Abstract
The aims of this study were to assess antibiotic resistance pheno-and genotypes in foodborne, clinical, and environmental Listeria isolates, as well as to elucidate the horizontal gene transfer potential of detected resistance genes. A small fraction of in total 524 Listeria spp. isolates (3.1%) displayed acquired antibiotic resistance mainly to tetracycline (n = 11), but also to clindamycin (n = 4) and trimethoprim (n = 3), which was genotypically confirmed. In two cases, a tetracycline resistance phenotype was observed together with a trimethoprim resistance phenotype, namely in a clinical L. monocytogenes strain and in a foodborne L. innocua isolate. Depending on the applied guidelines, a differing number of isolates (n = 2 or n = 20) showed values for ampicillin that are on the edge between intermediate susceptibility and resistance. Transferability of the antibiotic resistance genes from the Listeria donors, elucidated in vitro by filter matings, was demonstrated for genes located on transposons of the Tn916 family and for an unknown clindamycin resistance determinant. Transfer rates of up to 10−5 transconjugants per donor were obtained with a L. monocytogenes recipient and up to 10−7 with an Enterococcus faecalis recipient, respectively. Although the prevalence of acquired antibiotic resistance in Listeria isolates from this study was rather low, the transferability of these resistances enables further spread in the future. This endorses the importance of surveillance of L. monocytogenes and other Listeria spp. in terms of antibiotic susceptibility.
Keywords: Antibiotics, environmental, food, Listeria, resistance.
Introduction
Listeria monocytogenes is an emerging foodborne pathogen causing listeriosis and accounts after Salmonella for the second-most food-related deaths in the United States (Scallan et al. 2011). In the European Union, listeriosis even led by far to the most food-related deaths (n = 134) caused by bacteria in 2011, much more than salmonellosis (n = 56) (EFSA and ECDC 2013). A typical treatment of this invasive disease is the application of penicillin or ampicillin together with gentamicin (Swaminathan and Gerner-Smidt 2007); however, adaptations can be advisable for certain patients and clinical manifestations (recently reviewed by Allerberger and Wagner 2010). Regardless of an early antibiotic therapy, which is crucial for physical recovery of the patients (Mayrhofer et al. 2004), mortality rates are very high, ˜20% to 30% (Watson 2009). Next to L. monocytogenes, also the knowledge about transferable resistance genes present in other Listeria species is important, as L. innocua was recognized as a reservoir of antibiotic resistance (ABR) for L. monocytogenes (Bertrand et al. 2005). Listeria strains are frequently detected in food products due to their ubiquitary occurrence in the environment. In addition, growth and survival of these psychrotrophic bacteria are favored particularly due to the increasing demand for minimally processed foods and absence of preservatives. It was estimated that 99% of listeriosis cases occur due to foodborne transmission of the bacteria (Scallan et al. 2011).
Listeria spp. are often exposed to low levels of antibiotics, as these agents are used in large amounts both in human and animal medicine (Aarestrup 2012). This can lead to an increased selective pressure which favors emergence of ABR due to mutations or acquisition of mobile genetic elements. Thus, it was not surprising, when the first acquired ABR in L. monocytogenes was reported in 1990 (Poyart-Salmeron et al. 1990). Further multidrug-resistant Listeria strains due to acquisition of plasmids were sporadically detected (Quentin et al. 1990; Hadorn et al. 1993; Tsakris et al. 1997). A way to limit the spread of antimicrobial resistances is to reduce the application of these substances. This consideration led to the ban of antibiotics as feed additives in Switzerland already in 1999 (Arnold et al. 2004) and from January 2006 onward also in the European Union (Castanon 2007). In contrast, antimicrobial compounds are still administered as growth promoters, for example, in the United States, without control by veterinarians (Jones and Ricke 2003). An announcement of the Food and Drug Administration from April 2012 suggests stopping application of antibiotics for growth promotion on a voluntary basis (http://www.fda.gov).
