Background: NrdEF ribonucleotide reductases are found in many bacterial pathogens and function in vitro with either iron or manganese cofactors.
Results: Streptococcus sanguinis mutants lacking NrdEF or its manganese cofactor cannot grow aerobically or cause endocarditis.
Conclusion: The manganese cofactor is essential for virulence.
Significance: This work may explain why many NrdEF-containing bacteria require manganese for virulence and oxygen tolerance.
Keywords: Bacterial Metabolism, Bacterial Pathogenesis, Manganese, Ribonucleotide Reductase, Streptococcus, Infective Endocarditis
Abstract
Streptococcus sanguinis is a cause of infective endocarditis and has been shown to require a manganese transporter called SsaB for virulence and O2 tolerance. Like certain other pathogens, S. sanguinis possesses aerobic class Ib (NrdEF) and anaerobic class III (NrdDG) ribonucleotide reductases (RNRs) that perform the essential function of reducing ribonucleotides to deoxyribonucleotides. The accompanying paper (Makhlynets, O., Boal, A. K., Rhodes, D. V., Kitten, T., Rosenzweig, A. C., and Stubbe, J. (2014) J. Biol. Chem. 289, 6259–6272) indicates that in the presence of O2, the S. sanguinis class Ib RNR self-assembles an essential diferric-tyrosyl radical (FeIII2-Y•) in vitro, whereas assembly of a dimanganese-tyrosyl radical (MnIII2-Y•) cofactor requires NrdI, and MnIII2-Y• is more active than FeIII2-Y• with the endogenous reducing system of NrdH and thioredoxin reductase (TrxR1). In this study, we have shown that deletion of either nrdHEKF or nrdI completely abolishes virulence in an animal model of endocarditis, whereas nrdD mutation has no effect. The nrdHEKF, nrdI, and trxR1 mutants fail to grow aerobically, whereas anaerobic growth requires nrdD. The nrdJ gene encoding an O2-independent adenosylcobalamin-cofactored RNR was introduced into the nrdHEKF, nrdI, and trxR1 mutants. Growth of the nrdHEKF and nrdI mutants in the presence of O2 was partially restored. The combined results suggest that MnIII2-Y•-cofactored NrdF is required for growth under aerobic conditions and in animals. This could explain in part why manganese is necessary for virulence and O2 tolerance in many bacterial pathogens possessing a class Ib RNR and suggests NrdF and NrdI may serve as promising new antimicrobial targets.
Introduction
Ribonucleotide reductases (RNRs)3 perform the task, essential in nearly all organisms, of converting ribonucleotides to the deoxyribonucleotides needed for DNA replication and repair (Fig. 1A) (1). There are three classes of RNRs. These classes share a common structural fold in their large subunit (α), where nucleotide reduction occurs, and a conserved cysteine that must be oxidized to a thiyl radical to initiate nucleotide reduction. They are distinctive in their requirements for a metallo-cofactor that catalyzes this oxidation (Fig. 1A) and in their quaternary structures. The class II RNRs (NrdJ) have an α or α2 subunit structure and form the active thiyl radical through homolysis of the carbon cobalt bond of adenosylcobalamin (AdoCbl, Fig. 1C). The class III enzymes (NrdD, α2) require a glycyl radical for that purpose that is generated by a second protein or activase (NrdG, β2) that can function either stoichiometrically or catalytically with α2. NrdG uses an [4Fe4S]+ cluster and S-adenosylmethionine to generate the glycyl radical (Fig. 1A). The class Ia and Ib RNRs found in humans and many bacteria have an (α2)n(β2)m (n = 1 or 3 and m = 1 or 3) subunit structure, in which the thiyl radical on α is generated by a dimetal-tyrosyl radical (Y•) cofactor on a second subunit, β (2). Class Ia enzymes, typified by the Escherichia coli NrdA (α) NrdB (β) RNR, require a FeIII2-Y• cofactor to function in vitro and in vivo (3). More recently, class Ib (NrdEF, α/β) RNRs from several bacterial species, including E. coli, Bacillus subtilis, and Corynebacterium ammoniagenes, have been shown to contain a MnIII2-Y• cofactor in vivo (4–7). This discovery was especially interesting because it had previously been demonstrated that class Ib RNRs could self-assemble an active FeIII2-Y• cofactor in vitro upon addition of FeII and O2, as with class Ia enzymes (8). The key breakthrough in the discovery and in vitro characterization of the MnIII2-Y• cofactor was the identification of the essential role of NrdI (9), an unusual flavodoxin that is often encoded in the same operon with NrdE (α) and NrdF (β). This flavodoxin turned out to be an essential factor in the biosynthetic machinery, providing the missing oxidant required to oxidize the MnII2 form of NrdF to the active MnIII2-Y• cofactor (10). In this report, we focus on the metallation of class Ib and its interplay with class III RNR in Streptococcus sanguinis in an effort to determine their importance for virulence.
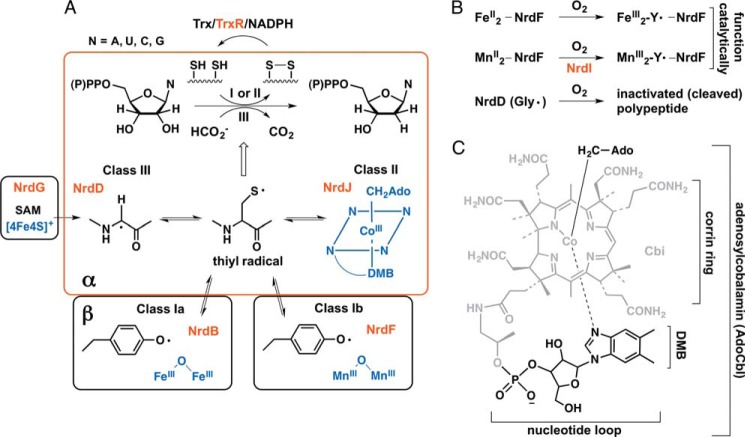
FIGURE 1.
All classes of RNRs catalyze the reduction of ribonucleotides to deoxyribonucleotides and require a metallo-cofactor to generate a thiyl radical that initiates this process. A, all RNRs have a structurally homologous α subunit (red) where nucleotide reduction occurs and is initiated by the thiyl radical. RNRs are classified based on the metallo-cofactors (blue) that oxidize the active site cysteine into the transient thiyl radical. This oxidation in the class I RNRs requires a second subunit, β (circled in black), designated NrdB for the Ia and NrdF for the Ib proteins. The oxidation uses AdoCbl for the class II RNR (NrdJ) and a glycyl radical for the class III RNR (NrdD). The glycyl radical is generated by an activating enzyme NrdG (black) that requires S-adenosylmethionine (SAM) and a [4Fe4S]+ cluster. In the class I and II RNRs, deoxynucleotide formation is accompanied by oxidation of two cysteines to a disulfide and consequently requires Trx, TrxR, and NADPH for multiple turnovers. NrdH-redoxin can function as a Trx in reduction of class Ib RNRs. In the class III RNRs, formate is the reductant. B, O2 is required to assemble the active MnIII2-Y• and FeIII2-Y• cofactors of NrdF (class Ib RNR), which then act catalytically with NrdE to produce deoxynucleotides. The glycyl radical cofactor of the class III RNR (NrdD) is inactivated by O2. C, structure of the AdoCbl cofactor used by the class II RNRs. Ado, adenosyl; AdoCbl, adenosylcobalamin; Cbi (in gray), cobinamide; DMB, dimethylbenzimidazole.
The three classes of RNRs differ with regard to their interaction with O2 (Fig. 1B). Both class Ia (NrdAB) and class Ib (NrdEF) enzymes require O2 to assemble the active dimetallo-cofactor. However, this requirement is catalytic in that once the active cofactor (FeIII2-Y• or MnIII2-Y•) is generated, it catalyzes many turnovers of nucleotides to deoxynucleotides. However, the cofactor in the class III RNRs (NrdDG) must be assembled under strictly anaerobic conditions as the [4Fe4S]+ cluster in NrdG is inactivated by O2 and the active glycyl radical generated by NrdG and S-adenosylmethionine reacts rapidly with O2 resulting in cleavage of the NrdD polypeptide chain and permanent enzyme inactivation (11). In class II (NrdJ) enzymes, O2 is neither required for activity nor detrimental to the enzyme. Interestingly, however, distinct anaerobic and aerobic pathways have been identified for synthesis of both the corrin ring and the nucleotide loop portions of the AdoCbl cofactor (Fig. 1C) (12, 13).
Based on genome sequence analysis, most bacterial species, and particularly those associated with humans, possess RNRs belonging to two or more of these classes or subclasses (14). These species include the important Gram-positive pathogens Bacillus anthracis, Bacillus cereus, Corynebacterium diphtheriae, Enterococcus faecalis, Staphylococcus aureus, coagulase-negative staphylococci, and streptococci, all of which possess class Ib and class III RNRs (14). The contribution of the class III RNR to anaerobic growth and virulence has been reported in two of these species, S. sanguinis (15) and S. aureus (16). The corresponding class Ib RNRs in these two species have been identified as essential for aerobic growth based on the inability of researchers to recover null mutants (17–19).
Despite recent advances in the field, there remain fundamental gaps in our understanding of class Ib RNRs, particularly in species that rely on this enzyme as their only aerobic RNR. In several of the species listed above, the identification of genes encoding components essential for RNR activity other than the α and β subunits is in question because the genome encodes two or more paralogs, with none of the genes located in proximity to nrdEF. Moreover, despite the finding in other species that class Ib RNRs isolated from their endogenous hosts contain manganese as a cofactor (5–7, 20, 21), the activity of the iron-cofactored enzyme in vitro begs the question of whether this form may function in vivo under certain environmental conditions. Finally, published reports that mutants of S. aureus and S. sanguinis lacking a functional class III (anaerobic) RNR exhibit reduced virulence in animals models of disease (15, 16) suggest that some infection sites may be sufficiently lacking in O2 to preclude the need for an aerobic RNR. In the case of S. sanguinis, which is an abundant inhabitant of the human mouth (22), the disease examined was infective endocarditis. This disease is an infection of heart valves or endocardial tissues with a mortality that often exceeds 20% (23). Because of the seriousness of this disease, the lack of a vaccine, and treatment regimens requiring sustained antibiotic administration or surgical intervention (23), a better understanding of bacterial factors required for disease causation is needed.
