Abstract
Escherichia coli and related enteric bacteria can survive under extreme acid stress condition at least for several hours. RpoS is a key factor for acid stress management in many enterobacteria. Although three rpoS-activating sRNAs, DsrA, RprA, and ArcZ, have been identified in E. coli, it remains unclear how these small RNA molecules participate in pathways leading to acid resistance (AR). Here, we showed that overexpression of ArcZ, DsrA, or RprA enhances AR in a RpoS-dependent manner. Mutant strains with deletion of any of three sRNA genes showed lowered AR, and deleting all three sRNA genes led to more severe defects in protecting against acid stress. Overexpression of any of the three sRNAs fully rescued the acid tolerance defects of the mutant strain lacking all three genes, suggesting that all three sRNAs perform the same function in activating RpoS required for AR. Notably, acid stress led to the induction of DsrA and RprA but not ArcZ.
Keywords: Acid resistance, Escherichia coli, RpoS, small noncoding RNA.
Introduction
Most pathogenic or nonpathogenic enterobacteria grow optimally at a pH range 5–9, and are therefore classified as neutrophilic bacteria. However, bacteria often face drastic pH changes along the gastrointestinal tract, such as gastric acid (pH 1.5 ˜ 2.5), alkaline secretion of pancreas (pH ˜ 9), and weak organic acid metabolites in intestine. The ability of enterobacteria to survive these conditions is critical for their colonization in host intestine or pathogenicity.
Escherichia coli can survive in extreme acidic condition (pH < 2.5) at least for several hours, and its acid resistance (AR) systems have thus been extensively studied (Lin et al. 1996). At least three discernible AR systems are responsible for protecting E. coli cells from acid stress (Foster 2004; Zhao and Houry 2010). Induction of each AR system varies depending on environmental conditions, such as growth/challenge media, growth phase, aerobic/anaerobic conditions, challenging pH, or exogenous amino acids. The AR1 system has been identified in cells grown oxidatively in rich media to the stationary phase, and shown to require rpoS and crp (Small et al. 1994; Castanie-Cornet et al. 1999). The glutamate-dependent AR2 and arginine-dependent AR3 systems include amino acid–dependent decarboxylase complexes comprising amino acid decarboxylases and countertransporters that exchange decarboxylation products for new extracellular amino acids. Recently, another AR system relying on L-glutamine, GadC, and glutaminase that releases gaseous ammonia to neutralize protons was reported (Lu et al. 2013).
The glutamate-dependent AR2 system plays a major role in AR and is most effective in relieving acid stress at very low pH (<2.5). The AR2 system contains two glutamate decarboxylases (GadA and GadB), and its countertransporter (GadC), which exchange γ-amino butyric acid (the decarboxylation product of glutamate) for new external glutamate (De Biase et al. 1999; Ma et al. 2012). Expression of gadA and gadBC operon is stimulated by the key activator, GadE (Ma et al. 2003a; Castanie-Cornet et al. 2010), and GadX activates gadE expression (Sayed et al. 2007; Sayed and Foster 2009). Transcription of gadX is primarily induced by the alternative sigma factor, RpoS (Tramonti et al. 2002). cAMP receptor protein (CRP) affects the AR2 system by repressing RpoS transcription (Ma et al. 2002, 2003b). Histone-like protein, H-NS, has been shown to directly suppress gadA and gadBC expression (Waterman and Small 2003; Giangrossi et al. 2005). Two-component systems, EvgA/S (Ma et al. 2004), PhoP/Q (Zwir et al. 2005), and TorS/R (Bordi et al. 2003), additionally regulate gadE transcription.
It is also known that three Hfq-interacting small noncoding RNAs, GadY, DsrA, and GcvB, participate in conferring AR to E. coli cells. GadY is encoded on the antisense strand between gadX and gadW, and positively regulates these genes by affecting processing of gadXW transcripts by RNase III (Opdyke et al. 2004, 2011; Tramonti et al. 2008). Overexpression of DsrA renders E. coli cells resistant to acid stress by inducing AR-related genes, such as hdeAB, gadAX, and gadBC (Lease et al. 2004), possibly through the actions of RpoS and/or H-NS, as DsrA is known to enhance rpoS translation and hns mRNA turnover (Sledjeski and Gottesman 1995; Sledjeski et al. 1996; Majdalani et al. 1998; Lease and Belfort 2000). GcvB, a regulator of genes involved in amino acid metabolism (Urbanowski et al. 2000; Pulvermacher et al. 2009a,2009b), enhances AR in a rpoS-dependent manner (Jin et al. 2009), although the underlying mechanism remains elusive.
In addition to AR, RpoS is crucial for general stress management and virulence in many pathogenic or nonpathogenic enterobacteria (Dong and Schellhorn 2010; Battesti et al. 2011). Three Hfq-interacting sRNAs, DsrA, RprA, and ArcZ, were reported to positively regulate rpoS expression (Sledjeski et al. 1996; Majdalani et al. 1998, 2001; Mandin and Gottesman 2010). Therefore, it is not difficult to speculate that these sRNAs would provide E. coli with AR by activating rpoS expression, but roles of these small RNA molecules in pathways leading to AR remain elusive due to the complexity of mechanisms of AR.
In this study, we showed the acquisition mechanisms of AR by E. coli cells through rpoS-activating sRNAs. The acquisition depends on RpoS and partially on GadX. Interestingly, DsrA and RprA, but not ArcZ, act as mediator molecules for the acid stress response. Our data should aid in improving our current understanding of the acquisition mechanisms of AR by E. coli cells through rpoS-activating sRNAs.
Experimental Procedures
Strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table1. MG1655 deletion mutants of rpoS, hfq, arcB, gadX, or gadW were obtained by P1 transduction using the deletion strains from the E. coli Keio strain collection (Baba et al. 2006). Knockout of sRNA genes was performed as described previously (Datsenko and Wanner 2000), using λRed-mediated recombination with pKD46 plasmid and kanamycin cassettes generated with polymerase chain reaction (PCR) using pKD13 as a template plasmid and the oligonucleotides listed in Supplementary Table 1. In case of the arcZ mutant, the-10 promoter mutation of arcZ was introduced to avoid unexpected effects on expression of the overlapped arcB gene because the kanamycin cassette sequence could be overlapped if the arcZ structural sequence is replaced with the kanamycin cassette. The kanamycin cassette for the arcZ mutant was generated as follows. A DNA fragment containing the region of positions −230 to −191 with respect to the transcription start of arcZ followed by the kanamycin resistance gene was prepared by PCR amplification using a ΔP10arcZP1/ΔP10arcZP2 primer pair. Another fragment carrying the −150 to +48 region of arcZ was prepared by PCR amplification using a ΔP10arcZP3/ΔP10arcZP4 primer pair. The two amplified PCR products were then linked using overlap extension PCR (Horton et al. 1989) with ΔP10arcZP1/ΔP10arcZP4 primer pair. This cassette was cloned into a pGEM-T vector (Promega, Tokyo, Japan), and site-directed mutagenesis of the −10 promoter region was performed. The mutated cassette containing the mutated-10 sequence (CTCGAG) was amplified using PCR. Kanamycin marked mutations were transferred into a desired strain background using bacteriophage P1 transduction (Silhavy et al. 1984). No expression of ArcZ in the arcZ mutant was confirmed by Northern blot (Fig. S4). To obtain MG1655Δ3 with deletion of all three rpoS-activating sRNA genes, the Flp recognition target (FRT)-flanked kanamycin cassette introduced into the first dsrA deletion strain was removed using Flp recombinase from pCP20 plasmid (Cherepanov and Wackernagel 1995), an additional rprA deletion was introduced by P1 transduction, the kanamycin cassette was removed again, and the arcZ promoter mutation was finally introduced by P1 transduction.
