Background: The cytochrome bc1 complex weakly associates with cytochrome oxidase (CcO) in the absence of the Rieske Rip1 subunit.
Results: The N-terminal domain of Rip1 enhances the stabilization of cytochrome bc1-CcO supercomplexes in yeast.
Conclusion: Induced stabilization of supercomplexes arises from bc1-dependent formation of CcO.
Significance: The late assembly intermediate of the bc1 complex can template the maturation of CcO even without cardiolipin.
Keywords: Bioenergetics/Electron Transfer Complex, Cardiolipin, Cytochrome Oxidase, Mitochondria, Mitochondrial Metabolism
Abstract
Yeast cells deficient in the Rieske iron-sulfur subunit (Rip1) of ubiquinol-cytochrome c reductase (bc1) accumulate a late core assembly intermediate, which weakly associates with cytochrome oxidase (CcO) in a respiratory supercomplex. Expression of the N-terminal half of Rip1, which lacks the C-terminal FeS-containing globular domain (designated N-Rip1), results in a marked stabilization of trimeric and tetrameric bc1-CcO supercomplexes. Another bc1 mutant (qcr9Δ) stalled at the same assembly intermediate is likewise converted to stable supercomplex species by the expression of N-Rip1, but not by expression of intact Rip1. The N-Rip1-induced stabilization of bc1-CcO supercomplexes is independent of the Bcs1 translocase, which mediates Rip1 translocation during bc1 biogenesis. N-Rip1 induces the stabilization of bc1-CcO supercomplexes through an enhanced formation of CcO. The association of N-Rip1 with the late core bc1 assembly intermediate appears to confer stabilization of a CcO assembly intermediate. This induced stabilization of CcO is dependent on the Rcf1 supercomplex stabilization factor and only partially dependent on the presence of cardiolipin. N-Rip1 exerts a related induction of CcO stabilization in WT yeast, resulting in enhanced respiration. Additionally, the impact of CcO stabilization on supercomplexes was observed by means other than expression of N-Rip1 (via overexpression of CcO subunits Cox4 and Cox5a), demonstrating that this is a general phenomenon. This study presents the first evidence showing that supercomplexes can be stabilized by the stimulated formation of CcO.
Introduction
The mitochondrial electron transfer chain (ETC)3 in Saccharomyces cerevisiae consists of Complex II and the two proton-pumping units Complex III (ubiquinol-cytochrome c reductase (bc1)) and Complex IV (cytochrome c oxidase (CcO)). These latter two complexes assemble into supercomplex structures, which appear to enhance the efficiency of electron transfer between complexes, thereby diminishing formation of reactive oxygen species arising from electron transfer steps (1). Because yeast cells lack Complex I, the two supercomplexes seen in yeast consist of the dimeric bc1 complex associated with one or two CcO monomeric complexes on either side of bc1 (1, 2). These trimeric III2IV1 and tetrameric III2IV2 supercomplexes in yeast mitochondria are resolved from the non-associated ETC complexes via blue native PAGE (BN-PAGE) (3, 4).
Single particle reconstructions of cryo-EM images of bovine and yeast ETC supercomplexes have shed light on the interface between CcO and bc1. Modeling of the EM projection maps predicts that yeast bc1 contacts CcO via Cob, Cyt1, Qcr6, Qcr7, Qcr8, and Qcr9 (5). In contrast, modeling of the bovine projection maps predicts a different interface involving the bc1 subunit cytochrome b (Cob), the Rieske subunit (Rip1), and subunit 11 (Qcr10) (6). A significant unresolved question is whether yeast and mammalian bc1 and CcO utilize different interfaces for interaction.
Recent attention has focused on the stability of the ETC supercomplexes. A series of factors, including Rcf1, Rcf2, and Aac2, have been reported to be important for stabilization (7–11). In addition, the inner membrane lipid cardiolipin (CL) has a key role in stabilizing the supercomplexes (12–14).
Biogenesis of the ETC complexes is dependent on coordinate steps of membrane insertion of mitochondrially encoded subunits and binding of proteins imported from the cytoplasm. The bc1 complex contains three subunits involved in electron transfer and proton pumping and seven to eight supernumerary subunits (15, 16). The essential catalytic core consists of cytochrome b (Cob), cytochrome c1 (Cyt1), and the Rieske iron-sulfur (FeS) protein Rip1. The assembly of bc1 in yeast proceeds in a modular pathway, with the mitochondrially encoded Cob seeding the assembly process (17, 18). A series of early Cob-containing core assembly intermediates exists, containing bc1 subunits Qcr7 and Qcr8 and assembly factors (17, 19–22). Additional subunits (Cor1, Cor2, Cyt1, Qcr6, and Qcr9) are added to form the late core intermediate lacking only Rip1 and the peripheral subunit Qcr10 (18, 19, 23). This nonfunctional late core intermediate exists loosely associated with CcO, and the addition of Rip1 and Qcr10 forms the active bc1 complex. Importantly for this study, mature bc1 forms stable associations with CcO.
The key event in the insertion of Rip1 into the late core assembly intermediate is its translocation across the inner membrane (IM) in a step mediated by the AAA-ATPase Bcs1 (18, 23, 24). The 215-residue Rip1 protein is imported into the mitochondrial matrix for insertion of its 2Fe-2S cluster (25). Bcs1 then translocates the C-terminal globular domain containing the FeS cluster across the IM in an ATP-dependent manner. Cells lacking Bcs1 resemble rip1Δ cells in the accumulation of the late core bc1 intermediate lacking Rip1 as well as Qcr10 (17).
We discovered a key Rip1 chaperone, Mzm1, which stabilizes Rip1 during the matrix maturation step, preventing misfolding (26–28). In the process of mapping the interaction interface between Mzm1 and Rip1, we constructed and expressed a series of Rip1 truncates in rip1Δ cells to probe interactions with Mzm1. We observed that Mzm1 interacted with the C-terminal 2Fe-2S domain of Rip1, but we also discovered that expression of a Rip1 truncate containing only the N-terminal 92 residues (designated N-Rip1) resulted in a surprising stabilization of the two bc1-CcO supercomplexes (28). After proteolytic removal of the matrix-targeting sequence, the remaining 61-residue N-terminal domain consists of an extended conformation with a 32-residue helix that spans the membrane. Neither the C-terminal domain (residues 93–215; C-Rip1) nor Rip1 lacking its transmembrane motif was able to enhance supercomplex abundance. The supercomplexes stabilized by N-Rip1 were unusual in that they contained Qcr10 yet lacked N-Rip1. Normally, Qcr10 is present only in cells in which Rip1 has been incorporated into bc1. N-Rip1 has a profound gain-of-function effect, but it failed to accumulate in rip1Δ cells (28).
