Background: PI4KA is a critical host factor for replication of hepatitis C virus in liver and a potential therapeutic target.
Results: PI4KA inhibitors prevent the maintenance of PtdIns(4,5)P2 pools during strong PLC activation.
Conclusion: PI4KA plays a critical role in maintaining plasma membrane phosphoinositide pools.
Significance: Safe pharmacological targeting of PI4KA is not feasible.
Keywords: Angiotensin II, G Protein-coupled Receptors, Phosphatidylinositol, Phosphatidylinositol Kinase, Phospholipase C
Abstract
Phosphatidylinositol 4-kinase type IIIα (PI4KA) is a host factor essential for hepatitis C virus replication and hence is a target for drug development. PI4KA has also been linked to endoplasmic reticulum exit sites and generation of plasma membrane phosphoinositides. Here, we developed highly specific and potent inhibitors of PI4KA and conditional knock-out mice to study the importance of this enzyme in vitro and in vivo. Our studies showed that PI4KA is essential for the maintenance of plasma membrane phosphatidylinositol 4,5-bisphosphate pools but only during strong stimulation of receptors coupled to phospholipase C activation. Pharmacological blockade of PI4KA in adult animals leads to sudden death closely correlating with the drug's ability to induce phosphatidylinositol 4,5-bisphosphate depletion after agonist stimulation. Genetic inactivation of PI4KA also leads to death; however, the cause in this case is due to severe intestinal necrosis. These studies highlight the risks of targeting PI4KA as an anti-hepatitis C virus strategy and also point to important distinctions between genetic and pharmacological studies when selecting host factors as putative therapeutic targets.
Introduction
Phosphatidylinositol 4-kinases (PI4Ks)3 have long been viewed as enzymes that produce PtdIns(4)P as an intermediate for PtdIns(4,5)P2 synthesis in the plasma membrane (PM). PtdIns(4,5)P2 is one of the most important regulatory lipids in the PM. It is a precursor of inositol 1,4,5-trisphosphate and diacylglycerol, generated by PLC enzymes, and of phosphatidylinositol 3,4,5-trisphosphate produced by PI3Ks. PtdIns(4,5)P2 also serves as regulator of ion channels and transporters and interacts with proteins that regulate both endocytosis and exocytosis (1). Therefore, PtdIns(4,5)P2 synthesis is essential for cells and so are the PI4Ks that produce PtdIns(4)P to be converted to PtdIns(4,5)P2. However, the identification of four distinct mammalian PI4Ks and subsequent studies on the biology of these enzymes clearly indicated that the different PI4K enzymes and PtdIns(4)P have functions unrelated to PtdIns(4,5)P2 synthesis (2–4). Yeast studies have shown that the Pik1 enzyme, the homologue of mammalian PI4KB, is important for secretion from the Golgi (5, 6), and the Golgi localization of the PI4KB enzyme, as well as the fluorescent reporters detecting PtdIns(4)P, firmly established the importance of this lipid in Golgi function in mammalian cells (7). Less clear is the role of the other type III PI4K, PI4KA. Yeast studies showed that its orthologue, Stt4, is important for the production of PM PtdIns(4,5)P2 (8), and it was also found that the PI4KA enzyme is responsible for maintenance of the PM PtdIns(4,5)P2 pool in mammalian cells (9, 10). The yeast homologue, Stt4, is localized to the PM and organized into signaling domains (11). Although the mammalian PI4KA is primarily located in the endoplasmic reticulum (2, 12), recent studies revealed that the EFR3 and TTC7 proteins together recruit the PI4KA enzyme to the PM (13). Part of the reasons why the functions of PI4KA have been so difficult to understand is the lack of specific inhibitors of its enzymatic functions. The only way to study the importance of the enzyme was to use siRNA-mediated gene silencing. This procedure takes several days, and assuming critical roles of these enzymes (both Pik1 and Stt4 mutant yeast strains are practically unviable), cells with efficient knockdown may be eliminated, and the surviving cells may have enough enzyme left to support essential functions. In addition, conclusions from siRNA studies may be confounded by functions prescribed to domains found outside the enzymatic modality, which may be involved in scaffolding or other protein-protein interactions, particularly in the context of infection (14).
An unexpected and much needed stimulus in PI4K research was brought about by the finding that PI4KA is an essential host factor for hepatitis C virus (HCV) replication, reported in several simultaneous studies using RNAi screens (15–19). In parallel studies, it was also shown that several small RNA enteroviruses reprogrammed the endoplasmic reticulum-Golgi trafficking machinery to serve their replication needs and used another PI4K, PI4KB, in the process (14, 20). These studies offered new possibilities to counter the establishment of viral replication machinery using PI4K inhibitors. The important feature of these studies was that viral replication was almost completely abolished by even partial PI4K knockdown, which elicited no apparent cellular toxicity in vitro (21). These findings have confirmed our previous conclusions that even small amounts of the enzyme can fulfill its “housekeeping” functions (9). Because of this difference for the enzyme for viral replication versus normal cellular functions, some viral studies have concluded that PI4Ks are not important for the cell, and they can be targeted without any problem as an antiviral therapeutic strategy. These conclusions started to be challenged by reports showing deleterious effects of PI4KA genetic inactivation (22).
In this study, we report on the characterization of a set of compounds that selectively inhibit PI4KA and interfere with HCV replication. We show that these compounds inhibit the synthesis of PtdIns(4)P in the PM and impair the maintenance of PtdIns(4,5)P2 levels under strong PLC activation. Curiously, the potency of these compounds to inhibit purified PI4KA in vitro and to inhibit PtdIns(4)P synthesis in the PM in cells shows significant variations raising questions about the ability of the compounds to reach the relevant cellular compartments despite similar chemistries. Importantly, the inhibitory effects on PtdIns(4)P in the PM and on PtdIns(4,5)P2 levels in PLC-stimulated cells were closely correlated. Toxicity studies in animals showed that the most potent small molecule inhibitors of PtdIns(4)P synthesis and PtdIns(4,5)P2 maintenance caused sudden death when applied at high doses with symptoms reminiscent of cardiovascular collapse. These may reflect the ability of the compound to inhibit PtdIns(4,5)P2 maintenance during Gq-coupled receptor signaling that is essential for maintaining vascular tone. Finally, genetic inactivation of the PI4KA enzyme in adult animals with a tamoxifen-induced conditional knock-out mouse caused a lethal gastrointestinal phenotype that was different from the acute drug-induced toxicity. These differences will require further studies to be fully understood but highlight the need for both types of approaches to anticipate the results of pharmacological interventions on the biology of whole animals.
EXPERIMENTAL PROCEDURES
Materials
Angiotensin II (human octapeptide) was from Bachem (Torrance, CA). Wortmannin was purchased from Calbiochem. All other chemicals were of the highest analytical grade. [γ-32P]ATP (6000 Ci/mmol) was purchased from PerkinElmer Life Sciences. myo-[3H]Inositol (30–80 Ci/mmol) was from Amersham Biosciences and American Radiolabeled Chemicals (St Louis, MO). The monoclonal anti-HA antibody (HA.11) was from Covance; the PtdIns(4)P antibody was from Echelon (Salt Lake City, UT; catalogue no. Z-P004), and the rabbit polyclonal anti-GST was from Millipore (Billerica, MA; catalogue no. AB3282).
Synthesis of PI4KA Active Compounds
A1 was synthesized as described in Ref. 23, and the synthesis of C1, F1, H1, and J1 was according to Ref. 24. The synthesis of M1 was described in Ref. 25, and G1 was synthesized according to the procedure detailed in Ref. 26.
Transfection of Cells for Microscopy
COS-7 cells or HEK293-AT1 cells (a HEK293 cell line stably expressing the rat AT1a angiotensin receptor) were used. Cells (50,000 cells/well) were plated onto 25-mm-diameter circular glass coverslips in 6-well plates, and plasmid DNAs (0.5–1 μg/well) were transfected with the PLCδ1-PH-GFP or Tubby domain-GFP fusion constructs (27) using the Lipofectamine 2000 reagent (Invitrogen) and Opti-MEM (Invitrogen) following the manufacturer's instructions.
Live Cell Imaging
After 20–24 h of transfection, cells were washed on glass coverslips with a modified Krebs-Ringer solution, containing 120 mm NaCl, 4.7 mm KCl, 1.2 mm CaCl2, 0.7 mm MgSO4, 10 mm glucose, 10 mm Na-Hepes, pH 7.4, and the coverslip was placed into a metal chamber (Atto, Invitrogen) that was mounted on a heated stage (35 °C or room temperature for HEK293-AT1 cells). Cells were incubated in 1 ml of the Krebs-Ringer buffer, and the stimuli were dissolved and added in 200 μl of warm buffer removed from the cells. Cells were examined in an inverted microscope. Confocal images were obtained with a Zeiss LSM 510-META laser confocal microscope (Carl Zeiss MicroImaging, Inc.) using a 63× oil-immersion objective equipped with an objective heater (Bioptech).
Measurements of Receptor-stimulated PtdIns(4,5)P2 Kinetics Using Tubby Domain Translocation
HEK293-AT1 cells were transfected with the Tubby domain-GFP construct. After 24 h, cells were mounted in the Atto chambers and imaged in a Zeiss LSM 510-META confocal microscope at room temperature. After stimulation with 100 nm AngII, the translocation of the GFP probe was monitored in time-lapse imaging. The translocation of the construct from the membrane to the cytosol was quantified by measuring cytosolic GFP intensity in regions of interest outside the nucleus and plotted against time using the Zeiss image processing software. Intensity curves were normalized to prestimulatory values, and they were averaged from a large number of cells in recordings obtained in several dishes in multiple independent experiments.
