Background: Circadian clock regulates various aspects of metabolism.
Results: Rhythmic acetylation of AceCS1 controls circadian oscillations in acetyl-CoA levels and in fatty acid elongation.
Conclusion: A previously unrecognized regulation of acetyl-CoA provides additional evidence to circadian regulation of metabolism.
Significance: Understanding of the role of acetyl-CoA in fatty acid elongation might provide therapeutic benefits for treating metabolic diseases and cancer.
Keywords: Acetyl Coenzyme A, Circadian Clock, Gene Regulation, Post-translational Modification, Signal Transduction, SIRT1
Abstract
The circadian clock regulates a wide range of physiological and metabolic processes, and its disruption leads to metabolic disorders such as diabetes and obesity. Accumulating evidence reveals that the circadian clock regulates levels of metabolites that, in turn, may regulate the clock. Here we demonstrate that the circadian clock regulates the intracellular levels of acetyl-CoA by modulating the enzymatic activity of acetyl-CoA Synthetase 1 (AceCS1). Acetylation of AceCS1 controls the activity of the enzyme. We show that acetylation of AceCS1 is cyclic and that its rhythmicity requires a functional circadian clock and the NAD+-dependent deacetylase SIRT1. Cyclic acetylation of AceCS1 contributes to the rhythmicity of acetyl-CoA levels both in vivo and in cultured cells. Down-regulation of AceCS1 causes a significant decrease in the cellular acetyl-CoA pool, leading to reduction in circadian changes in fatty acid elongation. Thus, a nontranscriptional, enzymatic loop is governed by the circadian clock to control acetyl-CoA levels and fatty acid synthesis.
Introduction
The circadian clock machinery is canonically described as a series of interconnected transcriptional and translational feedback loops (1, 2). Additional findings indicate that the clock relies on multiple levels of control, including post-transcriptional (3–5), post-translational (6), metabolic (7, 8), and transcription-independent pathways (9, 10). NAD+, a metabolite that acts as a critical coenzyme, has been shown to be an output of the circadian clock (7, 8). Moreover, fluctuations in NAD+ can also modulate the clock through NAD+-dependent deacetylation of histones, BMAL1, and PER2 by the SIRT1 deacetylase (11–13). SIRT1 has been shown to regulate several metabolic pathways and is implicated in controlling aging and inflammation through its deacetylase activity (14, 15). Interestingly, one of the proteins regulated by SIRT1-mediated deacetylation is acetyl-CoA synthetase 1 (AceCS1),3 a central enzyme involved in acetyl-CoA biosynthesis (16). Deacetylation of Lys-661 on AceCS1 by SIRT1 leads to activation of AceCS1 (16). Because SIRT1 activity, and the abundance of its cofactor NAD+, oscillate in a circadian manner (7, 11), we reasoned that AceCS1 acetylation, and in turn, acetyl-CoA abundance, may display circadian rhythmicity.
Acetyl-CoA, a metabolite that provides acetyl groups during the acetylation reaction, exists in two separate pools in the cell: a mitochondrial pool and a nuclear/cytosolic pool (17). The mitochondrial pool is derived mainly from the action of the enzyme pyruvate dehydrogenase and from fatty acid oxidation. The nuclear/cytosolic pool, responsible for protein acetylation and fatty acid synthesis, is produced by two enzymes: AceCS1 and ATP-citrate lyase (ACLY). Whereas ACLY uses citrate (produced during the tricarboxylic acid cycle) as a substrate for the production of acetyl-CoA, AceCS1 uses acetate. In mammals, acetate can be produced physiologically by the intestinal flora, alcohol metabolism, prolonged fasting, and histone deacetylation (18). In Saccharomyces cerevisiae, the homolog of AceCS1 (Acs2p), was shown to be the major source of acetyl-CoA (19). Importantly, Wellen et al. have reported that ACLY and AceCS1 are present in both the cytosol and the nucleus of mammalian cells, and that the loss of either of these proteins leads to a reduction in global histone acetylation (20). Moreover, reduction in histone acetylation upon loss of ACLY can be rescued by supplementing cells with acetate, supporting a critical role for AceCS1 in acetyl-CoA biosynthesis (20). In this study, we demonstrate a novel regulation of the enzymatic activity of AceCS1 by the circadian clock that results in the rhythmicity of fatty acid elongation.
EXPERIMENTAL PROCEDURES
Animals
The Clock-null mice and wild-type mice were from Dr. Steven Reppert. Clock/Clock mutant mice have been described (21). Mice housed in individual cages were entrained on a L12:D12 (12-h light:12-h dark) cycle for 2 weeks before analyses. Mice were sacrificed at specified circadian times, and livers were isolated. All research involving vertebrate animals was performed under a protocol approved by the Institutional Animal Care and Use Committee (IACUC). Animals were monitored on a daily basis by both the laboratory and University Lab Animal Resources (ULAR) veterinary staff for signs of distress, pain, and/or infection and were given ad libitum access to food and water. Cages were cleaned on a weekly basis and when visibly soiled to maintain a clean environment. All husbandry procedures and welfare policies were conducted according to the Guide for the Care and Use of Laboratory Animals, set forth by the Institute of Laboratory Animal Resources, Commission on Life Sciences, and National Research Council.
