SUMMARY
Cyclic di-GMP (c-di-GMP) is the prevalent intracellular signalling intermediate in bacteria. It triggers a spectrum of responses that cause bacteria to shift from a swarming motile phase to sessile biofilm formation. However, additional functions for c-di-GMP and roles for related molecules, such as c-di-AMP and c-AMP-GMP continue to be uncovered. The first usage of cyclic-di-nucleotide (c-di-NMP) signalling in the eukaryote domain emerged only recently. In Dictyostelid social amoebas, c-di-GMP is a secreted signal that induces motile amoebas to differentiate into sessile stalk cells. In humans, c-di-NMPs, which are either produced endogenously in response to foreign DNA or by invading bacterial pathogens, trigger the innate immune system by activating the expression of interferon genes. STING, the human c-di-NMP receptor, is conserved throughout metazoa and their closest unicellular relatives, suggesting protist origins for human c-di-NMP signalling. Compared to the limited number of conserved protein domains that detect the second messengers cAMP and cGMP, the domains that detect the c-di-NMPs are surprisingly varied.
Keywords: cyclic di-GMP, diguanylate cyclase, biofilm, cyclic di-AMP, diadenylate cyclase, stalk cell differentiation, Dictyostelium, STING, cGAS, 2′3′-cGAMP
INTRODUCTION
Nucleotide polymers are probably the oldest molecules of life, as proposed in the RNA-world hypothesis (1). The RNA building block ATP is the equally ancient carrier of cellular energy, while its cyclized form, cAMP is a carrier of sensory information in all domains of life. While the second messenger roles of cAMP and its sister molecule cGMP have been studied for over 50 years, the cyclic di-nucleotides c-di-GMP, c-di-AMP, c-AMP-GMP were more recently identified as information carriers in prokaryotes (2-4). As outlined in an excellent recent review (5), c-di-GMP has now far surpassed the prevalence and importance of cAMP as a second messenger in prokaryotes, while novel roles still emerge at a rapid rate. In this work, I will compare the recently discovered roles of c-di-nucleotides in eukaryotes with their established roles in prokaryotes and provide an overview of proteins that mediate synthesis, detection and degradation of cyclic di-nucleotides in the two domains.
Cyclic-di-nucleotide signalling in prokaryotes
C-di-GMP
C-di-GMP was identified in the late ‘80s as a cofactor that was required for activating the cellulose synthase of Gluconoacetobacter xylinus (2). However, it took an almost 15 year effort to determine how c-di-GMP was synthesized, detected and degraded. Cellulose synthase detects c-di-GMP with its associated PilZ domain, which is also present in other proteins that are directly regulated by c-di-GMP (6, 7). The synthetic enzyme, diguanylate cyclase is related to the type III nucleotidyl transferases, but contains a unique signature GGDEF sequence (8-11). The GGDEF domain proteins often harbour an EAL domain, which proved to be a 3′5′-phosphodiesterase (PDE) that hydrolyzes one of the phosphodiester bonds of c-di-GMP to form pGpG (8, 12). In addition, c-di-GMP can also be converted to GMP by a phosphohydrolase of the HD family, that also counts most of the cAMP and cGMP PDEs among its members. The c-di-GMP PDE is characterized by a signature HD-GYP sequence motif, that differs from the HD motif of the cAMP and cGMP PDEs (13).
While mechanisms and genes responsible for c-di-GMP metabolism were being identified, the abundance of GGDEF and EAL encoding sequences in emerging bacterial genome sequences suggested that c-di-GMP mediated signalling must be very widely used. While this was experimentally confirmed for c-di-GMP activation of cellulose synthase, cellulose production appeared to be only one aspect of a suite of c-di-GMP mediated responses that cause bacteria to shift from a swarming planktonic state to a sessile biofilm-associated lifestyle (14, 15, 11). The c-di-GMP induced synthesis of cellulose and other exopolysaccharides contributes to formation of the biofilm matrix, while c-di-GMP induced loss of flagella or other motility apparatus, formation of adhesive curli fimbriae or a stalk-like holdfast mark the developmental transition to a sessile life-style (16-18). For pathogenic bacteria, the planktonic state represents the virulent acute form of the infection, while the biofilm state represents the less virulent chronic stage, which is however more resistant to immune clearance and antibiotics.