Table 1.
Strains and plasmids used in this study.
| Name | Description | Source |
|---|---|---|
| Strains | ||
| MG1655 | E. coli MG1655 wild type | Laboratory stock |
| ΔarcZ | MG1655 ΔarcZ::kanR (promoter-10 mutant) | This study |
| ΔdsrA | MG1655 ΔdsrA::kanR | This study |
| ΔrprA | MG1655 ΔrprA::kanR | This study |
| MG1655Δ3 | MG1655 ΔdsrA ΔrprA ΔarcZ::kanR | This study |
| BW25113ΔrpoS | E. coli BW25113 ΔrpoS::kanR | Keio collection (Baba et al. 2006) |
| BW25113ΔarcB | E. coli BW25113 ΔarcB::kanR | Keio collection (Baba et al. 2006) |
| BW25113ΔgadX | E. coli BW25113 ΔgadX::kanR | Keio collection (Baba et al. 2006) |
| BW25113ΔgadW | E. coli BW25113 ΔgadW::kanR | Keio collection (Baba et al. 2006) |
| BW25113Δhfq | E. coli BW25113 Δhfq::kanR | Keio collection (Baba et al. 2006) |
| ΔrpoS | E. coli MG1655 ΔrpoS::kanR | This study |
| ΔarcB | E. coli MG1655 ΔarcB::kanR | This study |
| ΔgadX | E. coli MG1655 ΔgadX::kanR | This study |
| ΔgadW | E. coli MG1655 ΔgadW::kanR | This study |
| Δhfq | E. coli MG1655 Δhfq::kanR | This study |
| SG30013 | MG1655 lacX74 attRpoS750::LacZ, Reporter for transcription, translation, and degradation of RpoS | S. Gottesman (Wassarman et al. 2001) |
| Plasmids | ||
| pHMB1 | A derivative of pHM1 (Han et al. 2010), AmpR, IPTG-inducible transcription from immediate after EcoRI site, modified rnpB terminator (GAUUU to GGAGU) next to XbaI site. | This study |
| pArcZ | pHMB1 carrying arcZ | This study |
| pDsrA | pHMB1 carrying dsrA | This study |
| pRprA | pHMB1 carrying rprA | This study |
| pGEM3 | Cloning vector, AmpR | Promega |
| pGEM-T | Cloning vector, AmpR, Site-directed mutagenesis for arcZ | Promega |
| pG-ArcZ | pGEM3 carrying arcZ | This study |
| pBAD-myc-hisC | Control vector for pBAD-RpoS plasmid | Invitrogen |
| pBAD-RpoS | rpoS cloned in pBAD24, AmpR | Zhou and Gottesman (2006) |
| pKD46 | λRed recombination system under control of arabinose-inducible promoter, temperature-sensitive replication, AmpR | Datsenko and Wanner (2000) |
| pKD13 | Template plasmid for FRT-flanked kanamycin resistance cassette. | Datsenko and Wanner (2000) |
| pCP20 | FLP+, λ cI857+, λ PR Repts, AmpR, CmR, expression of site-specific Flp recombinase under control of heat-inducible promoter, temperature-sensitive replication. | Cherepanov and Wackernagel (1995) |
pG-ArcZ was constructed using the PCR-amplified DNA fragment containing the arcZ gene sequence from −107 to +208 relative to the transcription start site of ArcZ. For isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible overexpression of each sRNA, the pHMB1 plasmid was constructed by replacing the AatII site of pHM1 (Han et al. 2010) with a XbaI site and adding the modified rnpB terminator sequence (Kim et al. 1996) with mutations at the rne-dependent cleavage site (from GAUUU to GGAGU) adjacent to the XbaI site. ArcZ, DsrA, and RprA genes were amplified from MG1655 genomic DNA using PCR, and cloned into EcoRI and XbaI sites of pHMB1. The resulting plasmids express sRNAs from their native transcription start site and terminate transcription at the native terminator of each sRNA gene.
Protein identification
E. coli MG1655 cells containing control vector pGEM3 or pG-ArcZ were cultured overnight, diluted 1:100 into fresh lysogeny broth (LB) media, and grown for 17 h at 37°C. Cells were harvested, resuspended in phosphate buffered saline (PBS) buffer, and sonicated. After sonication, supernatant fractions were electrophoresed on a 10% sodiumdodecyl sulfate-polyacrylamide gel (SDS-PAGE). Protein bands of interest were isolated and digested in the gel with trypsin. Proteins were analyzed by electrospray hybrid quadrupole-time-of-flight tandem mass spectrometer (Q-TOF2 MS/MS).
RNA extraction and Northern blot analysis
Cells were grown overnight in LB broth with 100 μg mL−1 ampicillin if necessary. Overnight cultures were diluted 1:100 in fresh media and growth continued. For overexpression of sRNA, 1 mmol/L IPTG was added to cell cultures grown for 1 h after 1:100 dilution. Total cellular RNAs were extracted from culture at the desired time points by acidic hot phenol method, as described previously (Kim et al. 1996).
Northern blot analysis was carried out as described (Tezias et al. 2012). Briefly, 10 μg of total RNAs were fractionated on 7 mol/L urea, 5% PAGE, and electrotransferred onto a Hybond™-XL membrane (Amersham Biosciences, Little Chalfont, UK). Membranes were hybridized with 32P-labeled DNA probes in Rapid-Hyb buffer (Amersham Biosciences) according to the manufacturer's instructions. Hybridization signals were analyzed by using Image Analyzer FLA7000 (Fuji, Tokyo, Japan).
AR assay
Three colonies for each strain were pooled in LB. The overnight culture was diluted 1:100 in fresh LB and grown at 37°C. Exponentially growing cells were obtained from cultures grown for 2.5 h and stationary-phase cells from cultures grown for 12 h after dilution, respectively. If necessary, LB media containing 100 μg mL−1 ampicillin was used, and 1 mmol/L IPTG was added to diluted cultures grown for 1 h.