Reagents
All reagents used for HPLC-MS were from Sigma. Antibodies against total AceCS1 and ACLY were from Cell Signaling Technology; anti-BMAL1 (AB93806) and anti-tubulin were from Sigma. Anti-acetyl-AceCS1 was from the laboratory of Dr. John Denu as described in Ref. 16.
Cell Culture and Transfection
Mouse embryonic fibroblasts (MEFs) were cultured in DMEM supplemented with 10% FBS and antibiotics. Confluent MEFs were synchronized by treatment with 50% horse serum for 2 h. Control and AceCS1-knockdown mammary epithelial carcinoma cell lines were cultured in DMEM supplemented with 10% FBS and antibiotics. These cells were synchronized by treatment with 100 nm dexamethasone (Sigma) for 2 h. siRNA transfections were performed as described by Wellen et al. (20). ON-TARGETplus SMART pool siRNAs were from Dharmacon (mouse AceCS1 (L-065412-01-0010), mouse ACL (L-040092-01-0010), or a nontargeting control (D-001810-01-20)) and were transfected at a concentration of 20 nm using Lipofectamine RNAiMAX (Invitrogen). Stable knockdown of AceCS1 was achieved by using GIPZ lentiviral shRNAmir system (Thermo Scientific) according to the manufacturer's protocol. shRNA clone 4 (catalogue no. RMM4431-101266313) was the most effective clone in knocking down AceCS1 expression. Cells were selected by using puromycin.
Acetyl-CoA Measurements
We extracted and analyzed acetyl-CoA by modifying a previously reported method (26).. Briefly, cells grown in 15-cm dishes or 100 mg of liver tissue were harvested in water containing 5% trifluoroacetic acid and malonyl-CoA as an internal standard. After removal of debris and protein by centrifugation using 3-kDa cutoff filters, samples were loaded on a Sep-Pak C18 column and eluted using methanol. Samples were dried under N2 gas, resuspended in water containing 0.1% acetic acid, and analyzed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS).
Acetyl-CoA was analyzed using an Agilent 1100 series liquid chromatography coupled to an electrospray mass spectrometry detector (MSD Trap XCT, Agilent Technologies, Palo Alto, CA). Column was ZORBAX 300 Extend-C18 (2.1 × 150 mm 3.5 μm) maintained at 40 °C. Mobile phase A was methanol; mobile phase B was 5 mmol of di-n-butylammonium acetate (Tokyo Chemical Industry, Tokyo) + 5 mmol ammonium acetate buffer, pH 9.0. The elution gradient was as follows: 20% solvent A; 15 min linear gradient to 90% solvent B; 10–15 min 100% solvent B; post time was 15 min at 100% solvent A. The flow rate was 0.8 ml/min. Acetyl-CoA was detected in negative ionization mode with the source voltage set at 3500 V, skimmer at 40 V, and capillary exit at 151.8 V. Nebulizer pressure was set at 60 p.s.i., with drying gas (nitrogen) flow rate at 10 liters/min and drying temperature at 350 °C. Helium was used as a collision gas. Multiple reaction monitoring was used for the quantitative determination of acetyl-CoA, using malonyl-CoA as an internal standard. Extracted ion chromatograms for transitions with mass-to-charge ratios 808.1 > 408.1 and 852.1 > 808.1 were used for quantitation of acetyl-CoA and malonyl-CoA, respectively.
Fatty Acid Analyses
Cells were washed with ice-cold phosphate-buffered saline (PBS) and scraped into 1 ml of methanol/water (1:1, v/v) containing the d8-arachidonic acid (Cayman Chemicals, Ann Arbor, MI) as an internal standard. Protein concentrations were measured using the BCA protein assay (Pierce). Cells were sonicated and incubated with methanolic potassium hydroxide (1 m) for 1 h at 80 °C. The solutions were neutralized with glacial acetic acid (1–10 μl), and lipids were extracted with chloroform (2 volumes, 1.5 ml). The organic phases were collected and reconstituted in methanol (0.1 ml) for LC-MS analyses. Fatty acids were quantified using a 1100-LC system coupled to a 1946D-MS detector (Agilent Technologies) and equipped with an electrospray ionization interface. Fatty acids were separated on a reversed-phase XDB Eclipse C18 column (Agilent Technologies) eluted with a linear gradient (from 90% to 100% of methanol in water containing 0.25% acetic acid and 5 mm ammonium acetate in 2.5 min) at a flow rate of 1.5 ml/min with a column temperature of 40 °C. Electrospray ionization was in the negative mode, capillary voltage was set at 4 kV, and fragmentor voltage was 100 V. Nitrogen was used as drying gas at a flow rate of 13 liters/min and a temperature of 350 °C. Nebulizer pressure was set at 60 p.s.i. We used commercially available fatty acids as reference standards, monitoring deprotonated molecular ions [M-H]− in the SIM mode and d8-arachidonic acid (m/z 311.3) as internal standard.