Other developmental roles for c-di-GMP continue to emerge. In the predatory bacterium Bdellovibrio bacteriovorus c-di-GMP induces the transition from the axenic mode of feeding to invasion of prey bacteria (19). In the cyanobacterium Anabaena sp. c-di-GMP triggers differentiation of nitrogen-fixing heterocysts (20), while in Streptomyces coelicolor c-di-GMP regulates the formation of aerial hyphae (21).
The targets for c-di-GMP appeared to be numerous and are listed in Table 1, which also summarizes the entire repertoire of c-di-NMP synthetic and hydrolyzing enzymes and targets that are discussed below. As mentioned above, c-di-GMP binds to the PilZ domain of cellulose synthase, but the PilZ domain can also be part of other proteins, such as the Klebsiella transcription factor MrkH, which activates expression of fimbriae or the Pseudomonas aeruginosa protein Alg44, which is involved in biosynthesis of the extracellular polysaccharide alginate (22). Additionally, the Pilz domain can be expressed on its own, such as PlzA from the Lyme disease spirochaete Borrelia burgdorferi, which mediates c-di-GMP induced motility and infectivity of this pathogen (23). In addition to the PilZ domain, c-di-GMP has a steadily growing number of other targets, such as the transcriptional regulators FleQ (24), Vpst (25), CLP (26), Bcam1349 (27), which each interact with c-di-GMP in a distinctive, but unresolved manner. For CLP and Bcam1349, this may involve the intrinsic cNMP domain, the deeply conserved cAMP binding domain of the bacterial catabolite repressor, CRP, and eukaryote PKA (28). Another c-di-GMP target is the I-site of some GGDEF domains, which allosterically inhibits c-di-GMP synthesis by active DGCs. However, both the GGDEF and EAL domains are often degenerate, lacking DGC and PDE activity respectively. In three proteins, PelD, PopA and CdgG, the I-site of a defunct GGDEF domain acts as a c-di-GMP receptor, regulating protein function (29-31). In others, such as FimX and LapD, the substrate binding site of a defunct EAL domain functions as a c-di-GMP receptor (32, 33).
Table 1. Functional domains involved in the synthesis, detection and degradation of c-di-NMPs.
| ORGANISM/c-di-NMP/process | Protein name | Functional domain | Interpro/cd identifier | Ref. |
|---|---|---|---|---|
| BACTERIA | ||||
| c-di-GMP | ||||
| Synthesis | diguanylate cyclase (DGC) | GGDEF|Class III nucleotidyl cyclase | IPR000160|IPR001054 | (8) |
| Degradation | EAL | Diguanylate PDE, EAL domain | IPR001633 | (8) |
| HD-GYP | HD PDEase | IPR003607 | (13) | |
| Detection | PilZ | PilZ | IPR009875 | (7) |
| FleQ | unknown | (24) | ||
| Clp | unknown | (26) | ||
| Bcam1349 | unknown | (27) | ||
| FimX | EAL substrate binding site | IPR001633 | (55) | |
| PelD | RxxD motif in GGDEF domain | IPR000160 | (29) | |
| PNPase | unknown | (35) | ||
| Vpst | unknown | (25) | ||
| Vc2 Class I Riboswitch | GEMM | (36) | ||
| c-di-AMP | ||||
| Synthesis | diadenylate cyclase | DisA_N | IPR003390 | (3) |
| Degradation | YybT/GdpP | RecJ-like phosphoesterase | IPR001667 | (39) |
| Detection | DarR | GATase1_AraC_1 | cd03137 | (44) |
| c-AMP-GMP | ||||
| Synthesis | DncV (VC0179) | no previously identified domain | (4) | |
| Degradation | unknown | |||
| Detection | unknown | |||
| DICTYOSTELIA | ||||
| c-di-GMP | ||||
| Synthesis | diguanylate cyclase (DgcA) | GGDEF|Class III nucleotidyl cyclase | IPR000160|IPR001054 | (45) |
| Degradation | unknown | |||
| Detection | unknown | |||
| METAZOA | ||||
| 2′3′cGAMP | ||||
| Synthesis | Cyclic GMP-AMP synthase (cGAS) | MAB21 (male abnormal 21) | IPR024805 | (50) |
| Degradation | unknown | |||
| Detection | STING | STING-C | cd12146 | (50) |
This enumeration does not exhaust the variety of c-di-GMP targets (Table 1). The biofilm dispersal protein BcdA acts as a c-di-GMP sink by binding c-di-GMP (34). c-di-GMP also mediates signal dependent RNA processing by binding directly to RNA polynucleotide phosphorylase (PNPase) (35) and c-di-GMP regulates gene expression by binding with high affinity to riboswitches. Riboswitches are sequences in the 5′untranslated regions of bacterial mRNAs that fold into structures capable of binding small molecules, which then act to regulate gene transcription or translation (36, 37).