In this and the accompanying paper (24), we set out to characterize the activity of the class Ib RNR in S. sanguinis and the contribution of both RNRs to endocarditis virulence. We report the creation of S. sanguinis mutants deleted for nrdHEKF, nrdI, or trxR1 (Fig. 1A), and their characterization by growth studies and by heterologous complementation with the class II nrdJ gene from Lactobacillus leichmannii (25). We also report the testing of the nrdHEKF and nrdI mutants, along with an nrdD mutant lacking the class III RNR, in a rabbit model of endocarditis. Taken together, the data in this and the accompanying paper (24) identify the essential components required for class Ib RNR activity with MnIII2-Y• and demonstrate that this activity, but not that of NrdD, is indispensable for aerobic growth and disease causation in an animal model. These results could provide an explanation for previous findings indicating that manganese is essential for the virulence and O2 tolerance of S. sanguinis (26),4 S. aureus (28), and other species possessing a class Ib RNR (29–36). In addition, these results suggest that components of the class Ib pathway may serve as promising targets for new antimicrobials.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
A list of S. sanguinis strains used in this study is included in Table 1. All S. sanguinis mutants were derived from the SK36 background. Cells were grown at 37 °C in brain heart infusion (BHI; BD Biosciences) broth, supplemented with 1.5% (w/v) agar for plates. O2 concentrations were established using an AnoxomatTM Mark II programmable jar filling system (Advanced Instruments, Inc.), which replaced controlled amounts of air with an atmosphere of 10% H2, 10% CO2, and 80% N2 (w/w/w). For microaerobic incubations, gas concentrations were adjusted to 6% O2, 7% H2, 7% CO2, and 80% N2. Anaerobiosis was achieved in jars by creation of an atmosphere initially composed of 0.2% O2, 9.9% H2, 9.9% CO2, and 80% N2, with the remaining O2 removed by a palladium catalyst or by use of an anaerobic chamber (Coy Laboratory Products, Inc). The following antibiotics and concentrations were used: erythromycin (Erm; 10 μg/ml), kanamycin (Kan; 500 μg/ml), and spectinomycin (Spc; 200 μg/ml). E. coli strain DH10B (Invitrogen) was used for plasmid construction and was grown in Luria-Bertani medium for liquid culture or with 1.5% (w/v) agar added for surface plating. For antibiotic selection in E. coli, Spc was used at a concentration of 100 μg/ml.
TABLE 1.
Strains used in this study
Strain no. indicates the number used for strain designations in Figs. 4–8.
| Strain |
Description | Source or Ref. | |
|---|---|---|---|
| No. | Name | ||
| 1 | SK36 | WT isolate from human dental plaque; virulent in rat and rabbit models of infective endocarditis | 27, 43 |
| 2 | JFP36 | Ermr; SSA_0169::ermB | 48 |
| RNR-related mutants | |||
| 3 | JFP138 | Kanr; ΔnrdHEKF::aphA-3 | This study |
| 4 | JFP143 | Kanr; ΔnrdI::aphA-3 | This study |
| 5 | JFP150 | Kanr; ΔtrxR1::aphA-3 | This study |
| 6 | JFP141 | Spcr Cmr; nrdD::magellan2 SSA_0169::aad9; derived from 6-26 | 15 |
| Ssx_1668 | Kanr; ΔfmnG::aphA-3 | 19 | |
| Ssx_1683 | Kanr; ΔfmnI::aphA-3 | 19 | |
| Ssx_0813 | Kanr; ΔtrxR2::aphA-3 | 19 | |
| Homologously complemented mutants | |||
| JFP160 | Kanr Spcr; ΔnrdHEKF::aphA-3 SSA_0169::aad9 Phyper-spank lacZo nrdHEKF lacI | This study | |
| JFP161 | Kanr Spcr; ΔnrdI::aphA-3 SSA_0169::aad9 Phyper-spank lacZo nrdI lacI | This study | |
| JFP162 | Kanr Spcr; ΔtrxR1::aphA-3 SSA_0169::aad9 Phyper-spank lacZo trxB lacI | This study | |
| Heterologous expression strains | |||
| JFP127 | Spcr; SSA_0169::aad9 Phyper-spank lacZo nrdJ lacI | This study | |
| JFP130 | Spcr; SSA_0169::aad9 Phyper-spank lacZo nrdAB yfaE lacI | This study | |
| JFP139 | Kanr Spcr; ΔnrdHEKF::aphA-3 SSA_0169::aad9 Phyper-spank lacZo nrdAB yfaE lacI | This study | |
| 7 | JFP140 | Kanr Spcr; ΔnrdHEKF::aphA-3 SSA_0169::aad9 Phyper-spank lacZo nrdJ lacI | This study |
| 8 | JFP140c | Spontaneous mutant of JFP140; Kanr Spcr; ΔnrdHEKF::aphA-3 SSA_0169::aad9 Phyper-spank lacZo nrdJ lacI | This study |
| 9 | JFP157 | Kanr Spcr; ΔnrdI::aphA-3 SSA_0169::aad9 Phyper-spank lacZo nrdJ lacI | This study |
| 10 | JFP158 | Kanr Spcr; ΔtrxR1::aphA-3 SSA_0169::aad9 Phyper-spank lacZo nrdJ lacI | This study |
Plasmid and Strain Construction
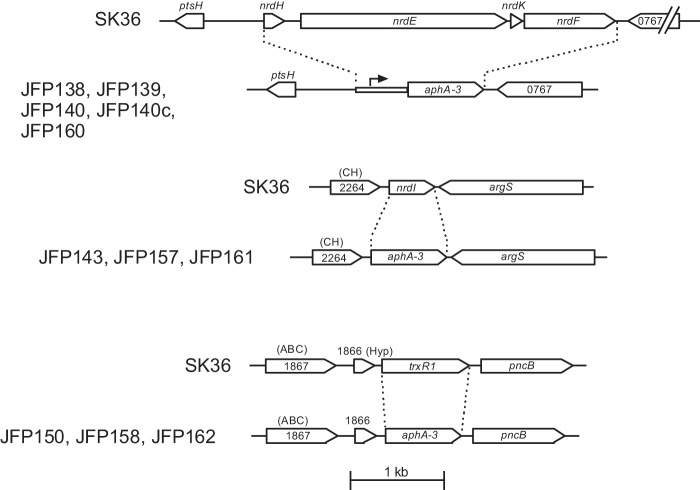
A list of all plasmids and oligonucleotides used in this study is included in supplemental Table S1. PCRs employed Phusion Taq or Platinum Pfx DNA polymerase. For allelic exchange mutagenesis, linear constructs were created by overlap PCR to replace SK36 genes with the aphA-3 gene (with or without its native promoter) encoding resistance to Kan, as described previously (Fig. 2) (19). Transformation of S. sanguinis with each construct was carried out as described previously (15) except that cells were grown anaerobically prior to and after transformation.
FIGURE 2.
Construction of mutants by allelic exchange. Dashed lines indicate sequence deletions. Strains containing each deletion are indicated to the left. The arrow indicates the native aphA-3 promoter. CH, conserved hypothetical protein; Hyp, hypothetical protein; ABC, putative ABC transporter permease. Four-digit numbers indicate locus tags. The nrdD locus and its mutagenesis have been illustrated previously (15).
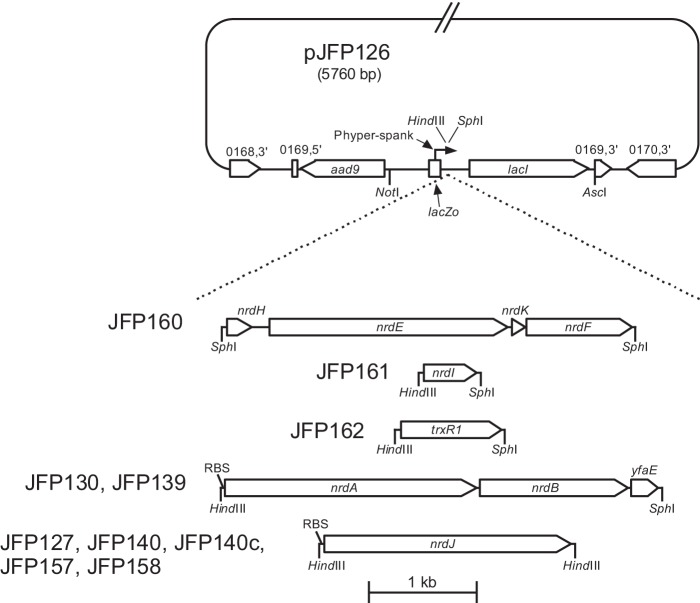
A suicide vector was constructed to allow for the insertion of complementing genes into an ectopic chromosomal site under the control of an inducible promoter. We began with plasmid pJFP96, which contains 1.35 kb of S. sanguinis DNA encompassing the SSA_0169 gene and flanking sequences, with the SSA_0169 gene interrupted by the aad9 gene encoding resistance to Spc.4 We first deleted HindIII and SphI restriction sites to facilitate downstream cloning reactions. The deletions were accomplished by digesting pJFP96 with HindIII and SphI (New England Biolabs), followed by treatment with T4 DNA polymerase (New England Biolabs) to create blunt ends, and ligating the blunt ends to re-circularize the plasmid using T4 DNA ligase (Invitrogen), resulting in creation of pJFP106. Next, an EcoRI to BamHI fragment of pDR111 (kindly provided by Dr. David Rudner, Harvard Medical School) containing the Phyper-spank lacZo lacI expression cassette was amplified by PCR using primers that replaced the EcoRI and BamHI restriction sites with NotI and AscI sites, respectively. The pDR111 fragment and pJFP106 were then digested with NotI and AscI and ligated together to create pJFP126 (Fig. 3). The following genes or operons were then PCR-amplified using flanking primers that introduced SphI and/or HindIII sites as follows: nrdHEFK, nrdI, and trxR1 from SK36; nrdAB yfaE from E. coli DH10B; and nrdJ from L. leichmannii (Fig. 3). The immediate source of the nrdJ gene was plasmid pSQUIRE, described previously (25). Predicted native ribosome-binding sites (RBSs) were included in the amplicons for S. sanguinis genes. For heterologous genes, the upstream primer was modified to add the sequence AGGAGAAATAA just upstream of the start codon. This sequence was chosen because it contained the most common RBS in the S. sanguinis SK36 genome (AGGAG) as predicted by the program RBSfinder (The Institute for Genomic Research), the most common spacer length (6 bp), and the actual sequence upstream from the rplX (ribosomal protein L24) gene. Inserts and pJFP126 were digested with SphI and/or HindIII, ligated, and electroporated into E. coli DH10B. Plasmid DNA was purified from DH10B cells using a Qiagen Mini Prep kit, and the inserts and flanking regions were sequenced (Retrogen) to confirm that the expected sequences were present. Transformation of S. sanguinis strains with each plasmid was carried out as described above. Spcr colonies were subjected to further purification by one or more rounds of growth in broth culture, sonication, and plating, and the resulting clones were verified to contain the expected structure, including lack of vector integration, by PCR and in most cases by DNA sequencing. Strain 6-26 containing a mini-transposon insertion in the nrdD gene (15) was made Spcr by transforming with plasmid pJFP96, described above, resulting in the creation of strain JFP141 (Table 1).
FIGURE 3.
Construction of complemented strains by allelic exchange. Dashed lines indicate sequence insertions. Strains containing each construct (without the pJFP126 vector backbone) are indicated to the left. The nrdAByfaE genes are from E. coli; nrdJ is from L. leichmannii. RBS, inserted ribosomal binding site identical to that upstream from the SK36 rplX gene. Other designations are as in Fig. 2. All regions except the pJFP126 vector backbone are drawn to scale.