There are relatively few epidemiological studies and thus only limited information on ABR prevalence and spread concerning Listeria spp. and many studies focused on local situations and special origins of Listeria isolates. Therefore, the aim of this study was the assessment of resistance pheno-and genotypes in foodborne, clinical, and environmental Listeria isolates from the different regions of Switzerland, as well as elucidating the horizontal gene transfer potential of detected ABR genes.
Material and Methods
Bacterial strains
Listeria strains were isolated from food for this study by application of ONE Broth-Listeria and Brilliance™ Listeria Agar (ready to use plates; both Oxoid, Pratteln, Switzerland). Other strains were obtained from own culture collections at ETH Zurich or provided by the food industry, hospitals, and veterinary clinics in Switzerland. In total 524 isolates were studied, 24 originated from poultry, 132 from meat products, 46 from dairy products, 18 from fish/seafood, 36 from production (swaps), five from processed food, eight from plant material, 100 from water, and 155 from humans. The Listeria strains of human clinical origin were provided anonymized as pure cultures from the culture collection of the Lausanne University Hospital (CHUV, Lausanne, Switzerland). The isolates were from 2006 (n = 33), 2007 (n = 54), 2008 (n = 151), 2009 (n = 142), 2010 (n = 68), 2011 (n = 72), and 2012 (n = 4) and comprised the species L. monocytogenes (n = 383), L. innocua (n = 50), L. ivanovii subsp. ivanovii (n = 5), L. ivanovii subsp. londoniensis (n = 20), L. welshimeri (n = 6), and L. seeligeri (n = 56). Four isolates were assigned to L. fleischmannii sp. nov. (Bertsch et al. 2013a). All strains were cultured on brain heart infusion (BHI) agar medium (Biolife Italiana S.r.l., Milano, Italy) at 37°C and stored at −80°C in BHI medium containing 33% glycerol.
DNA isolation and PCR assays
Chromosomal and extrachromosomal DNA from single bacterial colonies was isolated and purified as described before (Bertsch et al. 2013b). Sequencing analyses were performed by Microsynth AG (Balgach, Switzerland) or GATC Biotech AG (Konstanz, Germany). PCR assays with primers (purchased from Microsynth AG) used in this study are listed in Table S1. For templates up to 3 kb, PCR Master Mix (2X) with Taq polymerase (Fermentas, Le-Mont-Sur-Lausanne, Switzerland) was applied. Longer targets were amplified with either Phusion™ High-Fidelity DNA Polymerase (New England BioLabs, Allschwil, Switzerland) or GoTaq® Long PCR Master Mix (Promega, Duebendorf, Switzerland). L. monocytogenes isolates were further analyzed by application of multiplex PCR assays for rapid discrimination of the major serovars 1/2a, 1/2b, 1/2c, and 4b and for determination of the corresponding lineages (Table S1).
Antibiotic susceptibility
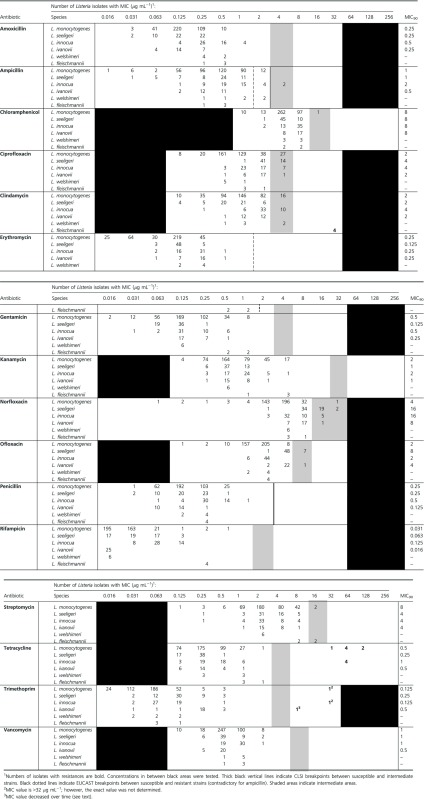
All isolates were screened for ABR phenotypes by broth microdilution testing according to the approved CLSI guidelines M45-A2 for L. monocytogenes (Jorgensen et al. 2010). Minimal inhibitory concentration (MIC) values values that were classified as intermediate or resistant (Table2) were tested three times. A microarray targeting >100 ABR genes of gram-positive bacteria (AMR+ve-2 ArrayTubes™; Alere Technologies GmbH, Jena, Germany), based on a previously published microarray (Perreten et al. 2005), was applied for genotypic resistance profiling, followed by PCR confirmation for known ABR genes (Table S1).