To assay AR, sample cells were harvested and resuspended M9 minimal media supplemented with 0.4% glucose. The suspended sample (˜108 cells mL−1 for exponential phase, ˜109 cells mL−1 for stationary phase) was diluted 1:10 or 1:100 with M9 minimal media supplemented with 0.4% glucose and 1.5 mmol/L glutamate, which was previously adjusted to pH 2.0 or 3.0 with concentrated HCl and incubated for 1 h at 37°C in a shaking incubator. Cells in 2 mL medium in a 14 mL test tube were grown with shaking at 250 rpm. pH changes were not observed during acid challenge of 1 h.
After acid exposure, cells were serially diluted in LB and plated on LB agar plates. The number of viable colonies was determined to calculate the colony-forming units (CFU mL−1). The survival rate was calculated based on the ratio of CFU of cells after acid exposure to CFU before the acid exposure. In parallel, cultures exposed to acid challenge were additionally diluted 1:10 serially in LB and 10 μL of each dilution was spotted onto LB plates.
β-galactosidase assay
Three colonies for each strain were pooled in LB, and the overnight culture was diluted 100-fold in fresh LB media–containing ampicillin (100 μg mL−1) and 1 mmol/L IPTG, if necessary. At the desired time points, the lacZ fusion reporter gene was assayed as described previously (Zhang and Bremer 1995; Lee et al. 2013). At least three independent experiments were performed.
Results
Overexpression of ArcZ leads to GadA accumulation in E. coli
ArcZ is a highly conserved sRNA in bacteria that was initially identified using genome-wide sRNA screening by independent research groups (Argaman et al. 2001; Wassarman et al. 2001). This sRNA is highly associated with Hfq protein in E. coli and Salmonella (Zhang et al. 2003; Sittka et al. 2008). Papenfort et al. (2009) characterized the function of ArcZ sRNA as a negative regulator of several mRNAs using microarray in Salmonella, whereas Mandin and Gottesman (2010) showed that ArcZ sRNA acts as a positive regulator of rpoS as well as a negative regulator of arcB in E. coli.
As an independent study of ArcZ function in E. coli, we initially examined the changes in protein expression patterns upon overexpression from multiple copies of arcZ on a multicopy plasmid. Bands displaying highly increased or decreased intensity were excised and identified by Q-TOF2 MS/MS. GadA/B, glutamate decarboxylases of the AR2 system, was highly induced, whereas the amount of TnaA, an alkali-inducible tryptophanase (Bordi et al. 2003), was significantly reduced (Fig. 1). This reciprocal change in the protein profile is similar to the case of GadY-overexpressed cells previously reported (Opdyke et al. 2004). GadY-overexpressed cells showed increased DnaK, GroEL, and GadA levels, and decreased TnaA expression. As a sRNA, GadY, positively regulates GadX, a transcriptional activator of gadA and gadBC operons, via base pairing with the 3′-UTR of gadX mRNA.
Figure 1.
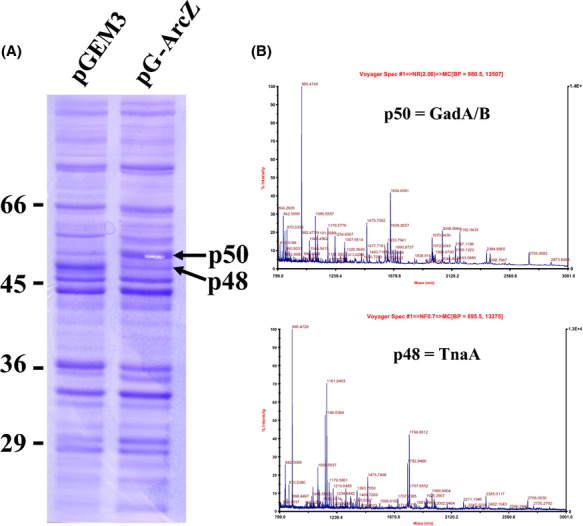
Protein expression profile of ArcZ overexpressing cells. (A) Equal amounts of total protein from Escherichia coli MG1655 cells containing pG-ArcZ and the pGEM3 vector, respectively, were subjected to electrophoresis on a 10% acrylamide SDS gel and compared. Increased or decreased protein bands are indicated with arrows. (B) Protein bands were excised and identified using Q-TOF2 MS/MS. As peptides containing the N-terminal end regions of p50 were not detected by Q-TOF MS/MS, GadA and GadB were indistinguishable so that p50 was assigned to GadA or GadB (GadA/GadB).
Effects of ArcZ on AR
As GadA/B induced by overexpression of ArcZ is a key enzyme in the AR2 system, we further examined the participation of ArcZ in AR of E. coli cells. For this purpose, we cloned the ArcZ sequence into the IPTG-inducible RNA expression vector, pHMB1, to effectively control the ArcZ expression. Exponentially growing E. coli cells were challenged at pH 2.0 in the presence of 1.5 mmol/L glutamate for 1 h, and their survival rates were determined. Our results showed that survival rates increase with the IPTG concentration (up to 5,000-fold at 1 mmol/L) (Fig. 2A). We also found that ArcZ levels were correlated with survival rates (Fig. 2B), supporting its contribution to acid tolerance in E. coli cells.
Figure 2.
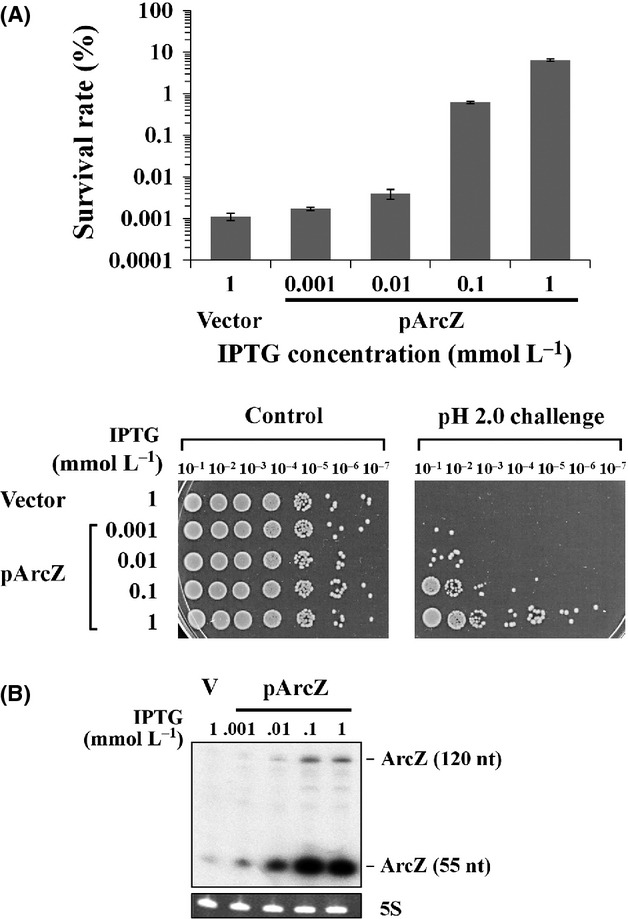
Effect of ArcZ overexpression on acid resistance of E. coli cells. (A) Survival rates of MG1655 transformed with IPTG-inducible RNA expression plasmid overexpressing ArcZ. Exponentially growing Escherichia coli cells were exposed to pH 2.0 for 1 h. Expression of ArcZ was increased with increasing IPTG concentrations. The RNA expression vector, pHMB1, was used as the control plasmid. Before and after acid exposure, cells were serially diluted in LB media and plated on LB agar plates. The survival rate was calculated based on the ratio of CFU of cells after acid exposure to CFU before acid exposure. Tenfold serial dilutions of cultures exposed to acid challenge were additionally spotted on LB agar and grown overnight. Cells containing the vector plasmid grown in the presence of 1 mmol/L were used as a control, instead of cells containing RNA expression plasmids grown at zero concentrations of IPTG. (B) IPTG-induced overexpression of ArcZ was analyzed by Northern blot analysis. A 32P-labeled antisense oligonucleotide for ArcZ was used as the probe.