Cells lacking Rip1 are stalled at the late core bc1 assembly intermediate, which loosely interacts with CcO as mentioned, yet these supercomplexes are largely destabilized by BN-PAGE conditions (12, 13). The stabilization of bc1-CcO supercomplexes in rip1Δ cells on BN-PAGE by expression of just the N-terminal domain of Rip1 provided an experimental system to address how Rip1 insertion into the late core assembly intermediate confers stabilization of the bc1-CcO supercomplexes. We show that the transient association of N-Rip1 with bc1 leads to a specific bc1-dependent accumulation of CcO, which triggers an enhanced abundance of bc1-CcO supercomplexes in WT yeast cells as well as yeast lacking the Bcs1 translocase or CL synthase.
MATERIALS AND METHODS
Yeast Strains and Vectors
Strains were all derivatives of BY4741 (MATa hisΔ1 leu2Δ0 met15Δ0 ura3Δ0) background. All single gene deletion strains were purchased from Open Biosystems and verified by PCR with specific primers. In addition, the crd1Δ strain was verified by tetrad dissection. To generate the rip1Δqcr10Δ strain, RIP1 was replaced with a LEU2 coding sequence in the qcr10Δ background. To generate the rip1Δcoa2Δ strain, COA2 was replaced with a Candida albicans URA3 coding sequence in the rip1Δ background. To generate the crd1Δqcr7Δ strain, QCR7 was replaced with a C. albicans URA3 coding sequence in the crd1Δ background. For Cor2-TAP purification, endogenous COR2 was replaced with COR2::TAP in the corresponding strains. Strains were grown in 2% dextrose or 2% galactose synthetic medium lacking relevant amino acids to maintain plasmid selection. The construction of plasmids expressing Rip1, N-Rip1, Rip1-Grx3, Cox4, and Cox5a was described previously (27–29). The RCF1 vector was a gift of Dr. Jared Rutter.
Preparation of Mitochondria
Mitochondria were isolated as described previously (28). Briefly, lyticase was used to create spheroplasts, which were subsequently ruptured by Dounce homogenization, and mitochondria were isolated by differential centrifugation. The total mitochondrial protein concentration was determined using the Bradford method (30).
SDS-PAGE and BN-PAGE Analysis
For SDS-PAGE, samples (10–30 μg of protein) were separated on 12 or 4–12% gradient polyacrylamide gels and transferred to nitrocellulose or PVDF membranes. Membranes were blocked prior to detection using 5% nonfat dry milk in PBS. BN-PAGE was performed using a Bis-Tris system (Invitrogen) with either 1% digitonin or 1% dodecyl maltoside. Chemiluminescent reagents with horseradish peroxidase-conjugated secondary antibodies were used to visualize proteins. Antibodies were either purchased, lab stocks, or generous gifts: anti-Myc (Roche Diagnostics); anti-porin (Molecular Probes); anti-peroxidase-antiperoxidase complex (PAP) (Open Biosystems); anti-Cox1, anti-Cox2, and anti-Cox3 (MitoSciences); anti-Rip1 and anti-Cor1/Cor2 (gifts from Dr. Bernard Trumpower); and anti-Cox13 (gift from Dr. Peter Rehling).
Cor2-TAP Purification
Cells were grown on synthetic galactose medium with auxotrophic selection, and mitochondria were isolated as described above. Mitochondria were lysed at 4 °C in lysis buffer (20 mm Tris (pH 7.4), 150 mm NaCl, 1 mm magnesium acetate, 1 mm imidazole, 2 mm CaCl2, 5 mm 2-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, Roche protease inhibitor mixture, and 1% digitonin) and clarified by centrifugation at 20,000 × g for 15 min. Solubilized mitochondrial proteins were incubated with calmodulin-agarose beads at 4 °C and then washed with buffer containing 20 mm Tris (pH 7.4), 150 mm NaCl, 1 mm magnesium acetate, 1 mm imidazole, 2 mm CaCl2, 5 mm 2-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, Roche protease inhibitor mixture, and 0.2% digitonin. The bound proteins were eluted by heating.
Mitochondrial Enzyme Assays
Cytochrome oxidase and bc1 enzymatic activities were measured using 10–20 μg of mitochondria in 40 mm potassium phosphate (pH 6.7) and 0.5% Tween 20 as reported previously (31, 32). Cytochrome oxidase activity was assessed by the oxidation of reduced cytochrome c at 550 nm, whereas bc1 was quantified by the reduction of oxidized cytochrome c at 550 nm.
In Vivo Mitochondrial Protein Translation Assay
Cells were grown overnight in selective medium containing 2% galactose and then re-inoculated the next day in 2% galactose. Mitochondrial gene products were labeled with [35S]methionine in whole cells in the presence of cycloheximide to inhibit cytoplasmic protein synthesis. Equivalent amounts of total cellular proteins were separated by SDS-PAGE. The gel was dried, and radiolabeled proteins were visualized by exposing autoradiography films at −80 °C.
Mitochondrial Respiration Assays
Cellular oxygen consumption was assessed in cells grown to stationary phase and then diluted to an A600 of 0.5 in 3 ml of 5% glycerol using a 5300A biological oxygen monitor (YSI Inc.).
