Abstract
Within- and between-host disease processes occur on the same timescales, therefore changes in the within-host dynamics of parasites, resources, and immunity can interact with changes in the epidemiological dynamics to affect evolutionary outcomes. Consequently, studies of the evolution of disease life histories, that is, infection-age-specific patterns of transmission and virulence, have been constrained by the need for a mechanistic understanding of within-host disease dynamics. In a companion paper (Day et al. 2011), we develop a novel approach that quantifies the relevant within-host aspects of disease through genetic covariance functions. Here, we demonstrate how to apply this theory to data. Using two previously published datasets from rodent malaria infections, we show how to translate experimental measures into disease life-history traits, and how to quantify the covariance in these traits. Our results show how patterns of covariance can interact with epidemiological dynamics to affect evolutionary predictions for disease life history. We also find that the selective constraints on disease life-history evolution can vary qualitatively, and that “simple” virulence-transmission trade-offs that are often the subject of experimental investigation can be obscured by trade-offs within one trait alone. Finally, we highlight the type and quality of data required for future applications.
There has been a great deal of interest in developing theory that bridges within- and between-host dynamics to study infectious disease evolution. Explicitly tracking dynamics at both scales is important because between-host parameters, such as transmission and virulence, are intimately tied to processes occurring at the within-host level (Ewald 1983; Bremermann and Pickering 1983; van Baalen and Sabelis 1995; Frank 1996; Antolin 2008). Further, these parameters are likely to vary over the course of an infection due to corresponding changes in the within-host interactions between parasites, host resources, and host immunity. The resulting infection-age-specific patterns of transmission and virulence are referred to as the disease’s life history (Day 2003; Mideo et al. 2008a).
A considerable body of work has been developed that nests within-host models into between-host frameworks, allowing for explicit interactions between these levels (Sasaki and Iwasa 1991; Day 2001; 2002; Gilchrist and Sasaki 2002; Ganusov et al. 2002; André et al. 2003; Alizon and van Baalen 2005; Gilchrist and Coombs 2006; André and Gandon 2006; Coombs et al. 2007; Alizon and van Baalen 2008; reviewed in Mideo et al. 2008a). However, these results remain somewhat unconnected with empirical research, meaning predictions made are often qualitative and not disease specific. Part of the reason for this is the difficulty that researchers face when attempting to connect models with data. For many pathogens of interest, the required information about the mechanistic details of the within-host dynamics is often not available. Indeed, even for very well-controlled laboratory systems, for which many sophisticated experimental approaches are often available, we still lack a complete understanding of the factors that determine the within-host infection dynamics (e.g., Levin et al. 1997; Grant et al. 2008; Mideo et al. 2008c; Reece et al. 2009).
These difficulties motivated the development of the so-called function-valued trait approach that is presented in a companion paper (Day et al. 2011). This approach captures the relevant evolutionary information about the within-host aspects of disease in a phenomenological way, through the use of statistical estimates of genetic covariance functions for disease life-history traits. This formulation alleviates the need to understand the mechanistic processes through which these disease life-history traits arise.
Here, we make use of two detailed datasets from experiments that measured within-host dynamics of rodent malaria infections across a number of parasite genotypes (Bell et al. 2006; Reece et al. 2008), allowing for a robust quantification of the requisite covariance functions. Using these data, our goal is to demonstrate how the theory developed in the companion article can be applied, and to encourage the collection of further data that might be used. Our analyses reveal some interesting and biologically significant aspects of disease life-history evolution. We provide an example of how epidemiological dynamics and patterns of genetic covariance can interact to generate patterns of disease life-history evolution. The patterns of genetic covariance depend importantly on the precise within-host interactions between parasites and host factors and the data clearly demonstrate that the direction of evolution of disease life-history traits (their “evolutionary trajectories”) can be qualitatively altered when parasites infect different types of hosts. For rodent malaria, specifically, this work also goes beyond previous empirical estimates of the genetic correlations between life-history traits (Mackinnon and Read 1999) by measuring how those correlations change over the course of infection and by emphasizing that genetic correlations within a single trait may constrain (or accelerate) evolution.