Quantitative Real-time RT-PCR
Each quantitative real-time RT-PCR was performed using the Chromo4 real-time detection system (Bio-Rad). 1 μg of mRNA was reverse-transcribed using an iScript cDNA synthesis kit (Bio-Rad). cDNA template was mixed with the primers to final concentrations of 200 nm and 10 μl of iQ SYBR Green Supermix (Bio-Rad), respectively. The reaction was first incubated at 95 °C for 3 min, followed by 40 cycles at 95 °C for 30 s and 60 °C for 1 min.
[14C]Acetate Incorporation into Histones and Lipids
Control and AceCS1-knockdown mammary epithelial carcinoma cell lines or MEFs were used for these experiments. Cells were treated with 10 μCi of [14C]acetate for 2 h and then were harvested in PBS and processed for extraction of histones or lipids. For histone extraction, cells were lysed in 0.8 n HCl, and 50 μl of acid-soluble fraction was used for scintillation counting. For incorporation into lipids, cells were extracted with methanol:water and chloroform, centrifuged, and 50 μl of the lipid containing lower phase was used for scintillation counting.
Data Analyses
Results are expressed as means ± S.E. of multiple experiments. Student's t tests were used to compare two groups. Statistical significance was detected at the 0.05 level.
RESULTS
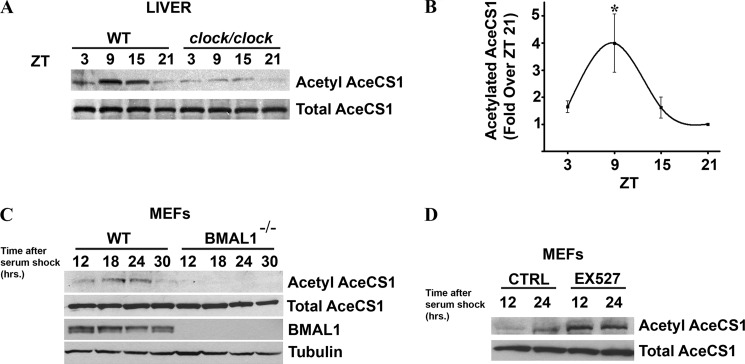
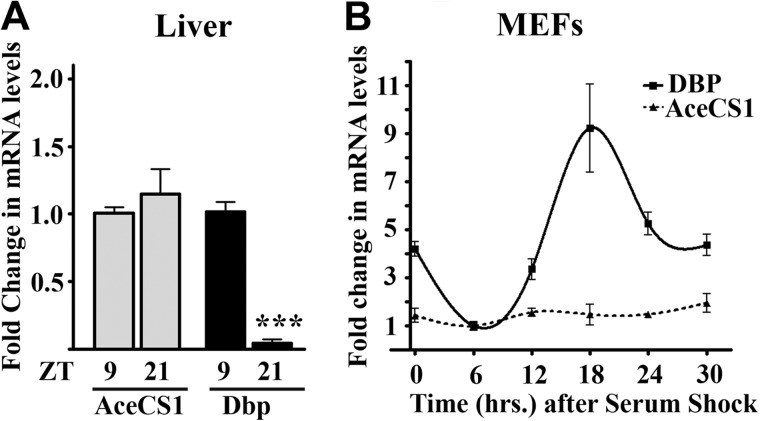
To determine whether acetylation of AceCS1 changes with the time of the day, liver extracts were prepared at different zeitgeber times (ZTs) from mice entrained in 12-h light:12-h dark cycle. Using an anti-acetyl-AceCS1 antibody, specific to the acetylated Lys-661 residue (16), we reveal that acetylation of AceCS1 oscillates in a circadian manner in the liver from wild-type (WT) mice (Fig. 1,A and B). The highest level of acetylation was observed at ZT9, whereas AceCS1 was mostly deacetylated at ZT21. Total levels of AceCS1 did not display circadian rhythmicity, either in protein levels (Fig. 1, A–D) or in mRNA levels (Fig. 2, A and B). Interestingly, the phase of oscillation of AceCS1 acetylation parallels that of BMAL1, another clock-related SIRT1 target (11, 12).
FIGURE 1.
AceCS1 acetylation is regulated by the circadian clock. A–C, mice entrained in 12-h light:12-h dark cycles were sacrificed at indicated times, and their liver was dissected out. Total lysates were prepared and resolved by SDS-PAGE. A, acetylated and total AceCS1 levels were detected by Western blotting using specific antibodies in WT and clock/clock livers. B, acetylated AceCS1 levels (normalized to total AceCS1 levels) were quantified by densitometry. *, p < 0.05 (versus ZT21); n = 4. Error bars, S.E. C, WT and Bmal1−/− MEFs were synchronized by serum shock. Total lysates were prepared at the indicated times after synchronization and resolved by SDS-PAGE followed by Western analysis. Acetylated and total AceCS1, BMAL1, and α-tubulin levels were detected by specific antibodies. D, WT MEFs were pretreated with 50 μm EX527 for 16 h and then synchronized by serum shock. 50 μm EX527 was also added to the medium during and after serum shock. Total lysates were prepared at indicated times, and acetylated and total AceCS1 levels were detected by Western blotting.
