), implying that divalent cations may be present in myroicolsin.
FIGURE 1.
Purification of myroicolsin from M. profundi D25 cultured in a gelatin-containing medium. A, gelatin zymography of the fermentation broth of strain D25. After electrophoresis, the samples on the gel at the left were stained with Coomassie Brilliant Blue. The gel on the right was washed with 2.5% (v/v) Triton X-100 at 4 °C three times, incubated with buffer B at 37 °C for 4 h, and stained with Coomassie Brilliant Blue. The bands of myroicolsin and myroilysin are indicated with arrows. B, purification of myroicolsin by ion-exchange chromatography on a DEAE-Sepharose Fast Flow column. ●, protein concentration; ○, relative activity; ——, NaCl concentration. C, purification and analysis of the present form of myroicolsin by gel filtration chromatography using FPLC (AKTA purifier, GE Healthcare). The samples were applied to a Sephadex G75 (10 × 300 mm) column and eluted with buffer B at a speed of 0.4 ml/min. The peaks of BSA (66 kDa), myroicolsin (56 kDa), and myroilysin (23 kDa) are indicated with arrows. D, analysis of the purity of myroicolsin by SDS-PAGE.
TABLE 1.
Physicochemical characteristics of myroicolsin
| Characteristics | Results |
|---|---|
| Length of DNA sequence (bp) | 2040 |
| No. of amino acid residues in mature enzyme sequence | 507 |
| No. of amino acid residues in signal sequence | 21 |
| No. of amino acid residues in N-propeptide | 74 |
| No. of amino acid residues in C-pro-secre-tail | 77 |
| Molecular mass (sequence) (Da) | 56,044.50 |
| Molecular mass (mass spectrometry) (Da) | 55,982.22 |
| Form in Tris-HCl buffer | Monomer |
| Isoelectric point (sequence) | 5.44 |
| Optimum temperature with type I collagen fiber (°C)a | 60 |
| Half-time at 60 °C (min)b | 10 |
| Optimum pH with type I collagen fiberc | 8.5 |
a The optimum temperature was determined by measuring the activities of myroicolsin with 5 mg of bovine insoluble type I collagen fiber in buffer B from 0 to 70 °C.
b Myroicolsin was incubated at 40, 50, and 60 °C. The collagenolytic activity was analyzed at intervals. The half-time was the time that it took to eliminate half of the activity of myroicolsin toward collagen at a given temperature.
c The optimum pH was determined by measuring the activities of myroicolsin with collagen fibers in Na2HPO4/KH2PO4 buffer from pH 5.0 to 7.0, Tris-HCl buffer from pH 7.0 to 9.5, NaHCO3/NaOH buffer from pH 9.5 to 11.0, and Na2HPO4/NaOH buffer from pH 11.0 to 13.0 at 37 °C.
TABLE 2.
Effects of metal ions and inhibitors on the collagenolytic activity of myroicolsin
| Metal ion | Relative activitya |
Metal ion | Relative activitya |
Inhibitor | Residual activityb | ||
|---|---|---|---|---|---|---|---|
| 2 mm | 4 mm | 2 mm | 4 mm | ||||
| % | % | % | % | % | |||
| Control | 100 | 100 | Ni2+ | 76.75 ± 1.65 | 54.16 ± 1.17 | Control | 100 |
| Ca2+ | 149.86 ± 3.23 | 192.49 ± 4.15 | Mn2+ | 73.29 ± 1.58 | 65.57 ± 1.41 | PMSF (2 mm) | 2.23 ± 0.88 |
| Sr2+ | 97.71 ± 2.11 | 90.92 ± 1.79 | Co2+ | 75.05 ± 1.62 | 50.23 ± 1.08 | EDTA (2 mm) | 29.59 ± 0.96 |
| Ba2+ | 99.87 ± 2.17 | 69.78 ± 1.83 | K+ | 71.31 ± 1.53 | 74.20 ± 1.60 | EGTA (2 mm) | 42.50 ± 2.17 |
| Li+ | 94.36 ± 2.03 | 80.11 ± 1.73 | Fe2+ | 66.57 ± 1.43 | 58.20 ± 1.25 | o-P (2.5 mm) | 78.79 ± 1.89 |
| Cu2+ | 79.47 ± 1.71 | 67.01 ± 1.44 | Sn2+ | 59.27 ± 2.10 | 40.01 ± 2.19 | ||
| Mg2+ | 78.61 ± 1.69 | 82.88 ± 1.79 | Zn2+ | 59.11 ± 1.27 | 24.19 ± 1.52 | ||
a Collagenolytic activity of myroicolsin toward 5 mg of bovine-insoluble type I collagen fibers was measured at 37 °C. The activity of myroicolsin without any metal ions was used as a control (100%).
b Myroicolsin was incubated with each inhibitor at 4 °C for 30 min, and the collagenolytic activities were measured at 37 °C. The activity of myroicolsin without any inhibitor was used as a control (100%).
Substrate Specificity of Myroicolsin
Various proteinaceous and synthetic substrates were used to determine the substrate specificity of myroicolsin. The results showed that myroicolsin had broad specificity toward several types of collagen, including types I, II, and IV (Table 3). Myroicolsin had high activity toward fish collagen. Myroicolsin was also active on gelatin, the degenerated form of collagen. In contrast, myroicolsin showed only slight activity toward casein or elastin (Table 3). These results indicated that myroicolsin is a collagenolytic protease that may play an important role in the degradation of marine collagens. For the hydrolysis of synthetic peptides, myroicolsin had the greatest activity toward N-succinyl-Ala-Ala-Pro-Arg-p-nitroanilide (AAPR; all peptide names abbreviated similarly). The hydrolysis of FAAF, AAPF, AAPK, and AAPL was also detectable. The tripeptides AAA, AAV, and GGG were poor substrates for myroicolsin (Table 3).
TABLE 3.
The substrate specificity of myroicolsin toward various proteins and synthetic peptides
| Substrate | Activitya | Substrate | Activity |
|---|---|---|---|
| units/mg | units/mg | ||
| Fish-insoluble collagen fiber | 7439.99 ± 181.2 | AAPRb | 5.54 ± 0.24 |
| Bovine-insoluble type I collagen fiber | 2027. 4 ± 97.6 | FAAF | 1.25 ± 0.24 |
| Mouse type IV collagen | 947.37 ± 32.4 | AAPF | 0.59 ± 0.03 |
| Bovine type II collagen | 715.27 ± 26.7 | AAPK | 0.55 ± 0.04 |
| Gelatin | 5464.03 ± 297.3 | AAPL | 0.50 ± 0.03 |
| Azocoll | 157.25 ± 4.3 | AAA | 0.15 ± 0.02 |
| Elastinorcein | 59.10 ± 2.8 | AAV | 0.11 ± 0.007 |
| Casein | 4.84 ± 0.09 | GGG | 0.04 ± 0.0001 |
a The values are specific activities (in units/mg) of myroicolsin as described under “Experimental Procedures.”
b AAPR, N-succinyl-Ala-Ala-Pro-Arg-p-nitroanilide. The other peptides are abbreviated in the same way as AAPR.