Western Blot Analysis
Six-ml overnight cultures were added to 14 ml of fresh BHI and incubated with 1–1000 μm isopropyl β-d-1-thiogalactopyranoside (IPTG; Fisher Scientific) for 3 h, harvested by centrifugation at 8600 × g for 10 min, washed once with 10 ml of PBS, and the cell pellet suspended in 1 ml of PBS. Cells were added to microcentrifuge tubes containing 0.1-mm silica beads (lysing matrix B, MP Biomedicals) and lysed using a bead beater (FastPrep-24, MP Biomedicals) at a setting of 6.5 m/s, twice for 1 min with 5 min cooling on ice in between. The lysate was centrifuged for 10 min, and the supernatant was transferred to a new tube. Total protein was quantified using a BCA protein assay (Thermo Scientific) with BSA as a standard, and equal amounts (15–20 μg) of protein of each sample were loaded into a 10 or 12% pre-cast Criterion SDS-polyacrylamide gel (Bio-Rad). Following electrophoresis, the proteins were transferred to PVDF (Millipore) or nitrocellulose (GE Healthcare) membranes at 100 V, 4 °C, for 1.5 h and blotted using rabbit antisera generated against NrdA and NrdB from E. coli strain MG1655, NrdF from S. sanguinis strain SK36, or NrdJ from L. leichmannii, the last two of which were generated for this study by Covance Research Products, Inc. Antisera were used at concentrations of 1:5000 (NrdA) or 1:10,000 (NrdB, NrdF, and NrdJ). Proteins were visualized with chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate, Pierce).
Activity Assay
Activity of L. leichmannii NrdJ was measured by monitoring conversion of [3H]CTP to [3H]dCTP. Lysates of JFP140c, prepared as described above, were assayed in duplicate following a previously published protocol with some modifications (37). All samples containing AdoCbl were kept in black microcentrifuge tubes. E. coli thioredoxin (Trx, 40 units/mg) and thioredoxin reductase (TrxR, 1400 units/mg) were isolated as described previously (38, 39). The assay mixture contained in a final volume of 140 μl the following: dATP (0.1 mm), NADPH (1 mm), E. coli TrxR (0.2 μm), E. coli Trx (20 μm), AdoCbl (20 μm), phosphatase inhibitor (Roche Applied Science), Hepes (25 mm, pH 7.5), EDTA (4 mm), MgCl2 (1 mm), and lysate (110 μg of total protein) at 37 °C. The reaction was initiated with [3H]CTP (0.5 mm, 4169 cpm/nmol), and aliquots (30 μl) were taken over 30 min and quenched by boiling for 2 min. The dCTP was quantitated by the method of Steeper and Steuart (40).
RNA and DNA Isolation
RNA was isolated using the Qiagen RNeasy mini kit from overnight cultures. Briefly, cells were grown overnight at 6% O2 in the presence of 100 μm to 1 mm IPTG, harvested by centrifugation, and lysed using 15 mg/ml of lysozyme (ICN), proteinase K (Invitrogen), and the bead beater, as described above. RNA was then purified using the RNA purification columns provided in the kit. After isolation, RNA was treated with a TURBO DNA-free kit (Ambion) to remove any contaminating DNA and electrophoresed on a 1% (v/v) formaldehyde gel in 1× MOPS buffer to confirm RNA integrity. DNA was isolated from cultures and rabbit homogenates using a Qiagen DNeasy Blood and Tissue kit, following the protocol for Gram-positive organisms. Briefly, the cells or tissues were harvested by centrifugation and digested using an enzymatic lysis buffer (20 mm Tris-Cl, 2 mm EDTA, 1.2% (v/v) Triton X-100), proteinase K, and a kit buffer. The lysates were then column purified, and the DNA was eluted with 10 mm Tris-Cl, 1 mm EDTA, pH 8.0.
RT-qPCR
Transcript levels were measured by RT-qPCR. Following RNA isolation, reverse transcription was carried out using SuperScriptIII (Invitrogen). cDNA was used as template with SYBR Green PCR Master Mix (Applied Biosystems) and primers specific for each gene. All qPCRs were performed in triplicate in a 7500 Fast Real Time PCR System (Applied Biosystems). Normalization was performed using the gapA gene (41), and the ΔΔCT method was used to compare expression in induced cultures relative to uninduced cultures (42). Control reactions without reverse transcriptase were used to confirm absence of genomic DNA contamination.
Growth Studies
Overnight cultures were started anaerobically (except in the case of JFP141, for which microaerobic conditions were used) and grown for 24 h at 37 °C in BHI containing the appropriate antibiotic(s) for each strain. Concurrently, tubes of 5-ml uninoculated BHI were incubated anaerobically or microaerobically at 37 °C. The following day, the overnight cultures were diluted and used to inoculate the BHI at a final dilution of 10−6. For some samples, one or more supplements including IPTG (1 mm), AdoCbl (10, 15, 50, or 150 nm; Sigma), dicyanocobinamide (Cbi; 5, 10, 20, 40, 80, 160, 320, or 640 nm; Sigma), or FeSO4 (6, 18, 50, 180, 1800, or 3000 μm; 99.999% purity; Alfa Aesar) were added at this time. Cultures were incubated at 37 °C anaerobically and in 6% O2 for 24 or 48 h. Bacterial densities in the inocula and the 24- and 48-h cultures were determined by dilution plating. Briefly, cultures were diluted 104-fold in PBS and sonicated at 50% power in a Biologics Model 150 sonicator to break up clumps and chains for 90 s, a time shown in trial experiments to maximize colony number. Cells were further diluted in PBS for spreading on BHI plates containing antibiotics using an Eddy Jet 2 spiral plater (IUL Instruments). Plates were incubated at 37 °C anaerobically or, for JFP141, in 6% O2, and colonies were counted 2 days later. Growth in low melting point agarose was as described previously (15), except that BHI was used in place of tryptic soy broth; 18-mm diameter glass tubes covered loosely with foil were used in place of Hungate tubes, and inocula consisted of 10−5 and 10−6 dilutions of 24-h cultures. Digital photographs of tubes were saved in TIF format, cropped, adjusted for size when necessary, and used without any other alterations.
Rabbit Endocarditis Model
Specific-pathogen-free male New Zealand White rabbits (2–4 kg; RSI Biotechnology) were used in an endocarditis model to assess virulence, as described previously.4 The protocol for these experiments was approved by the Institutional Animal Use and Care Committee and complied with all federal and institutional guidelines. All efforts were made to minimize suffering. Rabbits were sedated and anesthetized before surgery using bupivicaine at the incision site and ketamine, xylazine, glycopyrrolate, and buprenorphine subcutaneously and provided with buprenorphine as analgesic every 12 h following surgery until euthanasia. To create sterile heart valve vegetations, a catheter was inserted through the right carotid artery of each rabbit until it rested against or passed through the aortic valve. The catheters were then trimmed and sutured into place and the incision closed. Two days later, rabbits were inoculated via peripheral ear vein with a 0.5-ml mixture of test strains. The inocula were prepared by growing cultures overnight with antibiotics, followed by 10-fold dilution into BHI. After an additional 3 h of growth, these cells were harvested, washed with PBS, sonicated, and diluted to a final cell density of ∼107 cfu/0.5 ml. Remaining cells were enumerated by further dilution in PBS and spreading on BHI plates containing selective antibiotics using a spiral plater. About 20 h later, rabbits were sacrificed, and the vegetations from their aortic valves were removed, homogenized in PBS, sonicated, diluted in PBS, and plated for enumeration as for the inocula. Output values were normalized against inoculum ratios. Total DNA was isolated from the remaining portion of each homogenate as described above and used in qPCRs with primers specific for antibiotic resistance genes (ermB, aphA-3, or aad9) unique to each strain. As with RT-qPCR, SYBR Green PCR Master Mix was used, and the reactions were performed in triplicate. Standard curves were generated for each primer set using purified genomic DNA from each strain and were used to determine the quantity of DNA of each strain present in the vegetations from each animal. Results were then converted to genome equivalents.
RESULTS
Mutagenesis of S. sanguinis Genes Required for Ribonucleotide Reduction
To identify and characterize genes involved in S. sanguinis ribonucleotide reduction, a number of strains were constructed (Table 1). The most important deletion mutations are depicted in Fig. 2. We began by deleting the nrdHEKF operon and replacing it with the aphA-3 gene encoding Kan resistance to create strain JFP138. By homology, nrdE and nrdF encode the α and β subunits, respectively, of the S. sanguinis class Ib RNR, and NrdH is a glutaredoxin-like protein that acts to re-reduce the disulfide in NrdE (1). The nrdK gene encodes a hypothetical 36-amino acid protein of unknown function. The purification and biochemical properties of NrdH, NrdE, and NrdF are described in the accompanying paper (24) and confirm their functional assignments.
The nrdHEKF mutant was generated and passaged under anaerobic conditions. Loss of NrdF expression was confirmed by Western blot (data not shown). As would be expected for a strain whose only remaining RNR is a class III (anaerobic) enzyme (15, 43), the mutant could not be propagated in the presence of 6% O2, our usual microaerobic growth condition, which is also similar to dissolved O2 concentrations in venous blood (44). Indeed, this mutant routinely failed to grow in BHI broth under an atmosphere of as little as 1% O2 (data not shown). These results were also consistent with our previous finding that the nrdH, nrdE, and nrdF genes were among the 218 SK36 genes found to be essential in cells grown in BHI with 6% O2 (19). Interestingly, a nrdK mutant was recovered in that study, indicating that this gene is not essential.
It has been shown in E. coli, B. subtilis, and B. anthracis that a flavodoxin-like protein named NrdI is required along with O2 for formation of the NrdF-MnIII2-Y• cofactor in vitro (4, 7, 45, 46). Virtually all bacterial species analyzed to date that possess class Ib RNRs also contain at least one nrdI gene, and in several species, the gene is in the same operon as nrdE and nrdF (14). In SK36, there were three genes annotated as encoding NrdI or NrdI-like proteins, none of which were within or adjacent to the nrdHEKF operon (43). SSA_1668 and SSA_1683 were identified in our previous study as nonessential for growth in 6% O2, whereas SSA_2263 was found to be essential (19). We confirmed that the first two mutants grew normally in 6% O2 (data not shown). We then deleted the SSA_2263 gene under anaerobic conditions using the same strategy as described above for nrdHEKF, resulting in the creation of strain JFP143 (Fig. 2). This mutant had a growth phenotype similar to that of the nrdHEKF mutant. Based on data shown below and in the accompanying paper (24), which indicates that a recombinant SSA_2263 protein supported in vitro MnIII2-Y• assembly in NrdF, although the other two proteins did not (24), the nrdI designation was retained for SSA_2263. The products of the other two genes, SSA_1668 and SSA_1683, were both shown to bind FMN (24) and were named FmnG and FmnI, respectively.
NrdH, which reduces the disulfide formed concomitant with dNDP formation by NrdE in other bacterial species, has been shown to be reduced by TrxR in vitro (Fig. 1A) (47). Similar to the situation with NrdI, SK36 possesses two genes annotated as encoding TrxRs, SSA_0813 and SSA_1865, with the latter gene identified as essential under aerobic conditions (19). We confirmed that the SSA_0813 mutant grew normally in 6% O2 and used the same procedure described above to mutate the SSA_1865 gene, creating strain JFP150 (Fig. 2). As expected, JFP150 grew well anaerobically, but did not grow in the presence of 6% O2. The accompanying paper (24) showed that the SSA_1865 gene product acted as a reductant for NrdH, whereas the SSA_0813 gene product did not. The SSA_1865 gene was thus named trxR1, and the SSA_0813 gene, which was not examined further in this study, was named trxR2.