Measurements of Receptor-stimulated PtdIns(4,5)P2 Kinetics using FRET with the PLCδ1-PH Domain
HEK293-AT1 cells were transfected with the CFP- and YFP-tagged PLCδ1-PH constructs for 24 h. After transfection, cells were mounted in the Atto chambers and imaged in an Olympus IX70 microscope equipped with a Micromax 1024BFT (Princeton Instruments) camera, a Lambda-DG4 illuminator (Sutter), and a beam-splitter (Optical-Insights) that contained the filter set for recording YFP and CFP fluorescence. Data acquisition and processing were done with the MetaFluor software. The ratios of YFP to CFP for individual cells were calculated normalizing to their initial values before stimulation. Recordings from several cells in each dish and several dishes from separate experiments were pooled and averaged.
Analysis of myo-[3H]Inositol- or [32P]Phosphate-labeled Lipids
HEK293-AT1 cells plated on 12-well plates (at a density of 30,000 cells/ml) were labeled with myo-[3H]inositol (20 μCi/ml) in 1 ml of inositol-free DMEM supplemented with 2% dialyzed FBS for 24 h or with 2 μCi/ml o-[32P]phosphate for 3 h in phosphate-free DMEM supplemented with 2% dialyzed FBS. Cells were treated with various concentrations of the drugs (dissolved in DMSO) for 10 min and stimulated with angiotensin II (100 nm) for the indicated times or left unstimulated. Reactions were terminated by the addition of ice-cold perchloric acid (5% final concentration), and cells were kept on ice for 30 min. After scraping and freezing/thawing, the cells were centrifuged, and the cell pellet was processed to extract the phosphoinositides by an acidic chloroform/methanol extraction followed by thin layer chromatography (TLC) essentially as described previously (10, 28). TLC plates were sprayed with Enhance solution (PerkinElmer Life Sciences) and were subjected to autoradiography (in the case of 3H labeling) or were analyzed by a PhosphorImager (for the 32P-labeled samples). Autographic films were exposed several times to find the proper exposure for densitometric analysis.
Immunostaining for PM PtdIns(4)P and PtdIns(4,5)P2
A modified protocol (29, 30) was used. COS-7 cells were grown to ∼50% confluence on poly-l-lysine-coated 8-well test slides and then treated for 10 min with the indicated concentration of PI4K inhibitor in 0.1% DMSO/phenol red-free DMEM in a total volume of 25 μl at 37 °C, 10% CO2 in a humidified atmosphere. The cells were then rapidly fixed by the addition of a further 25 μl of PIPES buffer (137 mm NaCl, 2.7 mm KCl, 20 mm Na-PIPES, pH 6.8) containing 0.4% (w/v) formaldehyde and 0.4% (w/v) glutaraldehyde and incubated for 10 min at room temperature before placing the cells on ice for all subsequent steps. Autofluorescence was quenched by three 2-min washes in freshly prepared 1% (w/v) sodium borohydride in PIPES buffer. Cells were then stained for 30 min sequentially in ice-cold blocking buffer (PIPES buffer supplemented with 0.5% saponin and 5% normal goat serum) containing 1.25 μg/ml anti-PtdIns(4)P IgM and 50 nm GST-PH-PLCδ1, followed by 2.5 μg/ml rabbit polyclonal anti-GST in the continued presence of anti-PtdIns(4)P. After two rinses in ice-cold PIPES buffer, cells were stained for a further 30 min in ice-cold blocking buffer containing 5 μg/ml each of AlexaFluor647-conjugated anti-rabbit IgG and AlexaFluor555-conjugated anti-mouse IgM, as well as 5 units/ml AlexaFluor488-conjugated phalloidin (Life Sciences). Finally, cells were rinsed four times in ice-cold PIPES buffer, post-fixed for 10 min in ice-cold 4% (w/v) formaldehyde in PIPES buffer, rinsed three times in PIPES buffer, once in water, and mounted in ProLong Gold containing DAPI (Life Sciences).
Images were acquired on a Zeiss LSM 780 using a 20 × 0.8 NA air objective with the confocal pinhole fully opened to capture fluorescence from the entire depth of the cells. For quantitative analysis, the images were automatically analyzed by using an automated pipeline that used the open access package Cell Profiler (31) and by using a pipeline that used DAPI and phalloidin staining to identify and segment individual cells, as described previously (30). For each experiment, background fluorescence intensity (measured for cells whereby primary lipid probes were omitted) was subtracted. Individual intensity values were then normalized to the mean intensity value for the 0.1% DMSO control condition. These normalized intensity values were pooled to create the grand mean from independent experiments. Data were plotted in Prism 5 (Graphpad), and fit with the function shown in Equation 1,
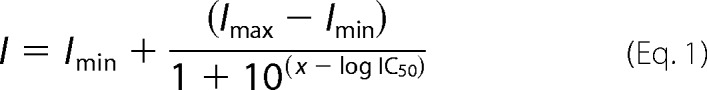 |
where I is the normalized mean pixel intensity, and x is log[inhibitor].
In Vitro PI Kinase and PIP 5-Kinase Measurements
Enzymes were prepared from COS-7 cells expressing the respective kinases epitope-tagged with an HA, FLAG, or Myc tag at their N termini. Proteins were immunoprecipitated from the cell lysates, and after several washes, their activity was measured on agarose beads. The activities of PI4Ks were measured as incorporation of radioactivity from [γ-32P]ATP into organic solvent-extractable material (32). The standard reaction mixture for PtdIns 4-kinase (50 μl final volume) contained 50 mm Tris/HCl, pH 7.5, 20 mm MgCl2, 1 mm EGTA, 1 μm PtdIns, 0.4% Triton X-100, 0.5 mg/ml BSA, 100 μm [γ-32P]ATP (2-μCi per reaction), and the enzyme. All assay components, except [γ-32P]ATP, were preincubated with inhibitors for 10 min at 30 °C. Inhibitors were dissolved in DMSO, which was also used in the control samples. Reactions were started by addition of [γ-32P]ATP, incubated for 10–30 min, and terminated by the addition of 3 ml of CHCl3/CH3OH/concentrated HCl (200:100:0.75 (v/v). Reactions were terminated, lipids extracted, and their activity measured by scintillation counting essentially as described previously (32).
The activity of PIP 5-kinases was measured as incorporation of [γ-32P]ATP into PtdIns(4)P. The kinase reaction was carried out in a 50-μl reaction volume containing 50 mm Tris, pH 7.5, 30 mm NaCl, 5 μCi of [γ-32P]ATP (50 μm final), 10 mm MgCl2, 67 μm PtdIns(4)P, and 133 μm phosphatidylserine. The reaction was initiated by adding ATP and carried out for 20 min. Reactions were terminated by addition of 100 μl of 1 m HCl and then extracted with 250 μl of CHCl3/MeOH (1:1) twice. Finally, lipids in organic phase were dried and quantified by scintillation counting.
Inhibition of HCV Replication
Compounds were assayed for activity against HCV using the genotype 1a, 1b (ET cell line), and 2a (Lunet cell line) subgenomic NS3-NS5B replicon model systems as described recently (33).
Conditional Knock-out Mice Studies
Cre-lox technology was used to generate a temporally controlled, conditional knock-out (cKO) of the Pi4ka gene. Standard gene targeting approaches were used to generate BA1 embryonic stem cells (hybrid C57BL/6 × 129/SvEv) heterozygous for the Pi4ka primary targeted allele (see Fig. 1 for details). An 8.95-kb region used to construct the targeting vector was subcloned from a C57BL/6 BAC clone. The 2-kb short homology arm and the 6-kb long homology arm were placed 5′ and 3′, respectively, of a loxP/FRT-flanked neomycin resistance cassette in the 3′–5′ orientation, together with the “target” region containing a further loxP element positioned 3′ to exon 48. Homologous recombination in neomycin-resistant ES cells was confirmed at the 5′ and 3′ ends by Southern blot analysis using NsiI-digested ES cell-derived genomic DNA and probes external to the homology arms, and the presence of the 3′ loxP element was confirmed by PCR (data not shown). Correctly targeted ES cell clones were subsequently injected into C57BL/6-derived blastocysts. Resultant male chimeras were crossed with C57BL/6 female mice carrying an Flp recombinase transgene to remove the neomycin resistance cassette, and the resulting mice heterozygous for the Pi4ka conditional allele were bred to wild-type mice to remove the Flp recombinase transgene. Standard breeding procedures were then used to generate study populations of mice homozygous for the Pi4ka conditional allele and carrying the R26MCM transgene. The latter transgene expresses the MerCreMer cDNA (34) from the Rosa26 locus (35) and enables tamoxifen-inducible, ubiquitous Cre recombinase-mediated deletion of the floxed region of conditional alleles in mice. Littermates homozygous for the conditional Pi4ka allele were used as controls.