Table 2.
Distribution of MICs among Listeria isolates with corresponding MIC90 values (for n > 20).
 |
In vitro conjugation experiments
Transferability of resistance determinants found in this study was analyzed on nitrocellulose membrane filters (0.45 μm; Millipore AG, Zug, Switzerland) and transconjugants were isolated as published before (Bertsch et al. 2013a). Streptomycin-resistant L. monocytogenes 10403S (Bishop and Hinrichs 1987) and rifampicin-resistant Ent. faecalis JH2-2 (Jacob and Hobbs 1974) were used as recipients and the transfer rate was calculated as transconjugants per donor.
Results and Discussion
Characterization of L. monocytogenes isolates
The majority (almost 90%) of the 383 L. monocytogenes isolates belonged to serovars 1/2a and 4b (Table 1). All isolates of serovars 1/2a and 1/2c (62.4%) corresponded to lineage 2, serovars 4b and 1/2b (37.6%) to lineage 1, respectively, and lineage 3 was not detected. It could be shown that lineage 2 was more common in foodborne isolates (73.4%) and lineage 1 in human isolates (51%) which is in accordance with a recent review (Orsi et al. 2011). The main serovar among foodborne isolates was 1/2a (69.1%), comparable to a study from Italy (Acciari et al. 2011) where 76.6% of L. monocytogenes strains from cheeses (n = 47) belonged to this serovar.
Table 1.
Serovar distribution of 383 Listeria monocytogenes isolates from this study.
| Serovars | Origin (%) | Food (%) | Human (%) | Environment (%) |
|---|---|---|---|---|
| Total number | 210 | 155 | 18 | |
| 1/2a | 228 (59.5) | 145 (69.1) | 74 (47.7) | 9 (50) |
| 1/2b | 28 (7.3) | 11 (5.2) | 17 (11) | – |
| 1/2c | 11 (2.9) | 9 (4.3) | 2 (1.3) | – |
| 4b | 116 (30.3) | 45 (21.4) | 62 (40) | 9 (50) |
Antibiotic resistance
The MIC distribution of the 16 tested antibiotics from Listeria isolates of species L. monocytogenes, L. seeligeri, L. innocua, L. ivanovii, L. welshimeri, and L. fleischmannii sp. nov. (Bertsch et al. 2013a) as well as MIC90 values (where growth of 90% of the isolates is inhibited) are presented in Table 2. As there are very few official susceptibility breakpoints available for L. monocytogenes (Jorgensen et al. 2010), partly inconsistent values are regularly adopted from other bacterial species like enterococci and staphylococci in many publications. In this study, we adhered strictly to only designating isolates as resistant whenever MICs of antibiotics were ≥3 twofold dilution steps higher than the majority of values and therefore clearly distinguishable. Classification as intermediate was based on a previous susceptibility study with different Listeria species (Troxler et al. 2000). We are, however, aware that the transition from intermediate to resistant is debatable in terms of classification, especially when no resistance genes can be detected. With our approach, all isolates were susceptible to amoxicillin, erythromycin, gentamicin, kanamycin, penicillin, rifampicin, and vancomycin. Intermediate values were obtained for ampicillin, chloramphenicol, ciprofloxacin, clindamycin, norfloxacin, ofloxacin, and streptomycin. Phenotypic resistances were detectable against clindamycin (n = 4), tetracycline (n = 11), and trimethoprim (n = 3). In total, 16 (3.1%) revealed resistance and 92 (17.6%) intermediate susceptibility to at least one tested antimicrobial substance among the 524 Listeria strains. Resistance was detected in four of 50 (8%) L. innocua, one of 25 (4%) L. ivanovii, seven of 383 (1.8%) L. monocytogenes, and in all four strains of L. fleischmannii sp. nov., which seem to be one single, persisting clone (Bertsch et al. 2013a). Grouped by their origin, 4.9% of all foodborne Listeria spp. isolates (n = 266) and 1.9% of the clinical isolates (n = 155; all L. monocytogenes; Table S2) displayed ABR, whereas none of the 105 environmental Listeria spp. strains (mainly from water) was resistant. The low value for all L. monocytogenes (1.8%) is