FIGURE 2.
AceCS1 mRNA levels do not oscillate. AceCS1 and Dbp (D site of albumin promoter (albumin D-box)-binding protein} gene expressions were quantified by quantitative PCR in WT liver, n = 10 (A) or serum-entrained WT MEFs, n = 3 (B). Gene expression was normalized to 18S ribosomal RNA. Expression at trough was set to 1. ***, p < 0.00001 versus Dbp expression at ZT9. Error bars, S.D. Dbp is a robustly oscillating transcript in most mammalian tissues and hence is used as a positive control for studying oscillation in gene expression.
To evaluate whether the circadian clock drives AceCS1 acetylation, we used clock/clock (c/c) mutant mice (21) and found that acetylation is indeed drastically reduced in the liver of these mutant mice (Fig. 1A). We further analyzed the oscillation in AceCS1 acetylation in cultured cells by using MEFs. WT and Bmal1−/− MEFs were synchronized by serum shock, and cells were harvested at different time intervals. Acetylated AceCS1 levels were rhythmic in the WT cells with a peak at 18–24 h after synchronization (Fig. 1C), paralleling BMAL1 acetylation profile in MEFs (7, 11). AceCS1 acetylation levels were almost undetectable in Bmal1−/− MEFs, whereas total protein levels of AceCS1 in both cell types remained unchanged and nonrhythmic. Thus, AceCS1 acetylation oscillates in a circadian manner both in vivo and in cultured cells. Next we validated the role of SIRT1 in circadian deacetylation of AceCS1 by using EX527, a direct pharmacological inhibitor of SIRT1 (22). Indeed, blocking SIRT1 functions leads to elevated and arrhythmic AceCS1 acetylation (Fig. 1D).
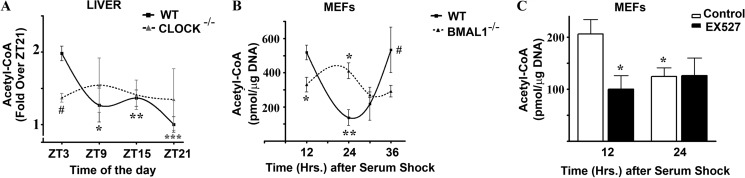
Because the acetylation status of AceCS1 controls its activity (16), we next sought to determine whether total cellular acetyl-CoA levels are also rhythmic. To do so, acetyl-CoA levels were measured by LC-MS/MS by using a modified version of the method described by Hayashi and Satoh (23). We found that acetyl-CoA levels were rhythmic in the liver of WT mice, with highest levels observed at ZT3 (Fig. 3A). This is in keeping with a scenario in which the peak of acetyl-CoA levels (ZT3) follows the peak of deacetylated (and hence, active) AceCS1 (ZT21). Next, we determined whether a functional circadian clock is important for the rhythmicity in the acetyl-CoA levels by analyzing the abundance of acetyl-CoA in the livers from Clock−/− mice. The peripheral tissues of Clock−/− mice have been shown to be arrhythmic (24). Consistent with a prominent role of the circadian clock machinery in regulating acetyl-CoA levels, there is no oscillation in the abundance of acetyl-CoA in the liver of Clock−/− mice (Fig. 3A). We then measured acetyl-CoA levels in cultured cells by synchronizing MEFs. WT MEFs displayed robust oscillation in the acetyl-CoA levels, with a peak at 12-h post-synchronization and trough at 24 h post-synchronization (Fig. 3B), in agreement with the cylic acetylation of AceCS1 (Fig. 1C). Because acetylation of AceCS1 in Bmal1−/− MEFs is low and noncyclic, we expected high and nonoscillating levels of acetyl-CoA in these cells, and this is in fact the case (Fig. 3B). Also, MEFs treated with EX527 displayed lower and nonoscillating levels of acetyl-CoA compared with untreated cells (Fig. 3C), paralleling the acetylation profile of AceCS1 (Fig. 1D). These results indicate that acetyl-CoA levels are rhythmic in mouse liver and in MEFs, that this rhythmicity is clock-controlled, and that the clock-driven acetylation of AceCS1 contributes to the cyclic abundance of acetyl-CoA.
FIGURE 3.
Circadian oscillation in Acetyl-CoA levels is controlled by the clock. Acetyl-CoA was extracted from mouse liver (A) or MEFs (B and C) harvested at indicated time points and analyzed by LC-MS/MS. A, relative abundance of acetyl-CoA in livers from WT and Clock−/− mice. ***, p = 0.00004 (WT ZT3 versus WT ZT21); **, p < 0.01 (WT ZT3 versus WT ZT 15); *, p < 0.05 (WT ZT3 versus WT ZT9); #, p < 0.05 (ZT3, WT versus CLOCK−/−); n = 4–7. Error bars, S.E. The total amount of acetyl-CoA (2200 pmol/μg DNA) was approximately 30% less in the Clock−/− liver at ZT3, compared with that in the WT liver (3200 pmol/μg DNA). B, acetyl-CoA levels in serum-entrained WT and Bmal1−/− MEFs. *, p < 0.05 (versus corresponding WT samples); **, p < 0.01 (versus WT 12 h); #, p < 0.05 (versus WT 24 h); n = 3. C, acetyl-CoA levels in serum-entrained WT MEFs treated with or without 50 μm EX527. *, p < 0.05 (versus control 12 h); n = 4.