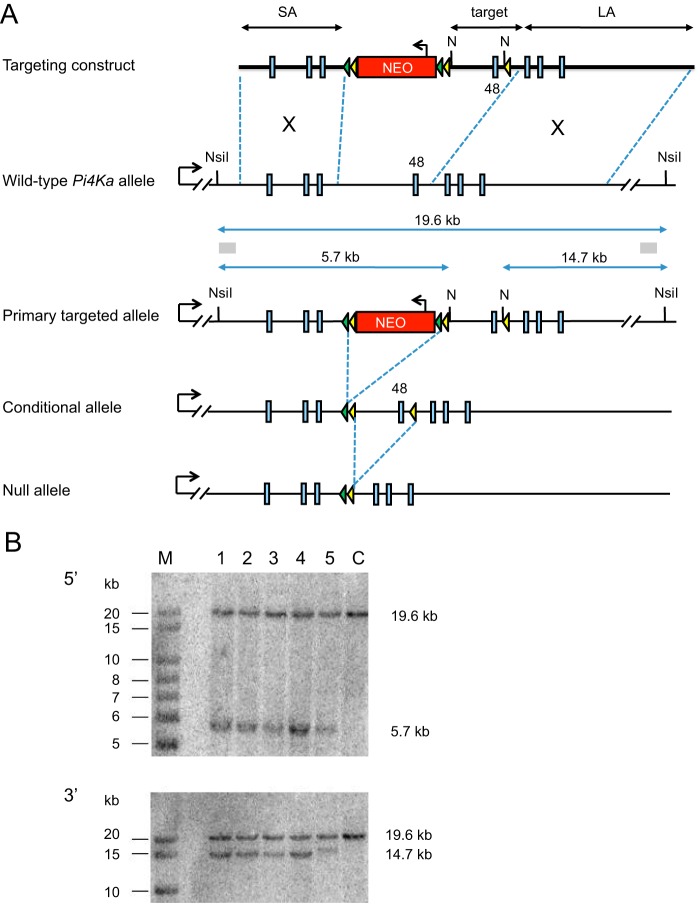
FIGURE 1.
Design and strategy of generating a conditional pi4ka allele. A, homologous recombination in ES cells was used to generate ES cells heterozygous for the Pi4ka primary targeted allele. The structure of the Pi4ka allele in the region of exon 48 is presented, with exons indicated as blue boxes. The structure of the targeting construct is shown with the short and long homology arms (SA and LA, respectively), the exon 48-containing target region, the neomycin selection cassette, and loxP and FRT elements (yellow and green arrowheads, respectively). The positions of the NsiI restriction sites (N), probes (gray boxes), and band sizes indicative of the wild-type and targeted Pi4ka allele used in the Southern blot screening strategy are indicated. The resultant primary targeted allele and the structure of the conditional and null alleles are shown following, respectively, Flp and Cre recombinase-mediated deletion. B, representative Southern blots using the 5′ and 3′ probes indicated in A. Genomic DNA from targeted ES cell clones (lanes 1–5), parental ES cell control DNA (C), and molecular size markers (M). Expected band sizes of the wild-type and targeted allele are indicated.
To induce MerCreMer recombinase activity, study mice were dosed orally once daily for 5 consecutive days with tamoxifen (1 mg/5 g body weight) dissolved in corn oil; control groups were dosed likewise with corn oil. A standard washout period of 17 days was applied to all recombinase activation studies to allow for any initial effect of dosing tamoxifen and corn oil to subside. All animal procedures were reviewed and approved by the GlaxoSmithKline Animal Care and Use Committee and were performed in accredited facilities in accordance with institutional guidelines and the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Research Council).
Animal Studies Using PI4KA Inhibitors
A 14-day oral toxicity study was performed in mice with the F1 compound. The compound was formulated as a solution in 30% solutol, 70% polyethylene glycol and administered to mice at a dose volume of 10 ml/kg/day. It was given to male mice (8/group) at doses of 0 (vehicle), 3, 10,4 20, and 404 mg/kg/day twice daily and 6 h apart for 74 or 14 days by oral gavage. These doses were based on the in vitro protein adjusted EC90 (0.16 μm) and a preliminary dose escalation study at doses of 3, 10, and 50 mg/kg (data not shown). All living animals were humanely euthanized upon completion of the study, and an exhaustive histopathology was performed, and sections were examined for any abnormalities.
RESULTS
Synthesis of Potent PI4KA-selective Compounds of Different Chemotypes
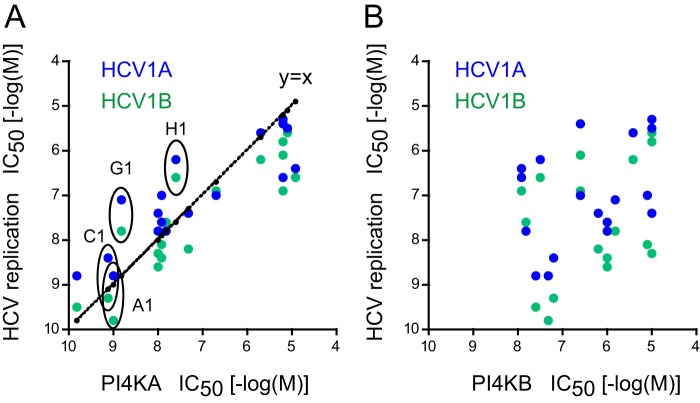
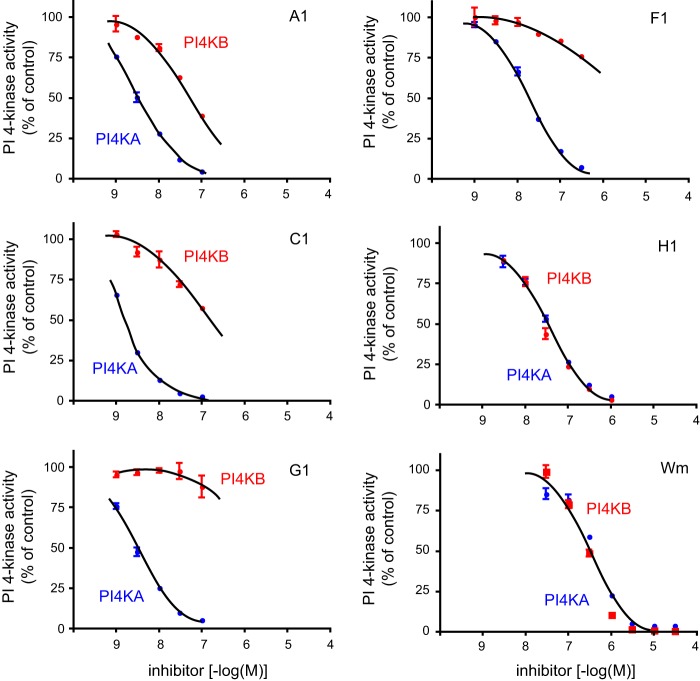
High throughput screening for inhibitors that inhibit HCV replication and also potently inhibit PI4KA has been the starting point in the generation of more selective PI4KA inhibitors (see Ref. 33 for more details). The first iteration of these compounds showed potent ATP-competitive inhibition of PI4KA but little selectivity against PI4KB and class I PI3Ks (33). Several rounds of structure-activity relationship studies using compounds with chemical substitutions in the original hits yielded inhibitors with high potency and selectivity (33). These were evaluated for their ability to inhibit PI4KA (using ADP-Glo assay) and HCV replication. These compounds were also tested against a whole set of PI kinases (Table 1) and subjected to KiNativ in situ kinase profiling to determine their effects on protein kinases. These analyses showed that several compounds had good selectivity and high potency against PI4KA with no effect on protein kinases. The antiviral effect showed very close correlation with the inhibitory potency against PI4KA (r2 = 0.758, p < 0.0001, and r2 = 0.797, p < 0.0001, for HCV1A and HCV1B, respectively) (Fig. 2A). In contrast, the potencies of the same compounds against PI4KB showed no correlation with HCV inhibition (Fig. 2B). A selected set of compounds (Fig. 3) was further tested in conventional PI4K lipid kinase assays based on PtdIns phosphorylation using 32P-labeled ATP and epitope-tagged PI4Ks expressed in COS-7 cells and immunoisolated. These experiments confirmed the IC50 values determined in the nonradioactive assays for PI4KA and PI4KB (Fig. 4 and Table 1). The compounds were also tested on the type II PI4Ks and on the three major forms of PIP 5-kinases (α, β, and γ) and were found to have no effect at a concentration of 10 μm (Table 1).
TABLE 1.
Inhibitory potencies of selected PI4KA inhibitors on PI3K and PI4K used in this study as measured with ADP-Glo or by [γ-32P]ATP in PI kinase assays (boldface)
IC50 values are expressed as −log (m). Note that larger numbers indicate more potent inhibition. ND means not determined in this study.
| Compound | PI4KA | PI4KB | PI3KA | PI3KB | PI3KG | PI3KD | PI4K2A | PI4K2B |
|---|---|---|---|---|---|---|---|---|
| A1 | 9.8/8.5 | 7.7/7.2 | 7.3 | 7 | 7.8 | 7.1 | <5 | <5 |
| C1 | 9.1/8.8 | 7.2/6.8 | 8.1 | 7.1 | 8.2 | 8.6 | <5 | <5 |
| F1 | 8.0/7.8 | 5.9/6.0 | 5.8 | 5.9 | 5.9 | 6.4 | <5 | <5 |
| G1 | 8.7/8.6 | 5.9/6.2 | 7.2 | 7.6 | 7.5 | 8.0 | <5 | <5 |
| H1 | 7.7/7.5 | 7.6/7.6 | 5.9 | 5.8 | 5.9 | 5.7 | <5 | <5 |
| Wm | 6.4 | 6.5 | ND | ND | ND | ND | <5 | ND |
FIGURE 2.