RESULTS
Stabilization of Qcr10-containing Supercomplexes Mediated by the N Terminus of Rip1
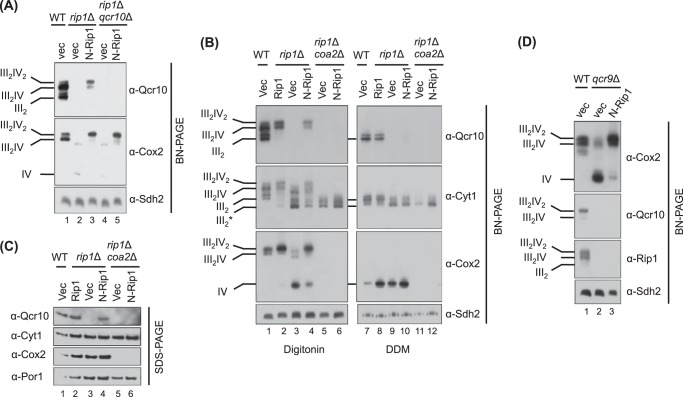
The association of Rip1 and Qcr10 with the late core bc1 assembly intermediate imparts stabilization to the bc1-CcO supercomplexes under BN-PAGE conditions. We demonstrated previously that expression of just the N-terminal domain of Rip1 (residues 1–92; designated N-Rip1) in rip1Δ BY4741 yeast cells induced stabilization of ETC supercomplexes without the Rip1 truncate being stably associated with the supercomplexes (28). In the absence of Rip1, CcO largely fractionated in a monomeric complex after BN-PAGE, although low abundance supercomplexes can be seen in digitonin-solubilized mitochondrial lysates in Fig. 1B (lane 3, compare anti-Cyt1 and anti-Cox2 blots). The presence of N-Rip1 induced a marked increase in the abundance of digitonin-solubilized bc1-CcO supercomplexes as visualized on BN-PAGE by immunoblotting with antisera to Cox2 (CcO subunit) or Qcr10 (bc1 subunit). This effect shown in Fig. 1A (lanes 1–3) is analogous to what we reported previously (28) and is shown for clarity and comparison. As with the supercomplexes in WT cells, the N-Rip1-induced ETC supercomplexes were stable with digitonin but not dodecyl maltoside solubilization (Fig. 1B, lane 4 versus lane 10). The induced stabilization of ETC supercomplexes is specific to the N-terminal half of Rip1; matrix targeting of the C-terminal Rip1 2Fe-2S domain is without effect (28). Although N-Rip1 was not stably associated with the supercomplexes, the final bc1 subunit Qcr10 was incorporated (Fig. 1A) (28). Qcr10 is typically unstable and rapidly degraded in rip1Δ cells. However, the expression of N-Rip1 in rip1Δ cells resulted in the stabilization of Qcr10 as seen by BN-PAGE (Fig. 1, A and B) and steady-state SDS-PAGE (Fig. 1C) despite the absence of stable N-Rip1 binding to bc1.
FIGURE 1.
N-Rip1-mediated stabilization of Qcr10 by bc1-CcO supercomplexes. A, BN-PAGE immunoblot of digitonin-solubilized mitochondria isolated from WT cells, from rip1Δ cells expressing a vector control (vec) and N-Rip1, and from rip1Δqcr10Δ cells expressing a vector control and N-Rip1. B and C, BN-PAGE immunoblot of either digitonin- or dodecyl maltoside (DDM)-solubilized mitochondria (B) and SDS-PAGE immunoblot of isolated mitochondria (C) from WT cells; from rip1Δ cells expressing Rip1, a vector control and N-Rip1; and rip1Δcoa2Δ cells expressing a vector control (vec) and N-Rip1. D, BN-PAGE immunoblot of digitonin-solubilized mitochondria from WT cells and from qcr9Δ cells expressing a vector control and N-Rip1. III2* denotes the late core intermediate of bc1. The succinate dehydrogenase subunit Sdh2 and porin were used as loading controls for BN-PAGE and SDS-PAGE, respectively.
To assess whether Qcr10 incorporation was responsible for the supercomplex stabilization, we tested the effect of N-Rip1 in rip1Δqcr10Δ cells. Expression of N-Rip1 in rip1Δqcr10Δ cells resulted in the stabilization of Cox2-containing ETC supercomplexes (Fig. 1A, lane 5), demonstrating that Qcr10 is not critical in supercomplex stabilization and therefore is unlikely to be a key stabilizing subunit at the supercomplex interface.
We next tested whether the N-Rip1-induced Qcr10 stabilization in rip1Δ cells was dependent on the presence of CcO. Coa2 is a key CcO assembly factor, and in its absence, CcO biogenesis is markedly impaired (29). The N-Rip1-mediated Qcr10 stabilization was abrogated in rip1Δcoa2Δ cells as seen by BN-PAGE (Fig. 1B, lane 6) and steady-state SDS-PAGE (Fig. 1C, lane 6). Qcr10 is stably associated with bc1 in cells lacking CcO as long as intact Rip1 is present (18). Thus, the observed N-Rip1-induced Qcr10 stabilization in rip1Δ (but not rip1Δcoa2Δ) cells suggests that Qcr10 stabilization is conditional and arises from the presence of ETC supercomplexes.
Role of Qcr9 in bc1-CcO Supercomplex Stabilization
Cells lacking the Qcr9 subunit of bc1 are deficient in Rip1 and also accumulate the late core assembly intermediate (33). The transmembrane helix of Rip1 is sandwiched between Qcr9 and Cob in the bc1 structure (16). It is unclear whether the destabilization of supercomplexes in qcr9Δ cells is due to the deficiency of Rip1 or the absence of both Rip1 and Qcr9. To assess whether N-Rip1 could stabilize supercomplexes of the late core intermediate and CcO in the absence of Qcr9, we expressed N-Rip1 in qcr9Δ cells. As with rip1Δ, the presence of N-Rip1 induced a marked stabilization of bc1-CcO supercomplexes on BN-PAGE (Fig. 1D). The induced stabilization of supercomplexes occurred without the stable association of N-Rip1. The absence of N-Rip1 was verified by using antisera specific to an N-terminal epitope in Rip1 as well as anti-Myc detection using an N-Rip1-Myc chimera (data not shown). In contrast to the observed stabilization of Qcr10 in rip1Δ cells, N-Rip1 conferred only minimal stabilization of Qcr10 and no stabilization of endogenous intact Rip1 in the qcr9Δ supercomplexes. Thus, Qcr9 has a key role in Qcr10 recruitment or stabilization and for the first time is shown to be nonessential for the stabilization of bc1-CcO supercomplexes.
Role of Bcs1 in N-Rip1-induced Stabilization of bc1-CcO Supercomplexes
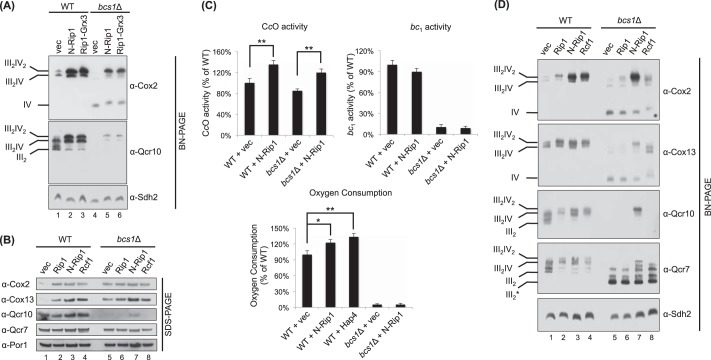
The insertion of intact Rip1 into the late core intermediate is dependent on the translocation of the 2Fe-2S domain of Rip1 across the IM by Bcs1 (24). The Rip1 translocation establishes the topology of the Rip1 transmembrane helix. Rip1 associates with Bcs1 through both its N- and C-terminal domains during translocation (24). To test the importance of Bcs1 for the observed N-Rip1 effect, we expressed N-Rip1 in bcs1Δ cells, which normally accumulate the same late core bc1 assembly intermediate as rip1Δ cells. Expression of N-Rip1 in bcs1Δ cells resulted in a similar increased abundance of bc1-CcO supercomplexes as seen in N-Rip1-containing rip1Δ cells (Fig. 2A, lanes 4 and 5). The presence of N-Rip1 led to a stabilization of steady-state Qcr10 levels in bcs1Δ cells (Fig. 2B, lane 7), and Qcr10 fractionated on BN-PAGE with the ETC supercomplexes (Fig. 2A, lane 5). Total CcO activity was augmented in N-Rip1-containing bcs1Δ cells (Fig. 2C), consistent with the enhanced abundance of Cox2-containing supercomplexes, but as expected, the cells were respiratory-deficient and showed minimal oxygen consumption due to the lack of a functional bc1 complex (Fig. 2C).