This vast, and probably only partially explored, versatility in c-di-GMP receptors forms a stark contrast with only two intracellular receptors for the more ubiquitous signals, cAMP and cGMP, i.e. the cNMP domain (28) and the GAF domain (38).
Cyclic-di-AMP and cyclic AMP-GMP
Bacteria use two other c-di-NMPs as second messengers, c-di-AMP and c-AMP-GMP, with again unique sets of metabolic enzymes and targets. The diadenylate cyclase, DAC or DisA_N, is part of the DNA scanning protein DisA that monitors Bacillus subtilis DNA for double strand breaks. The presence of Holliday junctions, the hallmark of partially repaired breaks, inhibits c-di-AMP synthesis by DAC. Further hydrolysis of c-di-AMP to pApA by YybT/GdpP prevents B. subtilis differentiation into spores (3, 39). DAC is a novel catalyst with little structural similarity to other mono- or di-nucleotidyl cyclases, and YybT/GdpP is a member the DHH domain phosphatases. The DAC domain was found to be present in most bacterial phyla and even in Archaea (3). Involvement of c-di-AMP in responses ranging from cell size regulation (40), bacterial cell growth (41), peptidoglycan cell wall homeostasis (42) to pathogenicity (43) in a range of bacterial species has since been uncovered. The first known target for c-di-AMP is the transcriptional repressor DarR (44), again binding c-di-AMP with sequences unrelated to previously known cNMP or c-di-NMP binding domains
The latest addition to the prokaryote c-di-NMPs is c-AMP-GMP. This hybrid molecule is synthesized from ATP and GTP by DncV, a cyclase of Vibrio cholerae, that is unrelated to any other nucleotidyl cyclases (4). In V.cholerae, c-AMP-GMP promotes intestinal colonization by down-regulating chemotaxis, but the DncV gene is also present in other proteobacteria and, like c-di-AMP and c-di-GMP, may regulate a range of cellular functions.
Cyclic-di-nucleotide signalling in eukaryotes
Dictyostelia
Until 2012, c-di-NMP signalling was considered to be confined to the prokaryote domain. DNA sequences with GGDEF domains were occasionally detected in the emerging genomes of disparate eukaryotes. However, in almost all cases, the sequences were very similar to prokaryote homologs and absent from close relatives of the eukaryote in question, indicating that they probably originated from contaminating bacterial DNA. The exception is a conserved GGDEF domain that is present in six genomes, sampled across the full genetic diversity of Dictyostelid social amoebas (45). Social amoebas chemotactically aggregate in response to nutrient stress to form multicellular aggregates which transform into fruiting structures. These structures consist of an aerial mas of spores that is supported by a column of stalk cells, encased in cellulose. Several species, such as D.discoideum have an intermediate light-sensitive migrating “slug” stage, which in nature serves to bring the organism to the top layer of the soil for optimal spore dispersal. Dictyostelids extensively use cAMP both in the traditional second messenger role and as a secreted first messenger for induction of chemotaxis and spore differentiation (46). Disruption of the DgcA gene, which encodes the D.discoideum GGDEF domain yielded dgcA- amoebas that form normal migrating slugs, but never initiate fruiting body formation. DgcA is expressed at the slug tip, where the stalk starts to form and was shown to synthesize c-di-GMP. However, unlike prokaryote c-di-GMP, D.discoideum c-di-GMP does not act as a second messenger, but is secreted to locally induce stalk cell differentiation (45). It is as yet unclear how cells detect and process the c-di-GMP signal, and whether c-di-GMP is hydrolysed. No EAL or HD-GYP domains are present in Dictyostelid genomes and only weak homologies are found to bacterial c-di-GMP binding proteins (Chen, Z. and Schaap, P., unpublished results). As a secreted signal, it may have entirely different targets, such as one of the 15 sensor histidine kinases or the 48 G-protein coupled receptors of D. discoideum.