similar to recent studies from France, where 2% of foodborne and environmental L. monocytogenes displayed acquired ABR (Granier et al. 2011) and about 1.3% of clinical isolates from humans, respectively (Morvan et al. 2010). The observation that ABR was most prevalent in L. innocua (8%) agrees with a previously published study (Walsh et al. 2001), where it was concluded that this might be due to a species-dependent ability to acquire resistances to antimicrobials. Intermediate values were most frequently found in L. seeligeri (48.2%), L. innocua (36%), and L. welshimeri (33.3%), and to a lesser extend in L. ivanovii (12%) and L. monocytogenes (11%). Only for L. monocytogenes occurrence of intermediate susceptibility varied depending on the isolation source and was much more prevalent among clinical isolates (25.8%; Table S2), compared with environmental samples (5.6%) and foodborne isolates (0.5%). The high number of isolates showing intermediate susceptibility values for ciprofloxacin (n = 49), clindamycin (n = 28), and norfloxacin (n = 28) (Table 2) is not surprising, as the natural population of several Listeria species was described as intermediate to these antibiotics before (Troxler et al. 2000). Intermediate susceptibilities against ciprofloxacin (34%) and clindamycin (27%) were, for example, also detected in L. monocytogenes isolates from a poultry cooking plant in the United States (Lyon et al. 2008).
Microarray analysis (Perreten et al. 2005) and a PCR assay (Table S1) revealed that all 11 phenotypic resistances to tetracycline were encoded by the tet(M) gene. In eight of these isolates (three clinical and three foodborne L. monocytogenes, as well as two foodborne L. innocua strains), this ribosomal protection gene was located on a conjugative transposon of the Tn916 family, indicated both by the presence of the transposon integrase gene int and by the detection of the circular transposon form by PCR (Table S1). No affiliation of the tet(M) gene with the Tn916 family could be detected for one foodborne L. monocytogenes and two foodborne L. innocua strains. Phenotypic resistance to clindamycin (MIC = 32 μg mL−1) in L. fleischmannii sp. nov. could not be linked to a resistance gene targeted on the microarray, as described before (Bertsch et al. 2013a). The only resistant L. ivanovii strain was isolated in 2008 from cheese and could be assigned to subspecies londoniensis. Its resistance to trimethoprim was encoded by the dfrA gene, which could be detected in two independent experiments both on a microarray and by PCR. However, the MIC decreased gradually from 8 to 0.25 μg mL−1 during storage at −80°C in 33% glycerol. Thus, dfrA could not be detected anymore after 1 year of storage. An explanation might be that the resistance gene was encoded on a transferable element which was unstable in the isolate, a phenomenon that was observed before in Listeria (Hadorn et al. 1993).
With application of the EUCAST clinical breakpoints for L. monocytogenes from 2012 (European Committee for Antimicrobial Susceptibility Testing), 20 isolates would be classified as resistant to ampicillin due to an MIC value >1 μg mL−1 (Table 2). Sticking to CLSI guidelines M45-A2 for L. monocytogenes (Jorgensen et al. 2010), where it is stated that resistance to ampicillin has not been described, we designated the two L. innocua strains with MICs >2 μg mL−1 (both isolated from fish) as intermediate (Table 2). Interestingly, fish had also been the source of L. monocytogenes strains with MICs >2 μg mL−1 for ampicillin in another study (Conter et al. 2009). As ampicillin is an important first-choice antibiotic for the treatment of listeriosis, we were testing these isolates for the presence of known genes encoding resistance to beta-lactam antibiotics (bla1, bla2, blaZ, and mecA) with the microarray hybridization assay (Perreten et al. 2005). However, none of these resistance genes was detected.