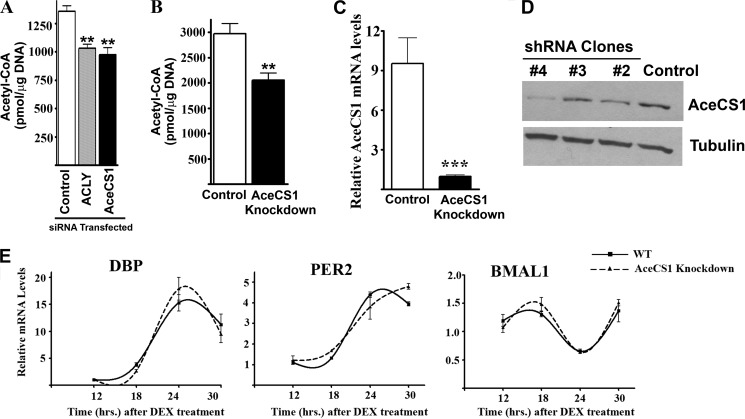
The relative contribution of ACLY and AceCS1 toward the intracellular abundance of acetyl-CoA is not fully understood. Blocking ACLY in cultured cells, either by RNAi (25) or by the specific inhibitor SB-204990 (26), has been shown to reduce the total cellular acetyl-CoA levels by ∼50%. Moreover, knockdown of ACLY in mouse liver by adenovirus-mediated RNAi caused ∼25% reduction in the hepatic acetyl-CoA levels (27). To determine the relative contribution of ACLY and AceCS1 on total cellular acetyl-CoA levels, we transiently knocked down ACLY and AceCS1 by siRNAs in cultured cells (Fig. 4A). Our results show that both ACLY and AceCS1 contribute significantly, and in a similar extent, to the total cellular acetyl-CoA pool. We reproducibly observed a reduction of acetyl-CoA levels by 24 or 28% upon the knockdown of ACLY or AceCS1, respectively (Fig. 4A). Furthermore, total acetyl-CoA levels were also reduced by 31% in a cell line where AceCS1 was stably knocked down (28) (Fig. 4B). AceCS1 mRNA levels were reduced by ∼90% in these cells (Fig. 4C). These results establish that AceCS1 is a major determinant of cellular acetyl-CoA.
FIGURE 4.
Regulation of acetyl-CoA levels by AceCS1. A, WT MEFs were transfected with siRNAs against ACLY, AceCS1, or a nontargeting control. Cells were harvested 3 days after transfection for acetyl-CoA measurement, **, p < 0.01, n = 3. Error bars, S.E. B and C, control and AceCS1-knockdown mammary epithelial carcinoma cell lines were used for acetyl-CoA measurement (B); **, p < 0.01, n = 8; AceCS1 gene expression by quantitative PCR (C); ***, p < 0.00001, n = 9. D, MEFs were infected with lentiviruses expressing shRNA against a nontargeting control or against AceCS1. Several clones were screened, and clone 4 (showing maximum down-regulation of AceCS1 protein levels) was selected for further experiments. E, control (WT) and AceCS1-knockdown MEFs were synchronized by dexamethasone, cells were harvested at indicated time points, and gene expression was analyzed by real-time quantitative PCR. Gene expression was normalized to actin mRNA levels.
Because acetyl-CoA levels could directly influence histone acetylation and thus, gene expression (20, 29, 30), we analyzed changes in circadian gene expression after knocking down AceCS1 in MEFs. Using a lentiviral shRNA against AceCS1, we generated a MEF cell line that expressed significantly lower levels of AceCS1 (Fig. 4D). When synchronized by dexamethasone, both control and AceCS1-knockdown MEFs displayed very similar, robust oscillation of core circadian gene expression (Fig. 4E). These results indicate that AceCS1 is not required for the regulation of circadian gene expression. Because the Km of histone acetyltransferases for acetyl-coA is relatively low (31), it is likely that modest fluctuations in acetyl-CoA levels might not be sufficient to alter histone acetylation and thus affect gene expression.