Gene Cloning and Sequence Analysis
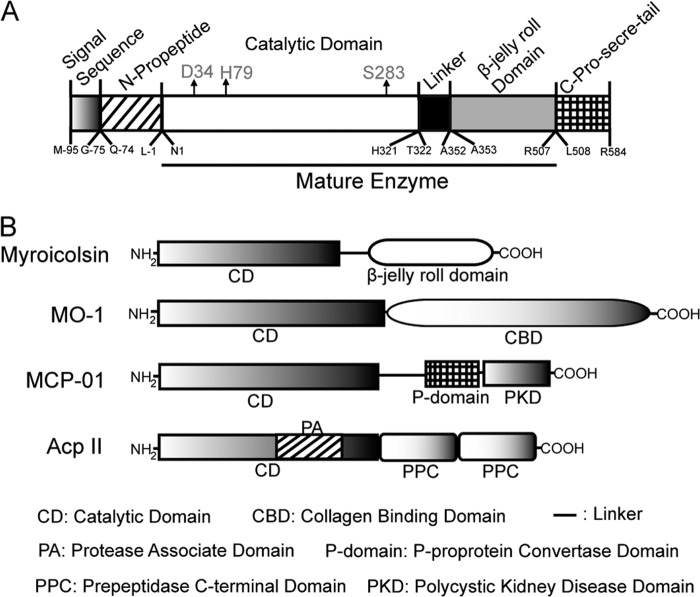
Based on the N-terminal amino acid sequence of myroicolsin and the conserved sequence in the catalytic domains of serine proteases, the complete gene of myroicolsin was cloned by a combination of PCR and thermal asymmetric interlaced PCR. The open reading frame of the whole gene is 2040 bp in length and was deduced to encode an S8 protease precursor of 679 amino acid residues. According to the results of sequence alignment and secondary structure prediction (PSIPRED version 3.3), we speculated that the precursor of myroicolsin contains a 21-residue signal sequence (Met−95–Gly−75), an N-propeptide (Gln−74–Leu−1), an S8 catalytic domain (Asn1–His321), a linker (Thr322–Ala352), a β-jelly roll domain (Ala353–Arg507), and a C-pro-secre-tail (Leu508–Arg584) (Table 1 and Fig. 2A). Based on the molecular mass of the purified myroicolsin and its N-terminal sequence, it could be deduced that the signal sequence, the N-propeptide, and the C-pro-secre-tail were cleaved spontaneously during enzyme maturation. The mature form of myroicolsin consists of 507 residues (Asn1–Arg507), including the catalytic domain, the linker, and the β-jelly roll domain (Fig. 2A). Myroicolsin exhibits the highest identity (46%) with a putative subtilisin-like protease (NCBI reference sequence: WP_002990379.1). Among characterized proteases, myroicolsin exhibits the highest identity (28%) with protease Kp43, an oxidatively stable alkaline protease of the S8 family (43). Moreover, myroicolsin has a domain architecture that is different from other reported subtilisin-like collagenolytic proteases (Fig. 2B). These results indicate that myroicolsin is a novel collagenolytic protease of the S8 family.
FIGURE 2.
Schematic diagrams of the domain structures of myroicolsin precursor (A) and structural comparison of myroicolsin and other characterized collagenolytic proteases of the S8 family (B). Myroicolsin, the protease studied here (AEC33275). The possible catalytic triad, Asp34, His79, and Ser283, is indicated by arrows in A. MO-1, the protease secreted by G. collagenovorans MO-1 (AB260948); MCP-01, the protease from Pseudoalteromonas sp. SM9913 (ABD14413); AcpII, the protease from A. collagenimarina AC40 (AB505451).
Functional Analysis of the C-pro-secre-tail, Linker, and β-Jelly Roll Domain
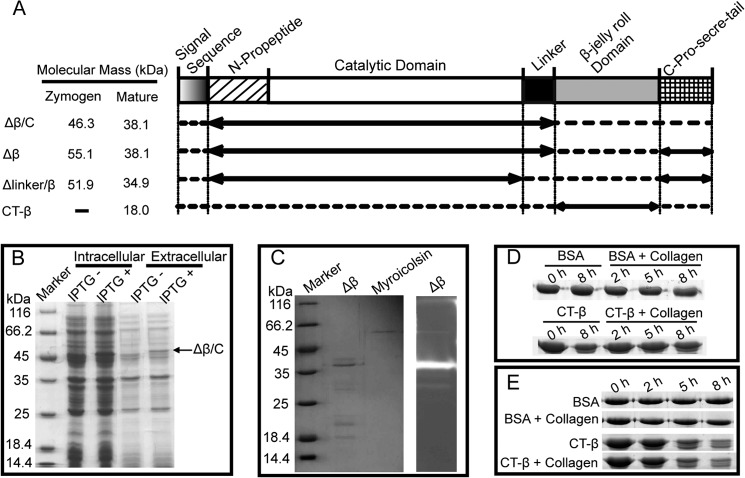
It is known that the N-propeptide of subtilisin-like proteases functions as an intramolecular chaperone in protein folding (44). To analyze the functions of the C-pro-secre-tail, the linker, and the β-jelly roll domain, four truncated mutants of myroicolsin were constructed (Fig. 3A). The recombinant mutants of myroicolsin were all expressed with a signal peptide of expression vector. The mutant Δβ/C, composed of the N-propeptide, the catalytic domain, and the linker, was expressed as an intracellular soluble protein with an apparent molecular mass of ∼46 kDa, consistent with the prediction from its sequence (46,466.27 Da) (Fig. 3, A and B). This recombinant Δβ/C protein could be secreted, and the extracellular form showed the same molecular mass (Fig. 3B), indicating that the N-propeptide was not cleaved from the extracellular form. This result suggested that the removed portion, including the β-jelly roll domain and the C-pro-secre-tail, is essential for the cleavage of the N-propeptide but not for the secretion and folding of the protein. Similar to the recombinant Δβ/C protein, Δβ could be secreted, and it was also able to remove the N-propeptide to become active (Fig. 3C), indicating that the β-jelly roll domain is not required for folding and maturation. Based on the results obtained for Δβ/C and Δβ, it was concluded that the C-pro-secre-tail may be involved in the removal of the N-propeptide during myroicolsin maturation. In contrast, recombinant Δlinker/β protein was insoluble, indicating that the linker may be required for protein folding. To analyze whether the β-jelly roll domain functions as a collagen-binding domain in mature myroicolsin, the recombinant β-jelly roll domain (the mutant CT-β) was expressed, and its collagen-binding ability was tested. The mutant CT-β showed minimal binding to insoluble collagen fiber at 0 or 25 °C (Fig. 3, D and E), and it became unstable after incubation at 25 °C for 5 h (Fig. 3E). Moreover, a comparison of the activities of myroicolsin and the mutant Δβ toward collagen indicated that the loss of the β-jelly roll domain had little effect on the collagenolytic activity of myroicolsin (Table 4), which suggests that the β-jelly roll domain plays a minor role in collagenolysis. These results suggested that myroicolsin has no C-terminal collagen-binding domain, unlike other characterized S8 collagenolytic proteases (25, 30).
FIGURE 3.
Expression and functional analysis of the truncated mutants of myroicolsin. A, schematic representation of the domain structures of myroicolsin and its mutants. The solid lines with arrows indicate the mutants' sequences, and the dotted lines indicate the deleted sequences. The predicted molecular masses from the amino acid sequence of each zymogen and mature protein are shown at the left. B, SDS-PAGE analysis of the overexpression of Δβ/C. The Δβ/C protein band is indicated by a black arrow. C, SDS-PAGE and gelatin zymography of Δβ. D and E, analysis of the collagen-binding ability of CT-β at 0 °C (D) and 25 °C (E). Collagen fibers (5 mg) were mixed with 20 μg of CT-β or BSA in 200 μl of buffer B and incubated at 0 or 25 °C. Samples were collected at 0, 2, 5, and 8 h and subjected to SDS-PAGE analysis. Collagen fibers incubated with BSA, CT-β incubated in buffer B, and BSA incubated in buffer B were used as the controls.