The most intriguing question about c-di-GMP function in Dictyostelia is whether it came in through the back door of lateral gene transfer (LGT), or whether it has deep origins in the first eukaryotes. Because a Dictyostelium amoeba consumes thousands of bacteria every day, LGT is not unlikely. One piece of evidence is suggestive of deep origins. The D.discoideum cellulose synthase, which is essential for stalk formation is more related to bacterial cellulose synthases than to other eukaryote (plant) enzymes. Although it does not have a PilZ domain, this domain, and its activation by c-di-GMP, may have been part of a signalling cassette that early eukaryotes “inherited” from bacteria, with the function of c-di-GMP in stalk formation changing over time.
Humans (and other animals)
The detection of viral or bacterial nucleic acids by cellular receptors is a critical step in the activation of the human innate immune system upon infection. The transmembrane protein STING (stimulator of interferon genes) is an important link in the pathway that leads from nucleic acid detection to expression of interferon genes. While STING itself does not interact with DNA, it was found to bind c-di-GMP derived from bacterial infections, and this interaction also induces interferon gene expression. c-di-GMP binds at a deep cleft between the monomers of the constitutively dimeric STING receptor (47).
The question how STING is activated by DNA was recently resolved by the finding that human cells synthesize the hybrid cyclic dinucleotide c-GMP-AMP using the enzyme cGAS, in response to the presence of cytosolic DNA (48, 49). This enzyme belongs to the nucleotidyl transferase family, which includes class III nucleotidyl cyclases, polyadenylate polymerase, DNA polymerase and oligoadenylate synthase (OAS1), with the latter being most similar to cGAS. cGAS harbours two N-terminal DNA binding domains, of which the second one is essential for activation of c-GMP-AMP synthesis by cytosolic DNA (48). c-GMP-AMP subsequently binds to STING to activate interferon gene expression.
Further studies showed that unlike c-di-AMP and c-di-GMP, in which both nucleotides are linked from the 3′OH of one to the 5′-phosphate of the other molecule, cGAS synthesizes a molecule in which the 2′OH of GMP is linked to the 5′-phoshate of AMP, and the 3′OH of AMP to the 5′-phosphate of GMP. This molecule is alternatively named 2′3′cGAMP (50) or c[G(2′,5′)pA(3′,5′)p] (51). 2′3′cGAMP binds to STING with 300-fold higher affinity than c-di-GMP or 3′3′cGAMP, however, its efficacy for interferon gene induction is only 30 times that of c-di-GMP. Structural studies revealed that 2′3′cGAMP sits deeper in the cleft between the STING dimers than c-di-GMP, and has three additional polar contacts with STING, explaining its higher affinity (50). Structural analysis of cGAS showed that it binds double stranded DNA in a non-specific manner by electrostatic interactions with the phosphodiester backbone. These interactions open the catalytic pocket of cGAS and reposition several catalytic residues. The formation of a 2′5′ linkage by cGAS is unusual for nucleotidyl cyclases, but similar to formation of 2′5′ oligoadenylates generated by OAS1 (52).
Detection of foreign DNA by cGAS and subsequent activation of STING represents a much broader mechanism for the detection of bacterial, viral and fungal infections than the direct interaction of STING with bacterial c-di-NMPs. It is therefore likely that the cGAS/STING system evolved independently to detect foreign DNA, and that STING activation by bacterial c-di-NMPs represents fortuitous use of an existing pathway.