FIGURE 2.
Enhancement of cellular respiration by N-Rip1 expression. A, BN-PAGE immunoblot of digitonin-solubilized mitochondria from WT cells and from bcs1Δ cells expressing a vector control (vec), N-Rip1, and the Rip1-Grx3 chimera. B and D, SDS-PAGE (B) and BN-PAGE (digitonin) (D) immunoblots of isolated mitochondria from WT cells and from bcs1Δ cells expressing a vector control, Rip1, N-Rip1, and Rcf1. The succinate dehydrogenase subunit Sdh2 and porin were used as loading controls for BN-PAGE and SDS-PAGE, respectively. C, CcO activity, bc1 activity, and total oxygen consumption of N-Rip1-expressing cells and a vector control. Specific enzymatic activities and oxygen consumption are represented as a percentage of the wild-type activity. *, p < 0.05; **, p < 0.01. The effect of N-Rip1 on bc1 activity in WT cells is not statistically significant.
We also investigated the specificity of N-Rip1 in inducing supercomplex stabilization. Overexpression of full-length Rip1 in bcs1Δ cells had only a minimal effect in inducing supercomplex stabilization relative to the effect with the N-Rip1 truncate (Fig. 2D, lane 6). Replacing the C-terminal 2Fe-2S domain of Rip1 with the heterologous globular domain from glutaredoxin-3 yielded a Rip1-Grx3 chimera (containing only residues 1–92 of Rip1) that was stably expressed and localized within the mitochondrial matrix (28). Expression of Rip1-Grx3 induced supercomplex stabilization in both rip1Δ cells (28) and bcs1Δ cells (Fig. 2A, lane 6). We demonstrated previously that the 32-residue N-terminal helical segment in Rip1-Grx3 is important for supercomplex stabilization (28). Thus, Bcs1 is not important to set the topology of N-Rip1 for the supercomplex stabilization. In addition, the effect of N-Rip1 on supercomplex stabilization exceeds the effect of the known supercomplex stabilization factor Rcf1 (Fig. 2D).
Enhancement of WT Cellular Respiration by Expression of N-Rip1
The N-Rip1 enhancement of CcO activity in bcs1Δ cells raised the question of whether N-Rip1 would stimulate respiration in WT cells. We therefore expressed N-Rip1 in WT BY4741 cells and tested its effect on supercomplex abundance and respiration. Strikingly, the presence of N-Rip1 (or Rip1-Grx3) resulted in a marked enhancement of the abundance of bc1-CcO supercomplexes in WT cells (Fig. 2A, lane 2, and Fig. 2D, lane 3). This marked effect of N-Rip1 in WT cells was not reproduced upon overexpression of full-length Rip1 (Fig. 2D, lane 2). The steady-state abundance of Cox2 and Cox13 was elevated relative to the vector-only control in the N-Rip1-containing WT cells with little change in the abundance of bc1 subunits (Qcr7 or Qcr10) (Fig. 2B, lane 3) or Cor1 and Cor2 (data not shown). CcO activity and oxygen consumption were elevated in the WT cells in the presence of N-Rip1, but no change was observed in bc1 enzymatic activity (Fig. 2C). The N-Rip1-induced enhancement of CcO in WT cells was dependent on the presence of bc1; expression of N-Rip1 in cor2Δ or qcr7Δ cells failed to elicit any CcO enhancement (data not shown).
Rip1-Grx3 Association with Supercomplexes
Although N-Rip1 is labile and not stably associated with the supercomplexes (28), the prediction is that it exerts its effect through associations with the ETC. The Rip1-Grx3 chimera is a stably expressed protein, unlike the isolated N-terminal domain. To test whether Rip1-Grx3 was associated with the bc1-CcO supercomplexes, we expressed the chimera in cells containing Cor2-TAP to permit facile affinity isolation of the bc1 complex. Mitochondria purified from these cells were solubilized with digitonin, and clarified lysates were affinity-purified with calmodulin beads to adsorb Cor2-TAP complexes. Affinity purification of bc1 from these WT cells resulted in the co-isolation of CcO and Rip1-Grx3 (Fig. 3A). Rip1-Grx3 was likewise co-purified with the late core bc1 assembly intermediate and CcO in Cor2-TAP rip1Δ cells expressing Rip1-Grx3 (Fig. 3B), but not in qcr7Δ cells (Fig. 3C), in which Cor2 is not associated in a bc1 assembly intermediate. Furthermore, the interaction of Rip1-Grx3 with the bc1-CcO supercomplex was abrogated in rcf1Δ cells (Fig. 3D), in which the tetrameric supercomplex is destabilized(9–11). Consistent with the lack of interaction of Rip1-Grx3 and the supercomplexes in rcf1Δ cells, neither Rip1-Grx3 nor N-Rip1 conferred stabilization of supercomplexes in rcf1Δ cells (data not shown).
FIGURE 3.
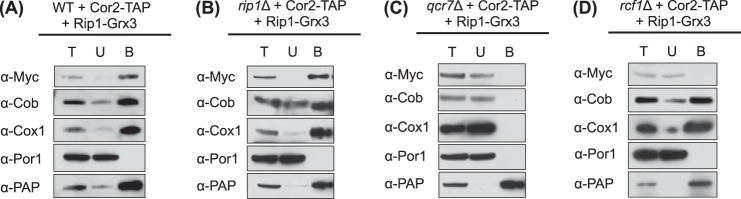
Rip1-Grx3 associates with bc1-CcO supercomplexes. Cor2-TAP purification was performed with digitonin-solubilized mitochondria isolated from the indicated strains expressing vector-borne Cor2-TAP and Rip1-Grx3, showing the total solubilized proteins (T), the unbound proteins (U), and the eluted bound proteins (B). The results in rip1Δ cells were similar with either vector-borne Cor2-TAP or with Cor2-TAP expressed from an integrant. The anti-Myc antibody was used to detect Rip1-Grx3, and the anti-peroxidase-antiperoxidase complex (PAP) antibody (α-PAP) was used to detect Cor2-TAP.
