Background: 13C hyperpolarization sensitively and non-destructively detects pyruvate-lactate exchanges in cancer cells.
Results: Combining 13C hyperpolarization with isotopomer analysis allowed many pyruvate-dependent pathways to be quantified simultaneously.
Conclusion: Monitoring H[13C]O3− production from hyperpolarized [1-13C]pyruvate yielded a quantitative readout of oncogene-regulated pyruvate dehydrogenase activity.
Significance: This approach might enable a broader quantitative assessment of metabolic activity in tumors.
Keywords: Cancer, Imaging, Metabolism, Nuclear Magnetic Resonance, Pyruvate, Anaplerosis, Hyperpolarization
Abstract
Metabolic reprogramming facilitates cancer cell growth, so quantitative metabolic flux measurements could produce useful biomarkers. However, current methods to analyze flux in vivo provide either a steady-state overview of relative activities (infusion of 13C and analysis of extracted metabolites) or a dynamic view of a few reactions (hyperpolarized 13C spectroscopy). Moreover, although hyperpolarization has successfully quantified pyruvate-lactate exchanges, its ability to assess mitochondrial pyruvate metabolism is unproven in cancer. Here, we combined 13C hyperpolarization and isotopomer analysis to quantify multiple fates of pyruvate simultaneously. Two cancer cell lines with divergent pyruvate metabolism were incubated with thermally polarized [3-13C]pyruvate for several hours, then briefly exposed to hyperpolarized [1-13C]pyruvate during acquisition of NMR spectra using selective excitation to maximize detection of H[13C]O3− and [1-13C]lactate. Metabolites were then extracted and subjected to isotopomer analysis to determine relative rates of pathways involving [3-13C]pyruvate. Quantitation of hyperpolarized H[13C]O3− provided a single definitive metabolic rate, which was then used to convert relative rates derived from isotopomer analysis into quantitative fluxes. This revealed that H[13C]O3− appearance reflects activity of pyruvate dehydrogenase rather than pyruvate carboxylation followed by subsequent decarboxylation reactions. Glucose substantially altered [1-13C]pyruvate metabolism, enhancing exchanges with [1-13C]lactate and suppressing H[13C]O3− formation. Furthermore, inhibiting Akt, an oncogenic kinase that stimulates glycolysis, reversed these effects, indicating that metabolism of pyruvate by both LDH and pyruvate dehydrogenase is subject to the acute effects of oncogenic signaling on glycolysis. The data suggest that combining 13C isotopomer analyses and dynamic hyperpolarized 13C spectroscopy may enable quantitative flux measurements in living tumors.
Introduction
Cancer cells display metabolic properties that distinguish them from surrounding, normal tissue. Tumor metabolism is regulated by oncogenes and tumor suppressors (1–4), suggesting that mutations in these genes orchestrate metabolic activities to support an environment conducive to malignancy (5, 6). In accordance with this view, mutations in a subset of metabolic enzymes, including fumarate hydratase, the succinate dehydrogenase complex, and isocitrate dehydrogenases-1 and -2 cause significant perturbations in intermediary metabolism and contribute directly to cancer (7–11). Metabolism is also highly sensitive to the effects of the tumor microenvironment, particularly hypoxia, which correlates with chemotherapeutic resistance and poor patient survival (12). Thus, the ability to measure metabolic flux in live tumors would potentially report clinically and prognostically valuable information.
Imaging of hyperpolarized 13C-enriched substrates is emerging as a promising technology for cancer diagnosis and monitoring because it enables the real-time detection of enzyme-catalyzed metabolic activities in tissues (13–18). Recently, imaging has also been shown to be capable of detecting malignant lesions in the human prostate, even diagnosing masses that were not positively identified by other methods (19). These studies revealed that exchanges between hyperpolarized pyruvate and lactate are readily detected in malignant cells and tend to be enhanced in tumors relative to nonmalignant tissue. Hyperpolarization involves the temporary redistribution of the populations of the available energy levels into a non-equilibrium state, enabling a massive gain in magnetic resonance signal. For 13C, this gain can exceed 10,000-fold (20), enabling detection of both a 13C-enriched substrate and some downstream metabolites with a temporal resolution of seconds. Hyperpolarized [1-13C]pyruvate is widely used to interrogate cancer metabolism, in part because the long T1 of this carboxyl carbon allows the hyperpolarized state to persist for multiple metabolic steps and also because pyruvate is located in a pivotal intersection of intermediary metabolism involving lactate, alanine, and oxidation in the tricarboxylic acid (TCA) cycle.
Cancer cells often display high rates of glycolysis and lactate secretion even when oxygen is plentiful (the Warburg effect). This oncogene-driven phenomenon contributes to the utility of tumor detection by uptake of the glucose analog [18F]fluoro-2-deoxyglucose (3). Abundant expression of lactate dehydrogenase (LDH) and the presence of a large lactate pool in tumor cells allows for rapid exchange between hyperpolarized [1-13C]pyruvate and [1-13C]lactate, making detection of [1-13C]lactate appealing for in vivo detection of cancer and for monitoring response to therapy (15, 21). However, cancer cells also oxidize pyruvate in the mitochondria, producing both energy and macromolecular precursors for cell growth (23). This is of particular interest because lung tumors, gliomas, and metastatic brain tumors have all been demonstrated to oxidize pyruvate in vivo in humans and mice (24–28). Therefore, assessment of both pyruvate/lactate exchanges and pyruvate oxidation in the mitochondria would provide a much more comprehensive view of cancer cell metabolism than lactate formation alone.
We previously used conventional 13C NMR spectroscopy to evaluate fluxes through competing metabolic pathways supplied by pyruvate, including LDH and the TCA cycle, in cultured cancer cells (29, 30). These same activities were detected in mouse and human tumors by infusing 13C-enriched glucose before surgery, extracting metabolites from surgically resected tumor tissue, and analyzing 13C enrichment patterns by NMR (26, 28). We also used hyperpolarized [1-13C]pyruvate to quantify flux into lactate (31). Here, we combined these methods to study two metabolically distinct cancer cell lines. First, we incubated cancer cells with thermally polarized [3-13C]pyruvate for several hours to produce steady-state labeling of metabolic intermediates. Next, using a selective excitation pulse to maximize detection of H[13C]O3− and [1-13C]lactate, we subjected cells to hyperpolarized [1-13C]pyruvate to measure flux into lactate and the TCA cycle. Combining the rate of pyruvate decarboxylation with steady-state isotopomer data provided a method to evaluate absolute flux rates through a variety of reactions associated with the TCA cycle.
EXPERIMENTAL PROCEDURES
Cell Culture Reagents and Basic Metabolism Experiments
Two cell lines, SF188-derived glioblastoma cells overexpressing human Bcl-xL (SFxL) and Huh-7 hepatocellular carcinoma cells were maintained in culture as described previously (30, 32, 33). Metabolic experiments were performed in Dulbecco's modified Eagle's medium (DMEM) prepared from powder lacking glucose, glutamine, phenol red, sodium pyruvate, and sodium bicarbonate. This basal medium was supplemented with 4 mmol/liter l-glutamine, 10% dialyzed fetal calf serum, 42.5 mmol/liter sodium bicarbonate, 25 mmol/liter HEPES, 10 units/ml penicillin, and 10 μg/ml streptomycin. Glucose and pyruvate were added as indicated for each experiment. To measure the rates of metabolite consumption/excretion in the medium, glucose, lactate, glutamine, and glutamate were measured using a BioProfile Basic 4 analyzer (NOVA Biomedical), and ammonia was measured using a spectrophotometric assay (Megazyme). For oxygen consumption assays, cells were harvested by trypsinization, suspended in fresh medium at a concentration of 108 cells/ml, and transferred to an Oxygraph water-jacketed oxygen electrode (Hansatech). The Akt inhibitor was Akt Inhibitor VIII (Calbiochem).
Pyruvate Decarboxylation Assay
Decarboxylation of [1-14C]pyruvate was measured essentially as described (34). Micro-bridges (Hampton Research) were placed into wells of a 24-well plate with one piece of 0.6 × 1 cm2 chromatography paper in each. Assay medium was prepared by supplementing DMEM (containing 10% fetal calf serum, 4 mm glutamine, and 6 mm sodium pyruvate) with 2.2 μCi of [1-14C]pyruvate. This medium was warmed to 37 °C and incubated for 2 h to remove any 14CO2 produced from spontaneous decarboxylation, then an aliquot was used to quantify radioactivity on a scintillation counter. This value was used to determine the specific activity of pyruvate, assuming a total pyruvate concentration of 6 mm. The specific activity ranged from 50 to 120 cpm/nmol of pyruvate. One million cells per well were then suspended in 370 μl of assay medium on ice. Each micro-bridge was moistened with 30 μl of 2 n NaOH, and the plate was sealed with adhesive film. Pyruvate metabolism was initiated by transferring the plate to a 37 °C water bath. After 15 min, metabolism was terminated by adding 50 μl of 20% trichloroacetic acid. The plate was re-sealed with adhesive film and incubated at 37 °C for another 60 min to release 14CO2 completely. Then the 14CO2-containing chromatography papers were collected for scintillation counts. Pyruvate oxidation rates were determined from the total specific activity in the reaction and were reported as nmol/106 cells/h. Wells containing culture medium but no cells were used to establish background levels of 14CO2. This level was subtracted from cell-containing wells.
