Background: 8-Halogenated guanine is a promutagenic lesion that promotes insertion of guanine opposite the lesion during DNA replication.
Results: 8-Bromoguanine forms Hoogsteen base pairing with G and Watson-Crick base pairing with C in the active site of polβ.
Conclusion: 8-Bromoguanine is well accommodated in the nascent base pair binding pocket of polβ.
Significance: Our structural studies provide insights into potential G to C mutations.
Keywords: DNA Damage, DNA Polymerase, DNA Repair, Enzyme Structure, Protein Structure, X-ray Crystallography
Abstract
8-Halogenated guanine (haloG), a major DNA adduct formed by reactive halogen species during inflammation, is a promutagenic lesion that promotes misincorporation of G opposite the lesion by various DNA polymerases. Currently, the structural basis for such misincorporation is unknown. To gain insights into the mechanism of misincorporation across haloG by polymerase, we determined seven x-ray structures of human DNA polymerase β (polβ) bound to DNA bearing 8-bromoguanine (BrG). We determined two pre-catalytic ternary complex structures of polβ with an incoming nonhydrolyzable dGTP or dCTP analog paired with templating BrG. We also determined five binary complex structures of polβ in complex with DNA containing BrG·C/T at post-insertion and post-extension sites. In the BrG·dGTP ternary structure, BrG adopts syn conformation and forms Hoogsteen base pairing with the incoming dGTP analog. In the BrG·dCTP ternary structure, BrG adopts anti conformation and forms Watson-Crick base pairing with the incoming dCTP analog. In addition, our polβ binary post-extension structures show Hoogsteen BrG·G base pair and Watson-Crick BrG·C base pair. Taken together, the first structures of haloG-containing DNA bound to a protein indicate that both BrG·G and BrG·C base pairs are accommodated in the active site of polβ. Our structures suggest that Hoogsteen-type base pairing between G and C8-modified G could be accommodated in the active site of a DNA polymerase, promoting G to C mutation.
Introduction
Chronic inflammation is closely associated with carcinogenesis (1, 2). One possible mechanism for inflammation-induced carcinogenesis involves DNA damage and mutation caused by reactive halogen species such as HOCl and HOBr, generated by the action of myeloperoxidase or eosinophil peroxidase on endogenous H2O2 and Cl−/Br− (Fig. 1A) (3, 4). The concentration of HOCl at the inflammation sites in vivo has been estimated to be ∼50 mm (5). Whereas reactive halogen species, potent oxidants that kill pathogens, play an important role in the host defense mechanism, they can also damage DNA especially under inflammatory conditions to generate various halogenated DNA lesions (5–7), with 8-halogenated G (haloG)2 such as 8-chloroguanine and 8-bromoguanine (BrG) being the major adducts (8–10). Recently, haloG has been detected in DNA of rat liver and in the urine of patients with hepatocellular carcinoma (10). Despite ∼1000-fold lower physiological concentration of Br− (100 μm) relative to that of Cl− (100 mΜ), the concentration of BrG is ∼5-fold higher than that of 8-chloroguanine under inflammatory conditions (10). The urine concentration of haloG was one tenth of that of 8-oxoguanine (oxoG) in healthy subjects, and half of that in diabetic patients, suggesting haloG as a potentially important lesion (10). Interestingly, 8-chloro-dGTP was efficiently hydrolyzed by hMTH1 (11), which hydrolyzes 8-oxo-dGTP to 8-oxo-dGMP. In addition, haloG was excreted to urine three times faster than oxoG in lipopolysaccharide-treated rats (10), suggesting the existence of an efficient DNA repair enzyme for haloG in cells.
FIGURE 1.
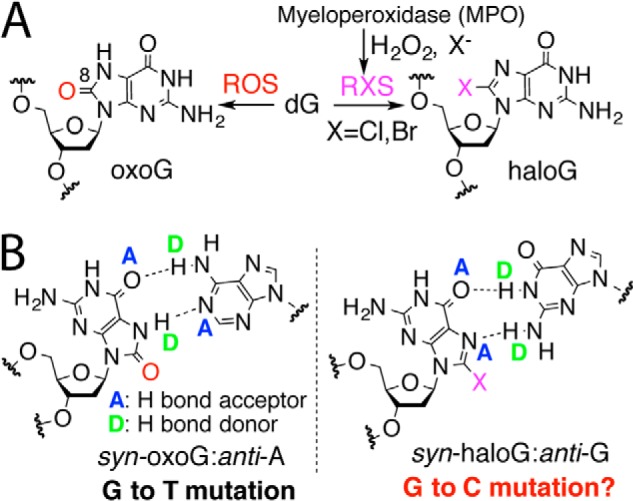
Hoogsteen base pairing of C8-modified guanine. A, generation of 8-haloG and 8-oxoG by reactive halogen species and reactive oxygen species, respectively, is shown. B, oxoG in syn conformation can form two H-bonds with A, causing G to T mutation. Similarly, haloG in syn conformation could form two H-bonds with G, potentially promoting G to C mutation. Note that H-bond motifs on the Hoogsteen edges of oxoG and BrG are different (AD [oxoG] versus AA [BrG]).
HaloG is a promutagenic lesion that facilitates misincorporation of G opposite the lesion during DNA replication in vitro (12, 13). Insertion efficiencies for G opposite templating BrG by DNA polymerases were similar to those for C, suggesting that persistence of haloG lesion in DNA could cause G to C transversion mutation. Currently, the mechanism underlying potential haloG-induced mutagenesis is largely unknown. We hypothesized that, like oxoG that induces G to T mutation by forming two H-bonds with anti-A using a H-bond donor and a H-bond acceptor on its Hoogsteen edge (14), haloG could promote G to C mutation by forming two H-bonds with anti-G using two H-bond acceptors on its Hoogsteen edge during DNA replication (Fig. 1B).
As an initial step to test our hypothesis, we wanted to solve x-ray structures of haloG-containing DNA bound to a human DNA polymerase β (polβ), which has been shown to insert a mixture of G and C opposite templating BrG in vitro, albeit with a ∼100-fold reduced insertion efficiency compared with insertion of C opposite G (13). Polβ is a DNA repair enzyme that fills short nucleotide gaps in DNA produced during base-excision DNA repair pathway (15). This enzyme belongs to X-family DNA polymerase and lacks an intrinsic proofreading 3′ → 5′ exonuclease activity. The protein contains an N-terminal lyase domain (8 kDa) and a C-terminal polymerase domain (31 kDa). The polymerase domain can be further divided into thumb, fingers, and palm subdomains typically observed in DNA polymerases.
Whereas several structures of haloG-containing nucleic acids have been published (16, 17), its protein-bound structure has not been reported. Here, we report seven x-ray structures of polβ bound to DNA containing BrG at varying stages of DNA replication. We determined a gapped binary complex structure of polβ bound to templating BrG, and two ternary complex structures of polβ with incoming nonhydrolyzable dGTP or dCTP analog paired with templating BrG. In addition, we determined four binary complex structures of polβ with BrG·C or BrG·G at varying positions of the enzyme active site. These x-ray structures provide structural basis for misincorporation of G opposite BrG by polβ and insights into potential haloG-induced G to C mutation.