Evolutionary origins of cGAS and STING
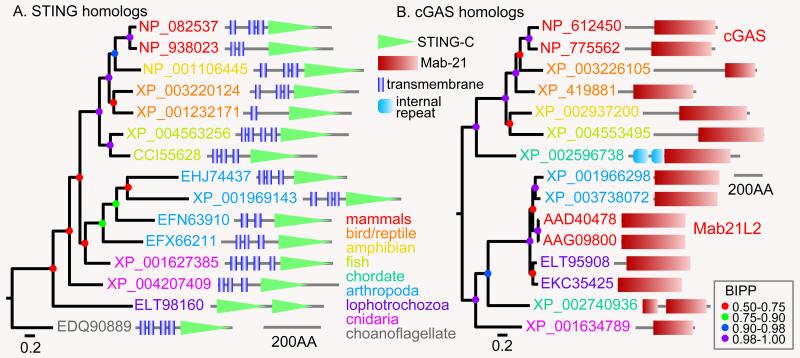
To detect co-evolution of cGAS and STING, we screened representative genomes of the major phyla or metazoa and their closest unicellular relative, the choanoflagellate Monosiga brevicollis, for homologs of the cGAS and STING genes. STING genes were found in all animal phyla, except Porifera, and also in M. brevicollis (Figure 1A). Alignment of the sequences with the structurally characterized human STING, showed that at least 9 out of 10 residues that are required for binding c-di-NMPs are conserved in all phyla, except in the lophotrochozoan Capitella telata (supplemental figure S1), which also has an aberrant functional domain architecture (Fig. 1A).
Figure 1. Phylogenetic analysis of STING and cGAS proteins.
The major vertebrate and invertebrate phyla were individually screened for homologs of human STING and cGAS proteins using BlastP. A few representative best hits for each phylum were selected, which were then used for a reverse screen of mammalian genomes. For STING, the reverse search yielded mouse and bat STING proteins. However, invertebrate cGAS homologs yielded mammalian Mab21 proteins as bidirectional hits. Protein sequences were aligned using M-coffee (53). After deletion of segments with poor consensus alignment, sequences were subjected to Bayesian inference for establishment of phylogenetic relationships between proteins (54). Analysis were run for 1 million generations under a mixed amino-acid model with rate variation between sites estimated by a gamma distribution. Bayesian inference posterior probabilities (BIPPs) of tree nodes are indicated by colored dots. Gene identifiers of the proteins are annotated with functional domain architectures and color-coded to represent the phyla from which they are derived. Corresponding species names are listed in the legends to supplemental figures 1 and 2.
The cGAS catalytic and DNA binding sequences are contained within the PFAM Mab21 domain, which was first detected in proteins involved in embryonic development. Blastp search with human cGAS detects Mab21 proteins throughout metazoa, but arthropod/lophotrochozoan Mab21 proteins are much more related to mammalian sensu stricto Mab21 proteins than to cGAS (Fig.1B). Sequence alignment (supplemental figure S2) shows that the vertebrate cGAS homologs share all residues required for DNA and substrate binding with human cGAS, while the invertebrate proteins lack most of the DNA binding residues and several of the substrate binding residues. Interestingly, conservation of essential residues in the chordates Branchiostoma floridae and Saccoglossus kowalevski is intermediate between cGAS and Mab21, with the proteins of the hemichordate S. kowalevski and the cephalochordate B. floridae being somewhat more similar to Mab21 and cGAS, respectively (Figs. S2 and 1B). This probably marks the emergence of cGAS in the chordate/vertebrate lineage.
Contrary to expectation, the comparative analysis shows that cGAS and STING did not evolve together and that metazoa and their protist ancestors detected c-di-NMPs long before they could synthesize 2′3′cGAMP. The function of this early metazoan c-di-NMP detection system presents an intriguing avenue for further study.
Concluding remarks
Similar to cAMP signalling, the rapidly expanding field of c-di-NMP signalling owes its existence to the original painstaking identification of a small biologically active molecule (2). Whereas molecular genetics and genomics are currently the most powerful tools to unravel gene function, they have limited ability to predict the repertoire of small molecules, that many of the cognate enzymes might synthesize. Our current understanding of particularly developmental signalling is therefore biased towards peptide signals that were discovered through genetics, leaving potentially important non-peptide mediated signalling undiscovered. The diverse functions of monomeric and polymeric nucleotides in information storage, energy transfer and signalling depend on the making and breaking of phosphodiester bonds. All life forms have a very large repertoire of enzymes to perform these functions and their potential for producing biologically active molecules may as yet be vastly underestimated. The expertise of dedicated biochemists, rather than geneticists, will however be needed to identify these molecules.