Gas Chromatography/Mass Spectrometry
Mass spectrometry experiments were performed essentially as described previously (30). Cultures of 80–90% confluent cells grown in 60-mm dishes were rinsed twice in phosphate-buffered saline then overlaid with DMEM-based medium containing 10% dialyzed fetal calf serum, 4 mm glutamine, and 6 mm pyruvate (either [1-13C]pyruvate or [3-13C]pyruvate; Cambridge Isotope Laboratories). At the end of the culture period, the medium was removed, and the cells were briefly rinsed in ice-cold normal saline. A cold solution of 50% methanol, 50% water was added, and the cells were lysed using repeated freeze-thaw cycles. After centrifugation to remove debris, the samples were evaporated and derivatized by trimethylsilylation (Tri-Sil HTP reagent; Thermo). An aliquot of 1–3 μl was injected onto an Agilent 6970 gas chromatograph networked to an Agilent 5973 Mass Selective Detector. Fragment ions m/z 334–338 and 465–471 were used to monitor enrichment in aspartate and citrate, respectively (30). The measured distribution of mass isotopomers was corrected for natural abundance of 13C as described (35).
NMR Spectroscopy of Extracted Metabolites
Cells were cultured to 80–90% confluence in eight 150-mm dishes. Freshly prepared DMEM with 10% dialyzed fetal calf serum, 0 mm glucose, 4 mm glutamine, standard concentrations of other amino acids, and 6 mm [3-13C]pyruvate was added to each dish, and the cells were cultured for 6 h, then either harvested immediately for extraction or collected for hyperpolarization experiments. For hyperpolarization, the cells were trypsinized, pelleted by centrifugation, and resuspended at a concentration of 1 × 108 cells/ml in fresh DMEM lacking glucose and pyruvate but containing 10% dialyzed fetal calf serum, 4 mm glutamine, and standard concentrations of other amino acids. This suspension was mixed thoroughly, then transferred to a 10-mm NMR tube that already contained hyperpolarized [1-13C]pyruvate so that the final concentration was 6 mm, precisely mimicking the nutrient availability of the steady-state labeling experiment. Acquisition of spectra was initiated immediately and proceeded for ∼3.5 min (see “Hyperpolarization, Shaped Pulse, and NMR Spectroscopy in Intact Cells” below for details on cell transfer and conditions for the hyperpolarization experiment). We previously reported that cells subjected to trypsinization and hyperpolarized [1-13C]pyruvate maintain rates of LDH flux similar to rates of adherent cells (31). To protect against depletion of oxygen from the medium during trypsinization and preparation of the cell suspension, the medium was mixed frequently, including immediately before transfer of the cells to the NMR tube, which occurred within a few seconds of introduction of [1-13C]pyruvate. Immediately after the hyperpolarization experiment, the cells were pelleted by centrifugation and frozen in liquid nitrogen. Frozen cell pellets were homogenized by sonication in 4 ml of 4% ice-cold perchloric acid. After centrifuging to remove debris, the acid-soluble material was neutralized with 8 n potassium hydroxide and centrifuged. Supernatants were subjected to lyophilization, reconstituted in deuterium oxide, and titrated to pH 7 for NMR analysis in a 3-mm tube.
NMR spectroscopy was performed on a Varian INOVA 14.1 T spectrometer (Agilent Instruments, Walnut Creek, CA) equipped with a 3-mm broadband probe with the observe coil tuned to 13C (150 MHz). Proton decoupling was performed using a standard WALTZ-16 pulse sequence. Carbon spectra were acquired under the following conditions: pulse flip angle 45°, repetition time 1.5 s, spectral width 35 kHz, number of data points 104,986, and number of scans 7,000–30,000, requiring 6–25 h. Free induction decays were zero-filled to 131,072 points and apodized with exponential multiplication. Peak areas were determined using ACD Labs SpecManager (Advanced Chemistry Development, Toronto, Canada). Glutamate isotopomer analysis to calculate FC2 (the fraction of acetyl-CoA labeled in position-2 with 13C) in Fig. 3 was performed using equations described in Malloy et al. (36). Relative flux values for the combined steady-state/hyperpolarization experiments in Table 1, including FC2 and ys (the rate of anaplerosis relative to citrate synthase flux), were calculated by tcaCALC (37).
FIGURE 3.
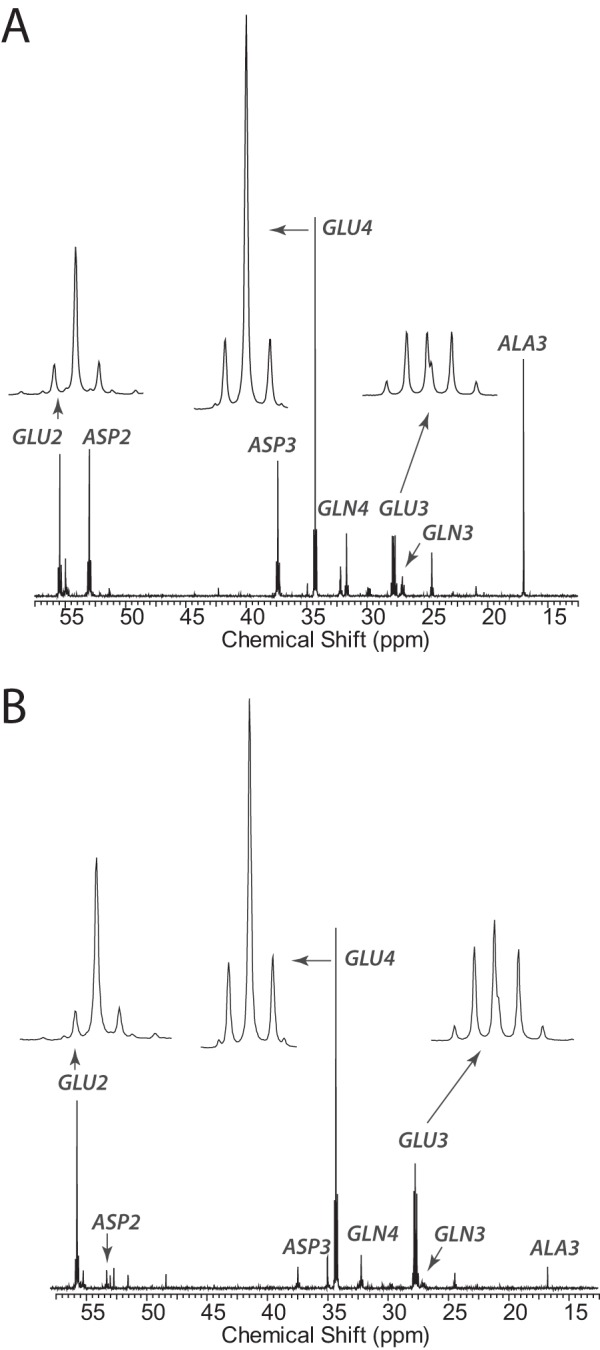
13C NMR steady-state analysis of pyruvate metabolism. A, carbon 13 NMR spectrum of metabolites extracted from SFxL cells after 6 h of culture in [3-13C]pyruvate. B, carbon 13 NMR spectrum of metabolites extracted from Huh-7 cells after 6 h of culture in [3-13C]pyruvate. GLU, glutamate; ASP, aspartate; GLN, glutamine; ALA, alanine.
TABLE 1.
Relative flux through various pathways in two different cancer cell lines as reported by a 13C isotopomer analysis of tissue glutamate
All fluxes are relative to a tricarboxylic acid cycle (TCAC) flux = 1.
| Fraction of acetyl-CoA derived from pyruvate (FC2) | PDH flux | Anaplerotic flux into the TCAC via PC (yPC) | PK flux | Anaplerotic flux of unlabeled substrates into the TCAC (yS) | |
|---|---|---|---|---|---|
| SFxL cells | 0.58 | 0.93 | 0.25 | 0 | 0.85 |
| 0.58 | 0.94 | 0.21 | 0 | 0.80 | |
| 0.93 | 0.56 | 0 | 0 | 0.45 | |
| Average | 0.70 | 0.81 | 0.15 | 0 | 0.70 |
| S.D. | 0.20 | 0.22 | 0.13 | 0 | 0.22 |
| Huh-7 cells | 0.61 | 1 | 1.18 | 0.27 | 0.33 |
| 0.62 | 0.96 | 0.90 | 0.07 | 0.32 | |
| 0.83 | 0.90 | 1.21 | 0.62 | 0.53 | |
| Average | 0.69 | 0.95 | 1.10 | 0.32 | 0.39 |
| S.D. | 0.12 | 0.05 | 0.17 | 0.28 | 0.12 |
| p value | 0.95 | 0.33 | 0.002 | 0.12 | 0.10 |
Hyperpolarization, Shaped Pulses, and NMR Spectroscopy of Intact Cells
A solution of [1-13C]pyruvic acid containing 15 mmol/liter OX63 radical was prepared for polarization. 8.6 μl of the solution was polarized for 2 h in an Oxford HyperSense dynamic nuclear polarization system (Oxford Instruments Molecular Biotools Ltd, Oxfordshire, UK) at 1.4 K with a microwave irradiation frequency of 94.125 GHz at 100 milliwatts. The frozen sample was dissolved with 4 ml of 15.3 mm sodium bicarbonate (heated to 190 °C and pressurized to 10 bar) in less than 10 s. An aliquot of the resulting solution was added to a 10-mm NMR tube (400 μl for 2 ml total volume or 200 μl for 1 ml total volume). The tube was placed in a Varian 10-mm broadband probe in a 9.4 tesla magnet equipped with a VNMRS console (Agilent). The NMR tube was directly attached by a thin tube to a syringe containing the suspension of cells. At time zero of the acquisition, either 1.6 or 0.8 ml of the cell suspension (for final total volumes of 2 or 1 ml, respectively) was added to the hyperpolarized solution, resulting in a final pyruvate concentration of 6 mm and a cell concentration of 1.0 × 108 cells/ml.