EXPERIMENTAL PROCEDURES
DNA Sequences
Oligonucleotides were purchased from Integrated DNA Technologies (IDT) or Midland Certified Reagent Co. (Midland, TX). They were purified by the manufacturer, and their sequences were confirmed by MALDI-TOF mass spectrometry. DNA substrates used for crystallographic studies consisted of a 16-mer template, a complementary 10-mer primer, and 5-mer downstream oligonucleotides (18). The template DNA sequence used for crystallization was 5′-CCGAC[BrG]TCGCATCAGC-3′. The upstream primer sequence was 5′-GCTGATGCGA-3′. The downstream oligonucleotide sequence was 5′-GTCGG-3′, and the 5′ terminus was phosphorylated. The DNA sequence almost identical to a published ternary complex structure (PDB ID 1BPY) was used to minimize sequence-dependent structural differences (19). The oligonucleotides were mixed and annealed to give a 1 mm mixture of gapped DNA as described (19).
Protein Expression and Purification
Polβ was expressed and purified from Escherichia coli with minor modifications of the method described previously (19, 20). The human polymerase β gene cloned into pET-30 (Novagen) was transformed into Rosetta2 (DE3) cells. Bacteria were grown in LB medium supplemented with 30 mg/liter kanamycin at 37 °C to an A600 = 0.6, and cooled to 28 °C for 30 min. Then the protein was induced by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. The culture was grown overnight at 28 °C, harvested by centrifugation (8000 × g for 10 min), resuspended in a lysis buffer (20 mm sodium phosphate, pH 7.5, 300 mm NaCl, 1 mm PMSF), and lysed by sonication for 2 min. The suspension was spun at 10,000 × g for 15 min, and a clear supernatant was loaded onto a HisTrap column (GE Healthcare) pre-equilibrated with lysis buffer and eluted by the gradient of 250 mm imidazole. After pooling the fractions containing the protein and extensive buffer exchange with 50 mm Tris, pH 7.5, 2 mm DTT, and 100 mm NaCl, the concentrate was applied to Mono S cation exchange column (GE Healthcare). Then the His tag was cleaved by incubating a mixture of factor Xa and thrombin overnight at 4 °C. To further separate from minor contaminants, human polymerase β was purified by size exclusion chromatography on a Superdex 75 column (GE Healthcare) pre-equilibrated with 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 200 mm NaCl. After purification, polβ was buffer-exchanged, concentrated to 20 mg/liter, and stored at −80 °C as described.
Protein-DNA Co-crystallization
The binary polβ complex containing templating BrG in a single-nucleotide gapped DNA was prepared under conditions similar to those described previously (19). Polβ was complexed with a single-nucleotide gapped DNA containing a 16-mer template (5′-CCGAC[BrG]GCGCATCAGC-3′), a complementary 10-mer primer (5′-GCTGATGCGC-3′), and a 5-mer downstream oligonucleotide (5′-pGTCGG-3′). The resulting polβ-DNA complex was used to obtain binary and ternary complex crystals in the absence or presence of an incoming nucleotide, respectively. The ternary polβ-DNA complex co-crystals with nonhydrolyzable dGTP or dCTP analog paired with templating BrG in a single-nucleotide gap at the active site were grown in a solution containing 50 mm imidazole, pH 7.5, 14–23% PEG3400, and 350 mm sodium acetate as described previously (21). Crystals were cryo-protected in mother liquor supplemented with 12% ethylene glycol and were flash-frozen in liquid nitrogen.
Data Collection and Refinement
Diffraction data were collected at 100 K using either a Rigaku MicroMax-007 HF microfocus x-ray generator with R-Axis IV++ imaging plate area detector or the beamline 5.0.3 Advanced Light Source at Berkeley Center for Structural Biology. All diffraction data were processed using HKL 2000 (22). The structures of the binary polβ complex with templating BrG in a single-nucleotide gapped DNA and the ternary complex of polβ with templating BrG paired with dGTP or dCTP analog were solved by molecular replacement with polβ with a single-nucleotide gapped DNA (PDB code 1BPX) as the search model. The model was built using COOT and refined using PHENIX software (23, 24). MolProbity was used to make Ramachandran plots (25).
RESULTS
Structure of a Single-nucleotide Gapped Binary Complex of Polβ with Templating BrG
We determined the x-ray structure of a single-nucleotide gapped binary complex of polβ bound to BrG-containing DNA. The structure of the gapped binary complex (PDB ID 4M2Y, see Table 1 for refinement statistics), refined to 2.3 Å resolution, is very similar to that of a published gapped binary structure (PDB ID 1BPX, r.m.s.d. = 0.689 Å) (Fig. 2A) (19). Protein is in an open conformation, with α-helix N containing Asn-279 and Arg-283, the minor groove recognition motifs, being located ∼10 Å from templating BrG.
TABLE 1.
Data collection and refinement statistics
| Parameters | Gapped binary (4M2Y)a | BrG·C N binary (4NLN) | BrG·G N binary (4NLZ) | BrG·C ternary (4NLK) | BrG·G ternary (4M47) | BrG·C N-1 binary (4NM1) | BrG·G N-1 binary (4NM2) |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P21 | P21 | P21 | P21 | P21 | P21 | P21 |
| Cell constants | |||||||
| a (Å) | 54.554 | 54.955 | 54.414 | 50.807 | 55.175 | 50.749 | 54.731 |
| b | 79.271 | 79.075 | 79.438 | 79.706 | 79.267 | 80.114 | 80.202 |
| c | 54.915 | 54.912 | 54.798 | 55.683 | 55.2944 | 55.272 | 55.109 |
| α (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| β | 105.48 | 106.59 | 105.37 | 107.83 | 107.80 | 107.52 | 106.60 |
| γ | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| Resolution (Å)b | 20–2.27 (2.31–2.27) | 20–2.26 (2.30–2.26) | 20–2.68 (2.73–2.68) | 20–2.49 (2.54–2.49) | 20–2.37 2.41–2.37) | 20–2.42 (2.46–2.42) | 20–2.55 (2.59–2.55) |
| <I/σ> | 18.4 (3.0) | 19.4 (2.26) | 8.5 (1.92) | 14.8 (2.18) | 19.6 (2.98) | 12.1 (1.91) | 20.6 (2.10) |
| Completeness (%) | 97.1 (94.8) | 98.7 (98.3) | 99.6 (99.5) | 99.9 (99.9) | 96.4 (95.4) | 100 (100) | 97.3 (85.4) |
| Rmergec (%) | 9.4 (38.5) | 8.7 (48.7) | 14.9 (50.7) | 12.2 (39.3) | 8.5 (32.6) | 10.9 (43.6) | 11.4 (49.9) |
| Redundancy | 4.7 (4.1) | 4.4 (4.2) | 4.2 (3.4) | 4.2 (3.6) | 2.7 (2.8) | 4.0 (3.6) | 6.7 (5.8) |
| Refinement | |||||||
| Rworkd/Rfreee (%) | 21.2/26.9 | 21.3/27.3 | 20.4/29.0 | 19.7/26.9 | 21.2/27.3 | 20.1/25.6 | 20.4/27.6 |
| Unique reflections | 20,228 | 20,837 | 12,597 | 14,578 | 17,813 | 16,215 | 14,626 |
| Mean B factor (Å2) | |||||||
| Protein | 28.4 | 37.5 | 22.2 | 31.5 | 42.3 | 24.8 | 40.7 |
| Ligand | 28.2 | 39.8 | 22.8 | 21.2 | 38.4 | 32.1 | 36.4 |
| Solvent | 25.1 | 36.1 | 16.1 | 29.1 | 36.6 | 23.2 | 40.1 |
| Ramachandran plot | |||||||
| Most favored (%) | 96.8 | 95.7 | 93.4 | 97.2 | 95.8 | 97.5 | 93.7 |
| Additional allowed (%) | 3.0 | 4.3 | 6.6 | 2.8 | 3.9 | 2.5 | 6.3 |
| r.m.s.d. | |||||||
| Bond lengths (Å) | 0.012 | 0.013 | 0.010 | 0.011 | 0.012 | 0.012 | 0.010 |
| Bond angles (°) | 1.651 | 1.842 | 1.664 | 1.659 | 1.623 | 1.748 | 1.627 |
a PDB ID codes.
b Values in parentheses are for the highest resolution shell.
c Rmerge = Σ|I−<I> /ΣI where I is the integrated intensity of a given reflection.
d Rwork = Σ|Fobs − Fcalc|/ΣFobs.
e Rfree = Σ|Fobs − Fcalc)|/ΣFobs, calculated using 5% of the data.