TABLE 4.
Substrate specificity of myroicolsin and mutant Δβ
| Substrate | Activity |
|
|---|---|---|
| Myroicolsin | Δβ | |
| units/mg | ||
| Bovine insoluble type I collagen fiber | 2741.8 ± 163.6 | 3802.2 ± 52.7 |
| Gelatin | 5507.2 ± 179.8 | 4813.5 ± 189.6 |
| AAPRa | 4.82 ± 0.08 | 4.20 ± 0.09 |
a N-succinyl-Ala-Ala-Pro-Arg-p-nitroanilide.
SEM and AFM Observation of Collagen Fiber Degradation by Myroicolsin
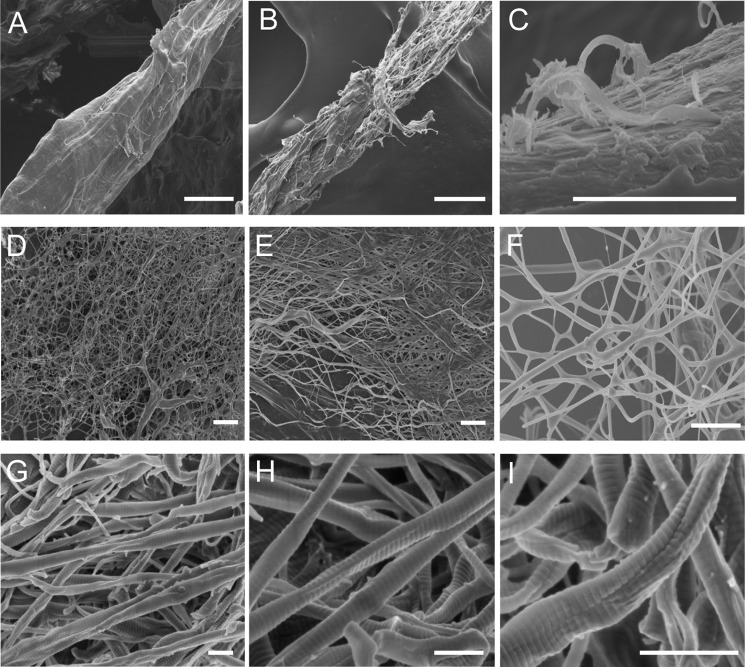
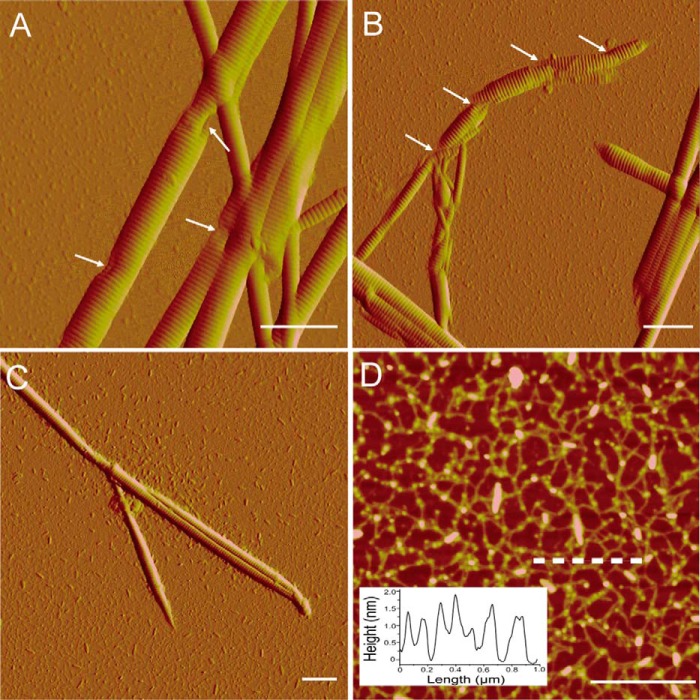
To study the collagenolytic mechanism of myroicolsin, the degradation of insoluble bovine type I collagen fibers by myroicolsin was observed by SEM and AFM. Under SEM, untreated collagen fibers showed a very compact structure (Fig. 4A). After incubation with myroicolsin at 30 °C for 1 h, the surface of the collagen fibers became irregular; fibril bundles were exposed, and some fibrils were released from the surface (diameter, 300 nm to 1 μm) (Fig. 4, B and C). Five hours later, the compact structure of the collagen fiber was totally devastated, and nearly all of the collagen fibers were dissociated into single fibrils (diameter, 100–500 nm) (Fig. 4, D–F). After treatment for 24 h, the sample was further dissociated into thinner fibrils or microfibrils (diameter, 50–200 nm). Remarkable indentation and breakage appeared on the surface of dispersed fibrils (Fig. 4, G–I).
FIGURE 4.
SEM observation of the time-dependent degradation of collagen fiber by myroicolsin. Collagen fibers (5 mg) were incubated with 5 μg of myroicolsin in 100 μl of buffer B at 30 °C with continuous mixing for 1 h (B and C), 5 h (D–F), and 24 h (G–I). The reaction performed without myroicolsin served as a control (A). Bars, 10 μm (A, B, and D), 2 μm (C, E, and F), and 250 nm (G–I).
To further investigate the degradation of collagen fibrils by myroicolsin, collagen incubated with myroicolsin at 30 °C for 48 h was observed by AFM. Consistent with SEM observations, the indentation and cleavage of fibrils were clearly observed under AFM (Fig. 5A). Moreover, the indentation could deepen (Fig. 5B) until the fibrils were further digested into microfibrils and oligomers of collagen monomers (Fig. 5C). In the digested mixture, abundant irregular arrangements of line-like structures were observed (Fig. 5D). The heights of these structures were ∼1.5 nm, which is in accord with the height of triple helix collagen monomers. The digestion of collagen monomers into small peptides and amino acids by myroicolsin was unobservable by AFM.
FIGURE 5.
AFM observation of the nanostructures of collagen fibrils and monomers released from collagen fibers by myroicolsin. Collagen fibers (5 mg) were incubated with 5 μg of myroicolsin in 100 μl of buffer B at 30 °C with continuous mixing for 48 h and then observed by AFM. A, disassembled collagen fibrils with obvious damages after treatment. The indentations are indicated with white arrows. B, further dissociation of collagen fibrils with remarkable indentation and breakage. C, small pieces of broken fibrils and oligomers of collagen monomers. D, height image of collagen monomers. Insets of D show the height analyses of the cross-section in the image indicated by a dashed line. D is the height image, and other panels are peak force error images. Bars, 1 μm.