A strain called 6-26 with a mutation in the nrdD gene encoding the α subunit of the class III (anaerobic) RNR was described previously (15). We modified this strain by inserting a Spcr gene cassette into the chromosome to facilitate antibiotic selection. The cassette was inserted into the SSA_0169 gene, resulting in the creation of strain JFP141 (Table 1). This chromosomal site was chosen because we established previously that insertion of aad9 or other antibiotic resistance genes into this locus had no effect on multiple phenotypes, including aerobic growth rate, anaerobic biofilm formation, genetic competence, and virulence for infective endocarditis in a rabbit model (48).
To ensure that the desired mutations were responsible for the O2-sensitive phenotype of the first three mutants created above, each was complemented with its deleted gene(s). The nrdHEKF operon and the nrdI and trxR1 genes, each with their native RBSs, were separately cloned into pJFP126, an IPTG-controlled expression plasmid, and then inserted into the same site in the SSA_0169 gene that was used for JFP141 creation (Fig. 3). Genes were placed under the control of an IPTG-inducible promoter system developed for B. subtilis and used previously in that species (7) but, to our knowledge, has never been shown to function in any Streptococcus. The aad9 gene and the lacI gene were also inserted into the chromosome, as indicated in Fig. 3. All three complemented mutants grew to high densities in 6% O2 in the presence of 100 μm IPTG, confirming the integrity of the original mutants (data not shown). An illustration of the nrdD locus and the construction of additional nrdD mutants and complementation constructs have been published previously (15).
Complementation of RNR Mutants with a Heterologous RNR
One of the goals of this and the accompanying paper (24) was to determine whether S. sanguinis NrdF functions with FeIII2-Y•, MnIII2-Y•, or either cofactor. The accompanying paper (24) establishes that S. sanguinis NrdI interacts with NrdF in vitro and that NrdI, but not its paralogs FmnG or FmnI, is required for MnIII2-Y• formation in vitro. This result, together with the finding that the nrdI mutant grows in the absence but not in the presence of O2, suggests that formation of the MnIII2-Y• is essential in aerobically grown cells. It could be argued, however, that NrdI might interact with more than one protein and that it has a role in some pathway other than ribonucleotide reduction that is essential for aerobic growth. To address this possibility, we sought to complement the nrdI mutant with a heterologous O2-tolerant RNR that would not require manganese or NrdI for activity. Restoration of growth in the presence of O2 by the heterologous RNR would indicate that the sole essential function of NrdI in S. sanguinis is to activate the class Ib RNR.
We used pJFP126 to place heterologous RNR genes into the same IPTG-inducible chromosomal expression locus that was used for the homologous complementation studies described above (Fig. 3). We first introduced into SK36 the nrdAB-yfaE operon from E. coli, encoding the α and β subunits of the class Ia, FeIII2-Y•-cofactored RNR, and a ferredoxin that plays a role in the maintenance and possibly synthesis of the FeIII2-Y•, respectively (49). As shown in Fig. 4A, we were able to demonstrate IPTG-inducible expression of NrdA by Western blot. Surprisingly, however, expression of NrdB was barely detectable above background (Fig. 4A), whereas the same antiserum produced strong bands when reacted with E. coli lysates (data not shown). This result was somewhat surprising because RT-qPCR analysis indicated 11.1 ± 0.9, 12.5 ± 1.1, and 12.7 ± 2.3 (mean ± S.D.) fold-induction for nrdA, nrdB, and yfaE, respectively, in cells incubated with 100 μm IPTG versus no-IPTG controls.5 The reason for this discrepancy between transcript and protein levels for NrdB has not been investigated further but may be due to our inclusion of a strong S. sanguinis RBS for translation of NrdA (see under “Experimental Procedures”), although we relied on native E. coli sequences for translation of NrdB and YfaE. We nevertheless decided to test for activity. Although our ultimate goal was complementation of an nrdI mutant, we began by deleting the nrdHEKF genes from this strain, which would indicate the activity of the heterologous RNR in the complete absence of the native class Ib RNR. Mutagenesis was performed as above, resulting in the creation of strain JFP139 (Table 1 and Figs. 2 and 3). Because of the poor expression of NrdB, it was not surprising that growth in this strain was indistinguishable from the noncomplemented nrdHEKF mutant as assessed by A660 or dilution plating. We have shown previously that BHI-grown SK36 cells contain more iron than manganese.4 Nevertheless, we considered that addition of FeII to the medium might increase the levels of free FeII in the cells and improve growth. However, supplementation with FeSO4 up to 180 μm had no significant effect on aerobic growth of JFP139 (data not shown). We therefore performed no further experiments with this strain or construct.
FIGURE 4.
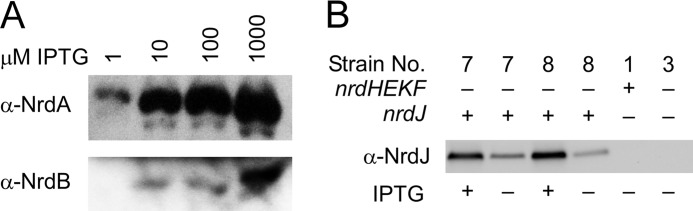
Analysis of heterologous RNR expression by Western blot. A, expression of NrdA and NrdB from E. coli in S. sanguinis strain JFP130 using antisera raised previously against the E. coli proteins. IPTG concentrations are indicated. B, expression of NrdJ from L. leichmannii in S. sanguinis strains JFP140 (strain 7) and JFP140c (strain 8) in the presence and absence of IPTG. The relevant genotype of each strain is indicated above. Strain numbers correspond to those in Table 1. −, no IPTG; +, IPTG was included at 100 μm.
We next introduced into SK36 the nrdJ gene, which encodes an O2-independent class II RNR from L. leichmannii that has been shown to employ AdoCbl (coenzyme B12; see Fig. 1C) as a cofactor (50) to create strain JFP127. Previous sequencing of the SK36 genome revealed a 70-kb gene cluster encoding an apparently complete anaerobic AdoCbl biosynthetic pathway and genes involved in the utilization of propanediol and ethanolamine (43). The latter two pathways are known to contain AdoCbl-requiring enzymes (51). Although no work has been done in S. sanguinis to examine the regulation of this pathway or to examine AdoCbl production, several of the pathway proteins were identified by proteomic analysis of SK36 cells grown in BHI and 6% O2 (43). In addition, all 28 contiguous AdoCbl biosynthetic genes, extending from SSA_0463 (cbiA) to SSA_0490 (cobS), as well as SSA_0510 and SSA_0512 (cobD and cobT, respectively), had low but detectable expression under the same growth conditions as assessed by microarray analysis (19). This result was also in agreement with our finding that expression of each of these genes was sufficient to produce Kan resistance from an integrated, promoterless aphA-3 gene, although many lowly expressed genes required the use of a cassette containing its own promoter to achieve Kan resistance (19).
The nrdHEKF genes were deleted from JFP127 as above, producing strain JFP140 (Table 1 and Figs. 2 and 3). NrdJ expression was then examined by Western blot, as shown in Fig. 4B. For ease of identification, each strain is identified in this and subsequent figures according to genotype and by an assigned strain number corresponding to that provided in Table 1. IPTG-inducible expression of NrdJ in JFP140 (strain 7) was evident, a result in agreement with RT-qPCR analysis, which indicated 24.6 ± 0.5-fold induction of nrdJ transcript levels in 100 μm IPTG versus the no-IPTG control. IPTG-inducible RNR activity was demonstrated in lysates of a derivative of strain 7 described below (Fig. 5). Based on results reported below, the nrdJ construct was also introduced into the nrdI and trxR1 mutants (Table 1 and Fig. 3).
FIGURE 5.
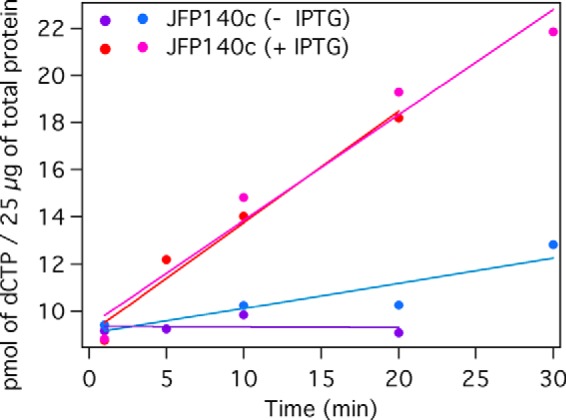
Characterization of NrdJ activity in S. sanguinis lysates. Amounts of [3H]dCTP produced by 25 μg of JFP140c (strain 8; Table 1) lysates prepared from uninduced ([minus]IPTG) and induced (+ IPTG, 1 mm) cultures are shown. Results from two independent experiments are shown.
Analysis of Mutant Growth in Broth Culture under Microaerobic and Anaerobic Conditions
We next wanted to characterize the effect of O2 on the growth of the strains outlined above. Given that class Ib RNRs require O2 for cofactor assembly whereas class III enzymes employ a glycyl radical that causes cleavage of the α subunit (NrdD) upon exposure to O2 (Fig. 1B) (11), this was a critical characteristic to study. During strain construction, cell growth was examined in two environments as follows: in an anaerobic chamber and in an atmosphere of 6% O2. Growth was assessed primarily by OD determination in broth cultures. Because we wanted a greater dynamic range than can be achieved by OD readings, we decided to measure cell density of broth cultures by dilution plating. One limitation of this assay is that O2 concentrations do not remain stable over time in still cultures of actively growing cells incubated with O2. S. sanguinis possesses a pyruvate oxidase that has been shown to consume O2 and pyruvate to form H2O2 and acetyl phosphate (52, 53), and we have shown in trial experiments that in BHI tubes incubated in a 6% O2 atmosphere, SK36 diminishes O2 levels to below the limit of detection (<0.1% using a PreSens Fibox 3 meter) in an overnight incubation (data not shown). In addition, we saw very little difference in trial experiments in the growth of mutants incubated in atmospheres containing O2 concentrations ranging from 1 to 6% (data not shown), suggesting that examination of growth at a concentration of O2 less than 1% might be needed to distinguish between strains. Finally, the potential for outgrowth of rare variants exists in broth culture. As described below, we therefore also employed an oxygen gradient assay that we (15) and others (54) have used previously. As described below, this assay results in the immobilization of cells within an O2 gradient that ranges from room air levels (21% O2) to standard anaerobiosis.
For the first assay, cells were cultured for 24 or 48 h in BHI broth in an anaerobic chamber or in jars containing 6% O2, and growth was measured by dilution plating (Fig. 6). WT cells (strain 1) grew well under both conditions, as expected. Under anaerobic conditions, growth of the nrdHEKF mutant (strain 3) was indistinguishable from that of WT at 24 or 48 h (Fig. 6A), but no growth of the nrdHEKF mutant occurred under 6% O2 at either time point, and by 48 h, no colonies were recovered (Fig. 6B). Both results were consistent with expectations, suggesting that O2 concentrations were sufficiently low under the anaerobic condition to allow NrdDG to function, whereas exposure to 6% O2 completely inactivated NrdD, leaving the nrdHEKF mutant with no mechanism for production of deoxyribonucleotides. Similarly, the nrdI and trxR1 mutants grew normally anaerobically but exhibited no growth in 6% O2. This result suggested that both gene products were necessary for growth in 6% O2.