FIGURE 2.
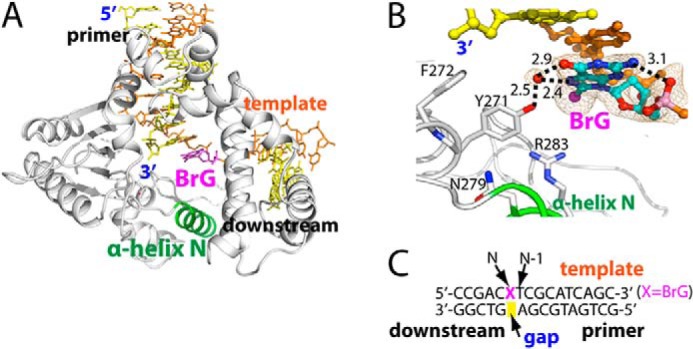
Structure of binary complex structure of polβ bound to DNA containing a single-nucleotide gap opposite templating BrG. A, overall structure of the binary gapped structure (PDB ID 4M2Y). Polβ is shown in white, and α-helix N containing the minor-groove recognition motif is shown in green. The template strand is shown in orange, and the primer and the downstream DNA are shown in yellow. BrG is shown in magenta. α-Helix N is in open conformation. B, active-site view of the gapped binary complex structure. An ordered water that bridges the OH of Tyr-271 and the Hoogsteen edge of BrG is depicted as a red sphere. Br is shown as a magenta sphere. Key H-bondings are indicated as dotted lines, where numbers in the panel indicate distances in Å. A 2Fo − Fc electron density map contoured at 1σ is shown around BrG and the ordered water. BrG is in syn conformation and forms an intramolecular H-bonding with its 5′-phosphate. C, DNA sequence used for crystallization of the gapped polβ complex. The positions of the gap, N, and N-1 are indicated.
The BrG gapped binary structure shows that, unlike G or oxoG, unpaired BrG at templating position preferentially adopts syn conformation. The syn-BrG is stabilized by an intramolecular H-bond between the N2 and the 5′-phosphate oxygen of BrG (Fig. 2B). In addition, an ordered water molecule stabilizes syn-BrG by bridging BrG and Tyr-271. The preferred syn conformation of BrG in the templating base position is in contrast with the previous observation that oxoG in the same position exists as a mixture of syn and anti conformers (14, 18), suggesting that size of C8-substituent may govern base conformation in the templating base position. The presence of syn-BrG in DNA triggers a local conformational change at the template DNA, where the BrG lesion moved ∼10 Å away from the position of G nucleotide residue seen in the published binary complex structure (PDB ID 1BPX) (19).
Ternary Structure of Polβ with Templating BrG Paired with an Incoming dCTP Analog
To elucidate structural features of polβ performing correct insertion opposite BrG, we obtained a pre-catalytic ternary structure of polβ in complex with an incoming nonhydrolyzable dCMPNPP (hereafter dCTP*) paired with templating BrG. The NH group in dNMPNPP replaces the bridging oxygen between Pα and Pβ, rendering the nucleotide analog resistant to dNMP transfer and hydrolysis. The use of nonhydrolyzable dNMPNPP thus enables the capture of ternary polβ structures bearing the critical coordination of 3′-OH of primer terminus to the catalytic metal ion (26). These nonhydrolyzable dNMPNPP nucleotides have been shown to have active-site coordination essentially identical to that of their natural nucleotides (e.g. PDB ID 2FMP and 2FMS (26)) and have been used in structural studies of various DNA polymerases (27–29).
The BrG·C ternary structure (PDB ID 4NLK) was refined to 2.5 Å resolution. The overall structure of the BrG·C ternary complex is very similar to that of published ternary structures with correct base pair, which assumes co-planar base pair conformation and closed protein conformation (PDB ID 2FMS, r.m.s.d. = 0.299 Å; PDB ID 2FMP, r.m.s.d. = 0.295 Å) (Fig. 3A) (26). Open-to-closed conformational activation of polβ for correct nucleotide incorporation typically involves the movement of α-helix N and the change in H-bonding interactions of Asn-279, Arg-283, and Tyr-271 with DNA (15). In the BrG·C ternary structure, the α-helix N moved ∼10 Å from the position in the gapped binary complex to sandwich the nascent BrG·C base pair between the primer terminus base pair and α-helix N. In addition, Asn-279, Arg-283, and Tyr-271 engage in H-bonding interactions with the minor groove edges of the incoming nucleotide, templating base, and primer terminus, respectively.
FIGURE 3.
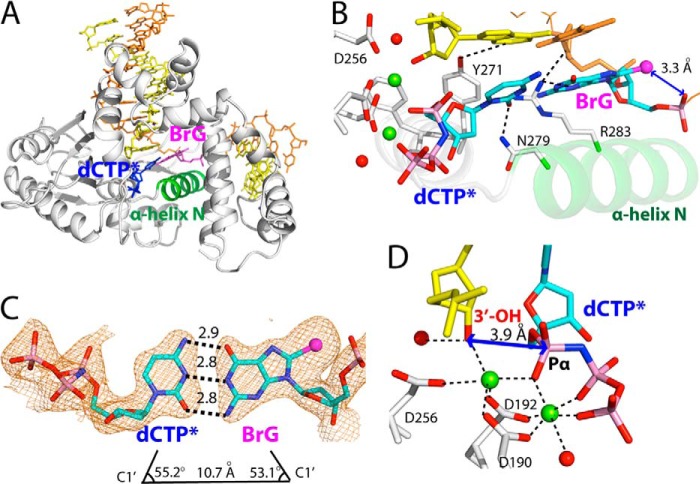
Pre-catalytic ternary structure of polβ inserting a nonhydrolyzable dCTP analog opposite templating BrG (PDB ID 4NLK). A, overall structure highlighting closed protein conformation and co-planar base pairing. B, active-site view of the BrG·C ternary structure. Metal ions are shown in green spheres, and ordered water molecules are shown in red spheres. BrG·dCTP* forms co-planar conformation, and α-helix N is in closed conformation. Asn-279 and Arg-283 interact with the minor groove edges of dCTP* and BrG, respectively. C, H-bonding interactions and geometry of the nascent BrG·dCTP* base pair. The distance between C1′(dCTP*) and C1′(BrG) and λ angles are indicated. A 2Fo − Fc map is contoured at 1σ around BrG and dCTP*. Numbers in the panel indicate distances in Å. D, close-up view of the BrG·C pre-catalytic complex. Distance between Pα of dCTP* and the 3′-OH of primer terminus is indicated. Note that the nucleotide-binding metal ion is hexa-coordinated, whereas the catalytic metal ion is penta-coordinated.