Biochemical Analysis of Collagen Fiber Degradation by Myroicolsin
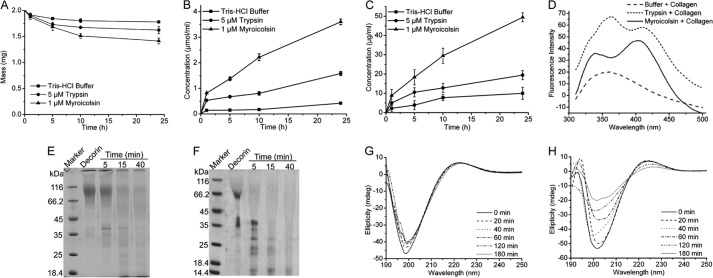
A series of biochemical experiments was performed to study the collagenolytic mechanism of myroicolsin. Collagen lost nearly 30% of its weight after incubation with 1 μm myroicolsin at 30 °C for 24 h (Fig. 6A). At the same time, more amino acids were released with increased treatment time (Fig. 6B). These results confirmed that insoluble collagen fibers were digested by myroicolsin to some extent. In addition, the released GAGs increased with treatment time, suggesting that the proteoglycans in collagen fibers were degraded (Fig. 6C). In vitro experiments showed that decorin, an important proteoglycan in collagen fibers, could be efficiently hydrolyzed by myroicolsin (Fig. 6F), further confirming that myroicolsin could release GAGs from collagen fibers. Because proteoglycans in collagen fibers are responsible for stabilizing interfibrillar organization (45), these results suggested that myroicolsin dissociated collagen fibers into fibrils by digesting the proteoglycans in collagen fibers. Collagen telopeptides are important covalent cross-links within and between collagen microfibrils (9). The cross-links between collagen telopeptides could also be hydrolyzed by myroicolsin to release pyridinolines and deoxypyridinolines (Fig. 6D), leading to the release of collagen monomers and microfibrils from fibrils. Moreover, CD spectra revealed that myroicolsin could destroy the triple helix structure of the collagen monomer (Fig. 6H), suggesting that the α1 and α2 chains in monomers could be released and further hydrolyzed into peptides and amino acids. Taken together, these biochemical results confirmed that myroicolsin could hydrolyze various cross-links within collagen fibers, fibrils, and microfibrils to release collagen monomers, which were further hydrolyzed into peptides and amino acids. In addition, as a control, the activity of trypsin on native collagen fibers was analyzed under the same conditions. When collagen fibers were treated with trypsin, less free amino acids and GAGs were released compared with myroicolsin treatment (Fig. 6, B and C). Trypsin could hydrolyze the proteoglycans (Fig. 6E) and telopeptides (Fig. 6D) between or within collagen fibrils, but it could not destroy the triple helix structure of the collagen monomer (Fig. 6G).
FIGURE 6.
The enzymatic degradation of the covalent cross-links in collagen fiber by myroicolsin. A, mass analysis of collagen fibers before and after incubation with 1 μm myroicolsin at 30 °C for 24 h. Collagen incubated with 5 μm trypsin or buffer B was used as a control. B and C, contents of amino acids (B) and GAGs (C) released from the degraded collagen. Collagen fibers (10 mg) were incubated with 1 μm myroicolsin, 5 μm trypsin, or buffer B at 30 °C for 24 h. The amino acid concentration in the supernatant of the digested mixture was quantitatively analyzed by the colorimetric ninhydrin method with l-leucine as the standard. The GAG concentration in the supernatant of the digested mixture was quantitatively analyzed by the dimethylmethylene blue assay. D, fluorescence spectra analysis of pyridinolines and deoxypyridinolines released from collagen telopeptides. Collagen fibers (20 mg) were incubated with 0.1 μm myroicolsin, 0.1 μm trypsin, or buffer B at 30 °C for 48 h. Pyridinolines and deoxypyridinolines in the supernatant of the digested mixture were detected on an FP-6500 spectrometer (Jasco, Japan) with an excitation wavelength of 295 nm at room temperature. E and F, SDS-PAGE analysis of decorin degradation by 0.5 μm trypsin (E) and 0.1 μm myroicolsin (F) at 30 °C. G and H, CD spectra of acid-soluble collagen incubated with 0.5 μm trypsin (G) and 0.1 μm myroicolsin (H) at 25 °C for 180 min. Error bars, S.D.
Cleavage Pattern of Bovine Type I Collagen by Myroicolsin
To study the cleavage pattern of bovine type I collagen by myroicolsin, the cleavage sites of collagen polypeptide chains α1 and α2 were analyzed by HPLC and mass spectrometry. When the collagen fibers were incubated with myroicolsin at 25 °C for 24 h, 40 released peptides were separated, and their molecular masses and sequences were identified (Table 5). Moreover, 22 peptides released from the collagen fibers by myroicolsin digestion at 50 °C for 10 h were also identified (Table 6). Based on the sequences of these identified peptides, the myroicolsin-mediated cleavage sites of polypeptide chains α1 and α2 of bovine type I collagens were determined (Fig. 7, A and B). Some of these cleavage sites were further confirmed with synthesized peptides (Fig. 7C). Based on the determined cleavage sites, the residue frequencies at P1 and P1′ sites were analyzed (Table 7). Myroicolsin showed different cleavage patterns against native collagen (25 °C) and denatured collagen (50 °C). In native collagen, the P1 position is often occupied by Gly, Arg, Pro, or Phe, and the P1′ position is almost always occupied by Gly. In denatured collagen, the P1 position is always a basic residue (Lys or Arg), and the P1′ position is still Gly.
TABLE 5.
Peptides released from bovine type I collagen fiber by myroicolsin at 25 °C
| Peak no. | MH+ observed | Mr (experimental) | Peptide sequence | Chain | Position |
|---|---|---|---|---|---|
| Da | Da | ||||
| 1 | 713.37 | 1424.71 | S↓TGISVPGPMGPSGPR↓G | α-1 | 172–186 |
| 2 | 989.97 | 1977.92 | Q↓GFQGPPGEPGEPGASGPMGPR↓G | α-1 | 199–219 |
| 3 | 732.83 | 1463.64 | F↓QGPPGEPGEPGASGPM↓G | α-1 | 201–216 |
| 4 | 887.92 | 1773.81 | F↓QGPPGEPGEPGASGPMGPR↓G | α-1 | 201–219 |
| 5 | 1064.53 | 2127.04 | G↓PAGAAGPAGNPGADGQPGAKGANGAP↓G | α-1 | 377–402 |
| 6 | 775.37 | 1548.72 | P↓SGPPGPKGNSGEPGAPGS↓K | α-1 | 423–440 |
| 7 | 806.37 | 1610.73 | S↓GEPGAPGSKGDTGAKGEP↓G | α-1 | 433–450 |