Frequency selective pulses were utilized to avoid depolarization of the pyruvate C1 resonance. Pulses were created in the PBox software provided with VNMRJ (Agilent). The pulse profile was designed to excite the pyruvate C2 resonance (205.9 ppm), lactate C1 (183.3 ppm), and H[13C]O3− (160.1 ppm) with a Gaussian shape of narrow bandwidth (350–500 Hz). Profiles were fit to a triple Gaussian function,
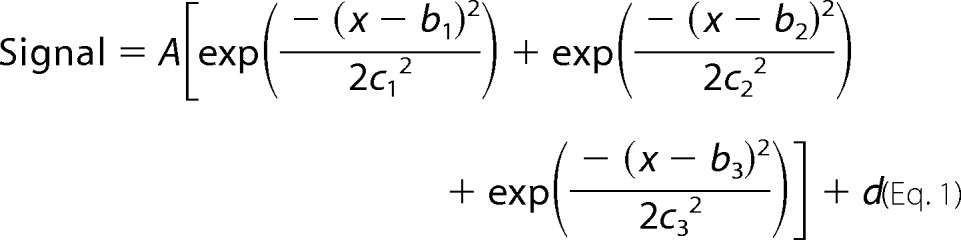 |
where A is the amplitude of excitation, b is the frequency location of each Gaussian shape, c is related to the width of the Gaussian shape such that the width at half-maximum height is 2(2ln(2))1/2c, and d is the signal amplitude in the absence of excitation; the maximum flip angle due to excitation is, therefore, inverse cosine(A + d). Selective pulses were applied every 2 s for a total of 100 13C spectra with an acquisition time of 1 s and a 1-s delay. Spectra were collected with a 32-kHz bandwidth and zero-filled before apodization and Fourier transformation. Using Bayesian Analysis software (38–41), the full set of 100 free induction decays were simultaneously fit to a five-resonance model with uncorrelated phase. Normalization of the peak areas was done using the pyruvate C2 resonance, and the lactate and bicarbonate areas were further adjusted to account for small differences in cell number among experiments.
In a separate set of experiments the decay of the signal due to radiofrequency depolarization was determined in the absence of cells using the selective pulse on a sample of hyperpolarized pyruvate. Each experiment began with 15 1° excitations with a 2-s repetition time followed by 15 selective pulses. The effective T1 was determined from the first set of pulses, and this value was used to extract the additional decay due to radiofrequency depolarization in the second set of 15 pulses. Both pulse profiles were tested with their corresponding sample volumes.
Modeling
The matrix shown in Scheme 1 was used to model flux as detected with hyperpolarized [1-13C]pyruvate, where ρi = 1/T1i. Pyr represents both extracellular and cytosolic [1-13C1]pyruvate, Lac represents total [1-13C1]lactate, MPyr is mitochondrial [1-13C1]pyruvate, and Bic is total H[13C]O3−. Because the cells were injected into a waiting sample of HP pyruvate, they experience an instantaneous increase in pyruvate concentration, and there was no need for a time-dependent delivery term. Export of lactate from the cells is not included because previous experiments showed this to be unnecessary to obtain accurate flux values (31). Inclusion of a mitochondrial pool of pyruvate was required to adequately model the initial delay detected in the H[13C]O3− time course. The model fits were done using 1000 sets of initial values as described previously (31). Radiofrequency depolarization rates were measured in cell-free phantom experiments, and T1 values were fit by the program.
SCHEME 1.
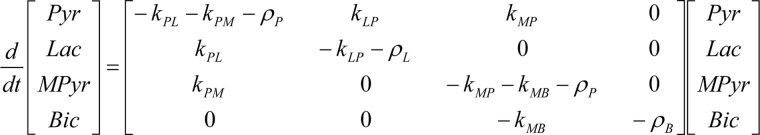
To establish absolute fluxes through other pathways associated with the TCA cycle, the rate of appearance of H[13C]O3− was used to reference all other TCA cycle fluxes. In SFxL cells, PDH4 flux was taken to be equivalent to total H[13C]O3− appearance as pyruvate cycling (PK) was inactive, and the contribution from oxidative reactions in the TCA cycle was assumed to be negligible (see “Results”). Because an isotopomer analysis yields metabolic fluxes relative to citrate synthase flux, absolute citrate synthase flux can be calculated as (absolute PDH flux)/(relative PDH flux). Remaining fluxes − the rate of anaplerosis via PC (yPC), the rate of entry of other unlabeled anaplerotic substrates into the cycle (yS), and flux through PK or other undefined pyruvate cycling pathways were obtained by multiplying the absolute citrate synthase flux by relative rates of these activities as derived from isotopomer analysis. Absolute cataplerosis was calculated by summing yPC and yS under steady state. Given that the parameter FC2 from isotopomer analysis in this experiment defines the fraction of acetyl-CoA that is derived from labeled pyruvate, the unlabeled fraction (FC0) from endogenous substrates is given by 1 − FC2. Thus, absolute fluxes from unlabeled substrates into the acetyl-CoA pool were calculated as (1 − FC2) × (absolute citrate synthase flux). In Huh-7 cells, a similar approach was taken, but additional calculations were used to account for the fact that PK was active. Due to symmetrization of 13C-labeled TCA cycle intermediates, the maximal amount of H[13C]O3− that can be produced from pyruvate cycling is half the total flux through that pathway. Thus, total H[13C]O3− production is equivalent to the sum of PDH flux and half of PK flux. The absolute PDH flux was calculated as (relative PDH flux from isotopomer analysis)/(total H[13C]O3− production). Other absolute fluxes were calculated as in SFxL cells.
RESULTS
Two Cancer Cell Lines with Distinct Pathways of Pyruvate Oxidation
We chose SFxL glioblastoma cells and Huh-7 hepatocellular carcinoma cells for this work. Both cell lines grow rapidly in culture (doubling time 22 ± 0.5 h in SFxL and 29 ± 0.1 h in Huh-7). Under the non-physiological conditions commonly used in mammalian cell culture (here, 10% fetal calf serum, 10 mm glucose, 4 mm glutamine, and room-air levels of oxygen), both cell lines consume glucose and glutamine rapidly and secrete lactate, glutamate, and ammonia into the extracellular space (Fig. 1A). Oxygen consumption was evident under these condition and persisted when glucose was replaced with 6 mm pyruvate, the same concentration used in experiments with hyperpolarized [1-13C]pyruvate (Fig. 1B). To determine the extent to which these cells could decarboxylate the C-1 position of extracellular pyruvate to CO2, cells were cultured with [1-14C]pyruvate, and 14CO2 was measured by scintillation counting. Both cell lines generated significant amounts of 14CO2 by this method, with Huh-7 cells producing it at a rate approximately three times higher than SFxL cells (Fig. 1C).
FIGURE 1.
Core metabolic activities in cancer cell lines with different routes of pyruvate oxidation. A, consumption and secretion of major nutrients in SFxL glioblastoma and Huh-7 hepatocellular carcinoma cells. Cells were cultured in complete medium containing 10 mm glucose and no pyruvate or no glucose and 6 mm pyruvate. Data are the average and S.D. of three cultures. B, oxygen consumption in both cell lines. C, oxidation of [1-14C]pyruvate to 14CO2 in both cell lines. Data are the average and S.D. of three cultures.
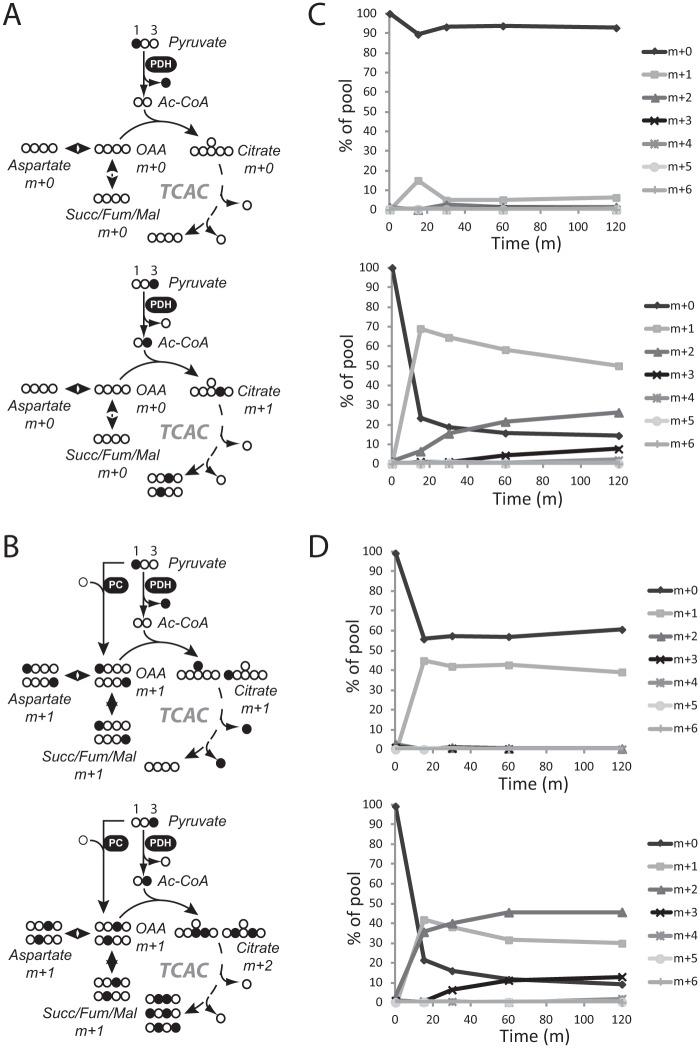
There are multiple mitochondrial pathways by which the carboxyl group of pyruvate can be released as CO2. First, PDH converts pyruvate to acetyl-CoA, releasing CO2 in the process (Fig. 2A). Alternatively, pyruvate can be carboxylated via PC to form oxaloacetate (OAA), and subsequent decarboxylation reactions involving either pyruvate cycling (42) or a forward turn of the cycle will release CO2 (Fig. 2B). To maximize detection of the various possible routes of pyruvate metabolism in SFxL and Huh-7 cells, each cell line was cultured in a large excess of pyruvate, with either [1-13C]pyruvate or [3-13C]pyruvate at a concentration of 6 mm in glucose-free medium. At several time points, the cells were freeze-clamped, metabolites were extracted, and mass spectrometry was used to determine 13C enrichment in TCA cycle intermediates. If PDH is active, but PC is not, citrate and other TCA cycle intermediates will be labeled by [3-13C]pyruvate but not by [1-13C]pyruvate (Fig. 2A). If both enzymes are active, then TCA cycle intermediates are labeled by both [3-13C]pyruvate and [1-13C]pyruvate (Fig. 2B). When cultured in [1-13C]pyruvate, Huh-7 cells demonstrated rapid and persistent 13C labeling in citrate and aspartate, but SFxL cells did not, indicating that only Huh-7 cells had high levels of PC activity (Fig. 2, C and D, and supplemental Fig. 1). When cultured in [3-13C]pyruvate, both cell lines transferred 13C to the TCA cycle (Fig. 2, C and D, and supplemental Fig. 1). However, citrate in SFxL cells was found to be labeled predominantly with one 13C (m+1), whereas m+2 and m+3 appeared only after a delay due to progression of citrate m+1 through complete turns of the cycle to acquire additional label from acetyl-CoA. In contrast, Huh-7 cells rapidly produced citrate m+2 and higher order labeling. These data demonstrate that SFxL cells possess PDH activity, whereas Huh-7 cells use both PDH and PC concurrently. These data are consistent with results from longer term cultures in which glucose instead of pyruvate was the source of 13C (29, 30).