The BrG·C ternary structure shows that the BrG·C base pair is well accommodated in the nascent base pair binding pocket of the enzyme. In the BrG·C ternary structure, BrG adopts anti conformation rather than syn conformation observed in the BrG gapped binary structure. The BrG and incoming dCTP* form co-planar a Watson-Crick base pair typically observed in structure with correct insertion (Fig. 3B) (30). The BrG·dCTP* is sandwiched between the primer terminus base pair and the α-helix N in closed conformation. The geometry of BrG·C base pair is essentially identical to that of G·C. The λ angles for BrG and dCTP* are 53.2° are 55.1°, respectively (Fig. 3C). The distance between C1′(dCTP*) and C1′(BrG) is 10.7 Å, which is similar to that observed in structure for correct insertion.
Our structure provides insight into the slower insertion efficiency for dCTP opposite BrG (13). Distance between the 3′-OH of primer terminus and Pα of incoming nucleotide (3.9 Å) is longer than that observed in correct insertion (3.4 Å), which would be suboptimal for nucleotidyl transfer. In addition, the 3′-OH of primer terminus is not well positioned for in-line attack on Pα of dCTP*. Furthermore, although the BrG·C ternary structure adopts co-planar Watson-Crick base pairing and closed protein conformation, the coordination sphere of the catalytic metal ion is not complete. In a ternary polβ complex structure with correct insertion, catalytic metal ion is complexed with three Asp residues (Asp-190, -192, and -256), Pα oxygen of the incoming nucleotide, the 3′-OH of primer terminus, and ordered water. In the BrG·C ternary structure, the ordered water molecule is not coordinated to catalytic metal ion. Combined effects of the longer 3′-OH-Pα(dCTP*) distance, nonideal trajectory for in-line attack, and incomplete coordination state of the catalytic metal ion would decrease the insertion efficiency for C opposite BrG.
Ternary Structure of Polβ with Templating BrG Paired with an Incoming dGTP Analog
To elucidate structural features of polβ misincorporating G opposite BrG, we solved the x-ray structure of a ternary complex of polβ with incoming nonhydrolyzable nucleotide analog dGTP* and templating BrG. The BrG·G ternary structure (PDB ID 4M47) was refined to 2.4 Å resolution (Fig. 4A).
FIGURE 4.
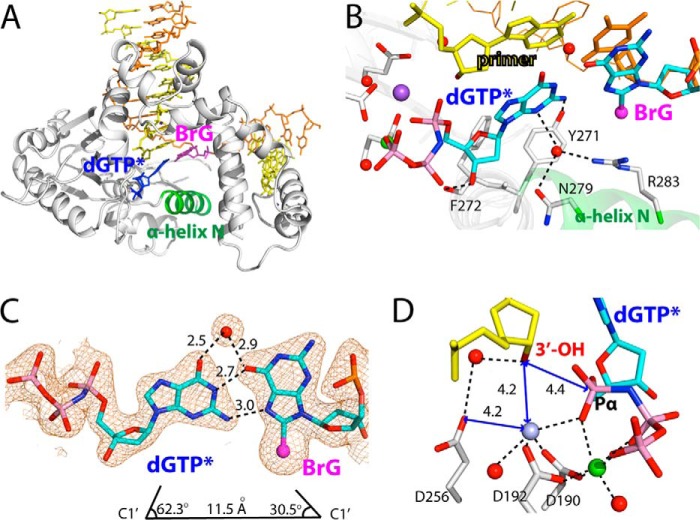
Pre-catalytic ternary structure of polβ inserting a nonhydrolyzable dGTP analog opposite templating BrG (PDB ID 4M47). A, overall structure. Protein is in intermediate conformation, and BrG·dGTP* forms a Hoogsteen base pair conformation. B, active-site view of the BrG·G ternary structure. BrG is in syn conformation and is paired with incoming dGTP* through its Hoogsteen edge. Note that H-bonding interactions of Asn-279, Arg-283, and Tyr-271 in this structure are completely different from those observed in the BrG·C ternary structure. C, H-bonding interactions and geometry of the nascent syn-BrG·anti-G base pair. A water molecule that bridges O6s of G and BrG is shown as a red sphere. The distance between C1′(dGTP*) and C1′(BrG) and λ angles are indicated. A 2Fo − Fc map is contoured at 1σ around BrG and dGTP*. D, close-up view of active-site metal ion binding site. Asp-256 and the 3′-OH of primer terminus are not liganded to the catalytic metal ion, and that distance between Pα of dGTP* and the 3′-OH of primer terminus is longer than that for correct insertion (3.4 Å). Note that Na+ ion occupies the catalytic Mg2+ ion site in this structure. Numbers in the panel indicate distances in Å.
The structure of the BrG·G ternary complex with mutagenic replication is very different from those of published polβ ternary complexes with a base pair mismatch, which typically showed staggered base pair conformation (Fig. 4A) (15, 28). In the BrG·G ternary structure, BrG adopts syn conformation and forms Hoogsteen base pairing with dGTP* in the nascent base pair binding pocket (Fig. 4B), which is reminiscent of published polβ-oxoG·A ternary structure with co-planar Hoogsteen base pair (31).
Although the geometry of the nascent mismatched syn-BrG·anti-G base pairing deviates considerably from that of a correct nascent base pair, with a longer C1′-C1′ distance (11.5 Å versus 10.5 Å) and significantly different λ angles (Fig. 4C), the Hoogsteen syn-BrG·anti-G base pair is stabilized by multiple H-bonds (Fig. 4B). The incoming dGTP* nucleotide forms two H-bonds with the Hoogsteen edge of syn-BrG. The base pair is further stabilized by an ordered water molecule that bridges O6 of both G and BrG.
Insertion of dGTP* into the enzyme active site is facilitated by the nucleotide minor-groove edge contacts to amino acid residues (Fig. 4B), with a H-bonding network significantly different from that of the published ternary complex structure (PDB ID 2FMS) (26), where Tyr-271 is H-bonded to a primer terminus base; Asn-279, an incoming nucleotide; and Arg-283, a templating base. In the BrG·G ternary structure, Tyr-271 is H-bonded to the minor-groove edge of N2 of dGTP*. Asn-279 and Arg-283, the minor-groove recognition motifs of polβ (32, 33), do not engage in direct H-bondings to the minor groove of the nascent base pair. Instead, Asn-279 is H-bonded to a bridging water molecule that interacts with N3 of dGTP*. Arg-283 is not H-bonded to the templating base. Instead, it is H-bonded to the bridging water molecule. In addition to these indirect minor-groove contacts, the H-bond between 3′-OH of dGTP* and the backbone carbonyl oxygen of Phe-272 contributes to stabilizing the insertion (Fig. 4A).
The two metal ion-binding site of the BrG·G ternary structure indicates that this complex requires further conformational change to reach a catalytically competent state (Fig. 4D). Apparently, the two metal ions, the nucleotide-binding and catalytic metal ions, are inserted in the active site of the enzyme. However, whereas the coordination sphere of the nucleotide-binding metal ion is complete, that of the catalytic metal ion is not. More specifically, Asp-190, Asp-192, the Pα oxygen of dGTP*, and a water molecule are coordinated to the catalytic metal ion, but Asp-256 and the 3′-OH of the primer terminus, typically involved in the completion of the coordination sphere of octahedral geometry, are not coordinated to it. Instead, Asp-256 is H-bonded to a bridging water molecule that interacts with 3′-OH of the primer terminus. The distance between the 3′-OH of the primer terminus and the Pα of dGTP* is longer (4.4 versus ∼3.5 Å) than the distance observed for correct insertion.