| 8 | 601.80 | 1201.59 | R↓GQAGVMGFPGPKG↓A | α-1 | 574–586 |
| 9 | 484.24 | 966.47 | E↓QGVPGDLGAPG↓P | α-1 | 660–670 |
| 10 | 605.31 | 1208.61 | G↓MPGERGAAGLPGP↓K | α-1 | 728–740 |
| 11 | 657.85 | 1313.67 | A↓GPAGPPGPIGNVGAPG↓P | α-1 | 844–859 |
| 12 | 767.37 | 1532.72 | P↓GPAGEKGAPGADGPAGAPG↓T | α-1 | 928–946 |
| 13 | 846.42 | 1690.83 | K↓SGDRGETGPAGPAGPIGPV↓G | α-1 | 1062–1080 |
| 14 | 874.93 | 1747.85 | K↓SGDRGETGPAGPAGPIGPVG↓A | α-1 | 1062–1081 |
| 15 | 910.45 | 1818.89 | K↓SGDRGETGPAGPAGPIGPVGA↓R | α-1 | 1062–1082 |
| 16 | 802.91 | 1603.80 | S↓GDRGETGPAGPAGPIGPV↓G | α-1 | 1063–1080 |
| 17 | 831.42 | 1660.82 | S↓GDRGETGPAGPAGPIGPVG↓A | α-1 | 1063–1081 |
| 18 | 866.94 | 1731.85 | S↓GDRGETGPAGPAGPIGPVGA↓R | α-1 | 1063–1082 |
| 19 | 774.40 | 1546.77 | G↓DRGETGPAGPAGPIGPV↓G | α-1 | 1064–1080 |
| 20 | 638.83 | 1275.65 | R↓GETGPAGPAGPIGPV↓G | α-1 | 1066–1080 |
| 21 | 780.91 | 1559.81 | R↓GETGPAGPAGPIGPVGAR↓G | α-1 | 1066–1083 |
| 22 | 626.29 | 1250.56 | P↓GPPGPPGPPSGGYD↓L | α-1 | 1183–1196 |
| 23 | 674.32 | 1346.62 | P↓GPPGPPSGGYDLSF↓L | α-1 | 1186–1199 |
| 24 | 548.75 | 1095.49 | P↓GPPSGGYDLSF↓L | α-1 | 1189–1199 |
| 25 | 804.41 | 1606.81 | A↓GARGSDGSVGPVGPAGPIG↓S | α-2 | 230–248 |
| 26 | 662.33 | 1322.65 | R↓GSDGSVGPVGPAGPIG↓S | α-2 | 233–248 |
| 27 | 633.82 | 1265.63 | G↓SDGSVGPVGPAGPIG↓S | α-2 | 234–248 |
| 28 | 1049.52 | 2097.06 | G↓FPGAPGPKGELGPVGNPGPAGPAG↓P | α-2 | 255–278 |
| 29 | 699.86 | 1397.68 | R↓GLVGEPGPAGSKGESG↓N | α-2 | 341–356 |
| 30 | 570.26 | 1138.51 | R↓GPNGDSGRPGEP↓G | α-2 | 431–442 |
| 31 | 690.86 | 1379.70 | G↓PRGFPGSPGNIGPAG↓K | α-2 | 447–461 |
| 32 | 628.67 | 1883.00 | G↓LPGIDGRPGPIGPAGARGEP↓G | α-2 | 468–487 |
| 33 | 1185.56 | 2369.10 | R↓GPSGPPGPDGNKGEPGVVGAPGTAGPSG↓P | α-2 | 608–635 |
| 34 | 501.24 | 1000.46 | R↓GEVGPAGPNGF↓A | α-2 | 713–723 |
| 35 | 606.81 | 1211.60 | G↓GPPGATGFPGAAGR↓T | α-2 | 779–792 |
| 36 | 653.85 | 1305.69 | R↓GLPGVAGSVGEPGPL↓G | α-2 | 881–895 |
| 37 | 488.71 | 975.41 | F↓GFDGDFYR↓A | α-2 | 1109–1116 |
| 38 | 524.23 | 1046.45 | F↓GFDGDFYRA↓D | α-2 | 1109–1117 |
| 39 | 460.20 | 918.39 | G↓FDGDFYR↓A | α-2 | 1110–1116 |
| 40 | 495.72 | 989.43 | G↓FDGDFYRA↓D | α-2 | 1110–1117 |
TABLE 6.
Peptides released from bovine type I collagen fiber by myroicolsin at 50 °C
| Peak no. | MH+ observed | Mr (experimental) | Peptide sequence | Chain | Position |
|---|---|---|---|---|---|
| Da | Da | ||||
| 1 | 748.77 | 1495.53 | K↓STGISVPGPMGPSGPR↓G | α-1 | 171–186 |
| 2 | 426.16 | 850.31 | R↓GFSGLDGAK↓G | α-1 | 268–276 |
| 3 | 768.92 | 767.91 | K↓GDAGPAGPK↓G | α-1 | 277–285 |
| 4 | 443.61 | 885.21 | R↓GSEGPQGVR↓G | α-1 | 361–369 |
| 5 | 550.17 | 1098.33 | K↓GAAGEPGKAGER↓G | α-1 | 586–597 |
| 6 | 717.52 | 2149.54 | K↓GDAGAPGAPGSQGAPGLQGMPGER↓G | α-1 | 709–732 |
| 7 | 882.82 | 1763.63 | R↓VGPPGPSGNAGPPGPPGPAGK↓E | α-1 | 882–902 |
| 8 | 904.37 | 2710.09 | K↓QGPSGASGERGPPGPMGPPGLAGPPGESGR↓E | α-1 | 984–1013 |
| 9 | 817.27 | 2448.79 | K↓GDRGETGPAGPPGAPGAPGAPGPVGPAGK↓S | α-1 | 1033–1061 |
| 10 | 988.33 | 1974.65 | K↓SGDRGETGPAGPAGPIGPVGAR↓G | α-1 | 1062–1083 |
| 11 | 780.78 | 1559.55 | R↓GETGPAGPAGPIGPVGAR↓G | α-1 | 1066–1083 |
| 12 | 418.64 | 835.25 | R↓GPAGPQGPR↓G | α-1 | 1084–1092 |
| 13 | 666.82 | 665.81 | R↓GIKGHR↓G | α-1 | 1105–1110 |
| 14 | 859.57 | 858.56 | R↓ADDANVVR↓D | α-1 | 1217–1224 |
| 15 | 768.74 | 2303.20 | R↓GPPGPPGKAGEDGHPGKPGRPGER↓G | α-2 | 131–154 |
| 16 | 420.63 | 839.25 | R↓GVVGPQGAR↓G | α-2 | 155–163 |
| 17 | 654.05 | 1959.13 | K↓GEPGAVGQPGPPGPSGEEGKR↓G | α-2 | 359–379 |
| 18 | 451.67 | 901.33 | R↓AGVMGPAGSR↓G | α-2 | 412–421 |
| 19 | 548.19 | 1094.37 | R↓GPRGDQGPVGR↓S | α-2 | 818–828 |
| 20 | 434.60 | 867.19 | R↓GPSGPQGIR↓G | α-2 | 995–1003 |
| 21 | 612.16 | 1222.31 | R↓GPAGPSGPAGKDGR↓I | α-2 | 1052–1065 |
| 22 | 596.75 | 1191.49 | R↓IGQPGAVGPAGIR↓G | α-2 | 1066–1078 |
FIGURE 7.
Analysis of cleavage pattern of bovine type I collagen by myroicolsin. A and B, the cleavage sites of the myroicolsin on bovine type I collagen polypeptide chains α1 (A) and α2 (B). LC-MS was used to separate the peptides released from collagen polypeptide chains by myroicolsin and to analyze their molecular masses. The sequences of the peptides were analyzed by Mascot MS/MS ion search tools and ExPASy tools. The identified peptide sequences released at 25 °C are underlined, and the cleavage sites are indicated by slashes. Boldface letters, peptide sequences released at 50 °C; #, cleavage sites. C, confirmation of the cleavage sites of collagen chains by myroicolsin with synthetic peptides. The cleavage sites are shown by arrows.
TABLE 7.
Specificity matrix of myroicolsin on bovine type I collagen fiber
| Amino acid | P1 |
Amino acid | P1′ |
||
|---|---|---|---|---|---|
| 25 °C | 50 °C | 25 °C | 50 °C | ||
| Gly | 23 | Gly | 37 | 31 | |
| Arg | 16 | 32 | Ser | 8 | 4 |
| Pro | 10 | Pro | 6 | ||
| Phe | 7 | Ala | 6 | 2 | |
| Ala | 6 | Glu | 2 | ||
| Ser | 6 | Ile | 2 | ||
| Val | 4 | Leu | 4 | ||
| Lys | 3 | 12 | Phe | 3 | |
| Gln | 1 | Thr | 3 | ||
| Glu | 1 | Lys | 3 | ||
| Leu | 1 | Gln | 3 | 1 | |
| Asp | 1 | Asp | 3 | 1 | |
| Met | 1 | Arg | 2 | ||
| Asn | 1 | ||||
| Met | 1 | ||||
| Val | 1 | ||||
A Model for Collagen Fiber Degradation by Myroicolsin
Based on our microscopic and biochemical analyses, a stepwise model for collagen fiber degradation by myroicolsin is proposed (Fig. 8). Myroicolsin first breaks the interfibrillar proteoglycan bridges, leading to the disassembly of the tight structure of collagen fibers and the exposure of collagen fibrils. Then telopeptides within collagen fibrils and microfibrils are hydrolyzed by myroicolsin, which accelerates the unfolding of the collagen structure. Thus, myroicolsin gains access to collagen monomers. Finally, the fibrillar structure of collagen is completely destroyed, and collagen monomers are degraded into peptides and free amino acids.