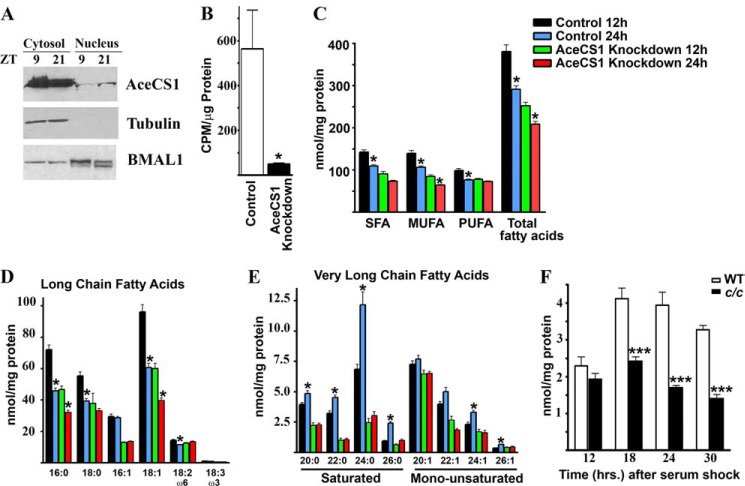
Acetyl-CoA is the carbon source for synthesis and elongation of fatty acids. Because AceCS1 is present predominantly in the cytosol (Ref. 20 and Fig. 5A) and the fatty acid synthesis is mostly dependent on cytosolic availability of acetyl-CoA, we explored whether fatty acid synthesis is under circadian control through the AceCS1-mediated oscillation in acetyl-CoA. Acetyl-CoA produced by AceCS1 has been shown to be utilized in lipid synthesis (16). To validate the role of AceCS1 in lipid synthesis, we measured the incorporation of 14C-labeled acetate into lipids in AceCS1-knockdown and control cell lines. There is a remarkable decrease in 14C incorporation into lipids in AceCS1-knockdown cells compared with the control cells (Fig. 5B).
FIGURE 5.
Regulation of fatty acid synthesis by AceCS1. A, WT mice were sacrificed at indicated time points, and liver was harvested. Cytosolic and nuclear extracts were fractionated. Total AceCS1, α-tubulin, and BMAL1 levels were detected by Western blotting using specific antibodies. B, control and AceCS1-knockdown mammary epithelial carcinoma cell lines were used for measurement of [14C]acetate uptake into lipids; *, p < 0.05, n = 3. C–E, LC-MS analysis of fatty acid levels in control and AceCS1-knockdown mammary epithelial carcinoma cell lines at indicated times after dexamethasone synchronization, n = 6. C, LC-MS analysis of fatty acid levels is shown at indicated times after dexamethasone synchronization. SFA, saturated fatty acids; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids. *, p < 0.01 versus corresponding 12-h sample, n = 6. D, long chain fatty acids; E, very long chain fatty acids;*, p < 0.01 versus corresponding 12-h sample, n = 6. Error bars, S.E. F, LC-MS analysis of saturated VLCFAs in WT and c/c MEFs is shown at indicated times after synchronization by serum shock. ***, p < 0.001 compared with the corresponding wild-type sample, n = 6.
To further understand the role of AceCS1 in lipid metabolism, we used a lipidomics approach. We analyzed the levels of fatty acids of varying length and unsaturation at two time points in synchronized control and the AceCS1-knockdown cultured cells. The overall levels of fatty acids were significantly reduced in the AceCS1-knockdown cells (Fig. 5C). Interestingly, most fatty acids demonstrated a trend where the levels were higher at 12 h post-synchronization compared with the 24-h time point. These fatty acids included saturated and monounsaturated long chain fatty acids (Fig. 5D). This could be because the fatty acids are either oxidized during this time period and/or they are being converted to very long chain fatty acids (VLCFAs). Supporting the latter scenario, we observed that the levels of VLCFAs are higher at the 24-h time point (Fig. 5E). Importantly, the change in VLCFA levels is absent in AceCS1-knockdown cells (Fig. 5E). These results suggest that the reduced level of acetyl-CoA in the AceCS1-knockdown cells leads to reduction in total fatty acid levels and also causes impaired elongation of long chain fatty acids into VLCFAs. To confirm that elongation of fatty acids is regulated by the circadian clock, we measured the levels of fatty acids in WT and c/c MEFs. Although there is a robust oscillation in VLCFAs in the WT MEFs, their levels are significantly lower and nonrhythmic in c/c MEFs (Fig. 5F). Our results demonstrate that the elongation of fatty acids, a process that requires acetyl-CoA, is under the control of the circadian clock machinery. These results also establish AceCS1 as an important contributor to fatty acid elongation.
DISCUSSION
Our findings provide evidence that AceCS1 functions as a circadian enzyme, thereby contributing to the cyclic cellular levels of acetyl-CoA. Rhythmicity in AceCS1 acetylation contributes to the oscillation of acetyl-coA levels and, in turn, regulates circadian fatty acid elongation. Acetate could also be converted to acetyl-CoA by the mitochondrial enzyme AceCS2. However, AceCS2 expression is significantly lower compared with AceCS1 in the mouse liver (32) and is almost undetectable in the MEFs (data not shown). Furthermore, in our experiments where cells were treated with [14C]acetate, knocking down AceCS1 is sufficient to reduce the acetate conversion to acetyl-CoA by ∼10-fold (Fig. 5B), confirming the prominent role of AceCS1 in these cells.