It is believed that polβ undergoes multiple conformational changes prior to catalyzing the nucleotidyl transfer (20, 34, 35), and the conformational reorganization appears to be metal coordination-dependent (32, 36–38). Our BrG·G ternary structure most likely represents an intermediate conformation where binding of the two metal ions to the insertion site has occurred, yet the full conformational change required for the chemical step has not occurred. This structure also suggests that, during nucleotide insertion into the active site of the enzyme, the coordination sphere for the nucleotide-binding metal ion completes first. The coordination sphere for the catalytic metal ion completes next, with coordination of Asp-256 and/or the 3′-OH to the catalytic metal ion potentially taking place at the final stages of conformational change to attain optimal geometry, which would facilitate formation of the closed conformation.
Comparison of active-site structures of the gapped binary, the BrG·C ternary, and the BrG·G ternary complexes shows that protein conformation in the BrG·G ternary structure is between those in the gapped binary and the BrG·C ternary structures (Fig. 5, A–D). Although the BrG·G ternary complex contains two metal ions and a Hoogsteen base pair, conformation of α-helix N is similar to that of the gapped binary structure with open conformation (Fig. 5D), suggesting that completion of coordination spheres of the two metal ions is required for the formation of closed protein conformation.
FIGURE 5.
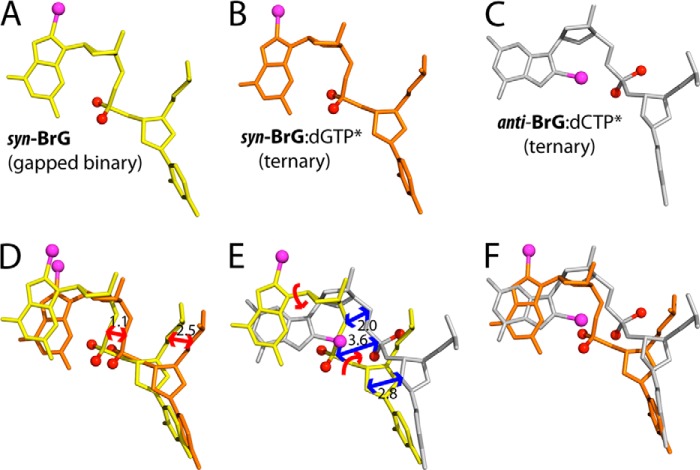
Comparison of active-site structures of polβ-BrG complexes. A, overlay of the gapped binary structure (gray) with the BrG·G ternary structure (green and blue). B, overlay of the gapped structure (gray) with the BrG·C ternary structure (yellow and red). C, overlay of the BrG·G ternary structure (green and blue) with the BrG·C ternary structure (yellow and red). D, overlay of the gapped binary, BrG·G ternary, and BrG·C ternary structures.
Our syn-BrG·anti-G ternary structure provides insights into previously observed ∼40-fold slower insertion of dATP opposite BrG relative to dGTP opposite BrG (13). Whereas dATP can form two H-bonds with syn-oxoG via Hoogsteen base pairing (18), it cannot form such H-bonds with syn-BrG due to mismatch between H-bond donors and acceptors (Fig. 1B); the Watson-Crick edge of dATP contains a H-bond donor and a H-bond acceptor, whereas the Hoosteen edge of BrG contains two H-bond acceptors. Therefore, formation of dATP·syn-BrG base pair in the nascent base pair binding pocket of the enzyme would be unfavorable.
Post-insertion Binary Structures of Polβ with BrG Paired with dC or dG at the N Position
To gain insights into post-insertion state of BrG·C and BrG·G, we obtained x-ray structure of binary complex of polβ bound to DNA containing templating BrG and primer terminus dC or dG at the N position, respectively. The BrG·C and BrG·G post-insertion structures were refined to 2.3 Å and 2.7 Å, respectively.
Interestingly, BrG·C at the N position of the BrG·C binary structure forms a staggered base pair conformation rather than co-planar conformation observed in the BrG·C ternary structure (Fig. 6, A and B). The overall structure of BrG·C post-insertion complex (PDB ID 4NLN) is very similar to that of published structure with a base pair mismatch (PDB 1TV9, r.m.s.d. = 0.329 Å) (39). The enzyme assumes an intermediate conformation, and BrG·C base pair is in staggered conformation. The BrG·C post-insertion structure shows an abasic site opposite templating BrG (Fig. 6B). Unlike anti-BrG in the BrG·C ternary structure, BrG is in the BrG·C binary structure is in syn conformation and does not form H-bonds with dC. The estranged dC forms two H-bonds with Arg-283 through its Watson-Crick edge (Fig. 6B).
FIGURE 6.
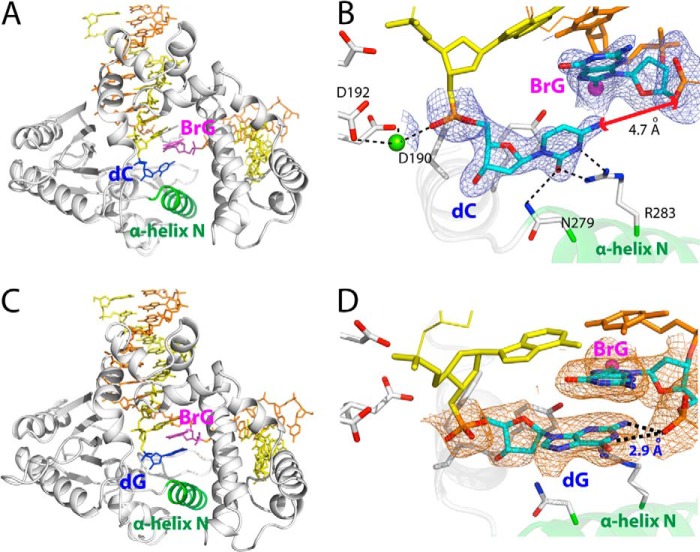
Post-insertion binary structures of polβ containing BrG·C or BrG·G at the N position. A, overall structure of the BrG·C nicked binary structure (PDB ID 4NLN). Protein is in open conformation, and BrG·C is in staggered conformation. B, active-site structure of the BrG·C post-insertion binary structure. BrG adopts syn conformation and does not form co-planar base pair conformation with primer terminus dC. A 2Fo − Fc map is contoured at 1σ around BrG and primer terminus dC. C, overall structure of the BrG·G post-insertion binary structure (PDB ID 4NLZ). Protein is in open conformation, and BrG·G is in staggered conformation. D, active-site structure of the BrG·G post-insertion binary structure. BrG adopts syn conformation and does not form co-planar conformation with primer terminus dG. Primer terminus dG stacks with BrG and is H-bonded to the 5′-phosphate of BrG.
In the BrG·G post-insertion structure (PDB ID 4NLZ), BrG·G forms staggered base pair conformation rather than co-planar conformation observed in the BrG·G ternary structure (Fig. 6, C and D). BrG in the BrG·G binary complex is in syn conformation and does not form Hoogsteen base pairing with primer terminus G. The staggered BrG·G base pair conformation is stabilized by an extensive π-π stacking interaction between BrG and the primer terminus dG. The distance between BrG and dG is ∼3.5 Å, which is similar to an average rise per base residue in B-form DNA (3.4 Å). In addition, the staggered BrG·G base pair is stabilized by two H-bonds formed between N2-H and N1-H of the primer terminus dG and the 5′-phosphate oxygen of BrG.
Post-extension Binary Structures of Polβ with BrG·C or BrG·G at the N-1 Position
To gain insight into base pair conformation of BrG·C and BrG·G at other positions of the enzyme active site, we determined two binary structures bearing BrG·C or BrG·G at the N-1 position. The BrG·C (N-1) and the BrG·G (N-1) binary complex structures were diffracted to 2.4 Å and 2.5 Å, respectively.