FIGURE 8.
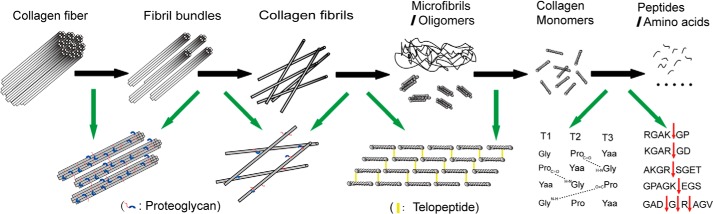
Schematic model for stepwise collagen degradation by myroicolsin. Black arrows indicate the process of collagen fiber and fibril hydrolysis by myroicolsin, and green arrows indicate the cross-links destroyed by myroicolsin in the corresponding step.
DISCUSSION
Collagenolytic serine proteases from deep sea bacteria are seldom studied. Thus far, only MCP-01 and AcpII have been reported (30, 31). In this paper, a collagenolytic serine protease myroicolsin, which is secreted by the deep sea sedimentary bacterium M. profundi D25, was purified and characterized. Myroicolsin had broad specificity to various collagens. Sequence analysis showed that myroicolsin is a novel protease of the S8 family with low identity (<30%) to characterized peptidases. In particular, myroicolsin is different from any other S8 protease in its domain structure. Although its precursor contains a signal sequence, an N-propeptide, a catalytic domain, a linker, a β-jelly roll domain, and a C-pro-secre-tail, the mature form only contains the catalytic domain, the linker, and the β-jelly roll domain. Therefore, the precursor of myroicolsin contains two propeptides. It is well known that bacterial subtilisin-like proteases require propeptides for folding and secretion (46, 47). Although the N-propeptide of myroicolsin may function as an intramolecular chaperone for protein folding as in other subtilisin-like proteases, our truncation mutation assay indicated that the C-pro-secre-tail is essential for the cleavage of the N-propeptide in myroicolsin maturation. Therefore, myroicolsin may mature from its precursor in a process similar to TK-SP (48).
Collagenolytic proteases, such as MCP-01 (30, 42) and MMP-1, -8, and -13 (49, 50), usually have a C-terminal domain that functions as a collagen-binding domain to mediate and facilitate collagen hydrolysis by the enzyme. However, there are also some collagenolytic proteases that do not need an additional domain for collagenolysis, such as MMP-12 (51) and the serine proteases from fiddler crab (16, 18). Although myroicolsin has a C-terminal β-jelly roll domain, our results showed that this domain had no collagen-binding ability and that the catalytic domain from myroicolsin could degrade collagen fibers with a similar efficiency as myroicolsin. Therefore, it seems that myroicolsin does not need a C-terminal collagen-binding domain for collagenolysis.
Although a number of subtilisin-like proteases from environmental and pathogenic microorganisms have been demonstrated to be collagenolytic proteases, the collagen degradation mechanisms of subtilisin-like collagenolytic proteases are still largely unknown. The S8 protease from Geobacillus collagenovorans MO-1 degrades type I and IV collagens into small pieces, suggesting that it digests collagens at multiple sites (25). Thirty-seven possible cleavage sites for MCP-01 on the chains of bovine type I collagen were determined by analyzing the N-terminal sequence of released peptides (30). It is still unclear how insoluble collagen fibers are degraded by subtilisin-like collagenolytic proteases. Our SEM and AFM observations showed that myroicolsin released fibrils from collagen fiber and that the fibrils were further degraded into collagen monomers. Biochemical assays indicated that this stepwise degradation of collagen fiber was achieved through the hydrolysis of the proteoglycans and telopeptides in collagen fibers and fibrils. Proteoglycans are primarily responsible for collagen fibrillogenesis, and they stabilize the cross-links between collagen fibrils within collagen fiber (10). Collagen telopeptides are non-triple-helical domains at the N and C termini of collagen monomers. Cross-links between neighboring telopeptides are essential for stabilizing the structure of mature collagen microfibrils (9, 52). Because of the hydrolysis of proteoglycans and telopeptides by myroicolsin, the complex hierarchical structure of collagen fiber was destroyed, and free collagen monomers were produced. Recently, the cysteine protease cathepsin K was reported to disassociate proteoglycans from collagen fiber prior to collagen degradation (53). The cleavage pattern of bovine type I collagen by myroicolsin was further analyzed. The cleavage sites of native collagen and denatured collagen are strikingly different, which may result from their different structures. Because of the atypical amino acid composition of collagen, Gly-Pro-Hyp is the most abundant triplet (10.5%). N-H(Gly)···O=C(Xaa) and Cα-H(Gly/Yaa)···O=C(Xaa/Gly) bonds hold three polypeptide chains together in a helical conformation and stabilize the triple helix structure of the collagen monomer (54). Therefore, the digestion of peptide bonds adjacent to Gly and Pro by myroicolsin at 25 °C should be related to the special triple helical structure of native collagen. Moreover, the breakdown of the cross-links between peptide chains can unfold the triple helix and enable myroicolsin to improve its collagen degradation efficiency by gaining access to additional cleavage sites. In contrast, when collagens are denatured at 50 °C, the triple helix of collagen monomers uncoils, and free polypeptide chains are released. The peptide bonds with P1 basic residues on the released polypeptide chains are then exposed and cleaved by myroicolsin, because basic residues are the favored P1 residues for myroicolsin.
Subtilisin-like proteases are usually nonspecific peptidases with a preference to cleave after hydrophobic residues because the S1 pocket of these proteases is composed of uncharged amino acids (11, 55, 56). However, our results indicated that although myroicolsin could cleave peptide bonds with hydrophobic residues at P1 positions, Arg was the most preferred P1 residue when synthetic peptides or denatured collagen was used as the substrate. Among serine proteases, trypsin-like proteases cleave the peptide bonds with basic residues at the P1 site because negatively charged groups are located at the bottom of their substrate-binding pocket (19). Therefore, it seems that myroicolsin exhibits a mixed type of P1 specificity, both trypsin-like and subtilisin-like.
In summary, our results show that myroicolsin from deep sea M. profundi D25 is a subtilisin-like collagenolytic protease with a novel domain structure and unique specificity. The mechanism of the myroicolsin-mediated degradation of insoluble collagen fiber has been revealed, and a model is proposed in Fig. 8. Our results provide more insight into the collagen degradation mechanism by subtilisin-like collagenolytic proteases. Because myroicolsin is secreted from a deep sea bacterium and insoluble collagen may be an important component of deep sea sediment PON, our results are also helpful for clarifying the process of deep sea sedimentary PON degradation.
The work was supported by National Natural Science Foundation of China Grants 91228210, 31290231, 31025001, and 31170055; Hi-Tech Research and Development Program of China Grants 2011AA090703 and 2012AA092103; China Ocean Mineral Resources R&D Association (COMRA) Special Foundation Grants DY125-15-T-05 and DY125-15-R-03; Special Fund of China for Marine-Scientific Research in the Public Interest Grant 201005032-6; and Program of Shandong for Taishan Scholars Grant 2008BS02019.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s)JF514144.