Inhibition of HCV replication by inhibitors of PI4KA. HCV replication was tested as described previously (33). A shows a tight correlation between the potencies of drugs against PI4KA and inhibition of HCV replication using two different HCV strains (r2 = 0.758, p < 0.0001, and r2 = 0.797, p < 0.0001, for HCV1A and HCV1B, respectively). B shows that no correlation was found with the PI4KB potencies and HCV replication with the same set of compounds (r2 = 0.149, p = 0.114, and r2 = 0.057, p = 0.341, for HCV1A and HCV1B, respectively). Compounds selected for future analysis are labeled in A.
FIGURE 3.
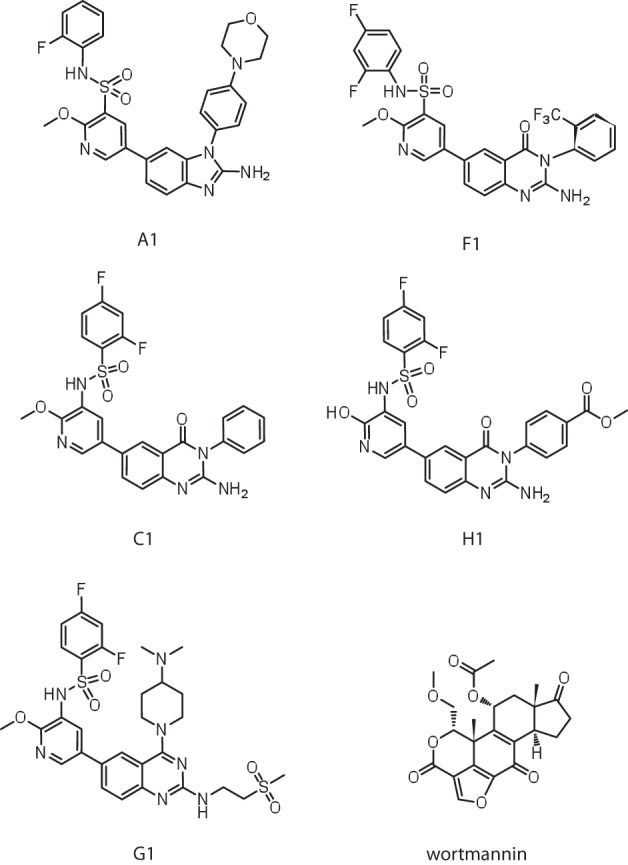
Structure of PI4KA inhibitors most characterized in this study.
FIGURE 4.
Inhibition of immunoisolated PI4KA and PI4KB by a selected set of PI4KA inhibitors in vitro. Enzymes were prepared from COS-7 cells expressing the respective kinases epitope tagged with an HA tag at their N termini. Proteins were immunoprecipitated from the cell lysates, and after several washes their activity was measured on agarose beads. The activities of PI4Ks were measured as incorporation of radioactivity from [γ-32P]ATP into organic solvent-extractable material. Activities were expressed as percent of DMSO-treated controls. Means ± range from duplicate determinations are shown from a representative experiment. Blue, PI4KA; red, PI4KB.
Inhibition of PI4KA Limits PtdIns(4)P Production in the PM
Next, we moved to intact cells to test the effects of a selected set of compounds on PtdIns(4)P levels using HEK293 cells stably expressing the AT1 angiotensin receptors (HEK-AT1). We used [3H]inositol labeling for 24 h to label the PtdIns(4)P and PtdIns(4,5)P2 pools to near equilibrium prior to adding the inhibitors for 10 min to see changes in the overall amounts of these lipids. These data showed that 10 min of treatment with the inhibitors potently decreased the levels of PtdIns(4)P with a negligible effect on PtdIns(4,5)P2, but the efficacy of the compounds showed significant differences. The most potent of these inhibitors was a compound we called A1, and the least potent was H1 with IC50 values ranging between 3 nm and 10 μm (Fig. 5, A and B and see below).
FIGURE 5.
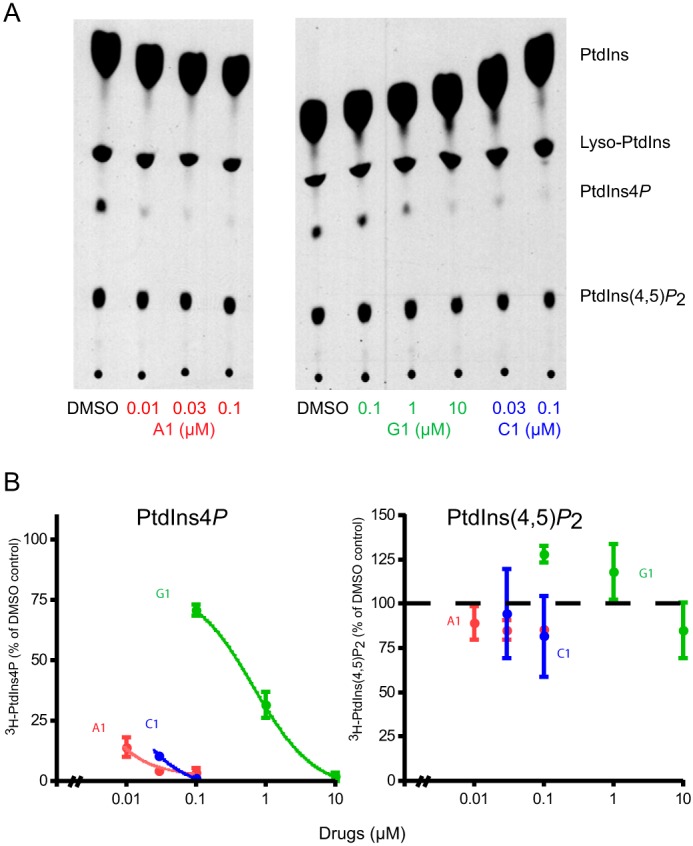
Effect of PI4KA inhibitors on PtdIns(4)P and PtdIns(4,5)P2 levels in [3H]inositol-labeled HEK293 cells. Cultured HEK293-AT1 cells were labeled with myo-[3H]inositol for 24 h and treated with the indicated concentrations of PI4KA inhibitors for 10 min. Cells were then denatured by the addition of 5% final perchloric acid, and labeled lipids were extracted and separated by TLC as detailed under “Experimental Procedures.” TLC plates were sprayed with ENHANCE and subjected to autoradiography. Different exposures were used to suit densitometry. A, TLC plates are shown from a representative experiment. B, results of densitometry from the same experiment performed in duplicate. Similar data were obtained in another experiment.
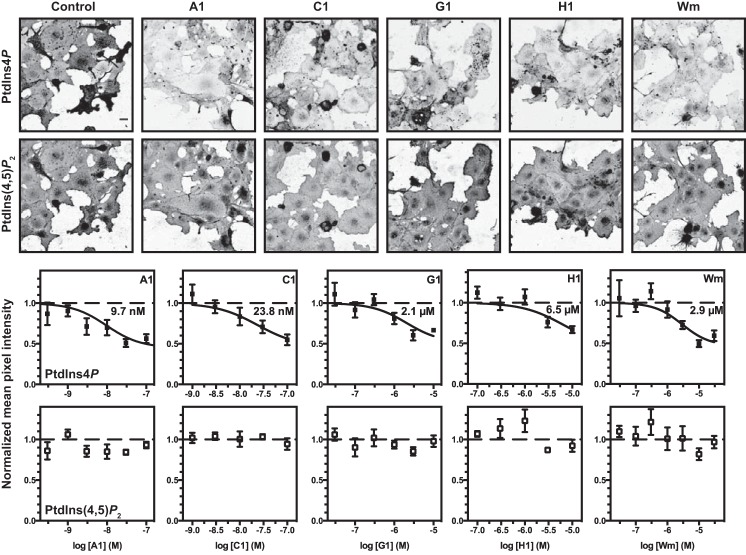
Because [3H]inositol labeling cannot distinguish between lipids found in the various compartments, we used quantitative immunofluorescence staining of cells treated with the compounds. We have previously identified specific fixation and permeabilization conditions that permit selective labeling of plasma membrane inositol lipids with antibodies or recombinant lipid-binding domains (29, 30). Note that relatively flat COS-7 cells do not usually exhibit a peripheral “ring” of plasma membrane but rather a continuous but uneven sheet-like appearance covering the entire cell. More rounded cells often appear brighter because they occupy a smaller footprint with more lateral membrane, especially in the case of the rounded mitotic cells present in most fields shown in Fig. 6. This is particularly apparent when using lower NA objectives without confocal optical sectioning, as shown in Fig. 6. Nonetheless, capturing the entire membrane-associated staining permits robust quantification (29, 30). As shown in Fig. 6, COS-7 cells stained with a PtdIns(4)P-specific antibody (29, 30) showed that this PM PtdIns(4)P pool was depleted with increasing inhibitor concentrations with a potency that was consistent with the 3H-labeling data (Fig. 6). Importantly, again, no effect was observed on PtdIns(4,5)P2 levels stained with the PLCδ1-PH domain in the same cells (Fig. 6).
FIGURE 6.
Effect of selected PI4KA inhibitors on plasma membrane PtdIns(4)P and PtdIns(4,5)P2 levels measured in fixed and immunostained COS-7 cells. Representative images of COS-7 cells treated for 10 min with 100 nm A1, 100 nm C1, 10 μm G1, 10 μm H1 or 30 μm Wm, as indicated, and then fixed and stained for PtdIns(4)P and PtdIns(4,5)P2. Graphs show the mean pixel intensity (normalized to vehicle controls) for the indicated concentration of each compound. The fitted IC50 value is indicated on each graph. Data are grand means ± S.E. of 4–5 independent experiments. The bar indicates 20 μm.