Notably, two isolates (0.4%) of this study were resistant to more than one antibiotic and therefore analyzed in more detail. The genetic basis of the novel transposon Tn6198 encoding tet(M) and dfrG, which was isolated from the clinical L. monocytogenes isolate TTH-2007, and of the novel plasmid pDB2011 carrying dfrD, erm(A), and spc, detected in the foodborne L. innocua strain TTS-2011, was published by our group elsewhere (Bertsch et al. 2013b,c2013c). Acquired multidrug resistance seems to be exceptional in Listeria (Morvan et al. 2010) and was only rarely reported from France (Poyart-Salmeron et al. 1990; Quentin et al. 1990), Greece (Tsakris et al. 1997), and Switzerland (Hadorn et al. 1993). In these four cases, resistances were mediated by partly similar plasmids (>35 kb in size). In a foodborne L. monocytogenes isolate (designated 04CEB563LM) from France, displaying resistance to tetracycline and trimethoprim, the tet(M) gene seems to be encoded on a member of the Tn916 family. However, the genetic basis of dfrD was not elaborated further (Granier et al. 2011).
Resistance to tetracycline due to the tet(M) gene was most prevalent among the isolates in this study, just as it was among Listeria spp. isolates in several studies before (Charpentier and Courvalin 1999). Reasons might be the extensive use of this antimicrobial worldwide, mainly in the past (Facinelli et al. 1993; Aarestrup 2012), selecting this resistance in enterococci and staphylococci, and spreading it to Listeria spp. by high-frequency transmissibility of tet(M) due to its presence on various Tn916-like conjugative transposons with a broad host range (Roberts and Mullany 2011). The detection of three different trimethoprim resistance genes (dfrA, dfrD, and dfrG) is remarkable and at the same time concerning, as there are very few reports of trimethoprim-resistant Listeria. This antibiotic compound is in the bactericidal drug combination trimethoprim/sulfamethoxazole (named co-trimoxazole), an option to treat listeriosis patients with intolerance to beta-lactams (Morvan et al. 2010). To the best of our knowledge, only in three L. monocytogenes isolates from France, a trimethoprim resistance gene (dfrD) could be identified so far. One of them is L. monocytogenes strain 04CEB563LM (see above). The dfrD gene was furthermore detected in the human clinical isolate U2A2348 (Morvan et al. 2010) and the environmental isolate BM4293, in which it was encoded on the 3.7 kb plasmid pIP823 (Charpentier and Courvalin 1997).
Horizontal gene transfer potential
Transferability of all resistance genes detected in this study was tested in vitro by bacterial matings on filters. Based on MIC values obtained with a broth microdilution test, donors and transconjugants were selected by adding appropriate amounts of antibiotics into the agar medium. Recipient L. monocytogenes 10403S is resistant to streptomycin, but susceptible to tetracycline, trimethoprim, and clindamycin. Recipient Ent. faecalis JH2-2 contains a nontransferable resistance to rifampicin and shows low MICs for tetracycline, trimethoprim, and clindamycin. All donors were susceptible to streptomycin and rifampicin. Successful filter mating experiments along with transfer rates are listed in Table 3. All five L. monocytogenes strains harboring tet(M) on a transposon of the Tn916 family transferred this transposon in similar frequencies. As expected, the strains containing tet(M) but lacking the int gene were unable to transfer the resistance gene.
Table 3.