FIGURE 6.
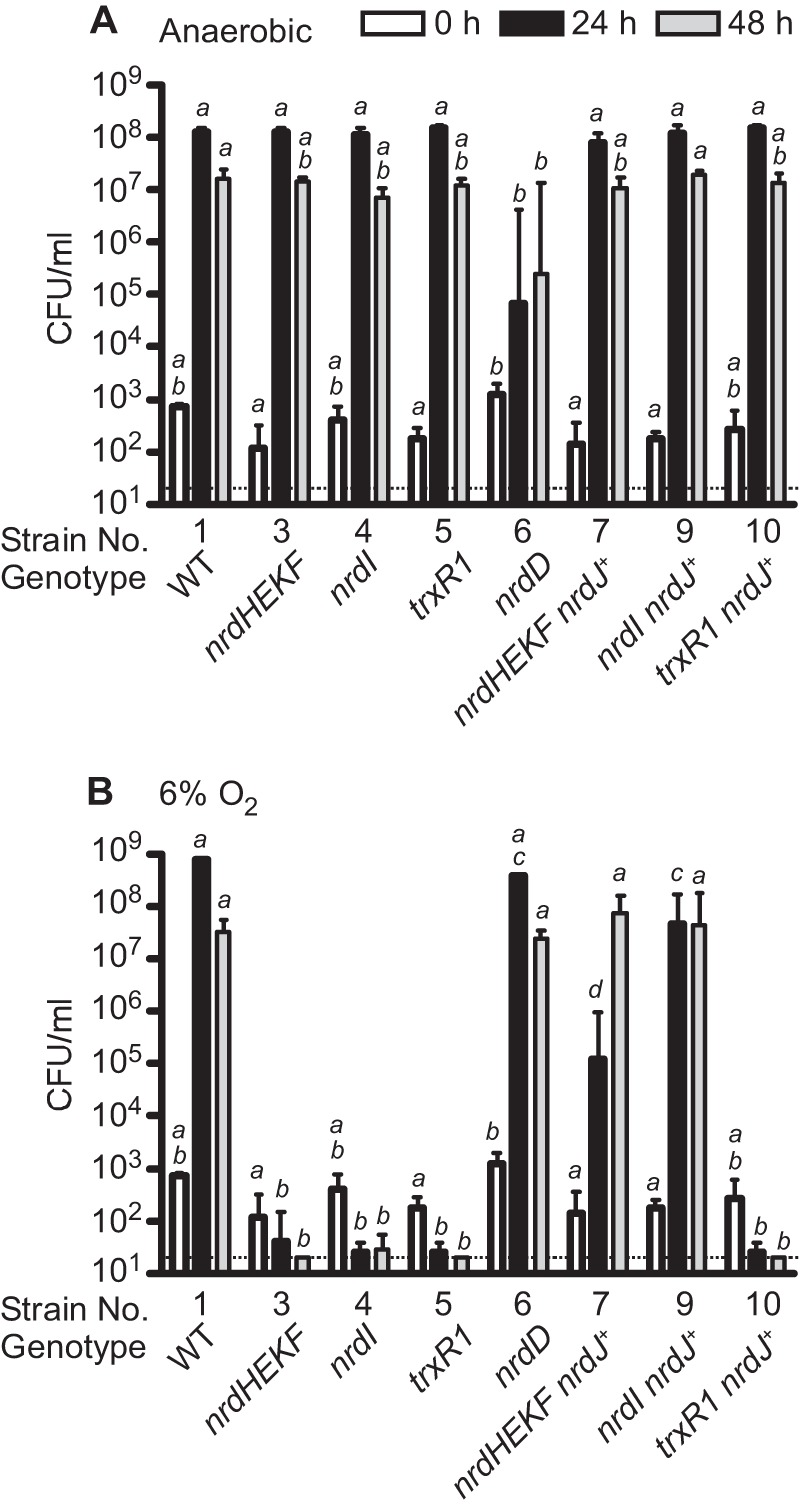
Growth of WT, mutant, and nrdJ-containing strains in BHI broth under standard anaerobic conditions (A) and in 6% O2 (B). Cultures were grown for the time indicated, and culture densities were then determined by dilution plating. Strain numbers correspond to those used in Table 1. Relevant genotypes are also provided. All strains containing nrdJ were cultured in the presence of 100 μm IPTG. Means and S.D. derived from log-transformed colony numbers are indicated. The dotted line indicates the limit of detection of the experiment (40 cfu/ml). Samples that do not share a letter across the same time point are significantly different (p < 0.05; analysis of variance with Tukey-Kramer post test).
The strain with a mutation in the nrdD gene encoding the α subunit of the anaerobic RNR was also examined. As expected, the nrdD mutant (strain 6) exhibited a very different phenotype from the other strains, growing indistinguishably from WT in 6% O2 (Fig. 6B), but reaching a density less than 0.1% that of WT at 24 h under anaerobic conditions (Fig. 6A). This result suggests that although tubes incubated anaerobically contained O2 levels sufficiently low to allow the anaerobic NrdDG enzyme to function in the nrdHEKF, nrdI, and trxR1 mutants, they also contained enough O2 to support limited growth of the nrdD mutant via NrdEF activity.
Complementation of Mutant Growth in Broth Cultures by a Class II RNR
We next examined strains into which the nrdJ class II RNR gene from L. leichmannii was introduced. As with their parental mutant strains, all three nrdJ-complemented mutants grew indistinguishably from WT under anaerobic conditions, as expected (Fig. 6A). In 6% O2, the nrdJ-complemented nrdHEKF mutant (strain 7) reached a density of ∼105 cfu/ml after 24 h and almost 108 cfu/ml after 48 h (Fig. 6B). Thus, although the introduction of NrdJ was not sufficient to restore growth to WT rates or densities, growth was dramatically increased compared with the parental nrdHEKF mutant. Given that no growth is possible without a functional RNR, this result strongly suggests that the exogenous NrdJ RNR can partially substitute for the missing class Ib (NrdEF) RNR. The nrdJ-complemented nrdI mutant (strain 9) grew to a density that was 1.2 logs less than WT at 24 h, but 6.3 logs greater than the parental nrdI mutant (Fig. 6B). The latter result suggested that the growth-limiting effect of the nrdI mutation was due to elimination of RNR activity rather than any other function NrdI might possess.
The nrdJ-complemented trxR1 mutant grew similarly to its trxR1 parent in 6% O2 (strains 10 and 5, Fig. 6B). This was not unexpected, as TrxR has been hypothesized to serve as the in vivo reductant for NrdJ (55), and TrxR1 was shown to support NrdJ activity in vitro (Fig. 5). The growth of this strain therefore indicated that there were no unexpected beneficial effects of the nrdJ (or lacI) genes on the anaerobic NrdDG that would allow it to function in the presence of O2. Ribonucleotide reduction by NrdJ provides the best explanation for restoration of aerobic growth by NrdJ in strains deleted for nrdHEKF or nrdI.
Growth of Mutant Strains in an O2 Gradient
We next wanted to examine growth in an O2 gradient, rather than the dichotomous 0 and 6% O2 conditions used in the broth assay. Our first interest was in determining whether the class Ib and class III RNRs function under mutually exclusive conditions or, instead, whether there is some concentration of O2 at which both enzymes are active. Second, Fig. 5B indicates that the nrdJ-complemented nrdI mutant (strain 9) grew to a level >2.5 logs higher than that of the nrdJ-complemented nrdHEKF mutant (strain 7) at 24 h. This suggested the possibility that we might be observing RNR activity from FeIII2-Y•-cofactored NrdEF in the nrdI mutant. This activity, although insufficient to support aerobic growth on its own, might work synergistically with NrdJ in strain 9 to improve growth relative to the NrdJ-complemented nrdHEKF mutant (strain 7). We wanted to examine this possibility more carefully under a range of O2 conditions.
An O2 gradient assay (15, 54) was therefore performed. Cells from 24-h cultures were diluted 105- or 106-fold into loosely covered glass tubes that contained 10 ml of molten BHI plus 1% low melting point agarose that had been preincubated for 2 days in an anaerobic chamber at 37 °C. The inoculated tubes were removed from the incubator and placed at room temperature to solidify and then removed from the anaerobic chamber and placed in a standard 37 °C incubator. At this temperature, the solidified agarose did not re-melt. Aerobic incubation was continued at 37 °C, creating an O2 gradient from top to bottom. Resazurin (56) was added to control tubes as a visual indicator of O2 levels (Fig. 7A).
FIGURE 7.

Colony formation by WT, mutant, and nrdJ-containing strains in an O2 gradient. Anaerobically preincubated tubes containing molten BHI and low melting point agarose were inoculated with 10−5 or 10−6 final dilutions of overnight cultures of the strains indicated, allowed to solidify, and then incubated at 37 °C in room air to create an O2 gradient. Strain genotypes and numbers corresponding to those used in Table 1 are provided. A, mutant strains. Resazurin, tubes containing 5 μg/ml resazurin. B, strains containing nrdJ. Cbi, cobinamide added to 320 nm final concentration. All strains containing the nrdJ gene were grown with 100 μm IPTG.
The WT strain grew throughout the tubes, as expected. In contrast, the nrdHEKF, nrdI, and trxR1 mutants all failed to form colonies in the same upper (oxygenated) region of each tube, which was also the only region in which the nrdD mutant exhibited colony formation (Fig. 7A). It thus appeared that any concentration of O2 sufficient to support colony formation via NrdEF activity (strain 6) was also sufficient to preclude colony formation via NrdDG (strains 3–5). Co-inoculation of tubes with the nrdHEKF and nrdD mutants (strains 3 and 6) resulted in colony formation throughout the tubes.
Interestingly, the nrdD mutant (strain 6) grew (albeit poorly) under anaerobic conditions in broth culture tubes (Fig. 6A), but it did not produce colonies in the “anaerobic” zone of the O2 gradient tubes (Fig. 7A). One explanation is that growth is occurring in the bottom portion of the gradient tubes but is not visible. The growth exhibited by the nrdD mutant in broth culture under anaerobic conditions in Fig. 6A was entirely undetectable by eye, even at 48 h (data not shown).