RpoS-dependent AR2 system activation by rpoS-activating sRNAs
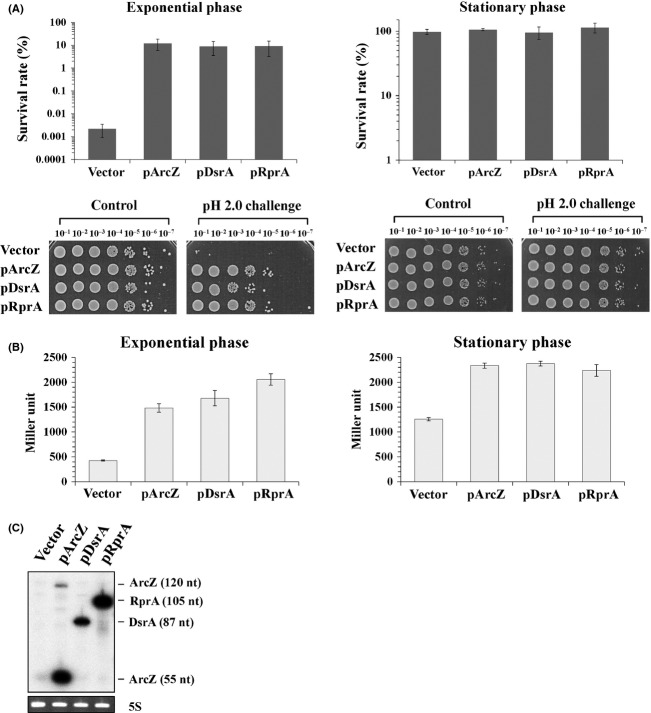
As ArcZ destabilizes antisense arcB mRNA and activates rpoS translation (Mandin and Gottesman 2010), we first tested whether enhanced AR observed with ArcZ overexpression is induced through arcB regulation (Fig. S1). However, upon expression of ArcZ in arcB mutant cells, enhanced AR was still observed, suggesting that arcB expression is not associated with ArcZ-induced AR. Therefore, we focused on whether ArcZ-induced translational activation of rpoS is responsible for AR. It was previously shown that the general stress response sigma factor RpoS plays a critical role in AR systems in commensal and pathogenic E. coli strains (Lin et al. 1996; Price et al. 2000). In addition to ArcZ, sRNAs such as DsrA and RprA are known to activate rpoS translation. Upon IPTG induction of DsrA or RprA from RNA expression plasmids, exponentially growing E. coli cells displayed significantly increased survival rates, similar to cells overexpressing ArcZ (Fig. 3A). Activation of rpoS translation by overexpression of all three sRNAs was confirmed using a rpoS-lacZ translational fusion (Fig. 3B). Overexpression of each sRNA led to a two-to fourfold increase in rpoS expression.
Figure 3.
Effect of overexpression of rpoS-activating sRNAs on acid resistance of Escherichia coli. (A) Acid resistance of E. coli cells overexpressing DsrA, RprA, and ArcZ, respectively, was examined. Survival rates of exponentially growing or stationary-phase cells after exposure to pH 2.0 for 1 h were determined. Overexpression of sRNAs was induced by 1.0 mmol/L IPTG. Values are averaged from at least three independent experiments. Tenfold serial dilutions of cultures exposed to acid challenge were additionally spotted on LB agar and grown overnight. (B) β-Galactosidase activities from cells containing a rpoS-lacZ translational fusion (SG30013) were measured upon sRNA overexpression at the exponential or stationary phase. (C) Overexpression of rpoS-activating sRNAs from the corresponding RNA expression plasmids was confirmed using Northern blot analysis. Antisense oligonucleotides for ArcZ, DsrA, and RprA were mixed, labeled with 32P, and used as probes.
We further examined the effects of rpoS-activating sRNAs on cells grown to the stationary phase. When stationary-phase cells were challenged to pH 2.0 in the presence of 1.5 mmol/L glutamate for 1 h, their survival rates were 100%, even in the absence of sRNA overexpression, suggesting that stationary cells have already acquired sufficient tolerance to the acid challenge. However, rpoS expression was still increased about twofold upon overexpression of any of the three sRNAs (Fig. 3B), suggesting that stationary-phase cells do not require further RpoS for AR under these conditions.
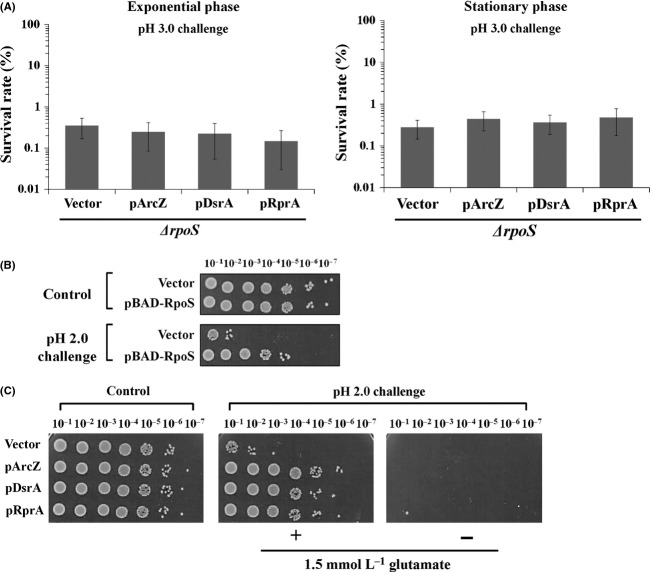
Next, we determined whether enhanced AR by the rpoS-activating sRNAs is achieved through RpoS in experiments with rpoS mutant cells. Unlike wild-type cells, rpoS mutant cells did not survive during acid challenge at pH 2.0, even with overexpression of rpoS-activating sRNAs, regardless of the growth phase (exponential growth or stationary) (Fig. S2). Subsequently, cells were challenged at pH 3.0 instead of pH 2.0. Notably, when rpoS-activating sRNAs were overexpressed, survival rates of rpoS mutant cells at the exponential and stationary phases were not increased (Fig. 4A), suggesting that the promotion of AR by these sRNAs is rpoS dependent. Moreover, overexpression of RpoS led to enhanced AR of exponentially growing E. coli cells (Fig. 4B), similar to that observed upon overexpression of rpoS-activating sRNAs. Our data clearly indicate that sRNAs confer AR in exponentially growing cells through upregulation of rpoS.