Combinations of donors and recipients applied in filter matings and obtained transfer rates.
| Listeria donor species | Resistance1 | Recipient2 | Selection3 | Transfer rates4 |
|---|---|---|---|---|
| L. monocytogenes | tet(M) + int | 10403S (STRR) | TET26 + STR26 | 2.5 × 10−8 to 5.7 × 10−6 |
| JH2-2 (RIFR) | TET26 + RIF13 | 9.4 × 10−8 to 1.6 × 10−7 | ||
| L. innocua | tet(M) + int | 10403S (STRR) | TET26 + STR26 | 2.3 × 10−8 to 1.1 × 10−5 |
| JH2-2 (RIFR) | TET26 + RIF13 | 5.4 × 10−8 to 2.4 × 10−7 | ||
| L. fleischmannii sp. nov.5 | CLIR | 10403S (STRR) | CLI13 + STR26 | 3.7 × 10−6 to 5.3 × 10−5 |
| JH2-2 (RIFR) | CLI13 + RIF13 | 5.1 × 10−7 to 8.4 × 10−7 |
Resistances to tetracycline [tet(M)] + integrase (int) and clindamycin (CLIR) were transferred.
Recipient Listeria monocytogenes 10403S is resistant to streptomycin (STRR) and Enterococcus faecalis JH2-2 to rifampicin (RIFR), respectively.
Selection occurred on agar medium supplemented with 26 μg mL−1 tetracycline (TET26), 26 μg mL−1 streptomycin (STR26), 13 μg mL−1 rifampicin (RIF13), and/or 13 μg mL−1 clindamycin (CLI13).
Transfer rates as transconjugants per donor.
Transfer of CLIR from L. fleischmannii sp. nov. DSM 24998T was mentioned before (Bertsch et al. 2013a).
The transfer rates of tet(M) both from L. monocytogenes and L. innocua donors to L. monocytogenes 10403S and Ent. faecalis JH2-2 were between 10−8 and 10−5 transconjugants per donor. Frequencies were higher (up to 10−5) in conjugation experiments between Listeria spp., whereas transfer to Ent. faecalis JH2-2 occurred less frequently (10−8 to 10−7). The differences in these values depending on the recipient have also been reported before (Facinelli et al. 1993), where tet(M) was transferred from L. innocua donors to both L. monocytogenes and L. innocua recipients with frequencies of 2 × 10−5 and to Ent. faecalis JH2-2 with frequencies of 10−7, respectively. The high-level clindamycin resistance detected in all four strains of L. fleischmannii sp. nov. was also transferred more frequently to Listeria recipients (up to 10−5) than to Enterococcus recipients (10−7). Transfer of Tn6198 from L. monocytogenes TTH-2007 to the same recipients occurred in frequencies between 10−9 and 10−6 (Bertsch et al. 2013b).
In conclusion, the relatively low number of ABR in all Listeria (3.1%) and L. monocytogenes strains (1.8%) from this work is in agreement with previous studies from Europe and does not implicate an apparent increase of ABR in this genus in this decade. However, next to the relatively common tet(M) gene, more detailed parallel studies revealed the presence of a presumably novel resistance to clindamycin (Bertsch et al. 2013a), of the novel conjugative transposon Tn6198 (Bertsch et al. 2013b), and of the novel, broad host-range, multidrug resistance plasmid pDB2011 (Bertsch et al. 2013c) among the 524 Listeria strains analyzed during this study. Even though no genes conferring resistance to ampicillin were detected in this study, the elevated MIC values of 4 μg mL−1 might warrant an increase in dosages for the successful treatment of listeriosis. The trend of this elevation was also observed in a huge study among clinical L. monocytogenes strains (n = 4816) from France over the last decades (Morvan et al. 2010). In contrast, we did not detect alarming MIC values for the other first-choice antibiotics amoxicillin and gentamicin.
The fact that all novel resistance elements were transferable and that MICs of ampicillin are at the edge of resistance reinforces the necessity of a broad surveillance of L. monocytogenes to detect changes in antibiotic susceptibility as early as possible. In addition, the other species of the genus Listeria must not be forgotten, as they can constitute reservoirs of antibiotic resistance.
Acknowledgments
This project was funded by the Competence Center Environment and Sustainability (CCES) directed by ETH Zurich. We are grateful to Vincent Perreten (University of Bern) for support during microarray hybridizations. For the provision of bacterial strains, we want to thank Roger Stephan (University of Zurich), Jacques Bille (CHUV Lausanne), Emmanuelle Roth (ALP Bern), and Rudolf Schneebeli (EAWAG Zurich).