Methods
EXPERIMENTAL DATA
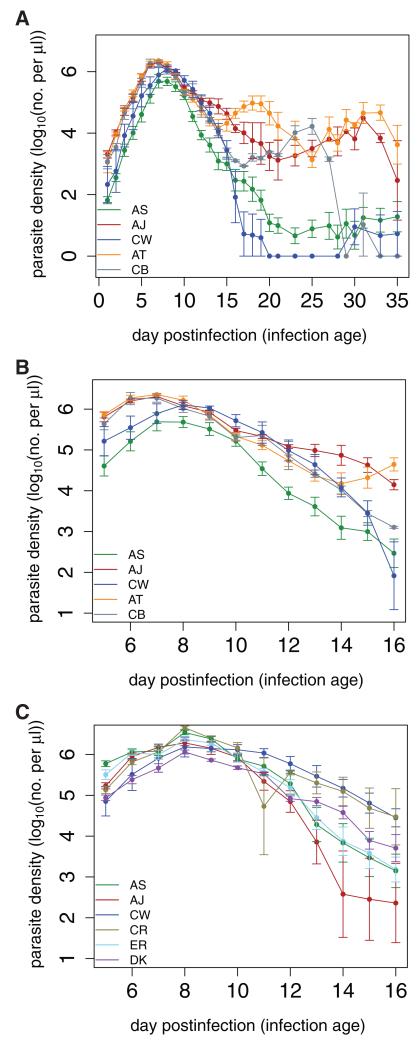
We use two previously published datasets of rodent malaria (Plasmodium chabaudi) infections in laboratory mice (Fig. 1). The first dataset is a composite of two independent experiments tracking P. chabaudi infections in 8-week-old female C57Bl/6J mice (Bell et al. 2006). Individual mice were inoculated with 106 parasites of a single genotype. The genotypes used were AS, AJ, AT, CB, and CW. This dataset provides 10 independent time series (i.e., 10 replicate infections in mice) for genotypes AS, AJ, and AT, and five time series for genotypes CB and CW. Asexual parasite densities were monitored over the first 35 days of infection using real-time quantitative PCR (see Bell et al. 2006, for details).
Figure 1. Dynamics of asexual parasite densities over the course of experimental infections in (A,B) female C57 mice or (C) male MF1 mice.
Data presented are means from infections with a particular genotype (5–10 mice per treatment) ± 1 standard error of the mean. Genotypes are denoted by different colored lines, as indicated by the legends. Note that (B) is the same dataset as in (A), but for only a subset of the days.
The second dataset also tracks the density of single genotype P. chabaudi infections; however, the hosts were 8 to 10-week-old male MF1 mice (Reece et al. 2008). Individual mice were inoculated with 106 parasites of one of the following genotypes: AS, AJ, CW, CR, ER, or DK (five replicate mice per treatment). Asexual parasite densities were monitored over days 5–16 postinfection using real-time quantitative PCR (see Reece et al. 2008, for more details). To better compare the evolutionary predictions from these hosts with those from the former, we also perform all analyses using only measurements from days 5–16 of the first dataset from C57 mice. In this way, we can directly compare evolutionary predictions from infections in different types of hosts, as well as from full and truncated datasets.
TRAIT DEFINITION
To explore models of evolutionary dynamics in the mouse-malaria system, we need to define how measures at the level of the host are related to both disease transmission and virulence. Malaria transmission occurs through specialized parasite forms known as gametocytes, but time series for gametocytes were not available for both datasets. For rodent malaria, there is some evidence that the dynamics of gametocyte densities track those of the asexual parasites, though they tend to be two or three orders of magnitude lower (e.g., Wargo et al. 2007). Assuming this is true, then defining transmission by a log-linear relationship captures at least one of the key aspects of natural malaria biology, namely that infectivity to mosquito vectors is a saturating function of (unlogged) gametocyte densities (Barnes and White 2005; Paul et al. 2007; Sinden et al. 2007). We therefore assume that transmission rate is proportional to the log10 of asexual parasite densities with a coefficient of 10−4 day−1. This translates, on average, to roughly one new transmission event every 3 to 30 days of infection (depending on the epidemiological scenario). These values fit with what has been estimated for malaria transmission in the field (e.g., Smith et al. 2007), although our modeling approach greatly simplifies the biology (see below).