Figure 4.
Role of RpoS in enhanced acid resistance by rpoS-activating sRNAs. (A) Survival rates of ΔrpoS mutant cells overexpressing DsrA, RprA, and ArcZ, respectively, after exposure to pH 3.0 were determined. As ΔrpoS cells did not survive during acid challenge at pH 2.0, cells were challenged at pH 3.0 instead. Overexpression of sRNAs was induced by 1.0 mmol/L IPTG. (B) Wild-type MG1655 cells overexpressing RpoS were exposed to pH 2.0. Overexpression of RpoS was induced from pBAD-RpoS by 0.002% arabinose in exponentially growing cells. Acid resistance was evaluated by analysis of colony-forming abilities of serial 10-fold dilutions of the cultures. (C) Glutamate dependency of acid resistance. Exponentially growing wild-type MG1655 cells were exposed to pH 2.0, and their colony-forming abilities analyzed by 10-fold serial dilution.
As hfq mutant cells showed extreme sensitivity to acid challenge as much as rpoS mutant cells did in our experimental conditions (Fig. S2) and overexpression of sRNAs in hfq mutant cells did not increase AR (G. Bak and Y. Lee, unpublished results), a further study on hfq has not been done.
As GadA/B, glutamate decarboxylases of the AR2 system, was highly accumulated upon overexpression of ArcZ, external glutamate that is an essential component of the AR2 system was included in AR assay. We examined whether AR induced by rpoS-activating sRNAs is increased when glutamate is omitted in acid challenge media. Enhanced AR was not observed in the absence of glutamate, indicating that rpoS-activating sRNAs act through the glutamate-dependent AR2 system (Fig. 4C).
Role of GadX in promotion of AR by rpoS-activating sRNAs
To identify the downstream molecules participating rpoS-activating sRNA-mediated AR, we assessed the involvement of GadX in this process. GadX is known to be a transcriptional activator of the gadE gene encoding the key transcriptional activator for gadA and gadBC genes (Masuda and Church 2003; Tramonti et al. 2006; Sayed et al. 2007; Sayed and Foster 2009). Transcription of gadX is not only primarily induced by RpoS in cells cultured in neutral rich media at the stationary phase, but also increased under acidic pH conditions (Ma et al. 2002, 2003b). To ascertain whether GadX is involved in the AR-acquiring process by rpoS-activating sRNAs, we examined the survival rates of gadX mutant cells overexpressing sRNAs. Upon challenge of exponentially growing cells at pH 2.0, enhanced AR was significantly decreased in the absence of gadX (Fig. 5), suggesting a role in enhanced AR. However, overexpressed sRNAs conferred some AR to gadX mutant cells, in contrast to rpoS mutant cells that showed no survival regardless of sRNA expression. Our findings support the existence of RpoS-dependent and GadX-independent pathways for AR enhancement by rpoS-activating sRNAs.
Figure 5.
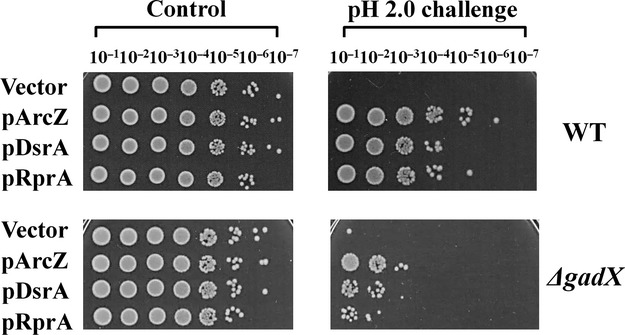
Role of GadX in enhanced acid resistance by rpoS-activating sRNAs. Expression of each rpoS-activating sRNA was induced with 1 mmol/L IPTG in gadX mutant cells, and exponentially growing cells exposed to pH 2.0. Acid resistance was evaluated based on the colony-forming abilities of serial 10-fold dilutions of the cultures. The results are representative of at least three independent experiments.
AR in the absence of rpoS-activating sRNAs
We examined the contribution of each rpoS-activating sRNA to AR using mutant strains lacking these sRNAs. Each mutant strain showed a three-to fivefold reduction in survival rates of stationary-phase cells upon acid challenge at pH 2.0 (Fig. 6A and B). When MG1655Δ3, a mutant strain lacking all three sRNA genes, was challenged, the survival rate was drastically reduced by about 1,000-fold (Fig. 6C). As all three rpoS-activating sRNAs are highly expressed in the stationary phase (Argaman et al. 2001), maintenance of the appropriate cellular levels of these sRNAs appears essential for E. coli cells to acquire full AR. Next, we examined the effects of overexpression of individual rpoS-activating sRNAs on AR defects of MG1655Δ3 cells. Upon overexpression of each sRNA in the MG1655Δ3 strain, AR at the stationary phase was fully recovered to the wild-type level and up to the level of wild-type cells overexpressing rpoS-activating sRNA at the exponential phase (Fig. 7), suggesting that the three sRNAs play the same role in AR.
Figure 6.