FIGURE 2.
Pyruvate rapidly supplies the TCA cycle in both SFxL and Huh-7 cells. A, predicted labeling of TCA cycle intermediates when PDH is active and PC is inactive in cells labeled with [1-13C]pyruvate (top) or [3-13C]pyruvate (bottom). Filled symbols are 13C. B, predicted labeling of TCA cycle intermediates when both PDH and PC are active in cells labeled with [1-13C]pyruvate (top) or [3-13]pyruvate (bottom). C, time course of citrate mass isotopomers in SFxL cells labeled with [1-13C]pyruvate (top) or [3-13]pyruvate (bottom). D, time course of citrate mass isotopomers in Huh-7 cells labeled with [1-13C]pyruvate (top) or [3-13]pyruvate (bottom). Abbreviations: TCAC, TCA cycle; Succ/Mal/Fum, succinate/malate/fumarate; OAA, oxaloacetate.
To establish conditions that would support glutamate isotopomer analysis for determining relative flux values of pyruvate-dependent metabolic pathways, we cultured large populations of SFxL and Huh-7 cells in a DMEM-based medium containing 6 mm [3-13C]pyruvate and 4 mm glutamine and 10% fetal calf serum. After 6 h of culture, the cells were flash-frozen, and metabolites were extracted for NMR analysis. The resulting 13C NMR spectra had excellent signal-to-noise and showed well-resolved multiplets in glutamate carbons 2, 3, and 4 (Fig. 3), indicating that a complete isotopomer analysis would be supported by these culture conditions. In this labeling scheme, the large singlet in glutamate C4 reflects PDH activity, and the additional multiplets in carbons 2, 3, and 4 indicate that OAA was labeled with 13C. Isotopomer analysis of spectra from each cell line revealed that under these conditions, [3-13C]pyruvate was a major source of the acetyl-CoA used in the TCA cycle of both cell lines (70% in SFxL and nearly 60% in Huh-7).
A Selective Pulse Technique to Maximize Detection of Pyruvate Oxidation
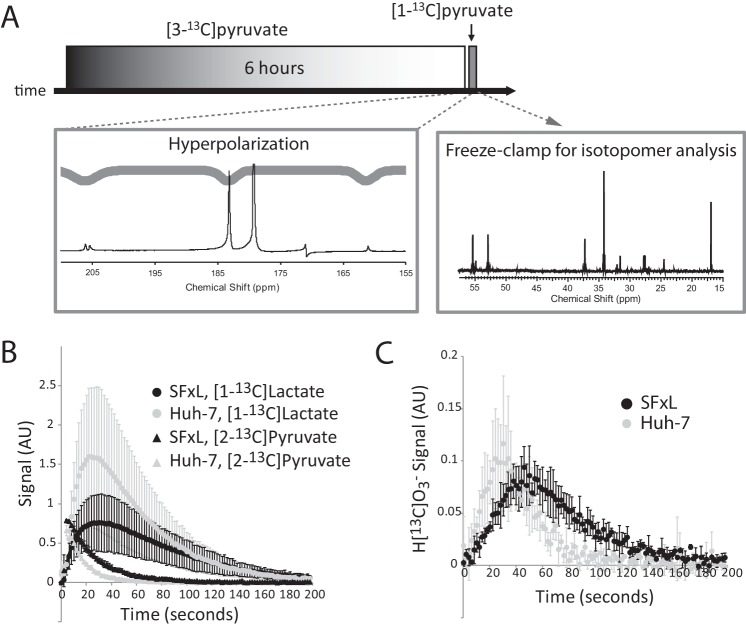
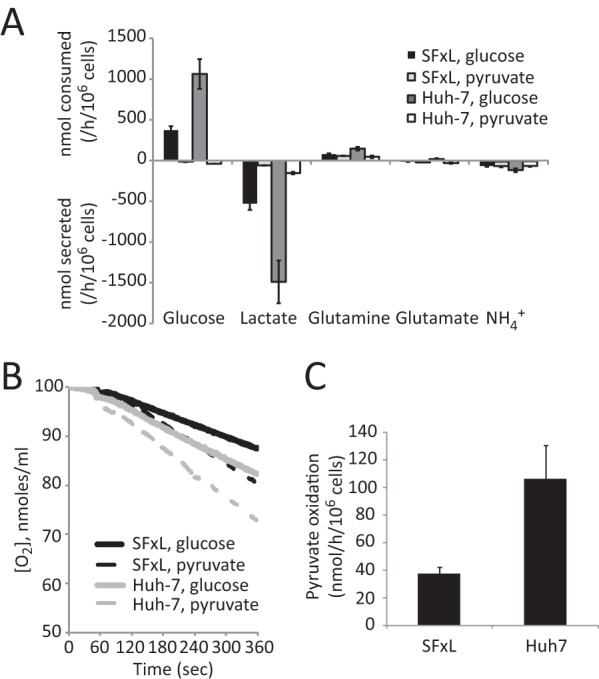
Most experiments involving hyperpolarized [1-13C]pyruvate in cancer cells report 13C exchanges between pyruvate and lactate but do not report production of carbon dioxide or bicarbonate. Having determined that SFxL and Huh-7 cells oxidize extracellular pyruvate within a few minutes, we tested whether this activity could be detected by 13C NMR using hyperpolarized pyruvate. A bolus of 6 mm hyperpolarized [1-13C]pyruvate was introduced to a suspension of SFxL cells, and 13C NMR spectra were continuously acquired for several minutes. When a non-selective excitation pulse was applied, a summation of spectra revealed labeling of lactate C1 (Fig. 4A), as previously reported for these cells (31). Labeling in alanine C1 and in C1 of pyruvate hydrate was also detected, but H[13C]O3− was not detected (Fig. 4A). Next, a frequency-selective excitation pulse was developed to optimize detection of H[13C]O3−. This shaped pulse selectively excites [1-13C]lactate and H[13C]O3− while avoiding excitation of [1-13C]pyruvate, thereby minimizing destruction of the hyperpolarized precursor (43). The pulse also excites [2-13C]pyruvate, which appears as a small doublet at 206 ppm because of natural abundance 13C at C2 (Fig. 5A). This resonance was used to monitor the abundance of hyperpolarized pyruvate in the absence of C1 excitation. When cells were analyzed under identical conditions using the new frequency-selective pulse, both [1-13C]lactate and H[13C]O3− were detected in individual scans as well as in summed spectra (Fig. 4B). This allowed a comparison of the dynamic labeling of [1-13C]lactate and H[13C]O3− (Fig. 4C). The time-dependent appearance of H[13C]O3− lagged behind the appearance of [1-13C]lactate, consistent with the expected faster kinetics of LDH relative to the various decarboxylation pathways, all of which require transport of pyruvate into the mitochondria.
FIGURE 4.
A selective pulse technique to detect oxidation of hyperpolarized pyruvate. A, summed spectra of a suspension of SFxL cells cultured with hyperpolarized [1-13C]pyruvate. NMR spectroscopy used a non-selective pulse. B, summed spectra of an identical suspension of SFxL cultured with hyperpolarized [1-13C]pyruvate but analyzed with a shaped pulse designed to excite [1-13C]lactate and [13C]bicarbonate. This pulse also excites [2-13C]pyruvate at 206 ppm. C, time course of [1-13C]lactate and [13C]bicarbonate signal in a suspension of SFxL cells analyzed using the shaped pulse.
FIGURE 5.
Two-stage analysis of pyruvate metabolism in cancer cells. A, overview of experimental scheme, detailed under “Results.” An example spectrum (sum of 50 free induction decays, approximately 3 min of culture time) and triple-Gaussian pulse Mz-profile corresponding to the hyperpolarized [1-13C]pyruvate experiment are shown on the bottom left. A representative 13C NMR spectrum from extracted metabolites, showing the effect of the 6-h culture in [3-13C]pyruvate is shown in the bottom right. B, average and S.D. for hyperpolarization experiments (n = 3) show the time course of detection of [1-13C]lactate and the natural abundance signal from carbon 2 of pyruvate in SFxL cells and Huh-7 cells. C, time course of the average and S.D. (n = 3) for H[13C]O3− signal in SFxL and Huh-7. AU, arbitrary units.