The amino acid sequence of this protein can be accessed through NCBI Protein Database under NCBI accession number AEC33275.
- PON
- particulate organic nitrogen
- MMP
- mammalian matrix metalloproteinase
- SEM
- scanning electron microscopy
- AFM
- atomic force microscopy
- GAG
- glycosaminoglycan
- PKD
- polycystic kidney disease
- C-pro-secre-tail
- Pro secretion system C-terminal sorting domain.
REFERENCES
- 1. Aluwihare L. I., Repeta D. J., Pantoja S., Johnson C. G. (2005) Two chemically distinct pools of organic nitrogen accumulate in the ocean. Science 308, 1007–1010 [DOI] [PubMed] [Google Scholar]
- 2. Brunnegård J., Grandel S., Ståhl H., Tengberg A., Hall P. O. (2004) Nitrogen cycling in deep-sea sediments of the Porcupine Abyssal Plain, NE Atlantic. Prog. Oceanogr. 63, 159–181 [Google Scholar]
- 3. Sarkar S., Pramanik A., Mitra A., Mukherjee J. (2010) Bioprocessing data for the production of marine enzymes. Mar. Drugs 8, 1323–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Exposito J. Y., Valcourt U., Cluzel C., Lethias C. (2010) The fibrillar collagen family. Int. J. Mol. Sci. 11, 407–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prockop D. J., Kivirikko K. I. (1995) Collagens. Molecular biology, diseases, and potentials for therapy. Annu. Rev. Biochem. 64, 403–434 [DOI] [PubMed] [Google Scholar]
- 6. Kadler K. E., Baldock C., Bella J., Boot-Handford R. P. (2007) Collagens at a glance. J. Cell Sci. 120, 1955–1958 [DOI] [PubMed] [Google Scholar]
- 7. Kadler K. E., Holmes D. F., Trotter J. A., Chapman J. A. (1996) Collagen fibril formation. Biochem. J. 316, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brodsky B., Persikov A. V. (2005) Molecular structure of the collagen triple helix. Adv. Protein Chem. 70, 301–339 [DOI] [PubMed] [Google Scholar]
- 9. Orgel J. P., Irving T. C., Miller A., Wess T. J. (2006) Microfibrillar structure of type I collagen in situ. Proc. Natl. Acad. Sci. U.S.A. 103, 9001–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scott J. E., Orford C. R., Hughes E. W. (1981) Proteoglycan-collagen arrangements in developing rat tail tendon. An electron microscopical and biochemical investigation. Biochem. J. 195, 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rawlings N. D., Barrett A. J., Bateman A. (2010) MEROPS. The peptidase database. Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kafienah W., Brömme D., Buttle D. J., Croucher L. J., Hollander A. P. (1998) Human cathepsin K cleaves native type I and II collagens at the N-terminal end of the triple helix. Biochem. J. 331, 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eisen A. Z., Henderson K. O., Jeffrey J. J., Bradshaw R. A. (1973) A collagenolytic protease from the hepatopancreas of the fiddler crab, Uca pugilator. Purification and properties. Biochemistry 12, 1814–1822 [DOI] [PubMed] [Google Scholar]
- 14. O'Farrell T. J., Guo R., Hasegawa H., Pourmotabbed T. (2006) Matrix metalloproteinase-1 takes advantage of the induced fit mechanism to cleave the triple-helical type I collagen molecule. Biochemistry 45, 15411–15418 [DOI] [PubMed] [Google Scholar]
- 15. Chung L., Dinakarpandian D., Yoshida N., Lauer-Fields J. L., Fields G. B., Visse R., Nagase H. (2004) Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 23, 3020–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsu C. A., Perona J. J., Fletterick R. J., Craik C. S. (1997) Structural basis for the broad substrate specificity of fiddler crab collagenolytic serine protease 1. Biochemistry 36, 5393–5401 [DOI] [PubMed] [Google Scholar]
- 17. Visse R., Nagase H. (2003) Matrix metalloproteinases and tissue inhibitors of metalloproteinases. Structure, function, and biochemistry. Circ. Res. 92, 827–839 [DOI] [PubMed] [Google Scholar]
- 18. Perona J. J., Tsu C. A., Craik C. S., Fletterick R. J. (1997) Crystal structure of an ecotin-collagenase complex suggests a model for recognition and cleavage of the collagen triple helix. Biochemistry 36, 5381–5392 [DOI] [PubMed] [Google Scholar]
- 19. Huber R., Bode W. (1978) Structural basis of the activation and action of trypsin. Acc. Chem. Res. 11, 114–122 [Google Scholar]
- 20. Robinson M. W., Corvo I., Jones P. M., George A. M., Padula M. P., To J., Cancela M., Rinaldi G., Tort J. F., Roche L., Dalton J. P. (2011) Collagenolytic activities of the major secreted cathepsin L peptidases involved in the virulence of the helminth pathogen, Fasciola hepatica. PLoS Negl. Trop. Dis. 5, e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takeuchi H., Shibano Y., Morihara K., Fukushima J., Inami S., Keil B., Gilles A. M., Kawamoto S., Okuda K. (1992) Structural gene and complete amino acid sequence of Vibrio alginolyticus collagenase. Biochem. J. 281, 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eckhard U., Schönauer E., Nüss D., Brandstetter H. (2011) Structure of collagenase G reveals a chew-and-digest mechanism of bacterial collagenolysis. Nat. Struct. Mol. Biol. 18, 1109–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoshida E., Noda H. (1965) Isolation and characterization of collagenases I and II from Clostridium histolyticum. Biochim. Biophys. Acta 105, 562–574 [DOI] [PubMed] [Google Scholar]
- 24. Gupta R., Beg Q. K., Lorenz P. (2002) Bacterial alkaline proteases. Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59, 15–32 [DOI] [PubMed] [Google Scholar]
- 25. Okamoto M., Yonejima Y., Tsujimoto Y., Suzuki Y., Watanabe K. (2001) A thermostable collagenolytic protease with a very large molecular mass produced by thermophilic Bacillus sp. strain MO-1. Appl. Microbiol. Biotechnol. 57, 103–108 [DOI] [PubMed] [Google Scholar]
- 26. Itoi Y., Horinaka M., Tsujimoto Y., Matsui H., Watanabe K. (2006) Characteristic features in the structure and collagen-binding ability of a thermophilic collagenolytic protease from the thermophile Geobacillus collagenovorans MO-1. J. Bacteriol. 188, 6572–6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim H. K., Ha Y. R., Yu H. S., Kong H. H., Chung D. I. (2003) Purification and characterization of a 33 kDa serine protease from Acanthamoeba lugdunensis KA/E2 isolated from a Korean keratitis patient. Korean J. Parasitol. 41, 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Windhorst S., Frank E., Georgieva D. N., Genov N., Buck F., Borowski P., Weber W. (2002) The major extracellular protease of the nosocomial pathogen Stenotrophomonas maltophilia. Characterization of the protein and molecular clonning of the gene. J. Biol. Chem. 277, 11042–11049 [DOI] [PubMed] [Google Scholar]