Surprisingly, in the BrG·C (N-1) structure (PDB ID 4NM1), protein assumes closed conformation rather than open conformation previously observed with polβ binary structures (Fig. 7, A and B). The α-helix N moved to engage in H-bonding interactions with the primer terminus base pair. BrG and dC form a co-planar Watson-Crick base pair rather than staggered base pair seen in the BrG·C (N) structure. A noteworthy observation from this closed binary structure is the presence of penta-coordinated metal ion near the primer terminus base. The metal ion is coordinated with the 5′-phosphate oxygen of primer terminus, monophosphate oxygen, a water molecule, Asp-190, and Asp-192. H-bondings and the base pair geometry of the BrG·C at the N-1 position are very similar to those of the normal Watson-Crick base pair (Fig. 7C).
FIGURE 7.
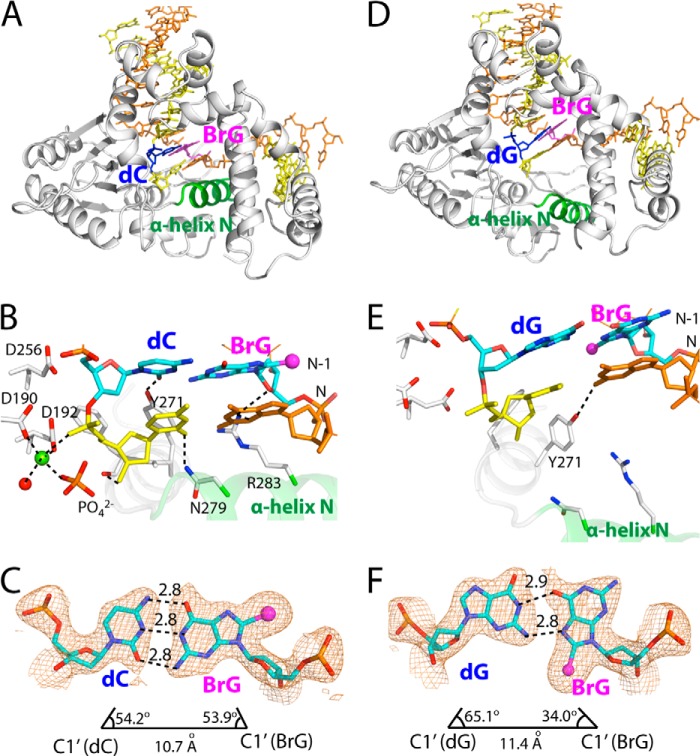
Post-extension binary structures of polβ containing BrG·C or BrG·G at the N-1 position. A, overall structure of the BrG·C post-extension binary structure (PDB ID 4NM1). Protein is in closed conformation, and BrG·C forms co-planar base pair. B, active-site view of the BrG·C post-extension complex. Asn-279, Arg-283, and Tyr-271 are H-bonded to minor groove edges of bases. Note that a penta-coordinated Mg2+ (shown in yellow-green) is present near primer terminus C. C, H-bondings and geometry of BrG·C at the N-1 position. The λ angles and C1′-C1′ distance are similar to those for normal Watson-Crick base pair. A 2Fo − Fc map is contoured at 1σ around BrG and dC. D, overall structure of the BrG·G post-extension binary structure (PDB ID 4NM2). Protein is in open conformation, and BrG·G is in co-planar conformation. E, active-site view of the BrG·C post-extension complex. Asn-279 and Arg-283 do not engage in H-bonding interaction with the primer terminus base pair. Note that a metal ion is not present near primer terminus base. F, H-bondings and geometry of BrG·G at the N-1 position. The C1′-C1′ distance is longer than that for the BrG·C base pair.
In the BrG·G (N-1) structure (PDB ID 4NM2), protein is in open conformation (Fig. 7D). BrG is in syn conformation and forms a co-planar Hoogsteen base pair with G (Fig. 7E). As seen in the BrG·G ternary structure, the O6 of BrG is H-bonded to the N1 of G, and the N7 of BrG is H-bonded to the N2 of G (Fig. 7F). However, the water-mediated H-bonding between O6 of BrG and O6 of G is lacking in this structure. The C1′-C1′ distance for BrG and G is ∼1 Å longer than that for correct base pair (11.4 Å versus 10.4 Å). The λ angles for BrG and G are similar to those seen for BrG·G ternary structure.
DISCUSSION
Comparison of our BrG·C and BrG·G ternary structures with published polβ ternary structures suggests that the coordination state of the catalytic metal ion dictates protein conformation of polβ ternary complex (35–38). Apparently, the coordination state of the catalytic metal ion varies among polβ ternary structures with different conformations. In polβ ternary structure with correct insertion, the catalytic metal ion adopts octahedral geometry by coordinating to the three catalytic Asp residues (Asp-190, -192, and -256), the 3′-OH of primer terminus, Pα oxygen of an incoming nucleotide, and an ordered water molecule. Protein in such structure generally adopts closed conformation. In our BrG·C ternary structure with correct insertion, although the coordination of a water molecule to the catalytic metal ion is lacking, the complex adopts closed protein conformation, indicating that coordination of water molecule to the catalytic metal ion is not required for the conformational activation of polβ. In polβ ternary structure with incorrect insertion, catalytic metal ion does not form octahedral coordination geometry and protein adopts intermediate conformation. In such a structure, Asp-256, the 3′-OH of primer terminus, and/or water molecule is not liganded to the catalytic metal ion, producing an incomplete coordination sphere of the catalytic metal ion. Published polβ ternary structure with dATP·dC (PDB ID 3C2L) or dATP·dG (PDB ID 3C2M) mismatch lacks the coordination of the primer terminus 3′-OH to the catalytic metal ion and shows intermediate protein conformation (28), indicating that the 3′-OH coordination to the catalytic metal ion is required for the conformational activation of polβ. In our BrG·G ternary structure, although the complex assumes Hoogsteen base pair conformation, coordination of Asp-256, primer terminus 3′-OH, and water molecule to the catalytic metal ion is lacking, and the complex adopts intermediate protein conformation. Taken together, these observations indicate that the coordination state of the catalytic metal plays an important role in conformational activation of polβ. Polβ appears to utilize the coordination state of catalytic metal ion as a kinetic checkpoint to discourage nucleotide misincorporation, which would be consistent with an induced-fit mechanism (19, 40), where an optimal conformation for nucleotidyl transfer is allowed for correct insertion, but not for incorrect insertion.
Structural differences between the BrG·C (N-1) binary structure with closed protein conformation and the BrG·G (N-1) binary structure with open protein conformation provide new insight into post-chemistry conformational change occurring during the catalytic cycle of polβ (Fig. 7, B and D). Currently, whereas pre-chemistry conformational change of polβ is relatively well understood (19, 35, 36, 41, 42), its post-chemistry conformational change is poorly understood due largely to the scarcity of polβ structure with post-chemistry conformational intermediate. Comparison of reported polβ binary structures with the BrG·G (N-1) and the BrG·C (N-1) binary structures suggests that the metal-ion coordination observed in the BrG·C (N-1) binary structure plays an important role in post-chemistry conformational reorganization and prevents closed-to-open conformational inactivation (Fig. 7B). In published polβ binary post-insertion structures with a correct base pair, metal-ion coordination similar to that observed near the primer terminus of our BrG·C (N-1) binary complex is lacking, and protein adopts open conformation. In polβ post-insertion binary structure with A·C or T·C mismatch (31), a single Mg2+ was observed, but this metal ion was not coordinated to amino acids. In our BrG·C (N-1) binary structure, the metal ion is coordinated to Asp-190, Asp-192, the 5′-phosphate oxygen of primer terminus, monophosphate oxygen, and water molecule. Interestingly, the metal ion and the monophosphate in the BrG·C (N-1) complex are similarly located where the nucleotide-binding metal ion and β-phosphate of an incoming nucleotide of the BrG·C ternary structure are. This suggests that the metal ion seen in the BrG·C (N-1) binary structure possesses coordination chemistry similar to that of the nucleotide-binding metal ion. As binding of nucleotide-binding and catalytic metal ions to the enzyme active site is believed to occur in discrete steps (30, 35–37, 42), release of the metal ions from the active site could occur in a stepwise fashion (42). Our BrG·C (N-1) binary structure with one metal-ion coordination state and closed protein conformation suggests that, during post-chemistry conformational reorganization, release of catalytic metal ion from the enzyme active site precedes that of nucleotide-binding metal ion and that the release of nucleotide-binding metal ion triggers a closed-to-open conformational inactivation. The BrG·C (N-1) binary structure would thus represent a close approximation of a post-chemistry conformational intermediate, where nucleotidyl transfer and catalytic metal-ion release have occurred, yet the release of nucleotide-binding metal ion and pyrophosphate from the enzyme active site has not occurred.