Combining Dynamic Measures of Flux with Steady-state 13C Isotopomer Analysis
If the sources of H[13C]O3− are known on a fractional basis from 13C isotopomer data, then it should be possible to use the rate of appearance of hyperpolarized H[13C]O3− to estimate fluxes through all pathways contributing to bicarbonate production. Because any decarboxylation reaction is essentially unidirectional, any ambiguities related to isotope exchange can be ignored. In the two cancer cell lines used here, hyperpolarized H[13C]O3− would be predominantly generated by PDH in SFxL cells, whereas PDH and PC could both contribute to H[13C]O3− production in Huh-7 cells. To obtain quantitative flux data for the two cell lines, a two-stage labeling experiment that combined isotopomer analysis with dynamic hyperpolarization data was performed (Fig. 5A). First, each cell line was incubated in DMEM containing 6 mm [3-13C]pyruvate, 4 mm glutamine, and 10% fetal calf serum for 6 h to achieve steady-state labeling of glutamate isotopomers. These same cells were then detached from culture dishes by brief exposure to trypsin and transferred to an NMR tube containing a solution of hyperpolarized [1-13C]pyruvate, also at a pyruvate concentration of 6 mm. The cells were transferred in DMEM containing 4 mm glutamine and 10% fetal calf serum such that they were exposed to the same nutrient milieu experienced during the 6-h incubation. The NMR tube was prepositioned in a Varian 10-mm broadband probe tuned to 13C. Serial excitation with the triple-Gaussian pulse was immediately initiated, and 13C NMR spectra were acquired until disappearance of the hyperpolarized 13C signal. At that point the cells were rapidly collected by centrifugation, frozen in liquid nitrogen, and extracted in acid to obtain the water soluble metabolites for isotopomer analysis. Dynamic labeling of both [1-13C]lactate and H[13C]O3− as well as the decay of signal from pyruvate C2 was apparent in both cell lines (Fig. 5, B and C).
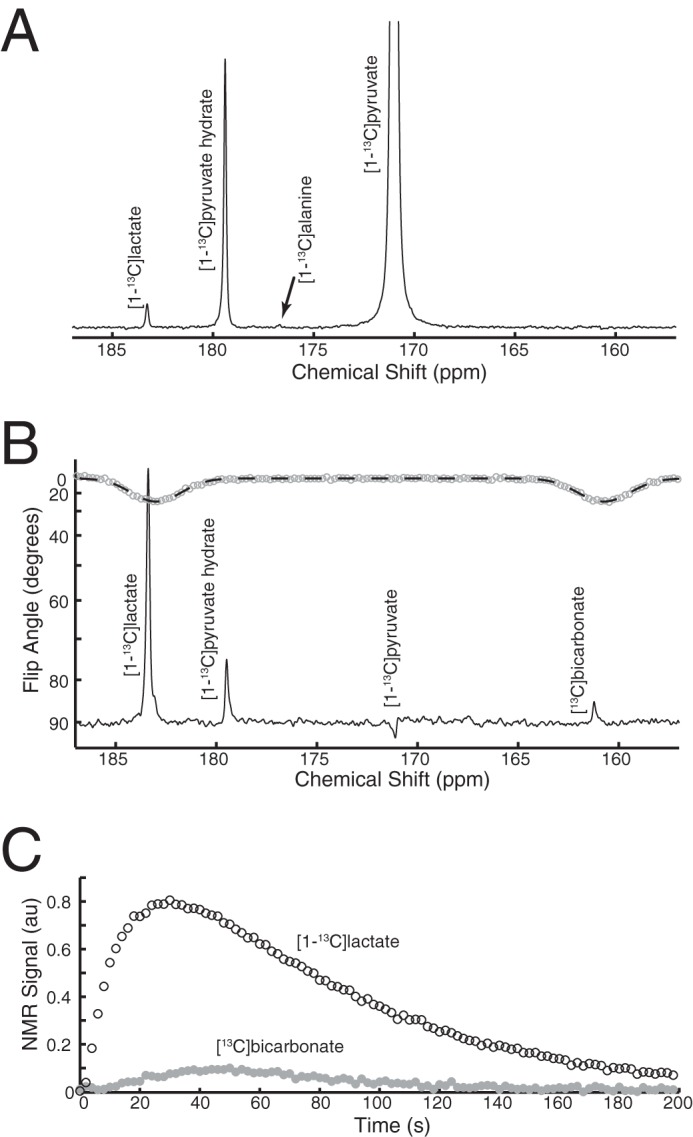
According to a previously established kinetic model (31), the initial LDH flux in these experiments was 190 ± 40 nmol/106 cells/h in SFxL and 380 ± 166 nmol/106 cells/h in Huh-7 (n = 3 for each cell line). Hyperpolarized H[13C]O3− appeared more rapidly on average in the Huh-7 cells, although the difference did not approach the increase in 14CO2 release measured in Fig. 1C. To determine the rate of appearance of H[13C]O3−, a multi-pool model was used to fit the hyperpolarization data. The model included exchanges between pyruvate and lactate, exchanges between cytosolic and mitochondrial pyruvate pools, and the conversion of mitochondrial pyruvate to H[13C]O3−. The fitting protocol generated 200 sets of randomized k values as starting values. Each starting value was chosen at random in the range of 0–2, with no other restrictions. Within each experiment (n = 3 for each cell line), the 50 best fits were essentially identical to each other. The top fit was used to calculate an overall rate of pyruvate decarboxylation (Fig. 6, A and B). When considering all six experiments, the rates of H[13C]O3− appearance in SFxL and Huh-7 cells were 38 ± 11 and 49 ± 20 nmol/h/106 cells, respectively. In SFxL cells, the averaged hyperpolarization data agreed closely with the 14CO2 experiment (39 versus 38 nmol/h/106 cells). In contrast, despite excellent fits generated by the model, the rate of formation of hyperpolarized H[13C]O3− in Huh-7 cells under-estimated the rate of 14CO2 measured in these cells (49 versus 100 nmol/h/106 cells).
FIGURE 6.
Kinetic analysis of pyruvate decarboxylation. Time courses for detection of signal for pyruvate carbons 1 and 2 (C1, C2), and H[13C]O3− in suspensions of cells treated with 6 mm hyperpolarized [1-13C]pyruvate. Results are shown for a typical experiment in SFxL cells (A) and Huh-7 cells (B). Rates of H[13C]O3− formation are derived from three independent experiments.
The data imply that hyperpolarization is unable to fully capture all decarboxylation reactions downstream of PC, possibly because of the time delay associated with these reactions and the rapid decay of the hyperpolarized signal. In contrast, the 14C experiments capture release of all 14CO2 from all reactions over the entire incubation period. To test the extent of PC-dependent 14CO2 formation, PC expression was silenced in SFxL and Huh-7 cells, and conversion of [1-14C]pyruvate to 14CO2 was re-measured. As expected, it was possible to suppress 14CO2 production by partially silencing PC expression, and this effect was much greater in Huh-7 cells (supplemental Fig. 2). We conclude that production of hyperpolarized H[13C]O3− from [1-13C]pyruvate in cancer cells primarily reflects PDH flux.
NMR spectroscopy of 13C-labeled metabolites extracted from each sample produced well resolved spectra, similar to those in Fig. 3. Analysis of glutamate isotopomers in these spectra revealed that essentially all of the acetyl-CoA interacting with the TCA cycle was either [2-13C]acetyl-CoA derived from [3-13C]pyruvate or was unlabeled (44). Therefore, the introduction of hyperpolarized [1-13C]pyruvate had a negligible effect on glutamate isotopomer labeling, as expected for the brief exposure of the cells to the hyperpolarized material. Isotopomer analysis was also used to calculate relative flux rates of numerous metabolic pathways interacting with the TCA cycle (36). Each pathway is normalized to citrate synthase flux given an arbitrary value of 1.0. As shown in Table 1, many of these activities were active in both cell lines. Relative PDH flux was similar in the two cell lines, consistent with the rapid labeling of citrate from [3-13C]pyruvate observed by mass spectrometry (Fig. 2). Anaplerotic flux from unlabeled sources (given the symbol, yS) was active in both cell lines, consistent with the previous report that both cell lines use glutamine as an anaplerotic precursor (30). However, flux through pyruvate carboxylase (yPC) was much higher in Huh-7 than SFxL. Pyruvate cycling (PK), another pathway that may produce CO2 from pyruvate C1 via either malic enzyme or phosphoenolpyruvate carboxykinase, was essentially inactive in SFxL cells but modestly active in Huh-7 cells. The high PDH flux and low yPC and pyruvate cycling (PK) in SFxL cells show that the great majority of pyruvate decarboxylation is PDH-dependent in these cells. On the other hand, PDH, yPC, and PK likely all contribute to bicarbonate production in Huh-7 cells.
Quantitative Models of Metabolic Flux from Single Extractions
Finally, we used information from both hyperpolarization and isotopomer analysis to develop quantitative models of metabolic flux in these two cell lines. Quantitative determination of one or more fluxes should make it possible to reference other reactions from the isotopomer analysis to it, thereby determining flux values throughout the network. We considered the potential routes by which H[13C]O3− could be formed from [1-13C]pyruvate. Each molecule of [1-13C]pyruvate metabolized by PDH generates one H[13C]O3−. By contrast, only a fraction of PC-dependent metabolism of [1-13C]pyruvate generates H[13C]O3−. PC converts [1-13C]pyruvate to [1-13C]OAA, and this may equilibrate with the malate, fumarate, and succinate pools, distributing up to half of the label to [4-13C]OAA. Citrate synthase will then transfer approximately half of the 13C originating on [1-13C]pyruvate to [6-13C]citrate and half to [1-13C]citrate. Release of 13C from [6-13C]citrate occurs at the isocitrate dehydrogenase step, but release from [1-13C]citrate does not occur until α-ketoglutarate dehydrogenase. Importantly, when a non-selective pulse is applied to either of these cell lines in the presence of hyperpolarized [1-13C]pyruvate, neither [6-13C]citrate nor [1-13C]citrate was observed (data not shown). Considering that both cell lines have appreciable citrate pools (Fig. 2), the lack of observable [13C]citrate suggests that forward cycling does not contribute significantly to the H[13C]O3− signal. Even the perfused liver, which contains abundant PC activity, produces only minimal amounts of observable hyperpolarized [6-13C]citrate (45). Pyruvate cycling may also contribute to decarboxylation of pyruvate at C1. These pathways involve the return of OAA to the pyruvate pool either through decarboxylation by phosphoenolpyruvate carboxykinase or by conversion to malate and decarboxylation by malic enzyme. Again, because of randomization of the 13C label between the 1 and 4 positions of OAA and malate, only a fraction of 13C can be released as H[13C]O3− on the first turn.