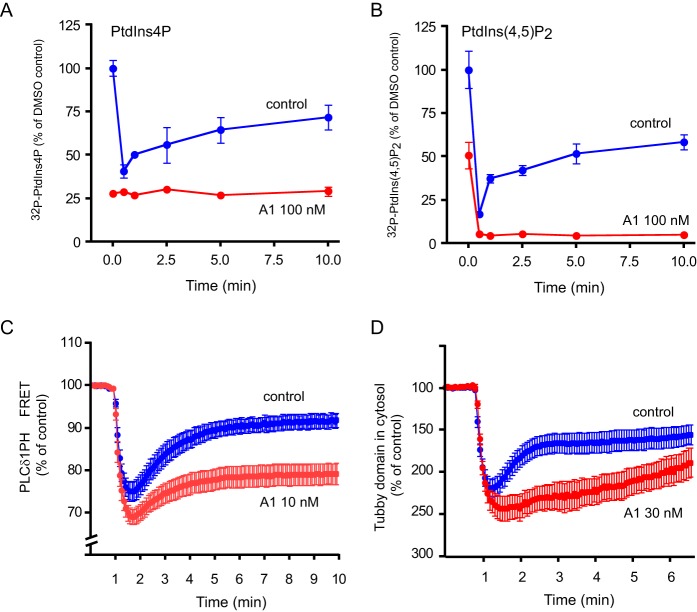
These data confirmed our earlier observation that PtdIns(4,5)P2 levels can be maintained in quiescent cells at a wide range of PtdIns(4)P concentrations (30). However, our previous studies also showed that wortmannin (Wm) or phenylarsine oxide treatment prevents the resynthesis of PtdIns(4,5)P2 during PLC activation (10, 30, 36). Although these inhibitors are not selective, their effect was attributed to inhibition of PI4KA in those studies. To test if selective PI4KA inhibitors indeed prevent the maintenance of PtdIns(4,5)P2 during PLC activation, we used HEK293-AT1 cells and stimulation with angiotensin II (AngII). We used several approaches to follow PtdIns(4,5)P2. First, we used cells labeled with [32P]phosphate for 3 h prior to stimulation with AngII, a condition we have previously used in these cells (10). Second, we used the PLCδ1-PH domain in FRET experiments and the Tubby domain in confocal studies to monitor PtdIns(4,5)P2 during stimulation. These protein domains are widely used as PtdIns(4,5)P2 reporters (37). As shown in Fig. 7A, in 32P-labeled cells pretreatment with 100 nm A1 for 10 min completely depleted PtdIns(4)P prior to AngII stimulation when compared with untreated control cells. As observed before, control cells showed a typical rapid depletion of PtdIns(4)P upon stimulation. There was no effect of AngII treatment on the already depleted PtdIns(4)P pools in cells pretreated with the A1 compound (Fig. 7A). As expected, stimulation with AngII rapidly depleted PtdIns(4,5)P2 followed by a partial replenishment of these pools in control cells (Fig. 7B). In contrast, AngII stimulation depleted PtdIns(4,5)P2 even more completely, and no replenishment was observed in A1-treated cells (Fig. 7B).
FIGURE 7.
Effect of PI4KA inhibitors on PtdIns(4)P and PtdIns(4,5)P2 levels in [32P]phosphate-labeled HEK293 cells or measured by PtdIns(4,5)P2-binding domains. A and B, cultured HEK293-AT1 cells were labeled with [32P]phosphate for 3 h and pretreated with the indicated concentrations of PI4KA inhibitors for 10 min before stimulation with AngII (10−7 m) for the indicated times. Reactions were stopped and labeled lipids analyzed as described in legend to Fig. 5. TLC plates were analyzed by a PhosphorImager and PtdIns(4)P (A) and PtdIns(4,5)P2 (B) were plotted as percent of activity obtained in control cells before AngII stimulation. Mann-Whitney test was used to compare the control and A1-treated groups, which showed a significant difference for the entire time period (p = 0.0022 for A and 0.0152 for B). Representative data are shown from one of two experiments performed in duplicate yielding identical results. C, FRET analysis of PtdIns(4,5)P2 changes using YFP- and CFP-tagged PLCδ1PH domains in HEK293-AT1 cells. See under “Experimental Procedures” for details. The ratios of YFP/CFP fluorescence were normalized to prestimulatory values. Note the truncated scale of the ordinate. Means ± S.E. of 23–25 cell recordings are shown obtained in five separate measurements (p < 0.0001 with Mann-Whitney test applied to the entire time period). D, using the Tubby domain-GFP translocation from the membrane to the cytosol as an index of PtdIns(4,5)P2 changes in the PM. HEK293-AT1 cells were transfected with the Tubby domain-GFP construct (27), and 24 h later cells were examined with confocal microscopy. Cells were pretreated with the inhibitors for 10 min before stimulation with AngII. Fluorescence changes in the cytosol were followed in time-lapse images in regions of interest placed outside the nucleus. Intensities were normalized to pre-stimulation values, and increases were plotted downward to better reflect the lipid changes in the membrane. Means ± S.E. of 10–13 cells are shown from three independent measurements from a representative experiment (p < 0.0001 with Mann-Whitney test applied to the entire time period).
The effects of the inhibitors were further tested on PtdIns(4,5)P2 kinetics using different approaches. The PH domain of PLCδ1 and the Tubby domain of the Tubby protein fused to fluorescent proteins have been used as probes for PtdIns(4,5)P2 in the membrane (27). To quantitate the lipid changes, we used both FRET analysis (in the case of the PH domain) (38) or increased intensity of fluorescence in the cytosol followed in confocal microscopy (in the case of the Tubby domain; note the inverted scale of the ordinate!). Both of these measurements showed the transient decrease in the level of PtdIns(4,5)P2 in the PM after AngII stimulation in control cells (Fig. 7, C and D, blue traces) and the severely impaired resynthesis of PtdIns(4,5)P2 in the PM in the A1-pretreated cells (Fig. 7, C and D, red traces). These results collectively showed that under acute and complete inhibition of PI4KA, cells are unable to maintain PtdIns(4,5)P2 levels against a robust PLC activation. However, PtdIns(4,5)P2 levels can be maintained in quiescent cells even when the PM PtdIns(4)P pools are largely reduced.
PI4KA Inhibitors Show a Wide Range of Potencies When Tested in Cellular Settings
Several PI4KA inhibitors of a similar chemotype were then tested to determine their ability to inhibit PtdIns(4,5)P2 synthesis during PLC activation. For this, we used HEK-AT1 cells labeled with [32P]phosphate for 3 h and stimulated with AngII for 10 min after a 10-min pretreatment with DMSO or varying concentrations of the selective PI4KA inhibitors. Because PtdIns(4,5)P2 levels return to about 75% of their initial value by 10 min of stimulation (see Fig. 7) and this replenishment of the pool is almost completely prevented by PI4KA inhibition (Fig. 7, B–D), this protocol provided a sensitive means to assess the potency of the inhibitors. As shown in Fig. 8, a large variation was observed between the potencies of the inhibitors to inhibit PtdIns(4,5)P2 resynthesis. The most potent inhibitor was A1 with an IC50 of about 3 nm, whereas the least active of these drugs was H1 with an IC50 of 30 μm. This analysis was then extended with additional compounds for which the IC50 for PI4KA and PI4KB inhibition was determined with the [32P]ATP-based PI4K assays. When the IC50 values for the in vitro kinase assays were plotted against the IC50 for PtdIns(4,5)P2 inhibition, it was apparent that, except for A1, most inhibitors were significantly less potent in the cellular assay. This finding was partially explained by the fact that even a fraction of the kinase activity can support PtdIns(4,5)P2 synthesis as concluded by earlier studies (10). However, it did not explain the fact that significant differences in cellular potencies were found within compounds that inhibited PI4KA in vitro with almost identical potencies (see J1, M1, and H1). This was not due to additional inhibition of PI4KB, because H1, which inhibited PI4KA and PI4KB equally in vitro and more potently than Wm, was less potent than Wm in the cellular assay. Part of the poor cellular potency must lie in the bioavailability of the compounds in the cellular compartment where they need to act, and indeed, the compounds that deviated the most from their in vitro potencies for PtdIns(4,5)P2 synthesis were also the outliers in their ability to inhibit HCV replication (see Fig. 1). Comparison of the potencies of the compounds on HCV replication and PtdIns(4,5)P2 inhibition also confirmed previous conclusions that viral replication is more sensitive to kinase inhibition, whereas PtdIns(4,5)P2 synthesis can be maintained even at a fraction of the total kinase activity.
FIGURE 8.