Although the association of Rip1-Grx3 with the supercomplexes is observed by co-immunoprecipitation, the interaction is destabilized by BN-PAGE conditions (28). Thus, Rip1-Grx3 is not observed in supercomplex bands on BN-PAGE.
Effect of CL on Supercomplex Stabilization
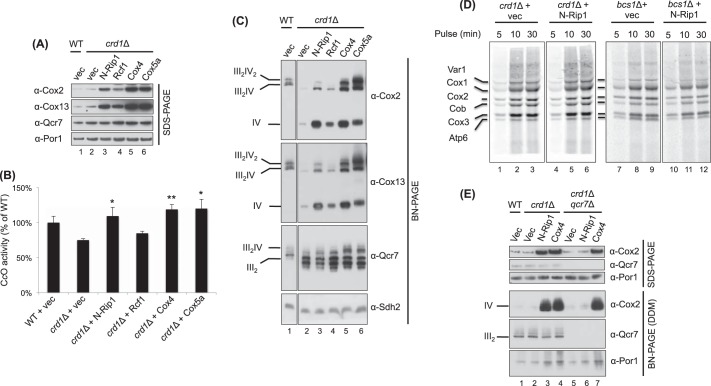
CL is a key stabilizing lipid for supercomplex formation (8, 13, 14). The yeast bc1-CcO supercomplexes have ∼50 molecules of bound CL in addition to other phospholipids (5, 14). Cells devoid of CL synthase (Crd1) have attenuated levels of trimeric and tetrameric supercomplexes (8, 13, 14). We therefore tested whether CL was important for the observed N-Rip1-induced enhancement of supercomplex abundance. Expression of N-Rip1 in crd1Δ cells resulted in a marked elevation in CcO subunits Cox2 and Cox13 by steady-state SDS-PAGE (Fig. 4A, lane 3) and CcO activity (Fig. 4B). This elevation of CcO subunit levels resulted in a modest enhancement of the abundance of the bc1-CcO supercomplexes on BN-PAGE, although a significant fraction of CcO remained in the monomeric complex on BN-PAGE in the absence of CL (Fig. 4C, lane 3). Although CcO levels and activity were enhanced, there was no change in the enzymatic activity of bc1 (data not shown) or steady-state levels of Qcr7 in crd1Δ cells in the presence of N-Rip1 (Fig. 4A, lane 3). N-Rip1 thus has a specific effect on CcO formation in crd1Δ cells.
FIGURE 4.
Elevated CcO levels restore supercomplexes in crd1Δ cells. A and C, SDS-PAGE (A) and BN-PAGE (digitonin) (C) immunoblots of isolated mitochondria from WT cells and from crd1Δ cells expressing a vector control (vec), N-Rip1, Rcf1, Cox4, and Cox5a. The succinate dehydrogenase subunit Sdh2 and porin were used as loading controls for BN-PAGE and SDS-PAGE, respectively. B, CcO activity of the strains used in A and C. CcO activities are presented as a percentage of the wild-type activity. The increase in CcO activity upon expression of N-Rip1, Cox4, or Cox5a in crd1Δ cells was statistically significant compared with crd1Δ cells expressing the vector control. *, p < 0.05; **, p < 0.01. D, in vivo mitochondrial protein synthesis was performed using the indicated strains and quenched by the addition of excess nonradioactive methionine. Samples were collected at the indicated times and analyzed by SDS-PAGE and autoradiography. E, SDS-PAGE and BN-PAGE (dodecyl maltoside (DDM)) immunoblots of isolated mitochondria from WT cells; from crd1Δ cells expressing a vector control, N-Rip1, and Cox4; and from crd1Δqcr7Δ cells expressing a vector control, N-Rip1, and Cox4.
Restoration of Supercomplex Formation in crd1Δ Cells by Elevated CcO Levels
We investigated the mechanism by which the expression of N-Rip1 in crd1Δ and bcs1Δ cells augmented CcO levels. The N-Rip1 truncate could enhance CcO levels by stimulating mitochondrial CcO subunit synthesis (Cox1, Cox2, and Cox3), by increasing the abundance of a CcO core subunit, or by decreasing the turnover rate of intact CcO (or an assembly intermediate). To test whether N-Rip1 stimulated mitochondrial CcO subunit synthesis, we used a mitochondrial protein translation assay, in which cells were labeled with [35S]methionine in the presence of cycloheximide to block translation in the cytoplasm. Only mitochondrial translation products, including Cox1, Cox2, and Cox3, are labeled in this protocol. The presence of N-Rip1 in either crd1Δ or bcs1Δ cells did not enhance the incorporation of [35S]methionine into Cox1, Cox2, or Cox3 relative to control cells at a series of pulse (Fig. 4D) or chase (data not shown) times.
Alternatively, CcO accumulation may be enhanced by increasing the abundance of one of the core CcO subunits. Cox5a, Cox6, and Cox8 are subunits that associate early in the maturation of Cox1, whereas Cox4 is added later (34–37). We tested whether overexpression of these peripheral subunits would mimic the effect of N-Rip1 in crd1Δ cells. Expression of the Cox4 and Cox5a subunits resulted in a dramatic enhancement of steady-state CcO subunits (Fig. 4A, lanes 5 and 6) as well as both trimeric and tetrameric bc1-CcO supercomplexes on BN-PAGE (Fig. 4C, lanes 5 and 6). In contrast, expression of either Cox6 or Cox8 had a positive but less dramatic effect relative to Cox4 or Cox5a (data not shown). The supercomplexes in crd1Δ cells were smaller in size relative to those in WT cells, as was reported previously (13). In addition to the observed CcO enhancement and supercomplex stabilization, CcO activity was elevated in the Cox4- and Cox5a-expressing crd1Δ cells (Fig. 4B). These data show clearly that elevated CcO levels can overcome the CL deficiency in stabilizing ETC supercomplexes. Likewise, elevated levels of Cox4 or Cox5a stimulated CcO formation in both WT and bcs1Δ cells (data not shown).