In SFxL cells, PC is much less active than PDH, and pyruvate cycling is inactive (Fig. 2 and Table 1). Thus, the rate of hyperpolarized H[13C]O3− formation in SFxL cells (38 ± 11 μmol/106 cells/h) is equivalent to the PDH flux. Using the relative flux data derived from isotopomer analysis, PDH flux was used to derive quantitative flux values for PC, anaplerosis/cataplerosis, citrate synthase/TCA cycle turnover, and the rates at which substrates other than labeled pyruvate feed the acetyl-CoA pool (Fig. 7A). In Huh-7 cells, PC and pyruvate cycling are active in addition to PDH. Assuming that both PDH and PC/pyruvate cycling can produce hyperpolarized H[13C]O3−, we used data from the isotopomer analysis to calculate the fraction of H[13C]O3− generated by PDH, converted this to a quantitative flux, and used this rate to determine the remaining rates in this network (Fig. 7B).
FIGURE 7.
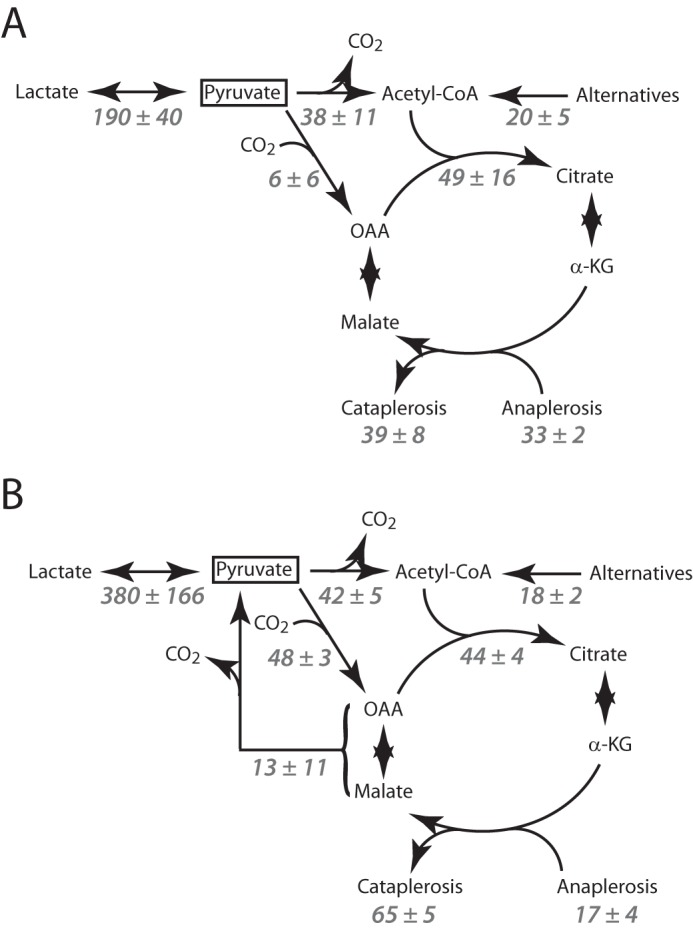
Quantitative models of pyruvate metabolism in SFxL cells (A) and Huh-7 cells (B). All flux values are in nmol/106/h. Each value is the average of three independent experiments ± S.D. α-KG, α-ketoglutarate.
Akt-mediated Glycolytic Flux Alters the Fate of Hyperpolarized [1-13C]Pyruvate
Because cancer cells typically have high rates of glycolysis, we next determined the effects of glucose on metabolism of [1-13C]pyruvate. We reasoned that hyperpolarization would provide a unique and dynamic view of the effects of glucose on multiple fates of pyruvate simultaneously. The presence of an active glycolytic flux was expected to alter pyruvate metabolism in at least two ways. First, glycolysis provides NADH that would result in a more physiological redox ratio than the glucose-free, high pyruvate conditions used in previous experiments and might stimulate the reduction of pyruvate to lactate by LDH. Second, the availability of a lactate pool would increase the opportunity for exchange reactions by which hyperpolarized 13C could be transferred from pyruvate to lactate (15). As expected, culture of SFxL cells with 10 mm glucose before and during exposure to 6 mm hyperpolarized [1-13C]pyruvate substantially increased the appearance of [1-13C]lactate (Fig. 8A). Peak lactate signal was ∼10 times higher in cultures containing glucose than in those lacking it (Fig. 7A). Unexpectedly, providing glucose also suppressed the appearance of H[13C]O3−. Although a small amount of this metabolite could be detected in summations of all spectra acquired over the course of a hyperpolarization experiment, time evolutions like those in Fig. 5C were no longer visible. The lack of time-dependent H[13C]O3− data made it impossible to determine specific flux rates as in the glucose-deprived cells.
FIGURE 8.
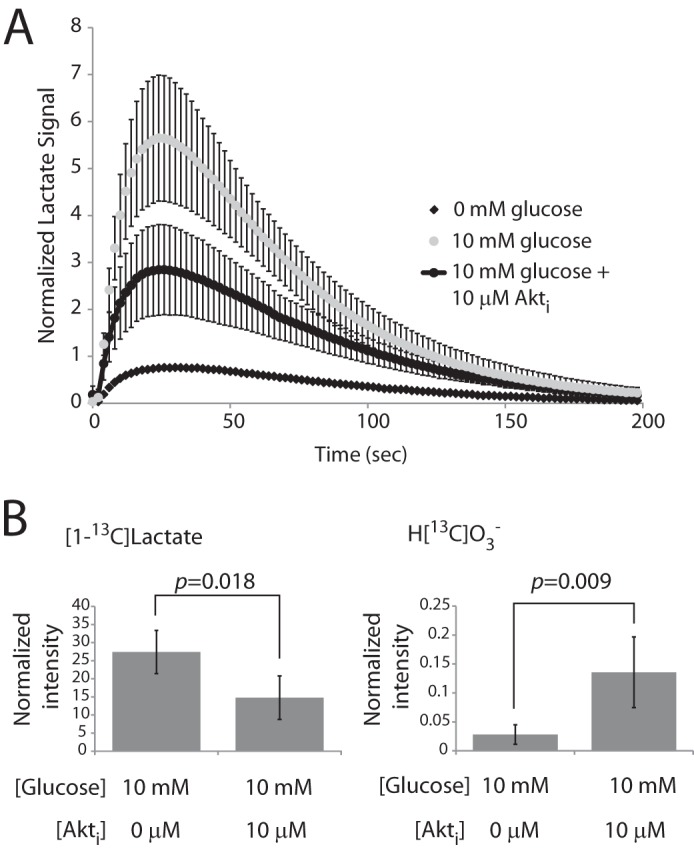
Akt-stimulated glycolysis acutely alters LDH and PDH flux. A, time evolution of [1-13C]lactate appearance in SFxL cells cultured in DMEM supplemented with 6 mm hyperpolarized [1-13C]pyruvate and 0, 10, or 10 mm glucose with 10 μm Akt inhibitor. The 0 mm glucose time course is the average of the experiments from Fig. 5B and is included here for reference. The other time courses are the average and S.D. for three independent experiments. B, normalized intensity for [1-13C]lactate and H[13C]O3− in 10 mm glucose with and without the Akt inhibitor. Data are the average and S.D. for several independent experiments (n = 5 for cultures containing glucose; n = 6 for cultures containing glucose and the Akt inhibitor).
Cancer cell glycolysis is largely under the control of oncogenic signaling. In SFxL cells, the kinase Akt enables cells to maintain high rates of lactate production and secretion (33). We, therefore, tested whether inhibiting Akt would alter the metabolism of hyperpolarized [1-13C]pyruvate. SFxL cells were cultured with a commercial inhibitor at doses that suppressed lactate secretion by ∼50% (33). This reduced the appearance of [1-13C]lactate but enhanced the production of H[13C]O3− (Fig. 8, A and B). These data demonstrate that metabolism of hyperpolarized [1-13C]pyruvate by both LDH and PDH is acutely and dramatically responsive to Akt-mediated effects on glycolysis.
DISCUSSION
We report here three advances in the quantitation of pyruvate metabolism in tumor cells. First, we developed an approach to maximize detection of bicarbonate in real time, making it possible to establish an accurate rate of PDH activity by monitoring formation of hyperpolarized H[13C]O3−. Second, by performing simultaneous steady-state labeling and isotopomer analysis, we used the rate of hyperpolarized H[13C]O3− formation to define a number of other rates related to pyruvate and TCA cycle metabolism. Third, we determined that Akt-driven effects on glycolysis suppress decarboxylation of hyperpolarized [1-13C]pyruvate in cancer cells.
Hyperpolarization has the capacity to report quantitative fluxes of enzyme-catalyzed reactions in tumors and has been used successfully to detect oncogene-driven LDH-catalyzed exchanges between pyruvate and lactate in vivo (16, 18). However, cancer cells both in culture and in solid tumors metabolize pyruvate in the mitochondria (28, 29). Mitochondrial pyruvate metabolism is important, because it delivers carbon to pathways involved in both the assimilation of biomass in tumors and in the energy-producing reactions in the TCA cycle and oxidative phosphorylation (23, 46). Furthermore, intravenous administration of [13C]glucose just before tumor resection has provided strong evidence that both PDH and PC are active in lung and brain tumors (24, 26, 28). Thus, hyperpolarization strategies designed to detect LDH without considering mitochondrial activity may miss many informative aspects of tumor metabolism.