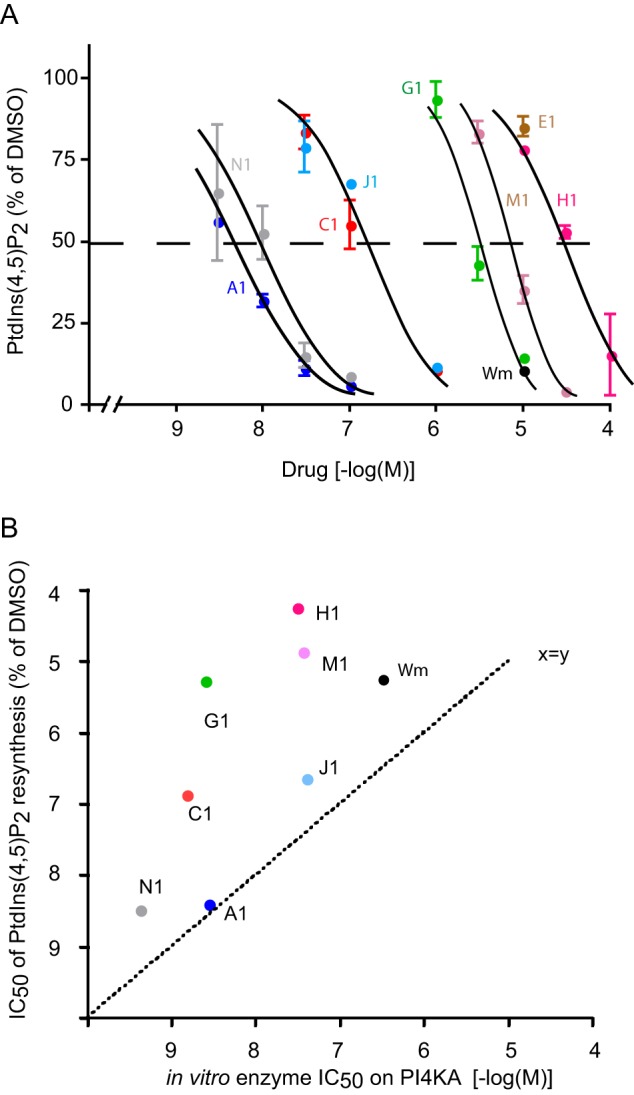
Comparison of IC50 values of PI4KA inhibitors for inhibiting PtdIns(4,5)P2 resynthesis in HEK293 cells and the PI4KA enzyme in vitro. A, HEK293-AT1 cells were labeled with [32P]phosphate for 3 h and pretreated with the indicated concentrations of PI4KA inhibitors for 10 min before stimulation with AngII (10−7 m) for an additional 10 min. Reactions were stopped, and labeled lipids were extracted and separated by TLC following quantification by a PhosphorImager. All PtdIns(4,5)P2 values were expressed as percent of the value from AngII-treated cell with DMSO pretreatment. This 100% value is about 60–70% of the initial pre-stimulatory PtdIns(4,5)P2 value in control cells (see Fig. 7B). Means ± S.E. are shown from 2 to 4 experiments performed in duplicate. Curve fitting and IC50 values were obtained as described under “Experimental Procedures.” B, comparison of the IC50 values determined in cells (from A) plotted on y axis, and in vitro kinase assays for PI4KA (from Fig. 4) plotted on x axis. Note the upward deviation of points (except for A1) from the x = y line indicating that the enzyme has to be inhibited almost completely to achieve inhibition of PtdIns(4,5)P2 resynthesis. It also shows that some of the drugs are less effective to reach the enzyme within the cell.
Cells Tolerate PI4KA Inhibition for Prolonged Periods without Significant PtdIns(4,5)P2 Depletion
Given the importance of PI4KA in the maintenance of plasma membrane PtdIns(4,5)P2 levels during robust agonist stimulation, we expected that cells would not tolerate prolonged treatments with PI4KA inhibitors. Also, we anticipated that cells growing in culture and subjected to an increased PLC activity and membrane remodeling would eventually get depleted in PtdIns(4,5)P2. However, surprisingly, HEK293 or COS-7 cells treated with A1 (30 nm) for 1 day showed no signs of change in their PtdIns(4,5)P2 levels or distributions as judged by expressed GFP-tagged PLCδ1PH or Tubby domain (data not shown). These results suggested that PI4KA may not be the sole source of PM PtdIns(4)P for PtdIns(4,5)P2 synthesis.
PI4KA Inhibitors Vary in Their Adverse Effects in Whole Animals
At this point only a 14-day oral toxicity study was performed in mice to test potential adverse effects of PI4KA inhibition in whole animals. It was performed with the F1 compound, which is the same chemotype as A1 but has been proven to have markedly better pharmacokinetic properties required to evaluate this chemotype in vivo. The compound was formulated as a solution in 30% solutol, 70% polyethylene glycol and administered to mice at a dose volume of 10 ml/kg/day. It was given to male mice (8/group) at doses of 0 (vehicle), 3, 10,4 20, and 404 mg/kg/day twice daily and 6 h apart for 74 or 14 days by oral gavage. These doses were based on the in vitro protein adjusted EC90 (0.16 μm) and a preliminary dose escalation study at doses of 3, 10, and 50 mg/kg (data not shown).
One mouse given 3 mg/kg/day and another mouse given 20 mg/kg/day were found dead on days 12 and 8, respectively, with no clinical signs noted. Four mice given 40 mg/kg/day were either found dead or were humanely euthanized in a moribund condition between days 11 and 14. Clinical signs for these mice included decreased activity, loss of skin elasticity, rough coat, eyes partially closed, hunched posture, cold to the touch, slow breathing, loose/watery feces, wet skin/fur, and staining in the urogenital/anal area. One animal had limited use of its forelimbs on day 12 prior to becoming moribund. All of these animals had slight body weight loss from day 1 to day 11 (0.78×) when compared with their pretest bodyweights. Clinical signs in animals that survived to terminal necropsy included loss of skin elasticity, loose/watery feces, decreased activity, rough coat, hunched posture, and brown/orange staining of the anal area. Animals treated with F1 at 40 mg/kg/day exhibited morbidity and mortality and test article-related changes in the stomach, thymus, spleen, and clinical pathology parameters. These included nonglandular gastric lesions present in all animals given 40 mg/kg/day characterized by combinations of erosion/ulceration, squamous epithelial cell hyperplasia and hyperkeratosis, lamina propria edema, a mixed inflammatory cell infiltrate, and occasional lamina propria vessels with medial degeneration/necrosis (data not shown). Individual mice at 40 mg/kg/day also had a gastric ulcer in the pylorus and multifocal foveolar cell necrosis in the glandular stomach. It is important to note that F1 does not cross the blood-brain barrier, which may explains the lack of CNS symptoms.
Several other potent PI4KA inhibitory compounds were then tested in animal studies for adverse effects. They all caused sudden death before any pathology could develop. These included C1, M1, and J1. These animals all appeared to die from a cardiovascular collapse. The toxicity of these compounds correlated with their ability to inhibit PtdIns(4,5)P2 resynthesis in the cellular assay. Because these compounds severely affected Gq signaling based on cellular data, it is expected that they cause pleiotropic effects in whole animals. These will require a lot more detailed analysis, including ruling out off-target effects.
Genetic Ablation of Pi4ka in Adult Animals Causes Lethality
To evaluate the importance of the kinase in whole animal models, cKO mice were generated by targeting exon 48 in the Pi4ka gene. Deletion of this exon generates a catalytically inactive truncated protein. (The targeting strategy is shown in Fig. 1, and a detailed description of the procedure is found under “Experimental Procedures.”) Homozygous mice were bred with a mouse line that carried a ubiquitous tamoxifen-inducible Cre transgene (34, 35) to generate homozygous conditional alleles and heterozygous inducible Cre transgenes. Mice were dosed orally once daily for 5 consecutive days with tamoxifen (or treated identically without tamoxifen) followed by a 17-day waiting period.
Pi4ka cKO mice were found moribund by day 7 post-tamoxifen treatment. These animals were humanely euthanized, and histopathology exhibited epithelial cell degeneration/necrosis (Fig. 9) with evidence of regeneration in the stomach, small intestine, and large intestine. Tamoxifen pharmacokinetics showed high levels in the gut and other organs, with little exposure in the CNS (data not shown). No gastrointestinal abnormalities were detected in animals in the corn oil (vehicle) control, tamoxifen-treated (not cKO), and cKO mice not treated with tamoxifen groups. These findings were similar to those reported in a recent cKO pi4ka mouse study (22).
FIGURE 9.