We consider virulence as the rate of disease-induced mortality in the host, and we assume that it is constant over the duration of the infection. We use this definition of virulence because our theory builds on previous theory for disease evolution, almost all of which equates virulence with disease-induced mortality (Day 2002). Our approach allows for time-dependent disease traits, but for simplicity we here assume that virulence is constant with respect to infection age for each genotype in this “proof-of-concept” example. While mortality does vary with infection age in P. chabaudi, genotypes tend to maintain their relative virulence rankings across all infection ages (i.e., a strain that is virulent at one infection age tends to be virulent at all infection ages). As a result, accounting for infection age dependence does not alter the qualitative results (Mideo et al, unpubl. data). Virulence was estimated using survival data from experimental infections. Specifically, we calculated a daily host mortality rate for each parasite genotype from the proportion of hosts surviving to the end of the experiment, under the assumption that the mortality rate is constant over time. Note that this procedure results in the virulence in C57 mice differing between the full and the truncated datasets because the duration of the experiment changes while the proportion of hosts surviving does not (because all host mortality occurred in the first eight days of infection).
MODELING APPROACH
The above laboratory experiments tracking parasite density provide an account of within-host disease dynamics of different parasite genotypes for the mouse-malaria system. The between-host disease dynamics, however, are more challenging to model because they involve potentially many aspects of parasite life history and host life history for which there are less empirical data. Here, we consider a simple SIR (Susceptible-Infected-Recovered) epidemiological model that assumes instantaneous mass action transmission. The model therefore does not account for all of the complexity of the malaria life cycle, such as the intricacies of sexual reproduction, or lags in between-host transmission due to development in the mosquito vector. Nevertheless it probably does capture some of the most important aspects of malaria transmission from the standpoint of disease life-history evolution. In particular, it captures the positive association between gametocyte levels within hosts and disease transmission (Robert et al. 1996; Collins and Jeffery 2003; Paul et al. 2007), under the assumption that gametocyte densities are proportional to asexual parasite densities. In principle, extending the epidemiological model to explicitly include vector dynamics poses no additional difficulty if such data were available.
Following the analyses of the theoretical examples in the companion paper (Day et al. 2011), we structure the analyses here into two parts: first, we study the evolution of transmission rate alone and, second, we study the joint evolution of transmission rate and virulence. Both analyses were performed on the two experimental datasets described above, and we take the perspective that the set of genotypes from each experiment represents a different parasite population.
Equation (10) from the companion paper describes the evolutionary dynamics of the within-host parasite density, as a function of infection age. Here, we use an analogous equation to model the evolutionary dynamics of transmission rate as a function of infection age, β(a). We use a discrete-time version of equation (10) in Day et al. (2011) because the data are collected in discrete time (i.e., each day during the infection) to account for the 24 hour life cycle of this parasite. In this case, the evolutionary dynamics of the average parasite transmission rate, denoted by the overbar, are given by
| (1) |
as a change per unit time. In the above expression, k is a combined measure of transmission and generation time of infections (Day et al. 2011). Roughly speaking, k is large when the product of the rate of generation of new infections and the generation time is large. q(s) describes the stable age distribution of infections, S is the number of susceptible hosts, and Gβ,β(a, s) describes any genetic autocovariance in transmission rates across infection ages. Specifically, Gβ,β(a, s) gives the autocovariance between transmission rate at age a and transmission at age s over the distribution of all parasite genotypes. Note that we have removed time dependencies of some of the parameters as compared with equation (10) of Day et al. (2011) because we assume the epidemiological dynamics is fast relative to evolution in the following examples.
The set of equations governing the joint evolutionary dynamics of the mean parasite transmission rate and mean virulence are given by the discrete-time analogues of equations 16 of the companion paper (Day et al. 2011):
| (2) |
| (3) |
where Gβ,v(a, s) and Gv,β(a, s), give the cross-covariances between transmission and virulence at infection ages a and s, with each matrix being the transpose of the other, and where σ(s) represents the reproductive value of an infection of age s. Gv,v is the autocovariance in virulence and, given that we have defined virulence as being constant across infection ages, each element of this matrix is identical and positive. Expressions for k and σ(s) are given in Appendix A.