Growth of nrdJ-complemented Strains in an O2 Gradient
We also examined growth of the nrdJ-complemented strains in the O2 gradient assay (Fig. 7B). The nrdJ gene had little effect on the growth of the trxR1 mutant (strain 10). Thus, NrdJ, like NrdEF, has very little activity in the absence of TrxR1. It was somewhat surprising that NrdJ appeared to support no growth in the upper aerobic portion of the tube (strain 7) given that in the broth culture study NrdJ supported detectable growth of the nrdHEKF mutant in 6% O2 (strain 7, Fig. 6B). One explanation for this result is that NrdJ provided limited RNR activity in 6% O2 that was enough to create a small anaerobic zone within the broth culture tube. This might have occurred, for instance, if the inoculum cells settled out of suspension and collected at the bottom of the (unshaken) culture tubes. Once a zone of anaerobiosis was established, it could presumably be extended by further growth. This would be consistent with the slowed growth of this strain in broth culture. It should also be noted that the level of growth exhibited by the nrdJ-complemented nrdHEKF mutant at 24 h in the 6% O2 broth culture (strain 7, Fig. 6B) was undetectable by eye. Thus, it is possible that some growth did occur in the aerobic zone in the O2 gradient assay, but it was undetectable. Finally, it should be noted that the highest level of O2 encountered in the broth culture was 6%, whereas the O2 gradients were formed under ambient atmosphere (∼21% O2).
As in the broth assay, the nrdJ-complemented nrdI mutant (strain 9) showed more growth than the nrdJ-complemented nrdHEKF mutant (strain 7). In the gradient experiment, it was possible to pinpoint the source of that growth. Examination of the strain 9 tube at high magnification reveals that there is a layer of small colonies just above the main boundary that is not present in the strain 7 tube. This may be further evidence of activity due to FeIII2-Y•-cofactored NrdEF. The larger isolated colonies may represent mutants that possess greater NrdJ activity, as hypothesized below.
Isolation of a Spontaneous Mutant of nrdJ-complemented Strain
One explanation for the incomplete complementation of the nrdI and nrdHEKF mutants afforded by the nrdJ gene (Fig. 7B) is that insufficient AdoCbl was available to support full NrdJ activity. This explanation is in agreement with the genome annotation, which shows that SK36 possesses the anaerobic rather than aerobic pathway for AdoCbl synthesis. In Salmonella enterica sv. typhimurium, which also possesses the anaerobic pathway, it has been shown that synthesis of the adenosyl cobinamide portion of AdoCbl is sensitive to inactivation by O2. It has also been shown that addition of AdoCbl or Cbi (Fig. 1C) restores the activity of AdoCbl-dependent enzymes that is normally lost under aerobic conditions (51). We therefore attempted to improve growth of the nrdJ-complemented nrdHEKF mutant (strain 7) in 6% O2 with addition of AdoCbl or Cbi. The former added to concentrations of 10, 15, 50, and 150 nm produced no effect, probably due to lack of an AdoCbl transporter in S. sanguinis. In initial studies, addition of Cbi to concentrations ranging from 5 to 80 nm also had no effect after 24 or 48 h in 6% O2, whereas concentrations of 160, 320, or 640 nm strongly inhibited growth. In one culture, however, the OD660 after 48 h in the presence of 320 nm Cbi was comparable with that of WT (data not shown). Cells from this culture were alternately passaged on plates and in broth with 320 nm Cbi. We confirmed that the nrdHEKF operon and NrdF protein were still absent as expected and that the phenotype was stable (data not shown). We concluded that we had likely isolated a spontaneous mutant of the nrdJ-complemented nrdHEKF mutant that was Cbi-resistant and that also grew better in the presence of O2 than its parent strain. This strain was named JFP140c (strain 8; Table 1).
The sequence of the nrdJ gene and promoter was identical in strain 8 and the original nrdJ-complemented nrdHEKF mutant (strain 7). Western blot analysis revealed similar expression levels of NrdJ in the two strains (Fig. 4B). Thus, the putative spontaneous mutation is not in NrdJ. The concomitant increase in growth and resistance to Cbi suggested that JFP140c acquired a mutation allowing increased conversion of Cbi to AdoCbl or formation of an AdoCbl analog. The first case could arise from increased expression of gene products involved in the assembly of the three major subcomponents of AdoCbl (57) (Fig. 1C). The second could involve a mutation that results in synthesis of an alternative base to dimethylbenzimidazole (Fig. 1C), resulting in a functional AdoCbl analog. Previous studies in Salmonella enterica, for example, found point mutations causing incorporation of adenine in place of dimethylbenzimidazole, resulting in formation of pseudo-B12 (58). An alternative explanation is that the spontaneous mutation in some way causes increased anaerobiosis or increased resistance of NrdD to O2. Although this possibility cannot be ruled out, it does not readily explain why added Cbi improves growth of strain 8 but inhibits growth of its parent strain 7.
Strain 8 was examined in the O2 gradient assay in the presence of IPTG alone or IPTG plus 320 nm Cbi. As shown in Fig. 7B, both tubes exhibited a zone of growth above the usual boundary, with the Cbi-supplemented tubes showing colony formation nearly to the tops of the tubes. Although additional studies could be undertaken to characterize AdoCbl production in S. sanguinis and thereby enhance NrdJ activity, the important point for this study is the demonstration that the limited complementation of the nrdHEKF mutant, and by extension the nrdI mutant, was likely due to low production of AdoCbl. Taken together, the results suggest that NrdJ partially complements RNR activity in O2-exposed cells that are deleted for nrdHEKF or nrdI and that the level of complementation can be improved by addition of an AdoCbl precursor.
Examination of Endocarditis Virulence in an Animal Model
We next wanted to test the activity of the S. sanguinis class Ib and class III RNRs in a mammalian host. Although S. sanguinis is an occasional cause of a wide variety of extra-oral infections, it is most often associated with infective endocarditis. This illness is thought to occur when bacteria are transported through the bloodstream to the heart of persons with pre-existing cardiac damage that has resulted in the deposition of platelets and fibrin to create sterile “vegetations” (23). Infective endocarditis is therefore most often modeled by introduction of an indwelling catheter past one of the heart valves of an animal to create a sterile vegetation that predisposes the animal to infection (59). To determine whether nrdHEKF and nrdI were required for disease causation, we tested the infectivity of the respective deletion mutants in the rabbit model of endocarditis. In addition, we re-examined the contribution of the nrdD gene to endocarditis virulence, since we had previously shown that the nrdD mutant that is the parent of strain 6 exhibited a mild (∼3-fold) reduction in virulence using a variation of this model.
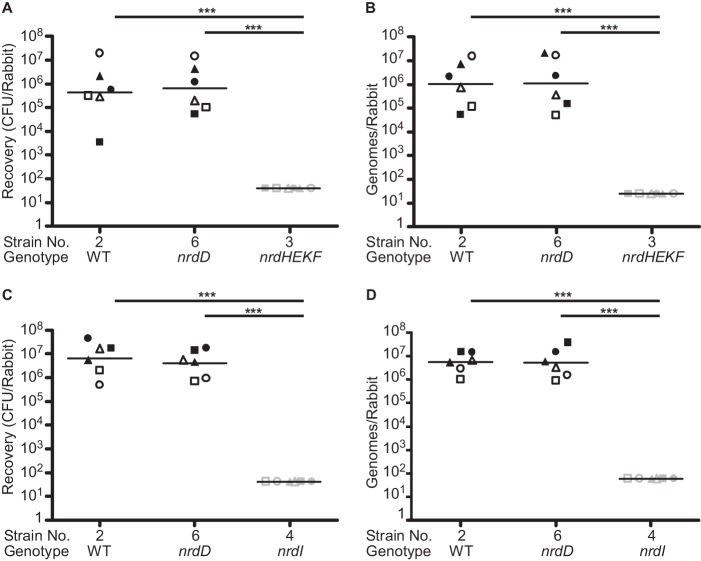
Rabbits were catheterized to create sterile vegetations and then co-inoculated with ∼107 cells of JFP36 (strain 2; an Ermr derivative of SK36 used previously for animal studies (26, 48, 60, 61)), the nrdD mutant (strain 6), and either the nrdHEKF mutant (strain 3) or the nrdI mutant (strain 4). Cell numbers in the mixed inoculum were confirmed by spreading on plates with the following selective antibiotics: Erm for WT; Spc for the nrdD mutant; and Kan for the nrdHEKF or nrdI mutants. Plates were incubated with 6% O2 for the nrdD mutant and anaerobically for all other strains. Twenty h after inoculation, rabbits were sacrificed, and vegetations were removed and homogenized. Half of each homogenate was plated on BHI with selective antibiotics for enumeration, as for the inoculum.
Fig. 8A indicates that there was no significant difference in recovery of the nrdD mutant versus WT from the six infected rabbits, but nrdHEKF mutant levels were below the limit of detection (∼40 cfu/rabbit) in the same rabbits. This suggested that nrdD was not required for infection, but the nrdHEKF operon was essential. We considered the possibility that nrdHEKF mutant cells could have died during sample processing due to O2 sensitivity. We therefore used the other half of each tissue homogenate for qPCRs with primer pairs specific for each antibiotic resistance marker. A standard curve was created by amplifying serial 10-fold dilutions of genomic DNA purified from each strain, allowing calculation of genome equivalents. The qPCR results (Fig. 8B) agreed remarkably well with the plating data for WT and the nrdD mutant. The results also confirmed the conclusions of the nrdHEKF mutant plating by indicating that the DNA from this strain was not detectable in the infected vegetations.
FIGURE 8.
Virulence of RNR-related mutants in a rabbit model of infective endocarditis. Rabbits were co-infected with the three strains indicated, and bacteria were recovered from infected vegetations by homogenization. Each experiment was performed on two occasions with three rabbits each, and the results are combined. When a particular symbol (e.g. a closed triangle) appears anywhere in A or B, it is referring to data from the same animal. The same is true for any symbol that appears in C or D. However, a given symbol used in A or B refers to a different animal than in C or D. Gray symbols represent the limit of detection for rabbits from which no bacteria or DNA were recovered. Horizontal lines indicate geometric means. Statistical significance was assessed by a repeated-measures analysis of variance with a Tukey-Kramer post test. ***, p < 0.001. Genotypes and strain numbers are as in Table 1. A and C, number of bacteria recovered by plating on selective antibiotics. B and D, number of genome equivalents determined by qPCR analysis of the antibiotic resistance genes used for selection in A and C.
An analogous experiment was performed with the nrdI mutant (strain 4) in place of the nrdHEKF mutant, and the results were the same, i.e. WT and nrdD mutant cells were isolated in large and equivalent numbers from all six rabbits, although nrdI mutant cells were not recovered from any of the same rabbits (Fig. 8C). Also, as with the nrdHEKF mutant, the nrdI mutant was not detected by qPCR in the vegetations from any of the six rabbits (Fig. 8D). It thus appears that NrdI is essential for growth and survival in the context of infective endocarditis.
DISCUSSION
We set out in this study to examine the role of the aerobic class Ib and anaerobic class III RNRs in the growth and virulence of the oral bacterium and infective endocarditis pathogen S. sanguinis. S. sanguinis is one of many pathogenic species that possess class Ib and class III enzymes as their sole RNRs.
We believe that S. sanguinis is an ideal organism to study the interplay of RNRs and pathogenesis. Previous work with strain SK36 produced the genome sequence (43), a comprehensive, ordered mutant library (19), an accurate list of essential genes (19), and an established animal disease model (15), all of which contributed to our ability to perform this work. In addition, intrinsic features of this organism were crucial. The high efficiency natural competence system of S. sanguinis (41) allowed us to confidently assess the essentiality of genes under aerobic conditions (19) and to readily produce transformants under anaerobic conditions. This is not a property shared by most other species. Indeed, class Ib RNR mutants could not be isolated in Streptococcus pyogenes, but low transformation frequencies in a control reaction made this result difficult to interpret (62). S. sanguinis is also one of a small number of human pathogens possessing genes for AdoCbl production, which allowed for the NrdJ complementation studies.