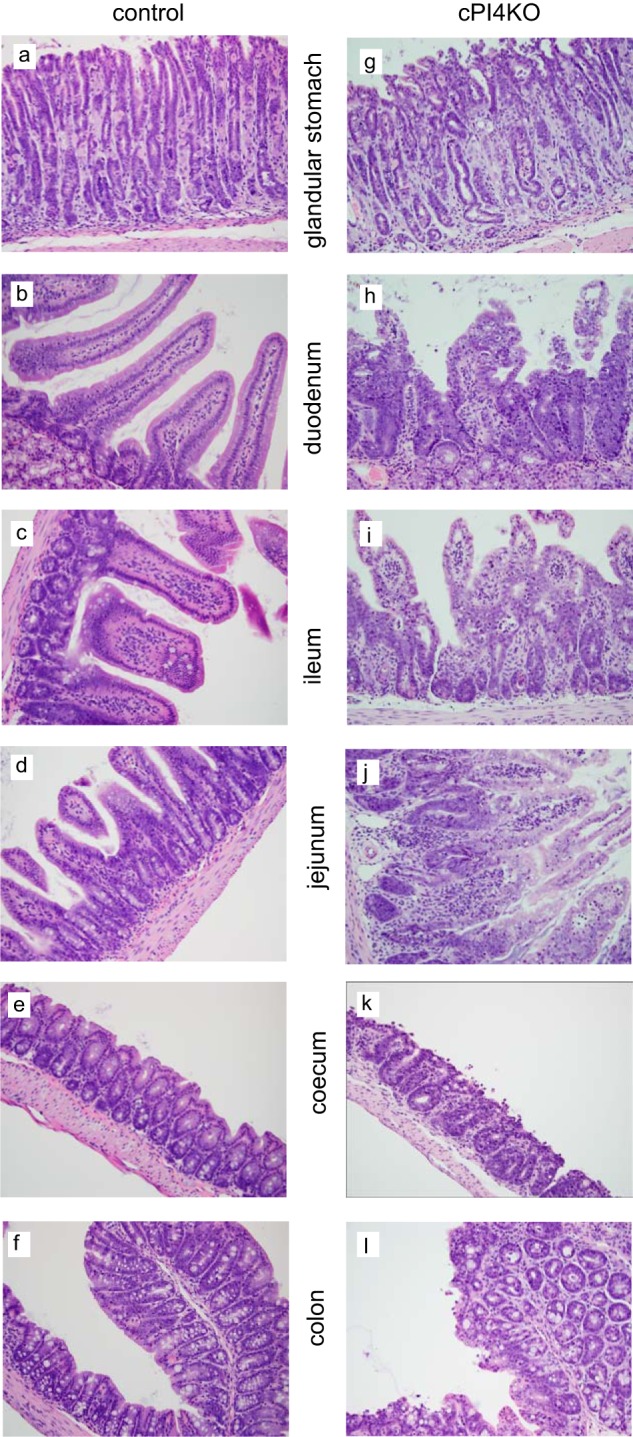
Histochemical analysis of selected segments of the GI track in wild-type and conditional pi4ka knock-out (cPI4KO) mice after tamoxifen treatment. Hematoxylin-eosin staining of the glandular stomach, duodenum, ileum jejunum, cecum, and colon. Panels a versus g, note the degeneration/necrosis of glandular pit, foveolar, and neck epithelial cells and lamina propria edema. Cellular debris is present within gastric pits in the cPI4KO animals. Panels b and c versus h and i, note the degeneration/necrosis of enterocytes on the villi and within intestinal crypts, crypt regeneration/hyperplasia (increased mitoses and cytoplasmic basophilia), and lamina propria edema in the cPI4KO animals both in the duodenum and ileum. Similar pathologies were observed in the jejunum, cecum, and colon (shown in panels d, e, and f versus panels j, k, and l).
DISCUSSION
These studies were designed to elucidate the effects of PI4KA inhibition in vivo and in vitro. Synthesis of selective PI4KA inhibitors has been motivated by the prospect that such inhibitors might be useful to fight HCV infection. RNAi studies identified PI4KA as a mandatory host factor in HCV replication, and those studies suggested that PI4KA down-regulation is reasonably tolerated by cultured cells (15, 17, 39, 40). In addition, it has been reported that HCV infection can induce the expression of PI4KA and increase PtdIns(4)P levels in humans (39). However, cellular studies using RNAi-mediated knockdown indicated that PI4KA is responsible for the maintenance of the PM PtdIns(4)P and PtdIns(4,5)P2 pools during strong PLC activation (10) and predicted that complete blockade of this enzyme may interfere with a number of signaling processes that require the presence of these lipids in the PM. The conclusion drawn from these studies was that cells tolerate a significant level of knockdown of PI4KA because even a small amount of enzyme is able to maintain its housekeeping functions, whereas HCV replication is strongly inhibited even by a 50% reduction of the level of this enzyme (15, 17, 39, 40). This has lent strong support to the notion that there is a therapeutic window for inhibiting HCV replication without affecting cellular functions.
A large number of compounds with various activities against PI4KA did indeed inhibit viral replication, with their antiviral potencies showing close correlation with their PI4KA inhibitory potencies. A selected set of potent PI4KA inhibitory compounds that were subjected to more detailed analysis in cultured cells strongly inhibited PtdIns(4)P synthesis, primarily affecting the PM pool of this lipid. Interestingly, the same treatment had very small if any effect on the PM PtdIns(4,5)P2 pools in quiescent cells. Only when cells were challenged with a strong PLC-activating stimulus did PtdIns(4,5)P2 levels decrease with a greatly diminished ability for these pools to resynthesize. Two recent studies have already noted the dissociation of PtdIns(4,5)P2 from PM PtdIns(4)P. One of these studies showed that PtdIns(4,5)P2 levels can recover after strong PLC activation even when the steady-state level of PtdIns(4)P is greatly reduced by an acutely PM-recruited PtdIns(4)P phosphatase, and only inhibition of PI4Ks impeded PtdIns(4,5)P2 recovery (30). Another study found that mouse embryo fibroblast cells obtained from a different conditional pi4ka knock-out mouse showed no signs of PtdIns(4,5)P2 depletion; in fact, these cells appeared to display increased PIP5K activity and accumulation of PtdIns(4,5)P2 in internal membranes (13). This latter feature is highly reminiscent of cells overexpressing PIP5Ks (41) or their activators, such as Arf6 (42). Notably, prolonged pharmacological blockade of PI4KA in this study did not yield the appearance of PtdIns(4,5)P2 in internal membranes in COS-7 cells, and the cells were able to maintain their PM PtdIns(4,5)P2 levels. These observations together suggest that there are additional mechanism(s) by which cells can supply PtdIns(4)P to the PM even when PI4KA is inhibited. Possible sources of PtdIns(4)P could be the endocytic compartments where the lipid is synthesized by type II PI4Ks and where the lipid can reach the membrane during vesicular trafficking as hinted by our previous study (43). Another alternative could be the production of PtdIns(4,5)P2 via PtdIns(5)P by the type II PIPKs, a possibility that needs further investigation.
At this stage, the animal studies were only designed to test the toxicity of PI4KA inhibitory compounds during oral administration. These animals displayed a variety of adverse effects in response to the inhibitor. Several animals showed sudden lethality, especially at the highest dose of treatment without any obvious pathology other than mild irritations documented in the upper GI tract. The sudden death suggested a cardiovascular insufficiency that can be caused by dehydration or a sudden loss of vascular tone. Although one cannot rule out off-target effects, one can speculate that strong activation of Gq-coupled receptors (such as those required to maintain vascular tone) may have caused a severe depletion of PtdIns(4,5)P2 levels in vascular smooth muscle cells. The inability to replenish PtdIns(4,5)P2 following stimulation would certainly be deleterious to maintenance of vascular tone or other aspects of cardiovascular homeostasis. Although these toxicity studies were not specifically designed to test these possibilities, they point to a direction that will have to be followed up in future in vivo studies. The animals dosed with a lower level of the compound remained alive throughout the studies and displayed moderate to severe GI abnormalities. This may be due to the highest exposure of these cells during oral administration or a specific role of PI4KA in gut homeostasis. Similarly, the cKO animals were dosed orally with tamoxifen providing highest exposure to the drugs or to Cre induction in the GI tract, and the most severe toxicities were observed in this tissue. The GI abnormalities observed here were similar to those observed in a recent study using a different cKO pi4ka mouse model (22). Interestingly, none of the cKO mice exhibited rapid lethality as was seen with some compounds, but rather they progressed to a moribund condition, suggesting significant differences in the pharmacological and genetic ablation of PI4KA on in vivo toxicity. More detailed studies are in progress to evaluate the efficiency of gene ablation in the various tissues and the specific consequences of the lack of the proteins. However, even these preliminary studies caution using only genetic ablation studies to determine safety of potential therapeutic targets.
Based on the results of the experiments in cultured cells, one would expect to see defects in many other tissues yielding a variety of symptoms. However, given the fact that cells can function with greatly reduced PI4KA levels unless very strongly stimulated via Gq-coupled receptors, it is most likely that these animals still maintained some PI4KA expression or activity in tissues other than the GI tract. For example, it is important to point out that the drugs used in theses studies do not cross the blood-brain barrier, and also, tamoxifen exposure of the brain was found to be minimal during this regime of tamoxifen administration (data not shown). More studies are in progress to address these questions with different experimental design.
In summary, this study showed that PI4KA is uniquely important in the maintenance of PtdIns(4,5)P2 levels during strong PLC activation, but it appears dispensable for normal cell growth and maintenance at least for a period of time. It also confirmed previous claims that a fraction of the enzyme or its activity is sufficient to support cellular functions, although HCV replication is more sensitive to inhibition of the enzyme. Although preliminary, the animal studies showed that the cells within the GI tract, especially those showing rapid turnover may require PI4KA for their normal functions. Additionally, acute inhibition of the enzyme with selective small molecules may also exert additional deleterious effects in addition to GI problems such as cardiovascular tone, possibly explaining the sudden death caused by these drugs. These results highlight the significant risks associated with pharmacological targeting of PI4KA. The compounds and animal models generated for these studies will greatly benefit further research on PI4KA to better understand the role of this important enzyme in normal physiology and disease.
Acknowledgments
Confocal imaging was performed at the Microscopy and Imaging Core of the National Institutes of Health, NICHD, with the kind assistance of Drs. Vincent Schram and James T. Russell.
This work was supported, in whole or in part, by National Institutes of Health grants from the Intramural Research Program of the Eunice Kennedy Shriver NICHD (to N. B., G. H., and T. B.).
Because of low exposures on day 1, the 10 mg/kg/day dose was raised to 40 mg/kg/day on days 8–13.
- PI4K
- phosphatidylinositol 4-kinase
- AngII
- angiotensin II
- GI
- gastrointestinal
- PI4KA
- phosphatidylinositol 4-kinase α
- PM
- plasma membrane
- PtdIns(4)P
- phosphatidylinositol 4-phosphate
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- Wm
- wortmannin
- PLC
- phospholipase C
- PH
- pleckstrin homology
- CFP
- cyan fluorescent protein
- HCV
- hepatitis C virus
- cKO
- conditional knock-out.