Fatty acid synthesis constitutes a major process that utilizes acetyl-CoA in all cells. We have reported a unique pathway by which the circadian clock regulates the abundance of acetyl-CoA, leading to a clock-driven control of fatty acid elongation. Abolishing the activity of AceCS1 causes a significant decrease in the cellular pool of acetyl-CoA and leads to dampening of oscillations in fatty acid synthesis. This transcription-independent pathway is based solely on cyclic enzymatic function, utilizing the NAD+-dependent SIRT1 deacetylase to control AceCS1 activity, and contributes to modulated biosynthesis of acetyl-CoA. Thus, our study adds another layer to important examples of transcription-independent control by the mammalian circadian clock (9, 10). These findings underscore that the circadian clock occupies a central position in controlling both NAD+ and acetyl-CoA levels in the cell, linking SIRT1 to fatty acid elongation. Increasing evidence reveals the links between the circadian clock and lipid metabolism (33–36). These may involve additional chromatin remodelers, such as HDAC3, whose recruitment to the genome and enzymatic output follow a circadian pattern. As many of the genes regulated by HDAC3 are involved in lipid metabolism, and loss of HDAC3 leads to increased de novo fatty acid synthesis and a fatty liver phenotype (33), future studies will need to explore its relationship with AceCS1 and the control by SIRT1. Our study has uncovered another level of interplay among the circadian clock, epigenetics, and metabolism. As de novo fatty acid synthesis is known to be increased in cancer (37) and obesity (38), our results might pave the way to future strategies for the use of sirtuin modulators (39) and chronotherapy in their treatment (40).
Acknowledgments
We thank Dr. Kathryn Wellen (University of Pennsylvania) for insightful discussions, Dr. Steven Reppert (University of Massachusetts Medical School) for providing Clock−/− mice, and Dr. Yukie Yoshii and Dr. Yasuhisa Fujibayashi (University of Fukui, Japan) for sharing the AceCS1-knockdown cell line.
Work in the Sassone-Corsi laboratory was supported by the National Institutes of Health, the Institut de la Sante et de la Recherche Medicale (France), and Sirtris Pharmaceutical Inc. Work in the Imhof laboratory was supported by the Bavarian-Californian Technology Center (BaCaTeC) and the Deutsche Forschungsgemeinschaft.
- AceCS1
- acetyl-CoA synthetase 1
- ACLY
- ATP-citrate lyase
- LC-MS/MS
- liquid chromatography coupled to tandem mass spectrometry
- MEF
- mouse embryonic fibroblast
- VLCFA
- very long chain fatty acid
- ZT
- Zeitgeber time.
REFERENCES
- 1. Hastings M. H., Reddy A. B., Maywood E. S. (2003) A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4, 649–661 [DOI] [PubMed] [Google Scholar]
- 2. Partch C. L., Green C. B., Takahashi J. S. (2013) Molecular architecture of the mammalian circadian clock. Trends Cell Biol. July 31, S0962–8924 10.1016/j.tcb.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Le Martelot G., Canella D., Symul L., Migliavacca E., Gilardi F., Liechti R., Martin O., Harshman K., Delorenzi M., Desvergne B., Herr W., Deplancke B., Schibler U., Rougemont J., Guex N., Hernandez N., Naef F. (2012) Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 10, e1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Menet J. S., Rodriguez J., Abruzzi K. C., Rosbash M. (2012) Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife 1, e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kojima S., Shingle D. L., Green C. B. (2011) Post-transcriptional control of circadian rhythms. J. Cell Sci. 124, 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mehra A., Baker C. L., Loros J. J., Dunlap J. C. (2009) Post-translational modifications in circadian rhythms. Trends Biochem. Sci. 34, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P. (2009) Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., Hong H. K., Chong J. L., Buhr E. D., Lee C., Takahashi J. S., Imai S., Bass J. (2009) Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O'Neill J. S., Reddy A. B. (2011) Circadian clocks in human red blood cells. Nature 469, 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O'Neill J. S., van Ooijen G., Dixon L. E., Troein C., Corellou F., Bouget F. Y., Reddy A. B., Millar A. J. (2011) Circadian rhythms persist without transcription in a eukaryote. Nature 469, 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L. P., Sassone-Corsi P. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirayama J., Sahar S., Grimaldi B., Tamaru T., Takamatsu K., Nakahata Y., Sassone-Corsi P. (2007) CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090 [DOI] [PubMed] [Google Scholar]
- 13. Asher G., Gatfield D., Stratmann M., Reinke H., Dibner C., Kreppel F., Mostoslavsky R., Alt F. W., Schibler U. (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 [DOI] [PubMed] [Google Scholar]
- 14. Chalkiadaki A., Guarente L. (2012) Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 8, 287–296 [DOI] [PubMed] [Google Scholar]