We define a matrix to describe the transitions between infections of different ages. Formally, this is similar to a Leslie matrix and has the general form
| (4) |
where n is the maximum age of infection and μ is the background host mortality rate. The long-term growth rate of the infected host class is described by the dominant eigenvalue of this matrix, λ, and we note that q(s) in the earlier expressions is its associated eigenvector.
Epidemiological dynamics
One of our main aims is to demonstrate how changes in the within-host dynamics can interact with changes in the epidemiological dynamics to affect evolutionary trajectories. Therefore our analyses focus on predicting disease life-history evolution in two different epidemiological settings: an expanding epidemic and an endemic situation.
In an expanding epidemic, the number of infected hosts is increasing. We assume in the analyses below that these dynamics have reached a stable infection age distribution quickly, relative to the speed of evolutionary change. At this stable age distribution, the number of infected hosts in each infection age class grows at a rate given by the eigenvalue of L with the largest magnitude, that is, the dominant eigenvalue. The distribution of hosts in each infection-age class is then given by the eigenvector of L associated with the dominant eigenvalue. We set the number of susceptible hosts in the population, S, to 1000, allowing us to then define L. We calculate λ and q(s) as the dominant eigenvalue and associated eigenvector of L. We are then able to calculate all of the other required quantities in equation (1) or (2) and (3). We note that our choice of S = 1000 is arbitrary in this proof of concept analysis and choosing larger values of S increases the strength of selection on transmission and vice versa.
In an endemic scenario, the number of infected hosts eventually stabilizes. Thus, in our analysis, we assume that these epidemiological dynamics have stabilized quickly relative to the speed of evolutionary change. Mathematically, this occurs when the dominant eigenvalue of L is equal to 1. For a given set of estimates of average transmission rate, , average virulence, , and mortality rate, μ, there is a unique value of S that satisfies this requirement. This value of S can be solved for explicitly (Appendix A). For all simulations, S in the endemic case was between 100 and 200. We can then define L and calculate q(s) as the eigenvector associated with λ = 1. All other required quantities in equation (1) or (2) and (3) can then be calculated.
STATISTICAL METHODS
We accounted for any error in the measured infection dynamics using parametric bootstrapping. The process started by fitting a semiparametric time series model to log10-density parasite dynamics of each mouse (Wood 2001). In this system, observation error is normally distributed on the log-scale (Mideo et al. 2008b; Miller et al. 2010), and we used general additive models (GAMs) to represent parasite densities. This approach enables a good statistical description of the time series trends where there is no a priori expectation of the dynamics. GAMs use a modified objective function that includes a likelihood term measuring fidelity to the data, as well as a term that penalizes excessive curvature (Wood 2001; 2006). The parameter that defines the relative weightings of these terms was estimated from generalized cross-validation (Wood 2001). For individual mice with less than 12 data points, the GAM models tended to overfit the data, as evidenced by a pronounced drop in the standard errors of residuals relative to other mice. To circumvent this problem, we used cubic polynomial for time series with less than 12 data points. A reanalysis of all data using cubic polynomials yielded no qualitative difference.
Uncertainty in the estimated auto- and cross-covariance matrices, as well as the expected evolutionary dynamics, was then calculated using parametric bootstraps of the mean parasite dynamics. For each bootstrap replicate, we generated an artificial dataset from each of the fitted GAMs (i.e., a single parametric bootstrap draw from each replicate mouse of each parasite genotype). A single mean bootstrap time series for each parasite genotype was generated by averaging over mouse-specific time series. The set of mean bootstrap time series from all parasite genotypes was then used to create a single bootstrap G matrix, as well as the accompanying evolutionary dynamics. Upper and lower 95% confidence bands for the G matrices and evolutionary dynamics were then calculated based on 10,000 of these bootstrap replicates.
In the experiment with C57 mice, two individuals died before day 8 postinfection. For these individuals, when considering only the truncated dataset, there were not enough data points to fit even cubic polynomials (i.e., there were three or fewer data points). These infections were therefore not included in generating the mean parasite dynamics or covariance matrices for the truncated dataset.
All analyses and simulations were done in the R statistical environment (version 2.10.1; The R Foundation for Statistical Computing, 2009, http://www.R-project.org), and the GAMs were fit using the mgcv library (Wood 2006).