- 29. Kim W. T., Kong H. H., Ha Y. R., Hong Y. C., Jeong H. J., Yu H. S., Chung D. I. (2006) Comparison of specific activity and cytopathic effects of purified 33 kDa serine proteinase from Acanthamoeba strains with different degree of virulence. Korean J. Parasitol. 44, 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao G. Y., Chen X. L., Zhao H. L., Xie B. B., Zhou B. C., Zhang Y. Z. (2008) Hydrolysis of insoluble collagen by deseasin MCP-01 from deep-sea Pseudoalteromonas sp. SM9913. Collagenolytic characters, collagen-binding ability of C-terminal polycystic kidney disease domain, and implication for its novel role in deep-sea sedimentary particulate organic nitrogen degradation. J. Biol. Chem. 283, 36100–36107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurata A., Uchimura K., Kobayashi T., Horikoshi K. (2010) Collagenolytic subtilisin-like protease from the deep-sea bacterium Alkalimonas collagenimarina AC40T. Appl. Microbiol. Biotechnol. 86, 589–598 [DOI] [PubMed] [Google Scholar]
- 32. Zhang X. Y., Zhang Y. J., Chen X. L., Qin Q. L., Zhao D. L., Li T. G., Dang H. Y., Zhang Y. Z. (2008) Myroides profundi sp. nov., isolated from deep-sea sediment of the southern Okinawa Trough. FEMS Microbiol. Lett. 287, 108–112 [DOI] [PubMed] [Google Scholar]
- 33. Chen X. L., Xie B. B., Bian F., Zhao G. Y., Zhao H. L., He H. L., Zhou B. C., Zhang Y. Z. (2009) Ecological function of myroilysin, a novel bacterial M12 metalloprotease with elastinolytic activity and a synergistic role in collagen hydrolysis, in biodegradation of deep-sea high-molecular-weight organic nitrogen. Appl. Environ. Microbiol. 75, 1838–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sankar S., Sekar S., Mohan R., Rani S., Sundaraseelan J., Sastry T. P. (2008) Preparation and partial characterization of collagen sheet from fish (Lates calcarifer) scales. Int. J. Biol. Macromol. 42, 6–9 [DOI] [PubMed] [Google Scholar]
- 35. Worthington Biochemical Co (1993) Worthington Enzyme Manual. Collagenase. Worthington Biochemical Co., Freehold, NJ [Google Scholar]
- 36. Abraham L. D., Breuil C. (1996) Isolation and characterization of a subtilisin-like serine proteinase secreted by the sap-staining fungus Ophiostoma piceae. Enzyme Microb. Technol. 18, 133–140 [DOI] [PubMed] [Google Scholar]
- 37. He H. L., Chen X. L., Li J. W., Zhang Y. Z., Gao P. J. (2004) Taste improvement of refrigerated meat treated with cold-adapted protease. Food Chem. 84, 307–311 [Google Scholar]
- 38. Chen X. L., Xie B. B., Lu J. T., He H. L., Zhang Y. (2007) A novel type of subtilase from the psychrotolerant bacterium Pseudoalteromonas sp. SM9913. Catalytic and structural properties of deseasin MCP-01. Microbiology 153, 2116–2125 [DOI] [PubMed] [Google Scholar]
- 39. Maciver B., McHale R. H., Saul D. J., Bergquist P. L. (1994) Cloning and sequencing of a serine proteinase gene from a thermophilic Bacillus species and its expression in Escherichia coli. Appl. Environ. Microbiol. 60, 3981–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y. G., Whittier R. F. (1995) Thermal asymmetric interlaced PCR. Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681 [DOI] [PubMed] [Google Scholar]
- 41. Farndale R. W., Buttle D. J., Barrett A. J. (1986) Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim. Biophys. Acta 883, 173–177 [DOI] [PubMed] [Google Scholar]
- 42. Wang Y. K., Zhao G. Y., Li Y., Chen X. L., Xie B. B., Su H. N., Lv Y. H., He H. L., Liu H., Hu J., Zhou B. C., Zhang Y. Z. (2010) Mechanistic insight into the function of the C-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 from deep sea Pseudoalteromonas sp. SM9913. Binding of the PKD domain to collagen results in collagen swelling but does not unwind the collagen triple helix. J. Biol. Chem. 285, 14285–14291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nonaka T., Fujihashi M., Kita A., Saeki K., Ito S., Horikoshi K., Miki K. (2004) The crystal structure of an oxidatively stable subtilisin-like alkaline serine protease, KP-43, with a C-terminal β-barrel domain. J. Biol. Chem. 279, 47344–47351 [DOI] [PubMed] [Google Scholar]
- 44. Eder J., Rheinnecker M., Fersht A. R. (1993) Folding of subtilisin BPN′. Role of the pro-sequence. J. Mol. Biol. 233, 293–304 [DOI] [PubMed] [Google Scholar]
- 45. Orgel J. P., Eid A., Antipova O., Bella J., Scott J. E. (2009) Decorin core protein (decoron) shape complements collagen fibril surface structure and mediates its binding. PLoS One 4, e7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y. J., Inouye M. (2008) The intramolecular chaperone-mediated protein folding. Curr. Opin. Struct. Biol. 18, 765–770 [DOI] [PubMed] [Google Scholar]
- 47. Tanaka S. I., Takeuchi Y., Matsumura H., Koga Y., Takano K., Kanaya S. (2008) Crystal structure of Tk-subtilisin folded without propeptide. Requirement of propeptide for acceleration of folding. FEBS Lett. 582, 3875–3878 [DOI] [PubMed] [Google Scholar]
- 48. Foophow T., Tanaka S., Angkawidjaja C., Koga Y., Takano K., Kanaya S. (2010) Crystal structure of a subtilisin homologue, Tk-SP, from Thermococcus kodakaraensis. Requirement of a C-terminal β-Jelly Roll domain for hyperstability. J. Mol. Biol. 400, 865–877 [DOI] [PubMed] [Google Scholar]
- 49. Manka S. W., Carafoli F., Visse R., Bihan D., Raynal N., Farndale R. W., Murphy G., Enghild J. J., Hohenester E., Nagase H. (2012) Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1. Proc. Natl. Acad. Sci. U.S.A. 109, 12461–12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Knäuper V., Cowell S., Smith B., López-Otin C., O'Shea M., Morris H., Zardi L., Murphy G. (1997) The role of the C-terminal domain of human collagenase-3 (MMP-13) in the activation of procollagenase-3, substrate specificity, and tissue inhibitor of metalloproteinase interaction. J. Biol. Chem. 272, 7608–7616 [DOI] [PubMed] [Google Scholar]
- 51. Taddese S., Jung M. C., Ihling C., Heinz A., Neubert R. H., Schmelzer C. E. (2010) MMP-12 catalytic domain recognizes and cleaves at multiple sites in human skin collagen type I and type III. Biochim. Biophys. Acta 1804, 731–739 [DOI] [PubMed] [Google Scholar]
- 52. Walton R. S., Brand D. D., Czernuszka J. T. (2010) Influence of telopeptides, fibrils and crosslinking on physicochemical properties of type I collagen films. J. Mater. Sci. Mater. Med. 21, 451–461 [DOI] [PubMed] [Google Scholar]
- 53. Panwar P., Du X., Sharma V., Lamour G., Castro M., Li H., Brömme D. (2013) Effects of cysteine proteases on the structural and mechanical properties of collagen fibers. J. Biol. Chem. 288, 5940–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cowan P. M., McGavin S., North A. C. (1955) The polypeptide chain configuration of collagen. Nature 176, 1062–1064 [DOI] [PubMed] [Google Scholar]
- 55. Perona J. J., Craik C. S. (1995) Structural basis of substrate specificity in the serine proteases. Protein Sci. 4, 337–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ballinger M. D., Tom J., Wells J. A. (1995) Designing subtilisin BPN′ to cleave substrates containing dibasic residues. Biochemistry 34, 13312–13319 [DOI] [PubMed] [Google Scholar]