- 15. Haigis M. C., Sinclair D. A. (2010) Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hallows W. C., Lee S., Denu J. M. (2006) Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Albaugh B. N., Arnold K. M., Denu J. M. (2011) KAT(ching) metabolism by the tail: insight into the links between lysine acetyltransferases and metabolism. ChemBioChem. 12, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shimazu T., Hirschey M. D., Huang J. Y., Ho L. T., Verdin E. (2010) Acetate metabolism and aging: an emerging connection. Mech. Ageing Dev. 131, 511–516 [DOI] [PubMed] [Google Scholar]
- 19. Takahashi H., McCaffery J. M., Irizarry R. A., Boeke J. D. (2006) Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Mol. Cell 23, 207–217 [DOI] [PubMed] [Google Scholar]
- 20. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vitaterna M. H., King D. P., Chang A. M., Kornhauser J. M., Lowrey P. L., McDonald J. D., Dove W. F., Pinto L. H., Turek F. W., Takahashi J. S. (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Napper A. D., Hixon J., McDonagh T., Keavey K., Pons J. F., Barker J., Yau W. T., Amouzegh P., Flegg A., Hamelin E., Thomas R. J., Kates M., Jones S., Navia M. A., Saunders J. O., DiStefano P. S., Curtis R. (2005) Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J. Med. Chem. 48, 8045–8054 [DOI] [PubMed] [Google Scholar]
- 23. Hayashi O., Satoh K. (2006) Determination of acetyl-CoA and malonyl-CoA in germinating rice seeds using the LC-MS/MS technique. Biosci. Biotechnol. Biochem. 70, 2676–2681 [DOI] [PubMed] [Google Scholar]
- 24. DeBruyne J. P., Weaver D. R., Reppert S. M. (2007) Peripheral circadian oscillators require CLOCK. Curr. Biol. 17, R538–R539 [DOI] [PubMed] [Google Scholar]
- 25. Hatzivassiliou G., Zhao F., Bauer D. E., Andreadis C., Shaw A. N., Dhanak D., Hingorani S. R., Tuveson D. A., Thompson C. B. (2005) ATP-citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8, 311–321 [DOI] [PubMed] [Google Scholar]
- 26. Chu K. Y., Lin Y., Hendel A., Kulpa J. E., Brownsey R. W., Johnson J. D. (2010) ATP-citrate lyase reduction mediates palmitate-induced apoptosis in pancreatic beta cells. J. Biol. Chem. 285, 32606–32615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Q., Li S., Jiang L., Zhou Y., Li Z., Shao M., Li W., Liu Y. (2010) Deficiency in hepatic ATP-citrate lyase affects VLDL-triglyceride mobilization and liver fatty acid composition in mice. J. Lipid Res. 51, 2516–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yoshii Y., Furukawa T., Yoshii H., Mori T., Kiyono Y., Waki A., Kobayashi M., Tsujikawa T., Kudo T., Okazawa H., Yonekura Y., Fujibayashi Y. (2009) Cytosolic acetyl-CoA synthetase affected tumor cell survival under hypoxia: the possible function in tumor acetyl-CoA/acetate metabolism. Cancer Sci. 100, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cai L., Sutter B. M., Li B., Tu B. P. (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaelin W. G., Jr., McKnight S. L. (2013) Influence of metabolism on epigenetics and disease. Cell 153, 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langer M. R., Fry C. J., Peterson C. L., Denu J. M. (2002) Modulating acetyl-CoA binding in the GCN5 family of histone acetyltransferases. J. Biol. Chem. 277, 27337–27344 [DOI] [PubMed] [Google Scholar]
- 32. Fujino T., Kondo J., Ishikawa M., Morikawa K., Yamamoto T. T. (2001) Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J. Biol. Chem. 276, 11420–11426 [DOI] [PubMed] [Google Scholar]
- 33. Feng D., Liu T., Sun Z., Bugge A., Mullican S. E., Alenghat T., Liu X. S., Lazar M. A. (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho H., Zhao X., Hatori M., Yu R. T., Barish G. D., Lam M. T., Chong L. W., DiTacchio L., Atkins A. R., Glass C. K., Liddle C., Auwerx J., Downes M., Panda S., Evans R. M. (2012) Regulation of circadian behaviour and metabolism by REV-ERBα and REV-ERB-β. Nature 485, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bugge A., Feng D., Everett L. J., Briggs E. R., Mullican S. E., Wang F., Jager J., Lazar M. A. (2012) Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Solt L. A., Wang Y., Banerjee S., Hughes T., Kojetin D. J., Lundasen T., Shin Y., Liu J., Cameron M. D., Noel R., Yoo S. H., Takahashi J. S., Butler A. A., Kamenecka T. M., Burris T. P. (2012) Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fritz V., Fajas L. (2010) Metabolism and proliferation share common regulatory pathways in cancer cells. Oncogene 29, 4369–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strable M. S., Ntambi J. M. (2010) Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit. Rev. Biochem. Mol. Biol. 45, 199–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cen Y., Youn D. Y., Sauve A. A. (2011) Advances in characterization of human sirtuin isoforms: chemistries, targets and therapeutic applications. Curr. Med. Chem. 18, 1919–1935 [DOI] [PubMed] [Google Scholar]
- 40. Sahar S., Sassone-Corsi P. (2009) Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer 9, 886–896 [DOI] [PubMed] [Google Scholar]