N-Rip1-induced CcO Formation Is Dependent on bc1
We tested whether the effect of Cox4 or N-Rip1 on stimulating CcO formation in crd1Δ cells was dependent on the presence of bc1. For this study, we compared crd1Δ cells with crd1Δqcr7Δ double null cells containing overexpressed Cox4 or N-Rip1. Qcr7 is a key bc1 subunit that stabilizes an early assembly intermediate of Cob, and in its absence, bc1 biogenesis is terminated at an early step (19, 38). The depletion of Qcr7 abrogated the effect of N-Rip1 on the accumulation of CcO in crd1Δ cells (Fig. 4E). However, the overexpression of Cox4 in crd1Δqcr7Δ cells led to the same marked elevation in CcO as seen in crd1Δ cells. Thus, N-Rip1 and Cox4 induce CcO accumulation through different mechanisms, with only the N-Rip1 effect being bc1-dependent.
DISCUSSION
The N-terminal domain of the catalytic bc1 subunit Rip1 has a dramatic effect in inducing the stabilization of supercomplexes consisting of mature CcO and the late core bc1 assembly intermediate (lacking Rip1 and Qcr10 subunits) on BN-PAGE (28). Such assemblies are normally destabilized by the conditions of BN-PAGE (12, 13). N-Rip1 induces the stabilization of ETC supercomplexes involving the late core intermediate that accumulates in rip1Δ, bcs1Δ, and qcr9Δ cells. The effect of N-Rip1 in inducing ETC supercomplex stabilization is specific for the N-terminal half of Rip1. Unlike N-Rip1, overexpression of neither the C-terminal 2Fe-2S domain nor full-length Rip1 had a positive effect on supercomplex abundance in bcs1Δ cells. The N-Rip1-induced stabilization effect is independent of Bcs1, suggesting that Bcs1-mediated partitioning is not required. Thus, the association of N-Rip1 with the late core intermediate, which likely requires a partitioning within the IM, is not dependent on Bcs1.
The effect of intact Rip1 in imparting stabilization of supercomplexes requires two steps: translocation of the FeS domain across the IM prior to insertion and its association with the late assembly intermediate. The use of N-Rip1 in the present studies dissociates the translocation step from the induced supercomplex stabilization step. The N-terminal Rip1 domain triggers supercomplex stabilization without being stably bound itself to the assembly.
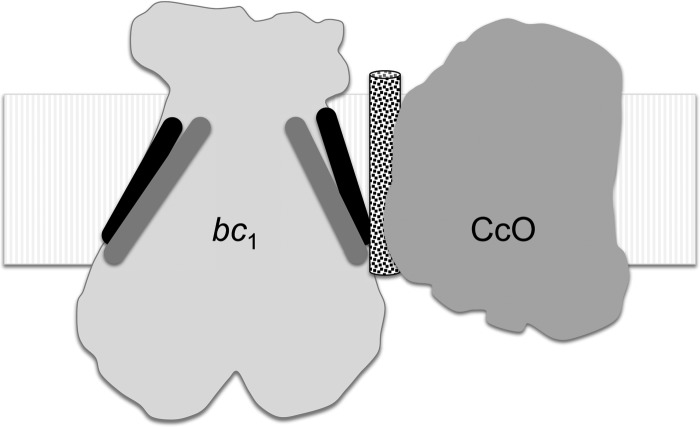
Our results demonstrate that ETC supercomplex formation can stabilize transient interactions of subunits within individual ETC complexes. The N-Rip1-mediated incorporation of Qcr10 into supercomplexes containing the late core bc1 intermediate is dependent on supercomplex formation. The CcO-dependent stabilization of Qcr10 suggests that, in yeast, Qcr10 is bound near the complex interface such that CcO stabilizes it (Fig. 5). Interestingly, supercomplex formation was also shown recently to stabilize the mammalian CcO subunit Cox7a2l (SCAF1) (39); SCAF1 was found to associate only with CcO in ETC supercomplexes.
FIGURE 5.
Schematic representation of the bc1-CcO supercomplex based on the cryo-EM reconstruction model of Dudkina et al. (6) of the bovine supercomplex. The approximate position of the Qcr10 subunit of bc1 is shown in the black cylinder, whereas the position of the helix of Rip1 is shown in the gray cylinder. As mentioned, the interface of bovine bc1 and CcO consists of bc1 subunits Cob, Rip1, and Qcr10, whereas the interface of yeast bc1 and CcO consists of bc1 subunits Cob, Cyt1, Qcr6, Qcr7, Qcr8, and Qcr9 (5, 6). Qcr10 is not present in the yeast bc1 structure (46). The stippled cylinder between bc1 and CcO is a representation of where Rcf1 may reside. No precise localization of Rcf1 is known from current structural studies of the supercomplex. The helix of Qcr9 packs on the outer surface of the Rip1 helix.
Expression of N-Rip1 increased the abundance of both tetrameric III2IV2 and trimeric III2IV1 bc1-CcO supercomplexes in bc1 mutants stalled at the late core assembly intermediate as well as in WT cells, in part through enhanced formation of CcO. This finding is the first example of increased supercomplex abundance arising from elevated CcO levels. This effect is readily apparent in WT yeast in the BY4741 background, but not in the W303 background, in which the basal levels of CcO are significantly higher. It should be noted that the restricted effect of N-Rip1 in BY4741 cells, and not W303 cells, is not related to the known mutant allele of Hap1 found in BY4741 cells (derived from S288c); expression of WT Hap1 in BY4741 cells failed to change the abundance of CcO or supercomplexes (data not shown).
A major observation of this study is that N-Rip1 facilitates CcO formation in a bc1-dependent manner. The induced CcO formation does not arise from stimulated mitochondrial translation of Cox1, Cox2, and Cox3. CcO biogenesis initiates with the mitochondrial synthesis and membrane insertion of the Cox1 subunit. Cox1 associates with a series of assembly factors and nuclear encoded subunits (Cox5a, Cox6, and Cox8) in early assembly intermediates, and Cox4 is added at a subsequent step (36). The observed enhancement of CcO formation in bcs1Δ and WT cells with Cox4 overexpression is consistent either with the availability of Cox4 being a rate-limiting step in CcO formation or with high Cox4 levels enhancing the stability of an early Cox1 assembly intermediate, allowing it to progress toward maturation rather than degradation. A likely scenario for the observed N-Rip1-induced formation of CcO is that the initial interaction of N-Rip1 with bc1 near the interface with CcO leads to enhanced binding of a CcO assembly intermediate whose maturation is limited in WT cells. The presence of a bc1 complex with a candidate-enhanced attachment interface may serve to template the full maturation of CcO, leading to elevated CcO levels. According to this model, Cox4 and Cox5a may be subunits that are limiting to CcO maturation, and the proposed enhanced interaction with bc1 may decrease the turnover rate of the Cox1 intermediate to which these subunits bind.