REFERENCES
- 1. Balla T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balla A., Balla T. (2006) Phosphatidylinositol 4-kinases; old enzymes with emerging functions. Trends Cell Biol. 16, 351–361 [DOI] [PubMed] [Google Scholar]
- 3. D'Angelo G., Vicinanza M., Di Campli A., De Matteis M. A. (2008) The multiple roles of PtdIns(4)P–not just the precursor of PtdIns(4,5)P2. J. Cell Sci. 121, 1955–1963 [DOI] [PubMed] [Google Scholar]
- 4. Minogue S., Waugh M. G. (2012) The phosphatidylinositol 4-kinases: don't call it a comeback. Subcell. Biochem. 58, 1–24 [DOI] [PubMed] [Google Scholar]
- 5. Walch-Solimena C., Novick P. (1999) The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1, 523–525 [DOI] [PubMed] [Google Scholar]
- 6. Hama H., Schnieders E. A., Thorner J., Takemoto J. Y., DeWald D. B. (1999) Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274, 34294–34300 [DOI] [PubMed] [Google Scholar]
- 7. D'Angelo G., Vicinanza M., Wilson C., De Matteis M. A. (2012) Phosphoinositides in Golgi complex function. Subcell. Biochem. 59, 255–270 [DOI] [PubMed] [Google Scholar]
- 8. Audhya A., Emr S. D. (2002) Stt4 PI4K localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2, 593–605 [DOI] [PubMed] [Google Scholar]
- 9. Balla A., Tuymetova G., Tsiomenko A., Várnai P., Balla T. (2005) A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-IIIα: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell 16, 1282–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balla A., Kim Y. J., Varnai P., Szentpetery Z., Knight Z., Shokat K. M., Balla T. (2008) Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIα. Mol. Biol. Cell 19, 711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baird D., Stefan C., Audhya A., Weys S., Emr S. D. (2008) Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J. Cell Biol. 183, 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wong K., Meyers ddR., Cantley L. C. (1997) Subcellular localization of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 272, 13236–13241 [DOI] [PubMed] [Google Scholar]
- 13. Nakatsu F., Baskin J. M., Chung J., Tanner L. B., Shui G., Lee S. Y., Pirruccello M., Hao M., Ingolia N. T., Wenk M. R., De Camilli P. (2012) PtdIns(4)P synthesis by PI4KIIIa at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 199, 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Altan-Bonnet N., Balla T. (2012) Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem. Sci. 37, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vaillancourt F. H., Pilote L., Cartier M., Lippens J., Liuzzi M., Bethell R. C., Cordingley M. G., Kukolj G. (2009) Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology 387, 5–10 [DOI] [PubMed] [Google Scholar]
- 16. Borawski J., Troke P., Puyang X., Gibaja V., Zhao S., Mickanin C., Leighton-Davies J., Wilson C. J., Myer V., Cornellataracido I., Baryza J., Tallarico J., Joberty G., Bantscheff M., Schirle M., Bouwmeester T., Mathy J. E., Lin K., Compton T., Labow M., Wiedmann B., Gaither L. A. (2009) Class III phosphatidylinositol 4-kinase α and β are novel host factor regulators of hepatitis C virus replication. J. Virol. 83, 10058–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tai A. W., Benita Y., Peng L. F., Kim S. S., Sakamoto N., Xavier R. J., Chung R. T. (2009) A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berger K. L., Cooper J. D., Heaton N. S., Yoon R., Oakland T. E., Jordan T. X., Mateu G., Grakoui A., Randall G. (2009) Roles for endocytic trafficking and phosphatidylinositol 4-kinase IIIα in hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 106, 7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trotard M., Lepère-Douard C., Régeard M., Piquet-Pellorce C., Lavillette D., Cosset F. L., Gripon P., Le Seyec J. (2009) Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 23, 3780–3789 [DOI] [PubMed] [Google Scholar]
- 20. Hsu N. Y., Ilnytska O., Belov G., Santiana M., Chen Y. H., Takvorian P. M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T., Cameron C. E., Ehrenfeld E., van Kuppeveld F. J., Altan-Bonnet N. (2010) Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141, 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tai A. W., Salloum S. (2011) The role of the phosphatidylinositol 4-kinase PI4KA in hepatitis C virus-induced host membrane rearrangement. PLoS One 6, e26300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaillancourt F. H., Brault M., Pilote L., Uyttersprot N., Gaillard E. T., Stoltz J. H., Knight B. L., Pantages L., McFarland M., Breitfelder S., Chiu T. T., Mahrouche L., Faucher A. M., Cartier M., Cordingley M. G., Bethell R. C., Jiang H., White P. W., Kukolj G. (2012) Evaluation of phosphatidylinositol-4-kinase IIIα as a hepatitis C virus drug target. J. Virol. 86, 11595–11607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Botyanszki J., Dickerson S. H., Leivers M. R., Li X., McFayden R. B., Redman A. M., Shotwell J. B., Xue J. (December 20, 2012) Benzimidazole derivatives as antiviral agents and their preparation. PCT International Patent Application WO 2012174312 A2
- 24. Banka A. L., Botyanszki J., Duan M. L., Leivers M. R., Shotwell J. B., Tallant M. D., Dickerson S. H., Tai V. W.-F., McFayden R. B., Redman A. M., Yu J., Li X., Garrido D. M., Catalano J. G., Adjabeng G. (June 28, 2012) Preparation of quinazolinone derivatives as antiviral agents. PCT International Patent Application WO 201287938 A1
- 25. Bengtsson M. L., Nikitidis G., Storm P., Bailey J. P., Griffen E. J., Arnould J.-C., Bird T., Geoffrey C. (May 18, 2006) Preparation of 5-heteroaryl thiazoles and their use as phosphoinositide 3-kinase (PI3K) inhibitors. PCT International Patent Application WO 2006051270 A1
- 26. Adams N. D., Burgess J. L., Michael G., Knight S. D., Newlander K. A., Ridgers L. H., Schmidt S. J. (December 24, 2008) Preparation of quinazoline derivatives as PI3 kinase inhibitors. PCT International Patent Application WO 2008157191 A2
- 27. Szentpetery Z., Balla A., Kim Y. J., Lemmon M. A., Balla T. (2009) Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol. 10, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakanishi S., Catt K. J., Balla T. (1995) A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositol phospholipids. Proc. Natl. Acad. Sci. U.S.A. 92, 5317–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hammond G. R., Schiavo G., Irvine R. F. (2009) Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns(4)P and PtdIns(4,5)P(2). Biochem. J. 422, 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hammond G. R., Fischer M. J., Anderson K. E., Holdich J., Koteci A., Balla T., Irvine R. F. (2012) PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 337, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J., Golland P., Sabatini D. M. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Downing G. J., Kim S., Nakanishi S., Catt K. J., Balla T. (1996) Characterization of a soluble adrenal phosphatidylinositol 4-kinase reveals wortmannin-sensitivity of type III phosphatidylinositol 4-kinases. Biochemistry 35, 3587–3594 [DOI] [PubMed] [Google Scholar]
- 33. Leivers A. L., Tallant M., Shotwell J. B., Dickerson S., Leivers M. R., McDonald O. B., Gobel J., Creech K. L., Strum S. L., Mathis A., Rogers S., Moore C. B., Botyanszki J. (2013) Discovery of selective small molecule type III phosphatidylinositol 4-kinase α (PI4KIIIα) inhibitors as anti-hepatitis C (HCV) Agents. J. Med. Chem. 10.1021/jm400781h [DOI] [PubMed] [Google Scholar]
- 34. Verrou C., Zhang Y., Zürn C., Schamel W. W., Reth M. (1999) Comparison of the tamoxifen regulated chimeric Cre recombinases MerCreMer and CreMer. Biol. Chem. 380, 1435–1438 [DOI] [PubMed] [Google Scholar]
- 35. Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 [DOI] [PubMed] [Google Scholar]
- 36. Várnai P., Balla T. (1998) Visualization of phosphoinositides that bind pleckstrin homology domains: calcium-and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Balla T., Varnai P. (2009) Visualization of cellular phosphoinositide pools with GFP-fused protein-domains. Curr. Protoc. Cell Biol. Chapter 24, Unit 24.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van der Wal J., Habets R., Várnai P., Balla T., Jalink K. (2001) Monitoring phospholipase C activation kinetics in live cells by FRET. J. Biol. Chem. 276, 15337–15344 [DOI] [PubMed] [Google Scholar]
- 39. Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M. S., Longerich T., Diehl S., Ramirez F., Balla T., Rohr K., Kaul A., Bühler S., Pepperkok R., Lengauer T., Albrecht M., Eils R., Schirmacher P., Lohmann V., Bartenschlager R. (2011) Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pollard T. D. (2007) Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 [DOI] [PubMed] [Google Scholar]
- 41. Rozelle A. L., Machesky L. M., Yamamoto M., Driessens M. H., Insall R. H., Roth M. G., Luby-Phelps K., Marriott G., Hall A., Yin H. L. (2000) Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10, 311–320 [DOI] [PubMed] [Google Scholar]
- 42. Brown F. D., Rozelle A. L., Yin H. L., Balla T., Donaldson J. G. (2001) Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szentpetery Z., Várnai P., Balla T. (2010) Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 8225–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]