Conflict of Interest
None declared.
Funding Information
This project was funded by the Competence Center Environment and Sustainability (CCES) directed by ETH Zurich.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. PCR assays with primers used in this study.
Table S2. Distribution of MICs among 155 clinical Listeria monocytogenes isolates with corresponding MIC90 values.
References
- Aarestrup F. Sustainable farming: get pigs off antibiotics. Nature. 2012;486:465–466. doi: 10.1038/486465a. [DOI] [PubMed] [Google Scholar]
- Acciari VA, Torresi M, Migliorati G, Semprini E, Giannatale Di P, Prencipe V. Characterisation of Listeria monocytogenes strains isolated from soft and semi-soft cheeses sampled in a region of Italy. Vet. Ital. 2011;47:15–23. [PubMed] [Google Scholar]
- Allerberger F, Wagner M. Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infect. 2010;16:16–23. doi: 10.1111/j.1469-0691.2009.03109.x. [DOI] [PubMed] [Google Scholar]
- Arnold S, Gassner B, Giger T, Zwahlen R. Banning antimicrobial growth promoters in feedstuffs does not result in increased therapeutic use of antibiotics in medicated feed in pig farming. Pharmacoepidemiol. Drug Saf. 2004;13:323–331. doi: 10.1002/pds.874. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Huys G, Yde M, D'Haene K, Tardy F, Vrints M, et al. Detection and characterization of tet(M) in tetracycline-resistant Listeria strains from human and food-processing origins in Belgium and France. J. Med. Microbiol. 2005;54:1151–1156. doi: 10.1099/jmm.0.46142-0. [DOI] [PubMed] [Google Scholar]
- Bertsch D, Rau J, Eugster MR, Haug MC, Lawson PA, Lacroix C, et al. Listeria fleischmannii sp. nov., isolated from cheese. Int. J. Syst. Evol. Microbiol. 2013a;63:526–532. doi: 10.1099/ijs.0.036947-0. [DOI] [PubMed] [Google Scholar]
- Bertsch D, Uruty A, Anderegg J, Lacroix C, Perreten V, Meile L. Tn6198, a novel transposon containing the trimethoprim resistance gene dfrG embedded into a Tn916 element in Listeria monocytogenes. J. Antimicrob. Chemother. 2013b;68:986–991. doi: 10.1093/jac/dks531. [DOI] [PubMed] [Google Scholar]
- Bertsch D, Anderegg J, Lacroix C, Meile L, Stevens MJA. pDB2011, a 7.6 kb multidrug resistance plasmid from Listeria innocua replicating in gram-positive and gram-negative hosts. Plasmid. 2013c;70:287–287. doi: 10.1016/j.plasmid.2013.06.001. . doi: 10.1016/j.plasmid.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Hinrichs DJ. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 1987;139:2005–2009. [PubMed] [Google Scholar]
- Castanon JI. History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 2007;86:2466–2471. doi: 10.3382/ps.2007-00249. [DOI] [PubMed] [Google Scholar]
- Charpentier E, Courvalin P. Emergence of the trimethoprim resistance gene dfrD in Listeria monocytogenes BM4293. Antimicrob. Agents Chemother. 1997;41:1134–1136. doi: 10.1128/aac.41.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Courvalin P. Antibiotic resistance in Listeria spp. Antimicrob. Agents Chemother. 1999;43:2103–2108. doi: 10.1128/aac.43.9.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conter M, Paludi D, Zanardi E, Ghidini S, Vergara A, Ianieri A. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 2009;128:497–500. doi: 10.1016/j.ijfoodmicro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J. 2013;11:3129. . doi: 10.2903/j.efsa.2013.3129. [Google Scholar]