It is likely that N-Rip1 transiently associates with bc1 near the interface and induces an interface alteration that favors docking by a Cox1 assembly intermediate. Rcf1 is predicted to reside at the interface of the bc1-CcO supercomplex (11) and may be close to where N-Rip1 associates to achieve its effect on supercomplex stabilization (Fig. 5).
These data are consistent with a model in which bc1 or its late stage assembly intermediate can template the maturation of CcO, which has not been observed previously. The N-terminal helix of Rip1 is not predicted to reside at the interface of bc1 and CcO based on the yeast cryo-EM image reconstruction model, but it is at the interface in the bovine reconstruction model (Fig. 5). Alternatively, these studies may suggest that the Rip1 N-terminal helix docks initially near the interface and may later move to its final docking site upon binding of Qcr10. Further refinement in the cryo-EM maps is needed to address the apparent differences in the interfaces of bovine and yeast bc1 and CcO complexes.
We have shown that supercomplex stabilization can be restored in cells lacking CL. We showed that increased formation of CcO is sufficient to enhance supercomplex stabilization in the absence of CL synthase (Crd1). ETC supercomplexes in yeast are destabilized on BN-PAGE in crd1Δ cells, although the assemblies have been reported to persist on clear native PAGE (13, 40). Thus, CL is not essential for the formation of the respiratory supercomplexes, but it is a key stabilizing factor for supercomplexes as well as the individual complexes (41, 42).
The abundance of ETC supercomplexes is enhanced to a greater extent in crd1Δ cells with the overexpression of the CcO peripheral subunit Cox4 or Cox5a rather than N-Rip1. The high levels of CcO are distributed in trimeric as well as tetrameric supercomplexes in the CL-deficient cells. It appears that, in the absence of CL, mass action of high CcO levels can restore both bc1-CcO supercomplexes. CL binding appears to neutralize positive charges that may destabilize supercomplexes due to charge repulsion in the absence of CL (43). In the absence of CL, something else may contribute to charge neutralization in bc1. It is intriguing that even with the elevation in CcO formation, the trimeric supercomplex persists and is not converted to the tetrameric species. An incomplete understanding exists as to what differentiates trimeric from tetrameric supercomplexes, other than Rcf1 and Cox13.
The dramatic elevation of CcO levels in crd1Δ cells containing either N-Rip1 or Cox4 (or Cox5a) arises from two distinct effects: the N-Rip1 effect on CcO formation is dependent on the presence of bc1, whereas the Cox4 effect is independent of bc1. It was reported previously that CcO subunits, including Cox4, were attenuated in abundance in crd1Δ cells cultured at 37 °C (44). However, at 30 °C, Cox4 levels were near normal. The striking induction of CcO formation by elevated expression of Cox4 or Cox5a in crd1Δ cells suggests that these subunits may still be limiting at 30 °C. We suggest that expression of either subunit may impart sufficient stability to a Cox1 assembly intermediate to allow its full maturation. One implication of this model is that cells may contain some level of a Cox1 assembly intermediate that is not matured and is subsequently degraded. This is suggested to occur in mammalian cells in the maturation of Cox1 (45).
Acknowledgments
We thank Drs. Hyung Kim and Mi-Young Jeong for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant ES03817 from NIEHS (to D. R. W.) and Training Grant T32 HL007576-25 (to J. L. F.).
- ETC
- electron transfer chain
- CcO
- cytochrome c oxidase
- BN-PAGE
- blue native PAGE
- CL
- cardiolipin
- IM
- inner membrane
- TAP
- tandem affinity purification tag
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Winge D. R. (2012) Sealing the mitochondrial respirasome. Mol. Cell. Biol. 32, 2647–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heinemeyer J., Braun H. P., Boekema E. J., Kouril R. (2007) A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J. Biol. Chem. 282, 12240–12248 [DOI] [PubMed] [Google Scholar]
- 3. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 4. Schagger H. (2001) Respiratory chain supercomplexes. IUBMB Life 52, 119–128 [DOI] [PubMed] [Google Scholar]
- 5. Mileykovskaya E., Penczek P. A., Fang J., Mallampalli V. K., Sparagna G. C., Dowhan W. (2012) Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J. Biol. Chem. 287, 23095–23103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dudkina N. V., Kudryashev M., Stahlberg H., Boekema E. J. (2011) Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl. Acad. Sci. U.S.A. 108, 15196–15200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dienhart M. K., Stuart R. A. (2008) The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol. Biol. Cell 19, 3934–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Claypool S. M., Oktay Y., Boontheung P., Loo J. A., Koehler C. M. (2008) Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Strogolova V., Furness A., Robb-McGrath M., Garlich J., Stuart R. A. (2012) Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell. Biol. 32, 1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y. C., Taylor E. B., Dephoure N., Heo J. M., Tonhato A., Papandreou I., Nath N., Denko N. C., Gygi S. P., Rutter J. (2012) Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 15, 348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vukotic M., Oeljeklaus S., Wiese S., Vögtle F. N., Meisinger C., Meyer H. E., Zieseniss A., Katschinski D. M., Jans D. C., Jakobs S., Warscheid B., Rehling P., Deckers M. (2012) Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 15, 336–347 [DOI] [PubMed] [Google Scholar]
- 12. Cruciat C. M., Brunner S., Baumann F., Neupert W., Stuart R. A. (2000) The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275, 18093–18098 [DOI] [PubMed] [Google Scholar]
- 13. Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schägger H. (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 [DOI] [PubMed] [Google Scholar]
- 14. Bazán S., Mileykovskaya E., Mallampalli V. K., Heacock P., Sparagna G. C., Dowhan W. (2013) Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J. Biol. Chem. 288, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xia D., Yu C. A., Kim H., Xia J. Z., Kachurin A. M., Zhang L., Yu L., Deisenhofer J. (1997) Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277, 60–66 [DOI] [PubMed] [Google Scholar]
- 16. Iwata S., Lee J. W., Okada K., Lee J. K., Iwata M., Rasmussen B., Link T. A., Ramaswamy S., Jap B. K. (1998) Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281, 64–71 [DOI] [PubMed] [Google Scholar]