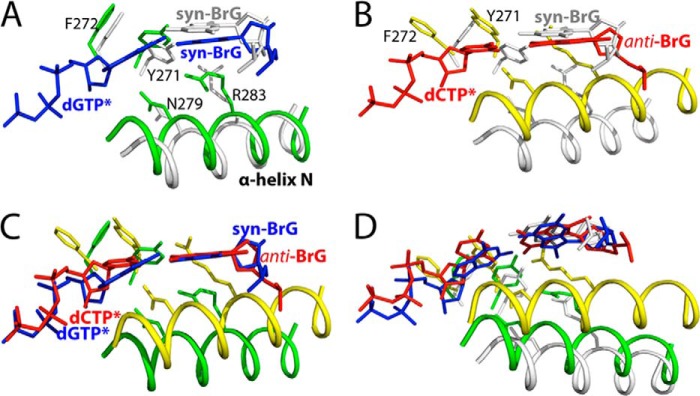
Polβ lacking an intrinsic proofreading exonuclease activity has shown to prevent nucleotide misincorporation by disallowing co-planar base pair conformation in the nascent base pair binding pocket (28, 30, 41). Mismatched base pairs such as A·C, T·C, and A·G have been shown to form nonplanar, staggered conformation in the enzyme active site (28, 41). The formation of Watson-Crick or Hoogsteen base pair conformation observed in the BrG·C and the BrG·G ternary structures and published oxoG·dATP and oxoG·dCTP ternary structures thus indicates that some 8-modified G can be tolerated in the enzyme active site (Fig. 8, A–C). Apparently, the nascent base pair binding pocket of polβ is plastic enough for accommodating both Hoogsteen and Watson-Crick base pairings. To accommodate a Hoogsteen syn-BrG·C base pair at the N position, template DNA containing BrG shifted ∼1 Å (Fig. 8, B and D). To accommodate a Watson-Crick anti-BrG·C base pair at the N position, the 5′-phosphodiester backbone of BrG underwent conformational reorganization, which prevented a steric clash between the 5′-phosphate of BrG and the bromine moiety (Fig. 8, C and E). The similar conformational reorganization of templating base and its phosphate backbone has been observed in polβ ternary structure with Hoogsteen syn-oxoG·dATP or Watson-Crick anti-oxoG·dCTP base pair (14, 18).
FIGURE 8.
Comparison of BrG conformation at the N position. Bromine moiety is shown in magenta sphere and the 5′-phosphate oxygen in red sphere. A, BrG in the gapped binary structure. B, BrG in the BrG·G ternary structure. C, BrG in the BrG·C ternary structure. D, overlay of BrGs in the gapped binary (yellow) and the BrG·G ternary (orange) structures. Numbers in the panel indicate distances in Å. Note that BrG in the BrG·G ternary structure shifted from the position in the gapped structure to accommodate BrG·G base pair. E, overlay of BrGs in the gapped binary (yellow) and the BrG·C ternary (gray) structures. F, overlay of BrGs in the BrG·G and the BrG·G ternary structures.
Our BrG·G ternary complex structure shows that the mutagenic Hoogsteen syn-BrG·anti-G base pairing is tolerated in the nascent base pair binding pocket of polβ, implying that BrG lesion could potentially promote G to C mutation by utilizing the two H-bond acceptors on its Hoogsteen edge (Fig. 1B). G to C transversion mutations comprise approximately 25% of mutations found in breast cancers (43), yet only a few mutagenic lesions promoting such mutations are known (44, 45), none of them utilizing the Hoosteen edge of the lesions in their mutagenic base pairings. Although further investigation will be required to evaluate potential haloG-induced mutagenesis, our studies suggest that a modified G with a bulky C8-substituent may adopt syn conformation and could facilitate G to C mutation by forming Hoogsteen base pairing using two H-bond acceptors on its Hoogsteen edge during DNA replication.
This work was supported by grants from a start-up fund from the College of Pharmacy at the University of Texas at Austin and the Robert Welch Foundation (Welch Grant F-1741).
- haloG
- 8-halogenated G
- BrG
- 8-bromoguanine
- oxoG
- 8-oxoguanine
- PDB
- Protein Data Bank
- polβ
- polymerase β
- r.m.s.d.
- root mean square deviation
- dNMPNPP
- 2′-deoxy-5′-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]nucleoside
- dCMPNPP
- 2′-deoxy-5′-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine.