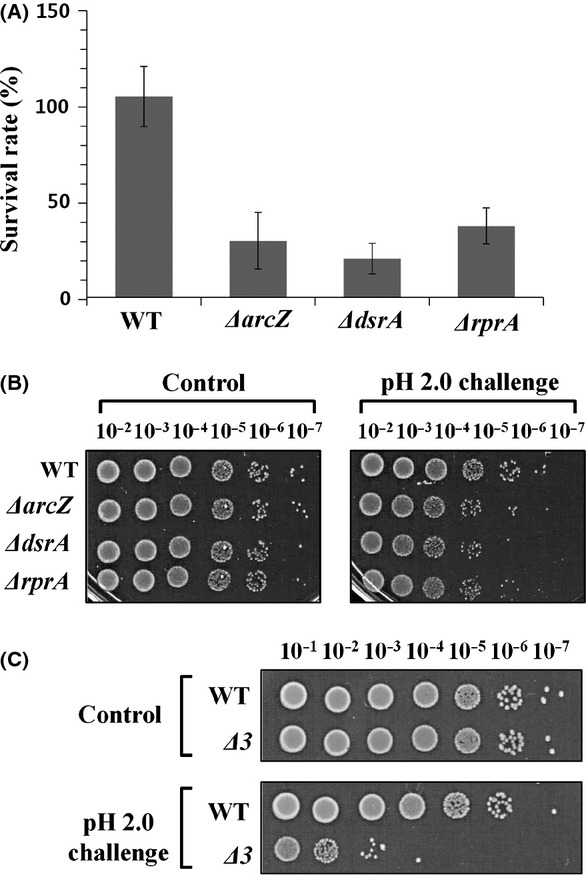
Acid resistance of cells lacking rpoS-activating sRNA genes. (A) Survival rates of Escherichia coli mutant cells with deletion of each rpoS-activating sRNA gene under conditions of acid shock (pH 2.0) were determined using stationary-phase cells (shown in linear scale). Values are averaged from at least three independent experiments. (B) Acid resistance of mutant cells was evaluated based on colony-forming ability. (C) Acid resistance of MG1655Δ3 cells lacking all three rpoS-activating sRNA genes was evaluated based on the colony-forming abilities of stationary-phase cells.
Figure 7.
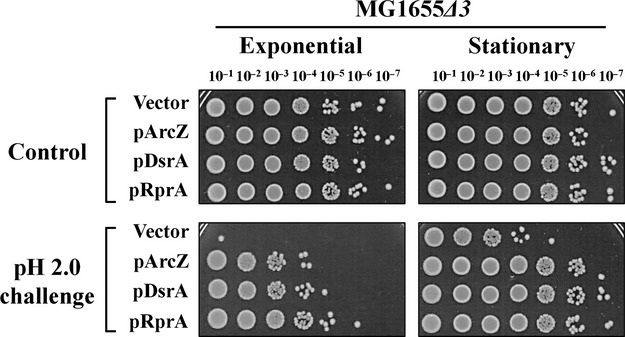
Recovery of acid resistance of mutant cells lacking all three rpoS-activating sRNA genes induced by single sRNAs. Each rpoS-activating sRNA was overexpressed in the triple mutant cells, and acid resistance evaluated based on colony-forming abilities of both exponentially growing and stationary-phase cells. Overexpression of sRNAs was induced by 1.0 mmol/L IPTG.
Induction of sRNA expression during acid challenge
All three rpoS-activating sRNAs are highly expressed in stationary phase, but it is not known whether they are induced during acid challenge. To examine their expression levels upon acid challenge, exponentially growing cells were subjected to moderate acid conditions of pH 5.0, as more harsh conditions may cause cell death. We observed increased expression of DsrA and RprA upon the pH shift, but not ArcZ (Fig. 8A). Expression of ArcZ was slightly reduced, probably due to the reduced growth rate because E. coli cells grew slowly when shifted to pH 5.0 (Fig. 8B).
Figure 8.
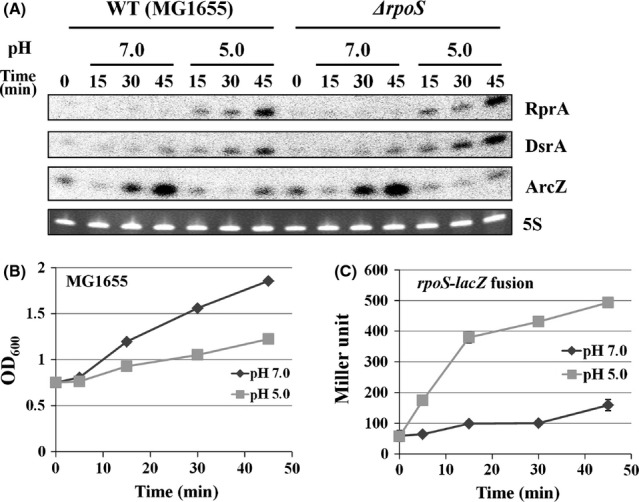
Induction of rpoS-activating sRNAs and activation of rpoS translation upon acid challenge. Overnight cultures of wild-type and ΔrpoS cells were diluted 1:100 in LB medium and grown for 2 h at 37°C. Cultures were split into two and either maintained or subjected to pH 5.0 (LB buffered by 100 mmol/L MES), then further incubated at 37°C. Aliquots were sampled at specific time intervals for Northern blot analysis (A), and for growth curves (B). Cells containing a rpoS-lacZ translational fusion (SG30013) was also shifted to pH 5.0 and aliquots were sampled for β-galactosidase assay (C).
As translational activation of rpoS upon acid exposure has been reported (Bearson et al. 1996; Battesti et al. 2011) in Salmonella typhimurium, we further examined whether exposure to low-pH triggers DsrA and RprA expression in rpoS mutant cells. DsrA and RprA were still induced in rpoS mutant cells (Fig. 8A). We also found that the shift of E. coli cells to pH 5.0 increased expression of rpoS-lacZ translational fusion (Fig. 8C), suggesting that acid-stimulated DsrA and RprA expression is responsible for increased translation of rpoS upon acid exposure.
Discussion
In this study, three rpoS-activating sRNAs, ArcZ, DsrA, and RprA, once they are expressed, were shown to play an equal role in conferring AR to E. coli cells through the AR2 system. This sRNA-mediated AR was totally dependent on RpoS and partially on GadX. Furthermore, DsrA and RprA, but not ArcZ, were induced upon exposure to acid. On the basis of these findings, we propose a model of acid stress response by rpoS-activating sRNAs (Fig. 9).
Figure 9.
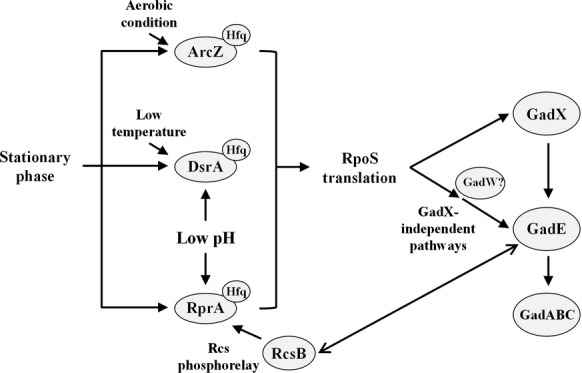
Model for acid resistance acquisition by rpoS-activating sRNAs in Escherichia coli cells. All three sRNAs are upregulated in the stationary phase. Expression of ArcZ and DsrA is induced under aerobic conditions and low temperatures, respectively. Expression of RprA is activated by Rcs phosphorelay. Expression of DsrA and RprA, but not ArcZ, is induced under acid stress. RpoS is activated by all three sRNAs in a Hfq-dependent manner. RpoS promotes gadX expression, and subsequently, GadX activates expression of GadE, which is a transcriptional activator for gadA and gadBC encoding key components in the AR2 system. RpoS-dependent, GadX-independent pathways of GadE induction also exist. RcsB, a response regulator of Rcs phosphorelay system, interacts with GadE to activate GadE.