Results
EVOLUTION OF TRANSMISSION RATE
The patterns of autocovariance in transmission rates across infection ages arising from the different datasets are presented in Figure 2 (left panels). To interpret these figures, it may be helpful to consider the genetic variance–covariance matrices common in evolutionary quantitative genetics studies. Such matrices quantify the extent to which selection acting on one trait will indirectly result in evolutionary changes to other traits (Lande, 1979; Lande and Arnold 1983), where a “trait” in this case is measured as a point estimate. To represent these matrices graphically, a set of traits would lie along both the x and y axes. Values along the diagonal would then represent the variance in a given trait, whereas off-diagonal values would represent covariances between traits. It is these covariances that describe the genetic relationships between traits and determine correlated effects of selection. In an analogous way, if a single trait is dependent on age (or, indeed, another continuous variable), then that trait can covary with itself across different ages (e.g., Kirkpatrick and Heckman 1989; Kirkpatrick et al. 1990; Pletcher and Geyer 1999). The axes in Figure 2 (left panels) represent transmission at different infection ages. The values along the diagonal represent the variance in transmission rate at a given infection age, whereas the off-diagonals represent the covariance between transmission rates at two different infection ages. It is these covariances that will determine whether, and how strongly, selection acting on transmission at one age will indirectly affect the evolution of transmission at other infection ages.
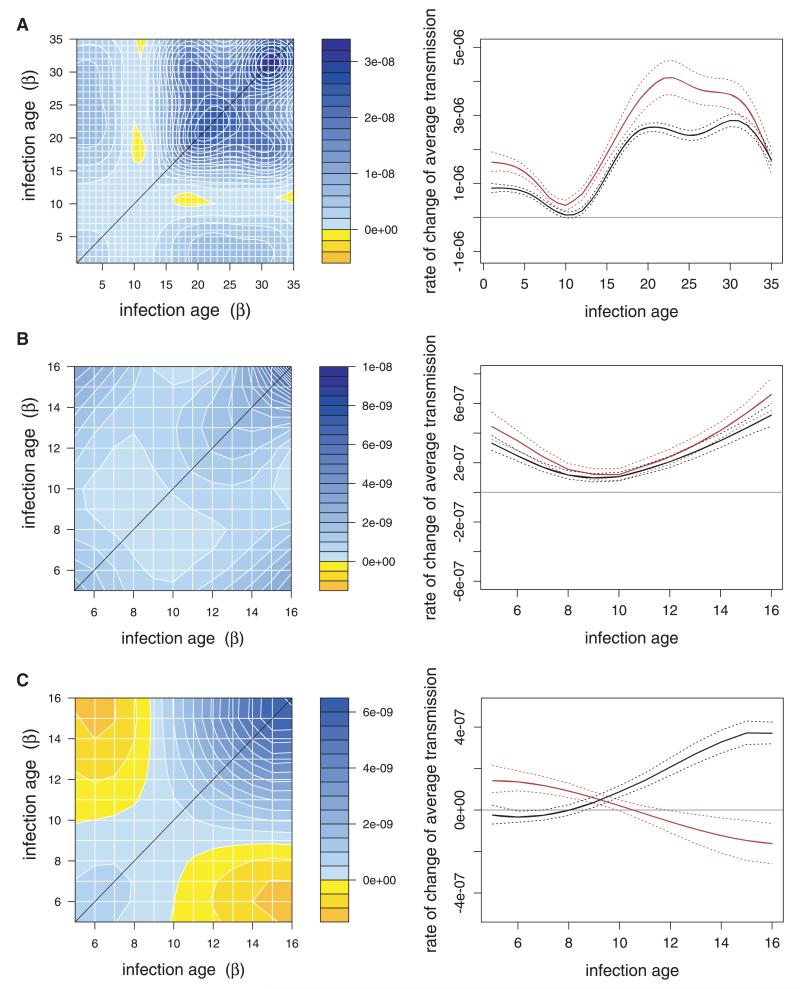
Figure 2. Autocovariance in transmission (Gβ,β) and predictions for the evolution of transmission rate for (A) parasites in C57 mice, given full infection data, (B) parasites in C57 mice, given truncated dataset, and (C) parasites in MF1 mice.