- Facinelli B, Roberts MC, Giovanetti E, Casolari C, Fabio U, Varaldo PE. Genetic basis of tetracycline resistance in food-borne isolates of Listeria innocua. Appl. Environ. Microbiol. 1993;59:614–616. doi: 10.1128/aem.59.2.614-616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier SA, Moubareck C, Colaneri C, Lemire A, Roussel S, Dao TT, et al. Antimicrobial resistance of Listeria monocytogenes isolates from food and the environment in France over a 10-year period. Appl. Environ. Microbiol. 2011;77:2788–2790. doi: 10.1128/AEM.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadorn K, Hächler H, Schaffner A, Kayser FH. Genetic characterization of plasmid-encoded multiple antibiotic resistance in a strain of Listeria monocytogenes causing endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 1993;12:928–937. doi: 10.1007/BF01992167. [DOI] [PubMed] [Google Scholar]
- Jacob AE, Hobbs SJ. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FT, Ricke SC. Observations on the history of the development of antimicrobials and their use in poultry feeds. Poult. Sci. 2003;82:613–617. doi: 10.1093/ps/82.4.613. [DOI] [PubMed] [Google Scholar]
- Jorgensen JH, Hindler JA, Bernard K, Citron DM, Cockerill FR, Fritsche TR, et al. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. Approved guideline–2nd edition. CLSI document M45-A2, vol. 30, no. 18. [DOI] [PubMed] [Google Scholar]
- Lyon SA, Berrang ME, Fedorka-Cray PJ, Fletcher DL, Meinersmann RJ. Antimicrobial resistance of Listeria monocytogenes isolated from a poultry further processing plant. Foodborne Pathog. Dis. 2008;5:253–259. doi: 10.1089/fpd.2007.0070. [DOI] [PubMed] [Google Scholar]
- Mayrhofer S, Paulsen P, Smulders FJ, Hilbert F. Antimicrobial resistance profile of five major food-borne pathogens isolated from beef, pork and poultry. Int. J. Food Microbiol. 2004;97:23–29. doi: 10.1016/j.ijfoodmicro.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Morvan A, Moubareck C, Leclercq A, Hervé-Bazin M, Bremont S, Lecuit M, et al. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chemother. 2010;54:2728–2731. doi: 10.1128/AAC.01557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi RH, Wiedmann HC, Bakker den M. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011;301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Perreten V, Vorlet-Fawer L, Slickers P, Ehricht R, Kuhnert P, Frey J. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J. Clin. Microbiol. 2005;43:2291–2302. doi: 10.1128/JCM.43.5.2291-2302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart-Salmeron C, Carlier C, Trieu-Cuot P, Courtieu AL, Courvalin P. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet. 1990;335:1422–1426. doi: 10.1016/0140-6736(90)91447-i. [DOI] [PubMed] [Google Scholar]
- Quentin C, Thibaut MC, Horovitz J, Bebear C. Multiresistant strain of Listeria monocytogenes in septic abortion. Lancet. 1990;336:375. doi: 10.1016/0140-6736(90)91916-x. [DOI] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 2011;35:856–871. doi: 10.1111/j.1574-6976.2011.00283.x. [DOI] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Troxler R, Funke A, Graevenitz von G, Wiedemann B, Stock I. Natural antibiotic susceptibility of Listeria species: L. grayi L. innocua L. ivanovii L. monocytogenes L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 2000;6:525–535. doi: 10.1046/j.1469-0691.2000.00168.x. [DOI] [PubMed] [Google Scholar]
- Tsakris A, Papa A, Douboyas J, Antoniadis A. Neonatal meningitis due to multi-resistant Listeria monocytogenes. J. Antimicrob. Chemother. 1997;39:553–554. doi: 10.1093/jac/39.4.553. [DOI] [PubMed] [Google Scholar]
- Walsh D, Duffy G, Sheridan JJ, Blair IS, McDowell DA. Antibiotic resistance among Listeria, including Listeria monocytogenes, in retail foods. J. Appl. Microbiol. 2001;90:517–522. doi: 10.1046/j.1365-2672.2001.01273.x. [DOI] [PubMed] [Google Scholar]
- Watson R. Listeriosis remains a cause for concern in Europe. BMJ. 2009;338:b319. doi: 10.1136/bmj.b319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. PCR assays with primers used in this study.
Table S2. Distribution of MICs among 155 clinical Listeria monocytogenes isolates with corresponding MIC90 values.