Supplementary Material
REFERENCES
- 1.Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618–618. [Google Scholar]
- 2.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 3.Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 4.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 7.Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 8.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, et al. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, et al. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol Lett. 2001;204:163–167. doi: 10.1111/j.1574-6968.2001.tb10880.x. [DOI] [PubMed] [Google Scholar]
- 10.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, et al. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, et al. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- 15.Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- 16.Romling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levi A, Jenal U. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol. 2006;188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Shea TM, Klein AH, Geszvain K, Wolfe AJ, Visick KL. Diguanylate cyclases control magnesium-dependent motility of Vibrio fischeri. J Bacteriol. 2006;188:8196–8205. doi: 10.1128/JB.00728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobley L, Fung RK, Lambert C, Harris MA, Dabhi JM, et al. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 2012;8:e1002493. doi: 10.1371/journal.ppat.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neunuebel MR, Golden JW. The Anabaena sp. strain PCC 7120 gene all2874 encodes a diguanylate cyclase and is required for normal heterocyst development under high-light growth conditions. J Bacteriol. 2008;190:6829–6836. doi: 10.1128/JB.00701-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran NT, Den Hengst CD, Gomez-Escribano JP, Buttner MJ. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J Bacteriol. 2011;193:3100–3108. doi: 10.1128/JB.01460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 23.Freedman JC, Rogers EA, Kostick JL, Zhang H, Iyer R, et al. Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol Med Microbiol. 2010;58:285–294. doi: 10.1111/j.1574-695X.2009.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Molecular Microbiology. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191:7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fazli M, O’Connell A, Nilsson M, Niehaus K, Dow JM, et al. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol. 2011;82:327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 28.Kopperud R, Krakstad C, Selheim F, Doskeland SO. cAMP effector mechanisms. Novel twists for an ‘old’ signaling system. FEBS Lett. 2003;546:121–126. doi: 10.1016/s0014-5793(03)00563-5. [DOI] [PubMed] [Google Scholar]
- 29.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, et al. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes & Development. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, et al. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro MV, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009;17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newell PD, Boyd CD, Sondermann H, O’Toole GA. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS biology. 2011;9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Q, Yang Z, Pu M, Peti W, Wood TK. Engineering a novel c-di-GMP-binding protein for biofilm dispersal. Environ Microbiol. 2011;13:631–642. doi: 10.1111/j.1462-2920.2010.02368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol. 2011;407:633–639. doi: 10.1016/j.jmb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 36.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schultz JE. Structural and biochemical aspects of tandem GAF domains. Handb Exp Pharmacol. 2009:93–109. doi: 10.1007/978-3-540-68964-5_6. [DOI] [PubMed] [Google Scholar]
- 39.Rao F, See RY, Zhang D, Toh DC, Ji Q, et al. YybT Is a Signaling Protein That Contains a Cyclic Dinucleotide Phosphodiesterase Domain and a GGDEF Domain with ATPase Activity. J Biol Chem. 2010;285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathogens. 2011;7:e1002217–e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehne FMP, Gunka K, Eilers H, Herzberg C, Kaever V, et al. Cyclic Di-AMP Homeostasis in Bacillus subtilis both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem. 2013;288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis sM in ss-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 2012;83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witte CE, Whiteley AT, Burke TP, Sauer J-D, Portnoy DA, et al. Cyclic di-AMP Is Critical for Listeria monocytogenes Growth, Cell Wall Homeostasis, and Establishment of Infection. mBio. 2013:4. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Li W, He ZG. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem. 2013;288:3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488:680–683. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaap P. Evolutionary crossroads in developmental biology: Dictyostelium discoideum. Development. 2011;138:387–396. doi: 10.1242/dev.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nature immunology. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 48.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J, Sun L, Chen X, Du F, Shi H, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Shi H, Wu J, Zhang X, Sun L, et al. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, et al. Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 55.Huang B, Whitchurch CB, Mattick JS. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J Bacteriol. 2003;185:7068–7076. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.