In the left column, x and y axes represent transmission rate at different infection ages. Values along the diagonal represent variance in transmission rate at a given infection age; values in the off-diagonals represent covariances between transmission rates at two different infection ages. For each pair of infection ages (each x–y combination) areas are either shaded blue or yellow to represent positive or negative covariances, respectively, between transmission rates at those ages. Values are in units of (per susceptible host per day)2. Note that the scales are different across the datasets. In the right column, red lines correspond to epidemic predictions, black lines correspond to endemic predictions. Solid lines represent mean predictions and dotted lines indicate confidence intervals on those predictions. Horizontal gray lines are where no evolutionary change is predicted; increased transmission is predicted above that line, and decreased transmission below it. Actual values represent change per day in transmission rate at different infection ages.
Qualitatively, the patterns of autocovariance across the three datasets are quite different and, consequently, so are the evolutionary predictions. For the full dataset in C57 mice, the autocovariance is nonnegative for most of the infection range; however, there are small areas of negative autocovariances roughly between infection ages (or days postinfection) 10 and 20, and between 10 and 35 (Fig. 2A). From the original data, this appears to be due to the fact that some genotypes reached peak parasite densities about two days later than others. This means that around infection age 10, those late-peaking genotypes have higher parasite densities, and consequently higher transmission rates, than the others with parasite densities that are already on the decline. Additionally, these same genotypes tend to have lower densities at later infection ages (e.g., genotype CW in Fig. 1A), resulting in the negative covariances between transmission at infection age 10 and later ages.
Responses to selection (or evolutionary trajectories) are determined by a combination of the strength of selection, genetic variation on which selection can act, and the genetic autocovariances (trade-offs) that constrain evolution by reducing other components of fitness (here, transmission at other infection ages). Predicted evolutionary responses (Fig. 2, right panels) reflect changes in the parasite population average transmission rate as compared to the average when all genotypes infect the host population at equal frequency. Equation (1) shows that the selection imposed under either epidemiological scenario will favor the evolution of increased transmission rates across all infection ages. Given the genetic covariances present in the full C57 data (Fig. 2A, left panel), the actual response to selection (Fig. 2A, right panel) will be constrained to some extent by the negative genetic covariances. In the epidemic case, most infections are young (Appendix B), making transmission at these ages evolutionarily most valuable. Furthermore, because there is an abundance of susceptible hosts in this case, the overall strength of selection for increased transmission is relatively large overall. As a result, we expect evolution to result in large increases in transmission rate at most infection ages. In the endemic case, the age distribution of infections shifts (Appendix B), making late transmission play an important role. In addition, the abundance of susceptible hosts is much reduced in this case, thereby reducing the overall strength of selection for increased transmission across infection ages. As a result, we predict an evolutionary increase in transmission at most infection ages, but the magnitude of the change is reduced as compared with the epidemic case. Also notice that, in both the epidemic and the endemic cases, very little evolutionary change in transmission at infection age 10 is predicted. This is due to the relative lack of genetic variance in transmission at this infection age, and the negative autocovariance between transmission at this age and transmission at ages 18–20. Evolutionary increases in transmission around ages 18–20 (for which there is substantial genetic variation) will tend to hamper evolutionary increases in transmission at infection age 10 because of this negative autocovariance, despite the fact that selection favors higher transmission rate at all infection ages (eq. 1).
In the truncated dataset with C57 mice, the autocovariance in transmission rates is nonnegative everywhere (Fig. 2B, left panel) because the dataset now does not contain the infection ages that negatively covaried with others. As a result, there is no genetic tradeoff between transmission rate at different infection ages. Nevertheless, we still predict relatively little evolutionary response to selection for increased transmission around infection age 10 because of the paucity of genetic variance for transmission at this age (Fig. 2B, right panel). Furthermore, the genetic variance in transmission rates across infection ages in the truncated dataset is relatively small compared with that at some infection ages in the full dataset (e.g., infection ages above 20). Consequently, the overall response of transmission to selection in the truncated dataset is predicted to be smaller. Finally, notice that the evolutionary response of transmission is again greater overall in the epidemic case than in the endemic case because the higher abundance of susceptible hosts increases the strength of selection.