These advantages allowed us to make several important and novel discoveries. First, we were able to establish the conditions under which the class Ib (NrdEF) and class III (NrdDG) RNRs support bacterial growth. By comparing mutants lacking each of these RNRs in the O2 gradient assay, we showed that there was little or no concentration of O2 at which both RNRs could support colony formation.
Next, we found that in the presence of the heterologous nrdJ gene, a nrdI mutant grew better than a nrdHEKF mutant in both growth assays. This result could be explained by several alternative models. In one model, NrdEF possesses partial activity in the absence of NrdI. The activity would be due to the presence of FeIII2-Y•-NrdEF (63), which in vitro forms readily without NrdI and possesses activity 30% that of MnIII2-Y•-cofactored NrdEF (24). The FeIII2-Y•-NrdEF, although insufficient to support continuous growth on its own, might work synergistically with NrdJ in strain 9 to improve growth in both assays relative to the NrdJ-complemented nrdHEKF mutant (strain 7). If this model is correct, it would provide the first evidence to date of in vivo activity of a FeIII2-Y•-cofactored class Ib RNR. A second model, suggested by a reviewer, is that in the absence of NrdEF, NrdI could diminish the activity of NrdJ or alter AdoCbl through its production of superoxide. NrdI, however, is used catalytically with its corresponding NrdFs in both E. coli and B. subtilis (4, 7), i.e. it is present at very low concentrations. Furthermore, in S. sanguinis H2O2, produced rapidly from O2⨪ in the presence of protons, is already present at high concentrations. Thus, we consider this alternative less likely. Distinguishing between these and other models warrants a more detailed analysis; these studies are underway.
The growth study results shed light on the virulence studies. Neither the nrdHEKF nor nrdI mutants grew appreciably in broth culture under 6% O2 or in the aerobic zone of the O2 gradient tubes, suggesting the complete inability of the S. sanguinis NrdDG enzyme to support growth in aerobic conditions. Likewise, the nrdD mutant grew indistinguishably from SK36 under these same conditions. We have recently shown that growth of S. sanguinis sodA (superoxide dismutase) and ssaB mutants in rabbit serum under 12% O2, the level present in arterial blood (44), closely mimics the results of virulence studies performed in the rabbit model.4 Neither the nrdI nor nrdHEKF mutants were recovered after 24 h of 37 °C incubation under these conditions (data not shown). Thus, it is not surprising that neither mutant showed any ability to cause endocarditis in this study.
We reported previously that an nrdD mutant (6-26) that was the progenitor of the mutant used here (JFP141) was less competitive than SK36 in a rabbit model (15). Although we cannot be certain why our results differed in that study, we believe this may be due to recent refinements in our rabbit model.4 For this study, we sonicated bacterial inocula and the cells recovered from rabbits to break up chains and clumps, which we did not do previously. We have found that this results in colony numbers that are more reproducible and more closely match expectations based on OD readings. We also used more favorable antibiotic resistance markers in this study. We introduced the Spcr determinant into the chloramphenicol-resistant 6-26 strain to create JFP141 (strain 6) because of previous difficulties using chloramphenicol as a selective agent (15). Also, rather than use SK36 as the virulent competitor strain, as in the previous study, we used an Ermr derivative, JFP36. In the previous experiment, 6-26 colonies were enumerated on plates with chloramphenicol, although plates without antibiotics were used for recovery of SK36 and 6-26, with the number of SK36 colonies determined indirectly by subtraction. We previously showed that both the Spc and Erm resistance markers are well expressed, producing indistinguishable plating efficiencies in the presence and absence of the corresponding antibiotics and that their use provides an accurate assessment of relative strain abundance (48). Considering that in our previous study the 6-26 mutant was recovered at a level 31% that of SK36 (15), either of the above changes to the model may account for this rather minor difference.
The results from this study were clear and convincing. Mutation of nrdD had no effect on endocarditis virulence, whereas deletion of nrdHEKF or nrdI completely abolished infectivity. We conclude that the cardiac vegetation is sufficiently aerobic to completely inactivate NrdD, so that NrdEF and the NrdI-dependent MnIII2-Y•-cofactor are essential for virulence.
A previous study from our group showed that the high affinity manganese transporter, SsaB, is essential for O2 tolerance and endocarditis virulence in S. sanguinis (26). This mirrored earlier findings showing that orthologs of this transporter are essential for endocarditis caused by other oral streptococci (29, 30) for pneumonia, bacteremia, and otitis media caused by Streptococcus pneumoniae (31–34), for bacteremia caused by S. pyogenes (35), and for mastitis caused by Streptococcus uberis (36). It is known that MnII contributes to O2 tolerance in part by serving as a cofactor for the superoxide dismutase SodA. However, we determined that SodA activity was reduced by only ∼23% in an S. sanguinis ssaB mutant growing under physiological conditions and that complete elimination of SodA activity by deletion of its gene produced a far milder defect in aerobic growth and virulence than mutation of ssaB.4 We therefore concluded that MnII has important functions required for S. sanguinis O2 tolerance and virulence apart from serving as a SodA cofactor. This is likely to be true for other manganese-requiring bacterial species as well.
Given its importance, it is surprising that after more than a decade of study there is still not a clear understanding of the mechanisms by which MnII contributes to O2 tolerance and virulence (64). It seems reasonable to suggest that a reduction in RNR activity due to manganese depletion, in concert with reduced SodA activity, may well account for the reduced virulence and O2 tolerance of the ssaB mutant and manganese transporter mutants of other streptococci. Studies to directly address this possibility in S. sanguinis are underway.
Finally, the finding of class Ib RNR essentiality, together with the absence of manganese-cofactored RNRs in eukaryotes, also suggests that essential components of the class Ib pathway may serve as promising targets for new antimicrobials. The results reported here suggest that any such drug is likely to be bactericidal when used to treat or prevent infective endocarditis. Given recent alarming increases in antibiotic resistance in streptococci, enterococci, and staphylococci (65–67), all of which are important endocarditis pathogens that employ a class Ib RNR, these results are particularly welcome.
Supplementary Material
Acknowledgments
We thank Nicai Zollar and Dr. Brian Bainbridge for assistance with this work, and Dr. David Rudner for providing pDR111.
This work was supported, in whole or in part, by National Institutes of Health Grants R56AI085195 (to T. K.), K12GM093857 (to Paul B. Fisher for D. V. R. and K. E. C.), and GM81393 (to J. S.).
This article was selected as a Paper of the Week.

This article contains supplemental Table S1 and additional references.
K. E. Crump, B. Bainbridge, S. Brusko, L. S. Turner, X. Ge, V. Stone, P. Xu, and T. Kitten, submitted for publication.
To our knowledge, this is the first demonstration that yfaE is co-transcribed with nrdA and nrdB.
- RNR
- ribonucleotide reductase
- AdoCbl
- adenosylcobalamin (coenzyme B12)
- BHI
- brain heart infusion
- Cbi
- dicyanocobinamide
- Erm
- erythromycin
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- Kan
- kanamycin
- RBS
- ribosome-binding site
- Spc
- spectinomycin
- Trx
- thioredoxin
- TrxR
- thioredoxin reductase
- Y•
- tyrosyl radical
- qPCR
- quantitative PCR.
REFERENCES
- 1. Cotruvo J. A., Stubbe J. (2011) Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu. Rev. Biochem. 80, 733–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jordan A., Reichard P. (1998) Ribonucleotide reductases. Annu. Rev. Biochem. 67, 71–98 [DOI] [PubMed] [Google Scholar]
- 3. Hofer A., Crona M., Logan D. T., Sjöberg B. M. (2012) DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cotruvo J. A., Jr., Stubbe J. (2010) An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry 49, 1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cox N., Ogata H., Stolle P., Reijerse E., Auling G., Lubitz W. (2010) A tyrosyl-dimanganese coupled spin system is the native metalloradical cofactor of the R2F subunit of the ribonucleotide reductase of Corynebacterium ammoniagenes. J. Am. Chem. Soc. 132, 11197–11213 [DOI] [PubMed] [Google Scholar]
- 6. Stolle P., Barckhausen O., Oehlmann W., Knobbe N., Vogt C., Pierik A. J., Cox N., Schmidt P. P., Reijerse E. J., Lubitz W., Auling G. (2010) Homologous expression of the nrdF gene of Corynebacterium ammoniagenes strain ATCC 6872 generates a manganese-metallocofactor (R2F) and a stable tyrosyl radical (Y•) involved in ribonucleotide reduction. FEBS J. 277, 4849–4862 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y., Stubbe J. (2011) Bacillus subtilis class Ib ribonucleotide reductase is a dimanganese(III)-tyrosyl radical enzyme. Biochemistry 50, 5615–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Atkin C. L., Thelander L., Reichard P., Lang G. (1973) Iron and free radical in ribonucleotide reductase. Exchange of iron and Mossbauer spectroscopy of the protein B2 subunit of the Escherichia coli enzyme. J. Biol. Chem. 248, 7464–7472 [PubMed] [Google Scholar]
- 9. Cotruvo J. A., Jr., Stubbe J. (2008) NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proc. Natl. Acad. Sci. U.S.A. 105, 14383–14388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cotruvo J. A., Jr., Stich T. A., Britt R. D., Stubbe J. (2013) Mechanism of assembly of the dimanganese-tyrosyl radical cofactor of class Ib ribonucleotide reductase: enzymatic generation of superoxide is required for tyrosine oxidation via a Mn(III)Mn(IV) intermediate. J. Am. Chem. Soc. 135, 4027–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. King D. S., Reichard P. (1995) Mass spectrometric determination of the radical scission site in the anaerobic ribonucleotide reductase of Escherichia coli. Biochem. Biophys. Res. Commun. 206, 731–735 [DOI] [PubMed] [Google Scholar]
- 12. Collins H. F., Biedendieck R., Leech H. K., Gray M., Escalante-Semerena J. C., McLean K. J., Munro A. W., Rigby S. E., Warren M. J., Lawrence A. D. (2013) Bacillus megaterium has both a functional BluB protein required for DMB synthesis and a related flavoprotein that forms a stable radical species. PLoS One 8, e55708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moore S. J., Warren M. J. (2012) The anaerobic biosynthesis of vitamin B12. Biochem. Soc. Trans. 40, 581–586 [DOI] [PubMed] [Google Scholar]
- 14. Lundin D., Torrents E., Poole A. M., Sjöberg B.-M. (2009) RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to GenBank. BMC Genomics 10, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paik S., Senty L., Das S., Noe J. C., Munro C. L., Kitten T. (2005) Identification of virulence determinants for endocarditis in Streptococcus sanguinis by signature-tagged mutagenesis. Infect. Immun. 73, 6064–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kirdis E., Jonsson I.-M., Kubica M., Potempa J., Josefsson E., Masalha M., Foster S. J., Tarkowski A. (2007) Ribonucleotide reductase class III, an essential enzyme for the anaerobic growth of Staphylococcus aureus, is a virulence determinant in septic arthritis. Microb. Pathog. 43, 179–188 [DOI] [PubMed] [Google Scholar]