RpoS is induced by variety of general stress conditions and aids in protecting cells against stress (Battesti et al. 2011). Acid shock is known to increase the translation and stability of RpoS in S. typhimurium and E. coli in LB media (Bearson et al. 1996; Heuveling et al. 2008; Battesti et al. 2011). We showed that a shift of exponentially growing E. coli cells to pH 5.0 not only increases the expression of translational rpoS-lacZ fusions but also induces DsrA and RprA. Therefore, acid stress appears to induce rpoS expression through induction of DsrA and RprA. ArcZ was not induced by the shift to pH 5.0, but activation of rpoS by ArcZ induced by other stimuli, such as aerobic conditions, can lead to AR of cells. As all three rpoS-activating sRNAs are abundant in the stationary phase (Argaman et al. 2001), high acid tolerance of stationary cells is mainly due to expression of RpoS induced by sRNAs. Cells depleted of any of the rpoS-activating sRNAs showed a three-to fivefold decreased AR at the stationary phase, suggesting that all three sRNAs are required for full AR. The three rpoS-activating sRNAs are triggered by different signals: ArcZ is highly expressed under aerobic conditions, DsrA is highly expressed at low temperatures and osmotic stress (Sledjeski et al. 1996; Majdalani et al. 2001), and RprA is induced by osmotic stress through Rcs phosphorelay (Majdalani et al. 2002; Majdalani and Gottesman 2005). Interestingly, RcsB protein, a response regulator of Rcs phosphorelay system, has been reported to interact with GadE to activate GadE (Castanie-Cornet et al. 2010). However, all three rpoS-activating sRNAs perform the same function in activating RpoS required for AR regardless of origin because overexpression of a single rpoS-activating sRNA can complement reduced acid tolerance caused by the absence of other sRNAs. Hfq is also essential for enhanced AR by rpoS-activating sRNAs (Fig. S2), possibly because sRNA-mediated activation of rpoS translation by these sRNAs is Hfq dependent (Battesti et al. 2011).
Among the known AR systems in E. coli, AR1 and AR2 require RpoS at least under certain conditions. While the mechanism underlying AR1 is not well understood, many regulators are involved in the AR2 pathway and function in different ways, depending on the growth conditions. Glutamate decarboxylases, GadA and GadB, key enzymes of the AR2 system, are induced by RpoS-dependent expression of GadX and subsequently GadX-dependent expression of GadE in stationary-phase cells grown in neutral LB media (Tramonti et al. 2002). On the other hand, exponentially growing cells have no acid tolerance at pH 2.0, possibly because GadA and GadB are not expressed during exponential phase (Ma et al. 2002; Tramonti et al. 2002). However, overexpression of rpoS-activating sRNAs can confer AR to exponentially growing cells through RpoS-GadX because direct overexpression of RpoS also promotes AR in E. coli cells. The finding that ΔgadX cells overexpressing sRNAs showed reduced AR, but were more acid tolerant than cells lacking RpoS supports the existence of RpoS-dependent, GadX-independent pathways for AR. Another AraC-family regulator, GadW, was reported to affect GadX-dependent activation of GadE (Ma et al. 2002, 2003b). However, in our experiments, gadW mutation did not affect AR promotion by sRNAs (Fig. S3), suggesting that gadW is not directly involved in sRNA-mediated AR. Although the RpoS-dependent, GadX-independent pathways remain to be demonstrated, they may subsequently involve activation of GadE, as overexpression of GadE leads to AR via expression of gadA and gadBC in gadX as well as rpoS mutant cells (Ma et al. 2003a). Thus, we conclude that enhanced AR by rpoS-activating sRNAs is conferred via a GadX-dependent or-independent RpoS-GadE circuit in both exponentially growing and stationary-phase cells.
Acknowledgments
This study was supported by the National Research Foundation of Korea (NRF) Grant by the Korea government (MSIP) (2010-0029167; 2011-0020322) and the Intelligent Synthetic Biology Center of Global Frontier Project funded by MSIP (2012M3A6A8054837). The authors would like to thank S. Gottesman and N. Majdalani for providing strain SG30013, and NBRP-E. coli at NIG for providing E. coli strains of the Keio collection.
Conflict of Interest
None declared.
Funding Information
This study was supported by the National Research Foundation of Korea (NRF) Grant by the Korea government (MSIP) (2010-0029167; 2011-0020322) and the Intelligent Synthetic Biology Center of Global Frontier Project funded by MSIP (2012M3A6A8054837).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Acid resistance of ΔacrB cells overexpressing ArcZ. Exponentially growing cells in the presence of 1 mmol/L IPTG were exposed to pH 2.0 for 1 h. Tenfold serial dilutions of cultures were spotted on LB agar and grown overnight. The results are representative of at least three independent experiments.
Figure S2. Acid resistance of Δhfq and ΔrpoS cells. Exponentially growing or stationary growing Δhfq and ΔrpoS cells were exposed to pH 2.0 or 3.0 for 1 h. Acid resistance was analyzed with serial 10-fold dilutions of acidchallenged cultures. The results are representative of at least three independent experiments.
Figure S3. Effects of GadW on acid resistance promoted by rpoS-activating sRNAs. Exponentially growing or ΔgadW cells were exposed to pH 2.0. Acid resistance was analyzed with serial 10-fold dilutions of acid-challenged cultures. Overexpression of sRNAs was induced by 1.0 mmol/L IPTG. The results are representative of at least three independent experiments.
Figure S4. Expression of ArcZ in wild-type and ΔarcZ backgrounds. Total RNA samples were extracted from wild-type MG1655 and ΔarcZ cells. Samples were taken from indicated time points after 1:100 dilution of the overnight culture. Expression of ArcZ sRNA was analyzed by Northern blot analysis. A 32P-labeled oligonucleotide specific to ArcZ was used as the probe.
Table S1. Oligonucleotide sequences used in this study.