REFERENCES
- 1. Coussens L. M., Werb Z. (2002) Inflammation and cancer. Nature 420, 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meira L. B., Bugni J. M., Green S. L., Lee C. W., Pang B., Borenshtein D., Rickman B. H., Rogers A. B., Moroski-Erkul C. A., McFaline J. L., Schauer D. B., Dedon P. C., Fox J. G., Samson L. D. (2008) DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J. Clin. Invest. 118, 2516–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weiss S. J., Test S. T., Eckmann C. M., Roos D., Regiani S. (1986) Brominating oxidants generated by human eosinophils. Science 234, 200–203 [DOI] [PubMed] [Google Scholar]
- 4. Gaut J. P., Yeh G. C., Tran H. D., Byun J., Henderson J. P., Richter G. M., Brennan M. L., Lusis A. J., Belaaouaj A., Hotchkiss R. S., Heinecke J. W. (2001) Neutrophils employ the myeloperoxidase system to generate antimicrobial brominating and chlorinating oxidants during sepsis. Proc. Natl. Acad. Sci. U.S.A. 98, 11961–11966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weiss S. J. (1989) Tissue destruction by neutrophils. New Engl. J. Med. 320, 365–376 [DOI] [PubMed] [Google Scholar]
- 6. Masuda M., Suzuki T., Friesen M. D., Ravanat J. L., Cadet J., Pignatelli B., Nishino H., Ohshima H. (2001) Chlorination of guanosine and other nucleosides by hypochlorous acid and myeloperoxidase of activated human neutrophils: catalysis by nicotine and trimethylamine. J. Biol. Chem. 276, 40486–40496 [DOI] [PubMed] [Google Scholar]
- 7. Valinluck V., Sowers L. C. (2007) Inflammation-mediated cytosine damage: a mechanistic link between inflammation and the epigenetic alterations in human cancers. Cancer Res. 67, 5583–5586 [DOI] [PubMed] [Google Scholar]
- 8. Suzuki T., Masuda M., Friesen M. D., Fenet B., Ohshima H. (2002) Novel products generated from 2′-deoxyguanosine by hypochlorous acid or a myeloperoxidase-H2O2-Cl- system: identification of diimino-imidazole and amino-imidazolone nucleosides. Nucleic Acids Res. 30, 2555–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stanley N. R., Pattison D. I., Hawkins C. L. (2010) Ability of hypochlorous acid and N-chloramines to chlorinate DNA and its constituents. Chem. Res. Toxicol. 23, 1293–1302 [DOI] [PubMed] [Google Scholar]
- 10. Asahi T., Kondo H., Masuda M., Nishino H., Aratani Y., Naito Y., Yoshikawa T., Hisaka S., Kato Y., Osawa T. (2010) Chemical and immunochemical detection of 8-halogenated deoxyguanosines at early stage inflammation. J. Biol. Chem. 285, 9282–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fujikawa K., Yakushiji H., Nakabeppu Y., Suzuki T., Masuda M., Ohshima H., Kasai H. (2002) 8-Chloro-dGTP, a hypochlorous acid-modified nucleotide, is hydrolyzed by hMTH1, the human MutT homolog. FEBS Lett. 512, 149–151 [DOI] [PubMed] [Google Scholar]
- 12. Sassa A., Ohta T., Nohmi T., Honma M., Yasui M. (2011) Mutational specificities of brominated DNA adducts catalyzed by human DNA polymerases. J. Mol. Biol. 406, 679–686 [DOI] [PubMed] [Google Scholar]
- 13. Efrati E., Tocco G., Eritja R., Wilson S. H., Goodman M. F. (1999) “Action-at-a-distance” mutagenesis: 8-oxo-7,8-dihydro-2′-deoxyguanosine causes base substitution errors at neighboring template sites when copied by DNA polymerase β. J. Biol. Chem. 274, 15920–15926 [DOI] [PubMed] [Google Scholar]
- 14. Batra V. K., Beard W. A., Hou E. W., Pedersen L. C., Prasad R., Wilson S. H. (2010) Mutagenic conformation of 8-oxo-7,8-dihydro-2′-dGTP in the confines of a DNA polymerase active site. Nat. Struct. Mol. Biol. 17, 889–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beard W. A., Wilson S. H. (2006) Structure and mechanism of DNA polymerase β. Chem. Rev. 106, 361–382 [DOI] [PubMed] [Google Scholar]
- 16. Pan B., Shi K., Sundaralingam M. (2006) Base-tetrad swapping results in dimerization of RNA quadruplexes: implications for formation of the i-motif RNA octaplex. Proc. Natl. Acad. Sci. U.S.A. 103, 3130–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ravelli R. B., Leiros H. K., Pan B., Caffrey M., McSweeney S. (2003) Specific radiation damage can be used to solve macromolecular crystal structures. Structure 11, 217–224 [DOI] [PubMed] [Google Scholar]
- 18. Batra V. K., Shock D. D., Beard W. A., McKenna C. E., Wilson S. H. (2012) Binary complex crystal structure of DNA polymerase β reveals multiple conformations of the templating 8-oxoguanine lesion. Proc. Natl. Acad. Sci. U.S.A. 109, 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sawaya M. R., Prasad R., Wilson S. H., Kraut J., Pelletier H. (1997) Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry 36, 11205–11215 [DOI] [PubMed] [Google Scholar]
- 20. Sawaya M. R., Pelletier H., Kumar A., Wilson S. H., Kraut J. (1994) Crystal structure of rat DNA polymerase β: evidence for a common polymerase mechanism. Science 264, 1930–1935 [DOI] [PubMed] [Google Scholar]
- 21. Beard W. A., Shock D. D., Batra V. K., Pedersen L. C., Wilson S. H. (2009) DNA polymerase β substrate specificity: side chain modulation of the “A-rule.” J. Biol. Chem. 284, 31680–31689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Batra V. K., Beard W. A., Shock D. D., Krahn J. M., Pedersen L. C., Wilson S. H. (2006) Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure 14, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao Y., Biertümpfel C., Gregory M. T., Hua Y. J., Hanaoka F., Yang W. (2012) Structural basis of human DNA polymerase η-mediated chemoresistance to cisplatin. Proc. Natl. Acad. Sci. U.S.A. 109, 7269–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., Wilson S. H. (2008) Structures of DNA polymerase β with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell 30, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clausen A. R., Murray M. S., Passer A. R., Pedersen L. C., Kunkel T. A. (2013) Structure-function analysis of ribonucleotide bypass by B family DNA replicases. Proc. Natl. Acad. Sci. U.S.A. 110, 16802–16807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Freudenthal B. D., Beard W. A., Shock D. D., Wilson S. H. (2013) Observing a DNA polymerase choose right from wrong. Cell 154, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krahn J. M., Beard W. A., Wilson S. H. (2004) Structural insights into DNA polymerase β: deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure 12, 1823–1832 [DOI] [PubMed] [Google Scholar]
- 32. Batra V. K., Perera L., Lin P., Shock D. D., Beard W. A., Pedersen L. C., Pedersen L. G., Wilson S. H. (2013) Amino acid substitution in the active site of DNA polymerase β explains the energy barrier of the nucleotidyl transfer reaction. J. Am. Chem. Soc. 135, 8078–8088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Freudenthal B. D., Beard W. A., Wilson S. H. (2013) DNA polymerase minor groove interactions modulate mutagenic bypass of a templating 8-oxoguanine lesion. Nucleic Acids Res. 41, 1848–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. (1994) Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science 264, 1891–1903 [PubMed] [Google Scholar]
- 35. Zhong X., Patel S. S., Tsai M. D. (1998) DNA polymerase β. 5. Dissecting the functional roles of the two metal ions with Cr(III) dTTP1. J. Am. Chem. Soc. 120, 235–236 [Google Scholar]
- 36. Freudenthal B. D., Beard W. A., Wilson S. H. (2012) Structures of dNTP intermediate states during DNA polymerase active site assembly. Structure 20, 1829–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang L., Arora K., Beard W. A., Wilson S. H., Schlick T. (2004) Critical role of magnesium ions in DNA polymerase β's closing and active site assembly. J. Am. Chem. Soc. 126, 8441–8453 [DOI] [PubMed] [Google Scholar]
- 38. Kirby T. W., DeRose E. F., Cavanaugh N. A., Beard W. A., Shock D. D., Mueller G. A., Wilson S. H., London R. E. (2012) Metal-induced DNA translocation leads to DNA polymerase conformational activation. Nucleic Acids Res. 40, 2974–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson S. J., Beese L. S. (2004) Structures of mismatch replication errors observed in a DNA polymerase. Cell 116, 803–816 [DOI] [PubMed] [Google Scholar]
- 40. Koshland D. E. (1958) Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 44, 98–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin P., Batra V. K., Pedersen L. C., Beard W. A., Wilson S. H., Pedersen L. G. (2008) Incorrect nucleotide insertion at the active site of a G:A mismatch catalyzed by DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 105, 5670–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., Wilson S. H. (2005) Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate. Structure 13, 1225–1233 [DOI] [PubMed] [Google Scholar]
- 43. Pfeifer G. P., Besaratinia A. (2009) Mutational spectra of human cancer. Hum. Genet. 125, 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stathis D., Lischke U., Koch S. C., Deiml C. A., Carell T. (2012) Discovery and mutagenicity of a guanidinoformimine lesion as a new intermediate of the oxidative deoxyguanosine degradation pathway. J. Am. Chem. Soc. 134, 4925–4930 [DOI] [PubMed] [Google Scholar]
- 45. Kino K., Sugiyama H. (2001) Possible cause of G·C → C·G transversion mutation by guanine oxidation product, imidazolone. Chem. Biol. 8, 369–378 [DOI] [PubMed] [Google Scholar]