- 17. Fey P. D., Endres J. L., Yajjala V. K., Widhelm T. J., Boissy R. J., Bose J. L., Bayles K. W. (2013) A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4, e00537–00512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang R., Lin Y. (2009) DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 37, D455–D458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu P., Ge X., Chen L., Wang X., Dou Y., Xu J. Z., Patel J. R., Stone V., Trinh M., Evans K., Kitten T., Bonchev D., Buck G. A. (2011) Genome-wide essential gene identification in Streptococcus sanguinis. Sci. Rep. 10.1038/srep00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cotruvo J. A., Stubbe J. (2011) Escherichia coli class Ib ribonucleotide reductase contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry 50, 1672–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin J. E., Imlay J. A. (2011) The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol. Microbiol. 80, 319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Socransky S. S., Manganiello A. D., Propas D., Oram V., van Houte J. (1977) Bacteriological studies of developing supragingival dental plaque. J. Periodontal Res. 12, 90–106 [DOI] [PubMed] [Google Scholar]
- 23. Thuny F., Grisoli D., Collart F., Habib G., Raoult D. (2012) Management of infective endocarditis: challenges and perspectives. Lancet 379, 965–975 [DOI] [PubMed] [Google Scholar]
- 24. Makhlynets O., Boal A. K., Rhodes D. V., Kitten T., Rosenzweig A. C., Stubbe J. (2014) Streptococcus sanguinis class Ib ribonucleotide reductase: high activity with both iron and manganese cofactors and structural insights. J. Biol. Chem. 289, 6259–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Booker S., Stubbe J. (1993) Cloning, sequencing, and expression of the adenosylcobalamin-dependent ribonucleotide reductase from Lactobacillus leichmannii. Proc. Natl. Acad. Sci. U.S.A. 90, 8352–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Das S., Kanamoto T., Ge X., Xu P., Unoki T., Munro C. L., Kitten T. (2009) Contribution of lipoproteins and lipoprotein processing to endocarditis virulence in Streptococcus sanguinis. J. Bacteriol. 191, 4166–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kilian M., Holmgren K. (1981) Ecology and nature of immunoglobulin A1 protease-producing streptococci in the human oral cavity and pharynx. Infect. Immun. 31, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kehl-Fie T. E., Zhang Y., Moore J. L., Farrand A. J., Hood M. I., Rathi S., Chazin W. J., Caprioli R. M., Skaar E. P. (2013) MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect. Immun. 81, 3395–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burnette-Curley D., Wells V., Viscount H., Munro C. L., Fenno J. C., Fives-Taylor P., Macrina F. L. (1995) FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect. Immun. 63, 4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paik S., Brown A., Munro C. L., Cornelissen C. N., Kitten T. (2003) The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J. Bacteriol. 185, 5967–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berry A. M., Paton J. C. (1996) Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect. Immun. 64, 5255–5262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marra A., Lawson S., Asundi J. S., Brigham D., Hromockyj A. E. (2002) In vivo characterization of the psa genes from Streptococcus pneumoniae in multiple models of infection. Microbiology 148, 1483–1491 [DOI] [PubMed] [Google Scholar]
- 33. Johnston J. W., Myers L. E., Ochs M. M., Benjamin W. H., Jr., Briles D. E., Hollingshead S. K. (2004) Lipoprotein PsaA in virulence of Streptococcus pneumoniae: surface accessibility and role in protection from superoxide. Infect. Immun. 72, 5858–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McAllister L. J., Tseng H.-J., Ogunniyi A. D., Jennings M. P., McEwan A. G., Paton J. C. (2004) Molecular analysis of the psa permease complex of Streptococcus pneumoniae. Mol. Microbiol. 53, 889–901 [DOI] [PubMed] [Google Scholar]
- 35. Janulczyk R., Ricci S., Björck L. (2003) MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect. Immun. 71, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith A. J., Ward P. N., Field T. R., Jones C. L., Lincoln R. A., Leigh J. A. (2003) MtuA, a lipoprotein receptor antigen from Streptococcus uberis, is responsible for acquisition of manganese during growth in milk and is essential for infection of the lactating bovine mammary gland. Infect. Immun. 71, 4842–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lohman G. J., Stubbe J. (2010) Inactivation of Lactobacillus leichmannii ribonucleotide reductase by 2′,2′-difluoro-2′-deoxycytidine 5′-triphosphate: covalent modification. Biochemistry 49, 1404–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chivers P. T., Prehoda K. E., Volkman B. F., Kim B. M., Markley J. L., Raines R. T. (1997) Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry 36, 14985–14991 [DOI] [PubMed] [Google Scholar]
- 39. Russel M., Model P. (1985) Direct cloning of the trxB gene that encodes thioredoxin reductase. J. Bacteriol. 163, 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Steeper J. R., Steuart C. D. (1970) A rapid assay for CDP reductase activity in mammalian cell extracts. Anal. Biochem. 34, 123–130 [DOI] [PubMed] [Google Scholar]
- 41. Rodriguez A. M., Callahan J. E., Fawcett P., Ge X., Xu P., Kitten T. (2011) Physiological and molecular characterization of genetic competence in Streptococcus sanguinis. Mol. Oral Microbiol. 26, 99–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 43. Xu P., Alves J. M., Kitten T., Brown A., Chen Z., Ozaki L. S., Manque P., Ge X., Serrano M. G., Puiu D., Hendricks S., Wang Y., Chaplin M. D., Akan D., Paik S., Peterson D. L., Macrina F. L., Buck G. A. (2007) Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 189, 3166–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Atkuri K. R., Herzenberg L. A., Niemi A.-K., Cowan T., Herzenberg L. A. (2007) Importance of culturing primary lymphocytes at physiological oxygen levels. Proc. Natl. Acad. Sci. U.S.A. 104, 4547–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cotruvo J. A., Jr., Stubbe J. (2012) Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: the class I ribonucleotide reductases as a case study. Metallomics 4, 1020–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Crona M., Torrents E., Røhr A. K., Hofer A., Furrer E., Tomter A. B., Andersson K. K., Sahlin M., Sjöberg B.-M. (2011) NrdH-redoxin protein mediates high enzyme activity in manganese-reconstituted ribonucleotide reductase from Bacillus anthracis. J. Biol. Chem. 286, 33053–33060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jordan A., Aslund F., Pontis E., Reichard P., Holmgren A. (1997) Characterization of Escherichia coli NrdH. A glutaredoxin-like protein with a thioredoxin-like activity profile. J. Biol. Chem. 272, 18044–18050 [DOI] [PubMed] [Google Scholar]
- 48. Turner L. S., Das S., Kanamoto T., Munro C. L., Kitten T. (2009) Development of genetic tools for in vivo virulence analysis of Streptococcus sanguinis. Microbiology 155, 2573–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu C. H., Jiang W., Krebs C., Stubbe J. (2007) YfaE, a ferredoxin involved in diferric-tyrosyl radical maintenance in Escherichia coli ribonucleotide reductase. Biochemistry 46, 11577–11588 [DOI] [PubMed] [Google Scholar]
- 50. Blakley R. L. (1965) Cobamides and ribonucleotide reduction: I. cobamide stimulation of ribonucleotide reduction in extracts of Lactobacillus leichmannii. J. Biol. Chem. 240, 2173–2180 [PubMed] [Google Scholar]
- 51. Roth J. R., Lawrence J. G., Bobik T. A. (1996) Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50, 137–181 [DOI] [PubMed] [Google Scholar]
- 52. Carlsson J., Edlund M. B., Lundmark S. K. (1987) Characteristics of a hydrogen peroxide-forming pyruvate oxidase from Streptococcus sanguis. Oral Microbiol. Immunol. 2, 15–20 [DOI] [PubMed] [Google Scholar]
- 53. Kreth J., Vu H., Zhang Y., Herzberg M. C. (2009) Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J. Bacteriol. 191, 6281–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sztukowska M., Bugno M., Potempa J., Travis J., Kurtz D. M., Jr. (2002) Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol. Microbiol. 44, 479–488 [DOI] [PubMed] [Google Scholar]
- 55. Booker S., Licht S., Broderick J., Stubbe J. (1994) Coenzyme B12-dependent ribonucleotide reductase: evidence for the participation of five cysteine residues in ribonucleotide reduction. Biochemistry 33, 12676–12685 [DOI] [PubMed] [Google Scholar]
- 56. O'Brien J., Wilson I., Orton T., Pognan F. (2000) Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 267, 5421–5426 [DOI] [PubMed] [Google Scholar]
- 57. Andersson D. I., Roth J. R. (1989) Mutations affecting regulation of cobinamide biosynthesis in Salmonella typhimurium. J. Bacteriol. 171, 6726–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Anderson P. J., Lango J., Carkeet C., Britten A., Kräutler B., Hammock B. D., Roth J. R. (2008) One pathway can incorporate either adenine or dimethylbenzimidazole as an α-axial ligand of B12 cofactors in Salmonella enterica. J. Bacteriol. 190, 1160–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Durack D. T., Beeson P. B., Petersdorf R. G. (1973) Experimental bacterial endocarditis. 3. Production and progress of the disease in rabbits. Br. J. Exp. Pathol. 54, 142–151 [PMC free article] [PubMed] [Google Scholar]
- 60. Callahan J. E., Munro C. L., Kitten T. (2011) The Streptococcus sanguinis competence regulon is not required for infective endocarditis virulence in a rabbit model. PLoS One 6, e26403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Turner L. S., Kanamoto T., Unoki T., Munro C. L., Wu H., Kitten T. (2009) Comprehensive evaluation of Streptococcus sanguinis cell wall-anchored proteins in early infective endocarditis. Infect. Immun. 77, 4966–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Roca I., Torrents E., Sahlin M., Gibert I., Sjöberg B.-M. (2008) NrdI essentiality for Class Ib ribonucleotide reduction in Streptococcus pyogenes. J. Bacteriol. 190, 4849–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hristova D., Wu C. H., Jiang W., Krebs C., Stubbe J. (2008) Importance of the maintenance pathway in the regulation of the activity of Escherichia coli ribonucleotide reductase. Biochemistry 47, 3989–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Papp-Wallace K. M., Maguire M. E. (2006) Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 60, 187–209 [DOI] [PubMed] [Google Scholar]
- 65. García-de-la-Mària C., Pericas J. M., Del Río A., Castañeda X., Vila-Farrés X., Armero Y., Espinal P. A., Cervera C., Soy D., Falces C., Ninot S., Almela M., Mestres C. A., Gatell J. M., Vila J., Moreno A., Marco F., Miró J. M., and Hospital Clinic Experimental Endocarditis Study Group (2013) Early in vitro and in vivo development of high-level daptomycin resistance is common in mitis group streptococci after exposure to daptomycin. Antimicrob. Agents Chemother. 57, 2319–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., Scheld M., Spellberg B., Bartlett J. (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 [DOI] [PubMed] [Google Scholar]
- 67. Bor D. H., Woolhandler S., Nardin R., Brusch J., Himmelstein D. U. (2013) Infective endocarditis in the U.S., 1998–2009: a nationwide study. PLoS One 8, e60033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.