References
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson SM, Jr Benjamin WH, Swords WE, Foster JW. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J. Bacteriol. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi C, Theraulaz L, Mejean V, Jourlin-Castelli C. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol. Microbiol. 2003;48:211–223. doi: 10.1046/j.1365-2958.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J. Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 2010;38:3546–3554. doi: 10.1093/nar/gkq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase D, Tramonti A, Bossa F, Visca P. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 1999;32:1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- Dong T, Schellhorn HE. Role of RpoS in virulence of pathogens. Infect. Immun. 2010;78:887–897. doi: 10.1128/IAI.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- Giangrossi M, Zattoni S, Tramonti A, De Biase D, Falconi M. Antagonistic role of H-NS and GadX in the regulation of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 2005;280:21498–21505. doi: 10.1074/jbc.M413255200. [DOI] [PubMed] [Google Scholar]
- Han K, Kim KS, Bak G, Park H, Lee Y. Recognition and discrimination of target mRNAs by Sib RNAs, a cis-encoded sRNA family. Nucleic Acids Res. 2010;38:5851–5866. doi: 10.1093/nar/gkq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuveling J, Possling A, Hengge R. A role for Lon protease in the control of the acid resistance genes of Escherichia coli. Mol. Microbiol. 2008;69:534–547. doi: 10.1111/j.1365-2958.2008.06306.x. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Jin Y, Watt RM, Danchin A, Huang JD. Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genomics. 2009;10:165. doi: 10.1186/1471-2164-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Park I, Lee Y. Mutational analysis of RNA structures and sequences postulated to affect 3′ processing of M1 RNA, the RNA component of Escherichia coli RNase P. J. Biol. Chem. 1996;271:19330–19337. doi: 10.1074/jbc.271.32.19330. [DOI] [PubMed] [Google Scholar]
- Lease RA, Belfort M. Riboregulation by DsrA RNA: trans-actions for global economy. Mol. Microbiol. 2000;38:667–672. doi: 10.1046/j.1365-2958.2000.02162.x. [DOI] [PubMed] [Google Scholar]
- Lease RA, Smith D, Mcdonough K, Belfort M. The small noncoding DsrA RNA is an acid resistance regulator in Escherichia coli. J. Bacteriol. 2004;186:6179–6185. doi: 10.1128/JB.186.18.6179-6185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Park H, Bak G, Kim KS, Lee Y. Regulation of transcription from two ssrS promoters in 6S RNA biogenesis. Mol. Cells. 2013;36:227–234. doi: 10.1007/s10059-013-0082-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Ma D, Chen Y, Guo Y, Chen GQ, Deng H, et al. L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 2013;23:635–644. doi: 10.1038/cr.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Richard H, Tucker DL, Conway T, Foster JW. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW) J. Bacteriol. 2002;184:7001–7012. doi: 10.1128/JB.184.24.7001-7012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 2003a;49:1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- Ma Z, Richard H, Foster JW. pH-Dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J. Bacteriol. 2003b;185:6852–6859. doi: 10.1128/JB.185.23.6852-6859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Masuda N, Foster JW. Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J. Bacteriol. 2004;186:7378–7389. doi: 10.1128/JB.186.21.7378-7389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Lu P, Yan C, Fan C, Yin P, Wang J, et al. Structure and mechanism of a glutamate-GABA antiporter. Nature. 2012;483:632–636. doi: 10.1038/nature10917. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N, Church GM. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 2003;48:699–712. doi: 10.1046/j.1365-2958.2003.03477.x. [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke JA, Fozo EM, Hemm MR, Storz G. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J. Mol. Biol. 2011;406:29–43. doi: 10.1016/j.jmb.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- Price SB, Cheng CM, Kaspar CW, Wright JC, Degraves FJ, Penfound TA, et al. Role of rpoS in acid resistance and fecal shedding of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2000;66:632–637. doi: 10.1128/aem.66.2.632-637.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV. Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiology. 2009a;155:106–114. doi: 10.1099/mic.0.023598-0. [DOI] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV. The small RNA GcvB regulates sstT mRNA expression in Escherichia coli. J. Bacteriol. 2009b;191:238–248. doi: 10.1128/JB.00915-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed AK, Foster JW. A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol. Microbiol. 2009;71:1435–1450. doi: 10.1111/j.1365-2958.2009.06614.x. [DOI] [PubMed] [Google Scholar]
- Sayed AK, Odom C, Foster JW. The Escherichia coli AraC-family regulators GadX and GadW activate gadE, the central activator of glutamate-dependent acid resistance. Microbiology. 2007;153:2584–2592. doi: 10.1099/mic.0.2007/007005-0. [DOI] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML, Enquist LW Cold Spring Harbor Laboratory. Experiments with gene fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl Acad. Sci. USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezias SS, Tsiftsoglou AS, Amanatiadou EP, Vizirianakis IS. Cloning and characterization of polyA-RNA transcripts encoded by activated B1-like retrotransposons in mouse erythroleukemia MEL cells exposed to methylation inhibitors. BMB Rep. 2012;45:126–131. doi: 10.5483/BMBRep.2012.45.2.126. [DOI] [PubMed] [Google Scholar]
- Tramonti A, Visca P, De Canio M, Falconi M, De Biase D. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 2002;184:2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramonti A, De Canio M, Delany I, Scarlato V, De Biase D. Mechanisms of transcription activation exerted by GadX and GadW at the gadA and gadBC gene promoters of the glutamate-based acid resistance system in Escherichia coli. J. Bacteriol. 2006;188:8118–8127. doi: 10.1128/JB.01044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 2008;70:965–982. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman SR, Small PL. Transcriptional expression of Escherichia coli glutamate-dependent acid resistance genes gadA and gadBC in an hns rpoS mutant. J. Bacteriol. 2003;185:4644–4647. doi: 10.1128/JB.185.15.4644-4647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bremer H. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 1995;270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- Zhao B, Houry WA. Acid stress response in enteropathogenic gammaproteobacteria: an aptitude for survival. Biochem. Cell Biol. 2010;88:301–314. doi: 10.1139/o09-182. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S. Modes of regulation of RpoS by H-NS. J. Bacteriol. 2006;188:7022–7025. doi: 10.1128/JB.00687-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, et al. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc. Natl Acad. Sci. USA. 2005;102:2862–2867. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Acid resistance of ΔacrB cells overexpressing ArcZ. Exponentially growing cells in the presence of 1 mmol/L IPTG were exposed to pH 2.0 for 1 h. Tenfold serial dilutions of cultures were spotted on LB agar and grown overnight. The results are representative of at least three independent experiments.
Figure S2. Acid resistance of Δhfq and ΔrpoS cells. Exponentially growing or stationary growing Δhfq and ΔrpoS cells were exposed to pH 2.0 or 3.0 for 1 h. Acid resistance was analyzed with serial 10-fold dilutions of acidchallenged cultures. The results are representative of at least three independent experiments.
Figure S3. Effects of GadW on acid resistance promoted by rpoS-activating sRNAs. Exponentially growing or ΔgadW cells were exposed to pH 2.0. Acid resistance was analyzed with serial 10-fold dilutions of acid-challenged cultures. Overexpression of sRNAs was induced by 1.0 mmol/L IPTG. The results are representative of at least three independent experiments.
Figure S4. Expression of ArcZ in wild-type and ΔarcZ backgrounds. Total RNA samples were extracted from wild-type MG1655 and ΔarcZ cells. Samples were taken from indicated time points after 1:100 dilution of the overnight culture. Expression of ArcZ sRNA was analyzed by Northern blot analysis. A 32P-labeled oligonucleotide specific to ArcZ was used as the probe.
Table S1. Oligonucleotide sequences used in this study.