Several important aspects of mitochondrial pyruvate metabolism are apparent from the data. First, combining isotopomer analysis with hyperpolarization revealed that essentially all decarboxylation of hyperpolarized [1-13C]pyruvate was the result of PDH activity rather than the effects of pyruvate carboxylation and subsequent CO2-releasing reactions. Second, the delayed formation of H[13C]O3− relative to [1-13C]lactate (e.g. Fig. 4C) is likely due to a delay associated with pyruvate transport, slower kinetics of PDH compared with LDH (47), and the greater number of steps needed to generate H[13C]O3− after transit of [1-13C]pyruvate through PC. However, the addition of more enzymatic steps to the model had no effect on the residual between the fit and the data, consistent with the idea that most of the hyperpolarized H[13C]O3− is produced by PDH in both cell lines. Second, there was a delay in H[13C]O3− formation in SFxL cells relative to Huh-7 cells. This delay was apparently due to multiple factors. The reduced initial LDH flux in SFxL is consistent with delayed entry of pyruvate via transporters on the cytoplasmic membrane. In addition, modeling of the hyperpolarization data revealed a substantial reduction in the rate constant for entry of pyruvate into the mitochondria in SFxL cells (data not shown), suggesting that Huh-7 cells may also have a higher capacity for mitochondrial pyruvate import.
These experiments are an extension of combining oxygen consumption measurements with isotopomer analysis data in perfused hearts and livers (45, 48). In those cases, isotopomer analysis was used to evaluate the amount of H[13C]O3− generated by PDH versus phosphoenolpyruvate carboxykinase. Simulating the data generated from a perfused organ ex vivo or in vivo is much more technically demanding than the cell suspensions used here. First, a pyruvate delivery function must be estimated to assess the starting concentration for the downstream reactions. This can be accomplished by using a non-metabolic marker such as 13C urea, which is co-injected with the pyruvate (49). The other complication is the presence of pyruvate in the vasculature that may not exchange with pyruvate in the target cells. In the methodology described here, cells were injected into the prepositioned hyperpolarized solution, enabling relatively simple kinetic models to be applied. Fitting the kinetic H[13C]O3− signal did not require differentiating intracellular pyruvate from extracellular pyruvate, although cytosolic and intramitochondrial pyruvate were considered as separate pools.
The addition of glucose to the culture medium dramatically altered [1-13C]pyruvate metabolism in SFxL cells. Exchanges between [1-13C]pyruvate and [1-13C]lactate were enhanced, whereas the appearance of H[13C]O3− was markedly suppressed. Enhanced labeling of [1-13C]lactate was expected, because glycolysis produces abundant lactate for participation in LDH-catalyzed exchanges with [1-13C]pyruvate (15). Furthermore, flux through glyceraldehyde-3 phosphate dehydrogenase in glucose-replete cells provides NADH for the LDH reaction. The findings indicate that LDH activity may be underestimated in experiments with hyperpolarized [1-13C]pyruvate unless glucose is provided. On the other hand, the suppression of pyruvate decarboxylation was unexpected. Because formation of hyperpolarized H[13C]O3− occurred exclusively through PDH in these experiments, the finding indicates that glycolysis constrains flow of [1-13C]pyruvate through the PDH reaction.
There are multiple potential explanations for this observation. One is that provision of glucose simply leads to substrate competition between pyruvate from glycolysis and hyperpolarized [1-13C]pyruvate, suppressing the appearance of H[13C]O3− from the latter. However, in these same cells glucose suppressed oxygen consumption relative to oxygen consumption on pyruvate (Fig. 1B). Thus, it is possible that suppressed H[13C]O3− production reflects a more fundamental inhibitory effect of glucose on oxidative metabolism. This might have broader implications for cancer cell metabolism. The Warburg effect refers to the historical observation that cancer cells preferentially convert pyruvate to lactate in the presence of oxygen, supposedly because of an intrinsic bottleneck limiting pyruvate oxidation (50). In contrast, our data demonstrate that glycolysis itself may constrain the cell ability to steer pyruvate toward PDH, at least when that pyruvate is provided as an exogenous substrate. Under the conditions used here, both anaerobic and aerobic pyruvate metabolism were acutely and reversibly impacted by the high glycolytic flux of SFxL cells. The lack of a fixed constraint on mitochondrial pyruvate metabolism recalls the Crabtree effect, in which a large glucose supply reversibly suppresses oxidative metabolism in yeast and cancer cells (22). Importantly, changes in pyruvate metabolism could be partially reversed in the presence of glucose by inhibiting Akt, the kinase that stimulates glycolysis in SFxL cells. This indicates that pyruvate handling is regulated by oncogenic signal transduction in cancer cells.
Translating approaches with 13C hyperpolarization to cancer patients poses significant challenges, including the need for appropriate pulse sequences, radiofrequency coils, and the ability to produce sufficient quantities of sterile hyperpolarized tracers for human studies (16). Most if not all of these challenges have been met, however, and successful proof-of-concept imaging to detect conversion of [1-13C]pyruvate to [1-13C]lactate has been demonstrated in human prostate cancer (19). Obtaining quantitative flux data from in vivo hyperpolarization, including quantitative rates of pyruvate decarboxylation, will require significantly more complicated modeling than what is used here. However, it may be possible to combine dynamic hyperpolarized 13C data with isotopomer analysis in solid tumors, because it is feasible and informative to obtain high quality 13C NMR spectra from tumors resected from patients infused intra-operatively with [13C]glucose (26). Importantly, the ability to observe definitive changes in flux is an essential component of metabolic analysis in any system, including tumors. For example, although conventional, steady-state 13C NMR can readily be used to detect major changes in the choice of metabolic pathway (e.g. an induction of PC relative to PDH), this approach would likely miss changes in absolute flux unless they are associated with differences in which pathways are active. The method described here solves that problem by providing a quantitative reference flux (pyruvate decarboxylation) to establish accurate rates across a broad network.
Supplementary Material
Acknowledgments
We thank members of the DeBerardinis laboratory and the Advanced Imaging Research Center at UT-Southwestern for critically evaluating the data.
This work was supported, in whole or in part, by National Institutes of Health Grant P41-EB015908, RR-02584, CA157996, and R37-HL34557. This work was also supported by a multi-investigator award from the Cancer Prevention and Research Institute of Texas (RP101243).

This article contains supplemental Figs. S1 and S2.
- PDH
- pyruvate dehydrogenase
- PK
- pyruvate kinase
- OAA
- oxaloacetate
- PC
- pyruvate carboxylase.
REFERENCES
- 1. Racker E., Resnick R. J., Feldman R. (1985) Glycolysis and methylaminoisobutyrate uptake in rat-1 cells transfected with ras or myc oncogenes. Proc. Natl. Acad. Sci. U.S.A. 82, 3535–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flier J. S., Mueckler M. M., Usher P., Lodish H. F. (1987) Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science 235, 1492–1495 [DOI] [PubMed] [Google Scholar]
- 3. Elstrom R. L., Bauer D. E., Buzzai M., Karnauskas R., Harris M. H., Plas D. R., Zhuang H., Cinalli R. M., Alavi A., Rudin C. M., Thompson C. B. (2004) Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899 [DOI] [PubMed] [Google Scholar]
- 4. Ying H., Kimmelman A. C., Lyssiotis C. A., Hua S., Chu G. C., Fletcher-Sananikone E., Locasale J. W., Son J., Zhang H., Coloff J. L., Yan H., Wang W., Chen S., Viale A., Zheng H., Paik J. H., Lim C., Guimaraes A. R., Martin E. S., Chang J., Hezel A. F., Perry S. R., Hu J., Gan B., Xiao Y., Asara J. M., Weissleder R., Wang Y. A., Chin L., Cantley L. C., DePinho R. A. (2012) Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. (2008) The biology of cancer. Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 6. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer. The next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 7. Parsons D. W., Jones S., Zhang X., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Siu I. M., Gallia G. L., Olivi A., McLendon R., Rasheed B. A., Keir S., Nikolskaya T., Nikolsky Y., Busam D. A., Tekleab H., Diaz L. A., Jr., Hartigan J., Smith D. R., Strausberg R. L., Marie S. K., Shinjo S. M., Yan H., Riggins G. J., Bigner D. D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dang L., White D. W., Gross S., Bennett B. D., Bittinger M. A., Driggers E. M., Fantin V. R., Jang H. G., Jin S., Keenan M. C., Marks K. M., Prins R. M., Ward P. S., Yen K. E., Liau L. M., Rabinowitz J. D., Cantley L. C., Thompson C. B., Vander Heiden M. G., Su S. M. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baysal B. E., Ferrell R. E., Willett-Brozick J. E., Lawrence E. C., Myssiorek D., Bosch A., van der Mey A., Taschner P. E., Rubinstein W. S., Myers E. N., Richard C. W., 3rd, Cornelisse C. J., Devilee P., Devlin B. (2000) Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287, 848–851 [DOI] [PubMed] [Google Scholar]
- 10. Niemann S., Müller U. (2000) Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat. Genet. 26, 268–270 [DOI] [PubMed] [Google Scholar]