- 17. Zara V., Conte L., Trumpower B. L. (2007) Identification and characterization of cytochrome bc1 subcomplexes in mitochondria from yeast with single and double deletions of genes encoding cytochrome bc1 subunits. FEBS J. 274, 4526–4539 [DOI] [PubMed] [Google Scholar]
- 18. Zara V., Conte L., Trumpower B. L. (2009) Evidence that the assembly of the yeast cytochrome bc1 complex involves the formation of a large core structure in the inner mitochondrial membrane. FEBS J. 276, 1900–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crivellone M. D., Wu M. A., Tzagoloff A. (1988) Assembly of the mitochondrial membrane system. Analysis of structural mutants of the yeast coenzyme QH2-cytochrome c reductase complex. J. Biol. Chem. 263, 14323–14333 [PubMed] [Google Scholar]
- 20. Gruschke S., Römpler K., Hildenbeutel M., Kehrein K., Kühl I., Bonnefoy N., Ott M. (2012) The Cbp3-Cbp6 complex coordinates cytochrome b synthesis with bc1 complex assembly in yeast mitochondria. J. Cell Biol. 199, 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kronekova Z., Rödel G. (2005) Organization of assembly factors Cbp3p and Cbp4p and their effect on bc1 complex assembly in Saccharomyces cerevisiae. Curr. Genet. 47, 203–212 [DOI] [PubMed] [Google Scholar]
- 22. Gruschke S., Kehrein K., Römpler K., Gröne K., Israel L., Imhof A., Herrmann J. M., Ott M. (2011) Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J. Cell Biol. 193, 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cruciat C. M., Hell K., Fölsch H., Neupert W., Stuart R. A. (1999) Bcs1p, an AAA-family member, is a chaperone for the assembly of the cytochrome bc1 complex. EMBO J. 18, 5226–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wagener N., Ackermann M., Funes S., Neupert W. (2011) A pathway of protein translocation in mitochondria mediated by the AAA-ATPase Bcs1. Mol. Cell 44, 191–202 [DOI] [PubMed] [Google Scholar]
- 25. Mühlenhoff U., Richter N., Pines O., Pierik A. J., Lill R. (2011) Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe-4S] proteins. J. Biol. Chem. 286, 41205–41216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Atkinson A., Khalimonchuk O., Smith P., Sabic H., Eide D., Winge D. R. (2010) Mzm1 influences a labile pool of mitochondrial zinc important for respiratory function. J. Biol. Chem. 285, 19450–19459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Atkinson A., Smith P., Fox J. L., Cui T. Z., Khalimonchuk O., Winge D. R. (2011) The LYR protein Mzm1 functions in the insertion of the Rieske Fe/S protein in yeast mitochondria. Mol. Cell. Biol. 31, 3988–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cui T. Z., Smith P. M., Fox J. L., Khalimonchuk O., Winge D. R. (2012) Late-stage maturation of the Rieske Fe/S protein: Mzm1 stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol. Cell. Biol. 32, 4400–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pierrel F., Khalimonchuk O., Cobine P. A., Bestwick M., Winge D. R. (2008) Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis facilitating the maturation of Cox1. Mol. Cell. Biol. 28, 4927–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 31. Capaldi R. A., Marusich M. F., Taanman J.-W. (1995) Mammalian cytochrome c oxidase: characterization of enzyme and immunological detection of subunits in tissue extracts and whole cells. Methods Enzymol. 260, 117–132 [DOI] [PubMed] [Google Scholar]
- 32. Ferramosca A., Conte A., Burri L., Berge K., De Nuccio F., Giudetti A. M., Zara V. (2012) A krill oil supplemented diet suppresses hepatic steatosis in high-fat fed rats. PLoS ONE 7, e38797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zara V., Conte L., Trumpower B. L. (2009) Biogenesis of the yeast cytochrome bc1 complex. Biochim. Biophys. Acta 1793, 89–96 [DOI] [PubMed] [Google Scholar]
- 34. Williams S. L., Valnot I., Rustin P., Taanman J.-W. (2004) Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1 or SURF1. J. Biol. Chem. 279, 7462–7469 [DOI] [PubMed] [Google Scholar]
- 35. Mick D. U., Wagner K., van der Laan M., Frazier A. E., Perschil I., Pawlas M., Meyer H. E., Warscheid B., Rehling P. (2007) Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26, 4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McStay G. P., Su C. H., Thomas S. M., Xu J. T., Tzagoloff A. (2013) Characterization of Cox1p assembly intermediates in Saccharomyces cerevisiae. J. Biol. Chem. 288, 26546–26556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McStay G. P., Su C. H., Tzagoloff A. (2013) Modular assembly of yeast cytochrome oxidase. Mol. Biol. Cell 24, 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee S. Y., Hunte C., Malaney S., Robinson B. H. (2001) The N-terminus of the Qcr7 protein of the cytochrome bc1 complex in S. cerevisiae may be involved in facilitating stability of the subcomplex with the Qcr8 protein and cytochrome b. Arch. Biochem. Biophys. 393, 215–221 [DOI] [PubMed] [Google Scholar]
- 39. Lapuente-Brun E., Moreno-Loshuertos R., Acín-Pérez R., Latorre-Pellicer A., Colás C., Balsa E., Perales-Clemente E., Quirós P. M., Calvo E., Rodríguez-Hernández M. A., Navas P., Cruz R., Carracedo Á., López-Otín C., Pérez-Martos A., Fernández-Silva P., Fernández-Vizarra E., Enríquez J. A. (2013) Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 [DOI] [PubMed] [Google Scholar]
- 40. Zhang M., Mileykovskaya E., Dowhan W. (2002) Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277, 43553–43556 [DOI] [PubMed] [Google Scholar]
- 41. Lange C., Nett J. H., Trumpower B. L., Hunte C. (2001) Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 20, 6591–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shinzawa-Itoh K., Aoyama H., Muramoto K., Terada H., Kurauchi T., Tadehara Y., Yamasaki A., Sugimura T., Kurono S., Tsujimoto K., Mizushima T., Yamashita E., Tsukihara T., Yoshikawa S. (2007) Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wenz T., Hielscher R., Hellwig P., Schägger H., Richers S., Hunte C. (2009) Role of phospholipids in respiratory cytochrome bc1 complex catalysis and supercomplex formation. Biochim. Biophys. Acta 1787, 609–616 [DOI] [PubMed] [Google Scholar]
- 44. Zhang M., Su X., Mileykovskaya E., Amoscato A. A., Dowhan W. (2003) Cardiolipin is not required to maintain mitochondrial DNA stability or cell viability for Saccharomyces cerevisiae grown at elevated temperatures. J. Biol. Chem. 278, 35204–35210 [DOI] [PubMed] [Google Scholar]
- 45. Weraarpachai W., Sasarman F., Nishimura T., Antonicka H., Auré K., Rötig A., Lombès A., Shoubridge E. A. (2012) Mutations in C12orf62, a factor that couples COX I synthesis with cytochrome c oxidase assembly, cause fatal neonatal lactic acidosis. Am. J. Hum. Genet. 90, 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lange C., Hunte C. (2002) Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 99, 2800–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]