- 11. Tomlinson I. P., Alam N. A., Rowan A. J., Barclay E., Jaeger E. E., Kelsell D., Leigh I., Gorman P., Lamlum H., Rahman S., Roylance R. R., Olpin S., Bevan S., Barker K., Hearle N., Houlston R. S., Kiuru M., Lehtonen R., Karhu A., Vilkki S., Laiho P., Eklund C., Vierimaa O., Aittomäki K., Hietala M., Sistonen P., Paetau A., Salovaara R., Herva R., Launonen V., Aaltonen L. A., and Multiple Leiomyoma Consortium (2002) Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata, and papillary renal cell cancer. Nat. Genet. 30, 406–410 [DOI] [PubMed] [Google Scholar]
- 12. Semenza G. L. (2010) Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29, 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Albers M. J., Bok R., Chen A. P., Cunningham C. H., Zierhut M. L., Zhang V. Y., Kohler S. J., Tropp J., Hurd R. E., Yen Y. F., Nelson S. J., Vigneron D. B., Kurhanewicz J. (2008) Hyperpolarized 13C lactate, pyruvate, and alanine. Noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 68, 8607–8615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brindle K. M., Bohndiek S. E., Gallagher F. A., Kettunen M. I. (2011) Tumor imaging using hyperpolarized 13C magnetic resonance spectroscopy. Magn. Res. Med. 66, 505–519 [DOI] [PubMed] [Google Scholar]
- 15. Day S. E., Kettunen M. I., Gallagher F. A., Hu D. E., Lerche M., Wolber J., Golman K., Ardenkjaer-Larsen J. H., Brindle K. M. (2007) Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat. Med. 13, 1382–1387 [DOI] [PubMed] [Google Scholar]
- 16. Kurhanewicz J., Vigneron D. B., Brindle K., Chekmenev E. Y., Comment A., Cunningham C. H., Deberardinis R. J., Green G. G., Leach M. O., Rajan S. S., Rizi R. R., Ross B. D., Warren W. S., Malloy C. R. (2011) Analysis of cancer metabolism by imaging hyperpolarized nuclei. Prospects for translation to clinical research. Neoplasia 13, 81–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Swisher C. L., Larson P. E., Kruttwig K., Kerr A. B., Hu S., Bok R. A., Goga A., Pauly J. M., Nelson S. J., Kurhanewicz J., Vigneron D. B. (2014) Quantitative measurement of cancer metabolism using stimulated echo hyperpolarized carbon-13 MRS. Magn. Reson. Med. 71, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu S., Balakrishnan A., Bok R. A., Anderton B., Larson P. E., Nelson S. J., Kurhanewicz J., Vigneron D. B., Goga A. (2011) [13C]Pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 14, 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nelson S. J., Kurhanewicz J., Vigneron D. B., Larson P. E., Harzstark A. L., Ferrone M., van Criekinge M., Chang J. W., Bok R., Park I., Reed G., Carvajal L., Small E. J., Munster P., Weinberg V. K., Ardenkjaer-Larsen J. H., Chen A. P., Hurd R. E., Odegardstuen L. I., Robb F. J., Tropp J., Murray J. A. (2013) Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13C]pyruvate. Sci. Transl. Med. 5, 198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ardenkjaer-Larsen J. H., Fridlund B., Gram A., Hansson G., Hansson L., Lerche M. H., Servin R., Thaning M., Golman K. (2003) Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 100, 10158–10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Golman K., Zandt R. I., Lerche M., Pehrson R., Ardenkjaer-Larsen J. H. (2006) Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 66, 10855–10860 [DOI] [PubMed] [Google Scholar]
- 22. Diaz-Ruiz R., Rigoulet M., Devin A. (2011) The Warburg and Crabtree effects. On the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta 1807, 568–576 [DOI] [PubMed] [Google Scholar]
- 23. Deberardinis R. J., Sayed N., Ditsworth D., Thompson C. B. (2008) Brick by brick. Metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 18, 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan T. W., Lane A. N., Higashi R. M., Farag M. A., Gao H., Bousamra M., Miller D. M. (2009) Altered regulation of metabolic pathways in human lung cancer discerned by 13C stable isotope-resolved metabolomics (SIRM). Mol Cancer 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fan T. W., Lane A. N., Higashi R. M., Yan J. (2011) Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics 7, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maher E. A., Marin-Valencia I., Bachoo R. M., Mashimo T., Raisanen J., Hatanpaa K. J., Jindal A., Jeffrey F. M., Choi C., Madden C., Mathews D., Pascual J. M., Mickey B. E., Malloy C. R., DeBerardinis R. J. (2012) Metabolism of [U-13C]glucose in human brain tumors in vivo. NMR Biomed. 25, 1234–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marin-Valencia I., Cho S. K., Rakheja D., Hatanpaa K. J., Kapur P., Mashimo T., Jindal A., Vemireddy V., Good L. B., Raisanen J., Sun X., Mickey B., Choi C., Takahashi M., Togao O., Pascual J. M., Deberardinis R. J., Maher E. A., Malloy C. R., Bachoo R. M. (2012) Glucose metabolism via the pentose phosphate pathway, glycolysis and Krebs cycle in an orthotopic mouse model of human brain tumors. NMR Biomed. 25, 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marin-Valencia I., Yang C., Mashimo T., Cho S., Baek H., Yang X. L., Rajagopalan K. N., Maddie M., Vemireddy V., Zhao Z., Cai L., Good L., Tu B. P., Hatanpaa K. J., Mickey B. E., Matés J. M., Pascual J. M., Maher E. A., Malloy C. R., Deberardinis R. J., Bachoo R. M. (2012) Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 15, 827–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C. B. (2007) Beyond aerobic glycolysis. Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheng T., Sudderth J., Yang C., Mullen A. R., Jin E. S., Matés J. M., DeBerardinis R. J. (2011) Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. U.S.A. 108, 8674–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harrison C., Yang C., Jindal A., DeBerardinis R. J., Hooshyar M. A., Merritt M., Dean Sherry A., Malloy C. R. (2012) Comparison of kinetic models for analysis of pyruvate-to-lactate exchange by hyperpolarized 13C NMR. NMR Biomed. 25, 1286–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang C., Sudderth J., Dang T., Bachoo R. M., Bachoo R. G., McDonald J. G., DeBerardinis R. J. (2009) Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 69, 7986–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., Thompson C. B. (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chuang J. L., Davie J. R., Wynn R. M., Chuang D. T. (2000) Production of recombinant mammalian holo-E2 and E3 and reconstitution of functional branched-chain α-keto acid dehydrogenase complex with recombinant E1. Methods Enzymol. 324, 192–200 [DOI] [PubMed] [Google Scholar]
- 35. Fernandez C. A., Des Rosiers C., Previs S. F., David F., Brunengraber H. (1996) Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31, 255–262 [DOI] [PubMed] [Google Scholar]
- 36. Malloy C. R., Sherry A. D., Jeffrey F. M. (1990) Analysis of tricarboxylic acid cycle of the heart using 13C isotope isomers. Am. J. Physiol. 259, H987–H995 [DOI] [PubMed] [Google Scholar]
- 37. Sherry A. D., Jeffrey F. M., Malloy C. R. (2004) Analytical solutions for 13C isotopomer analysis of complex metabolic conditions. Substrate oxidation, multiple pyruvate cycles, and gluconeogenesis. Metab. Eng. 6, 12–24 [DOI] [PubMed] [Google Scholar]
- 38. Bretthorst G. L. (1990) Bayesian analysis. I. Parameter estimation using quadrature NMR models. J. Magn. Reson. 88, 533–551 [Google Scholar]
- 39. Bretthorst G. L. (1990) Bayesian analysis. II. Signal detection and model selection. J. Magn. Reson. 88, 552–570 [Google Scholar]
- 40. Bretthorst G. L. (1990) Bayesian analysis. III. Applications to NMR signal detection, model selection, and parameter estimation. J. Magn. Reson. 88, 571–595 [Google Scholar]
- 41. Bretthorst G. L. (1991) Bayesian Analysis. IV. Noise and computing time considerations. J. Magn. Reson. 93, 369–394 [Google Scholar]
- 42. Burgess S. C., Hausler N., Merritt M., Jeffrey F. M., Storey C., Milde A., Koshy S., Lindner J., Magnuson M. A., Malloy C. R., Sherry A. D. (2004) Impaired tricarboxylic acid cycle activity in mouse livers lacking cytosolic phosphoenolpyruvate carboxykinase. J. Biol. Chem. 279, 48941–48949 [DOI] [PubMed] [Google Scholar]
- 43. Larson P. E., Kerr A. B., Chen A. P., Lustig M. S., Zierhut M. L., Hu S., Cunningham C. H., Pauly J. M., Kurhanewicz J., Vigneron D. B. (2008) Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J. Magn. Reson. 194, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeffrey F. M., Storey C. J., Sherry A. D., Malloy C. R. (1996) 13C isotopomer model for estimation of anaplerotic substrate oxidation via acetyl-CoA. Am. J. Physiol. 271, E788–E799 [DOI] [PubMed] [Google Scholar]
- 45. Merritt M. E., Harrison C., Sherry A. D., Malloy C. R., Burgess S. C. (2011) Flux through hepatic pyruvate carboxylase and phosphoenolpyruvate carboxykinase detected by hyperpolarized 13C magnetic resonance. Proc. Natl. Acad. Sci. U.S.A. 108, 19084–19089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Caro P., Kishan A. U., Norberg E., Stanley I. A., Chapuy B., Ficarro S. B., Polak K., Tondera D., Gounarides J., Yin H., Zhou F., Green M. R., Chen L., Monti S., Marto J. A., Shipp M. A., Danial N. N. (2012) Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 22, 547–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Curi R., Newsholme P., Newsholme E. A. (1988) Metabolism of pyruvate by isolated rat mesenteric lymphocytes, lymphocyte mitochondria, and isolated mouse macrophages. Biochem. J. 250, 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Merritt M. E., Harrison C., Storey C., Jeffrey F. M., Sherry A. D., Malloy C. R. (2007) Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proc. Natl. Acad. Sci. U.S.A. 104, 19773–19777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. von Morze C., Larson P. E., Hu S., Yoshihara H. A., Bok R. A., Goga A., Ardenkjaer-Larsen J. H., Vigneron D. B. (2012) Investigating tumor perfusion and metabolism using multiple hyperpolarized 13C compounds. HP001, pyruvate. and urea. Magn. Res. Imaging 30, 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Warburg O. (1956) On respiratory impairment in cancer cells. Science 124, 269–270 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.