The above results for infections in C57 mice differ in qualitatively interesting ways from the results for infections in MF1 mice (Fig. 2C). In MF1 mice there is a pronounced genetic trade-off between early and late transmission, as indicated by the negative autocovariance between transmission at infection ages 0 to 8 and transmission at infection ages 12–16 (i.e., the yellow areas in the Gβ,β plot, left panel). As a result, any evolutionary increase in transmission at early infection ages will tend to come at the expense of reduced transmission at late infection ages and vice versa, despite selection favoring increased transmission everywhere (eq. 1). Consequently, the qualitative evolutionary dynamics we predict (Fig. 2C, right panel) depend very strongly on the epidemiological dynamics. In the epidemic case, most infections are relatively young, making selection strongest at these infection ages. Thus, transmission at early infection ages is expected to evolve upwards at the expense of reduced transmission late in the infection. In the endemic case, however, the age distribution of infections is more uniform, making selection at later infections ages also important. In this case, the balance turns in the other direction, and transmission at late infection ages is predicted to evolve upwards at the expense of reduced transmission early in the infection.
EVOLUTION OF TRANSMISSION RATE AND VIRULENCE
To understand the evolutionary dynamics of transmission rates when virulence can also evolve, we have to consider the patterns of cross-covariance between the two traits (Fig. 3) in addition to the patterns of autocovariance in transmission (Fig. 2, left column). In Figure 3, the x-axis represents virulence at different infection ages, and the y-axis represents transmission at different infection ages. The values along the diagonal represent the covariance between transmission and virulence at a given infection age, whereas the off-diagonals represent the cross-covariance between transmission rate at a given infection age and virulence at a different age. The patterns of cross-covariance show that for each dataset a genetic trade-off between transmission and virulence emerges over some infection ages but not others, as indicated by the positive genetic covariances between the two for some infection age pairs (i.e., the darker blue areas of Fig. 3). Interestingly, there is even a negative genetic covariance between transmission at early infection ages and virulence in the dataset with MF1 mice (Fig. 3C), meaning that strains with high early transmission also tend to have lower virulence. This results from the fact that the genotype with the highest virulence has one of the lower transmission rates early on (see CR in Fig. 1C).
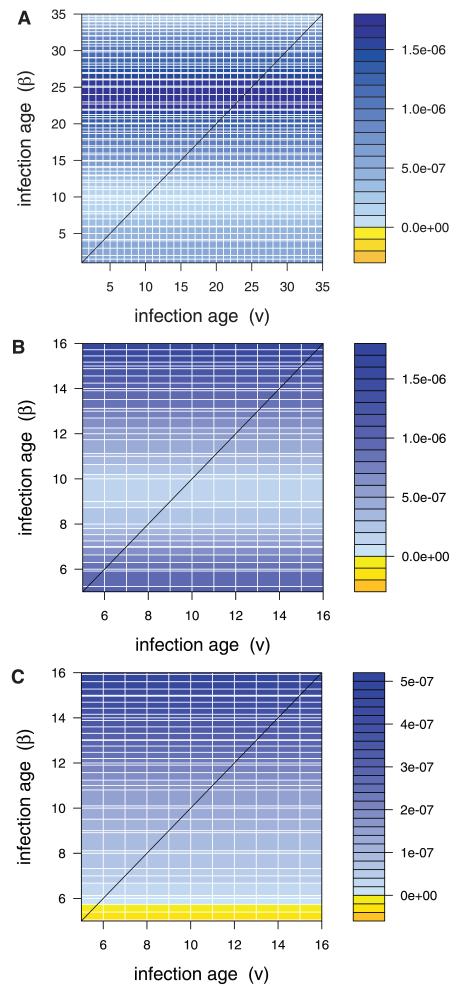
Figure 3. Cross-covariance between transmission and virulence, (Gβ,v), for (A) parasites in C57 mice, given full infection data, (B) parasites in C57 mice, given truncated dataset, and (C) parasites in MF1 mice.
The x-axis represents infection ages for virulence, and the y-axis represents infection ages for transmission. Values along the diagonal represent covariance between transmission rate and virulence at a given infection age; values in the off-diagonals represent cross-covariances between transmission rate at one infection age and virulence at another. For transmission at a given age of infection (i.e., a particular y-value), there is no variation in the value of the cross-covariance over the x-axis, because we assume that virulence is constant over age of infection. For virulence at a given age of infection (i.e., a particular x-value), there is variation over the y-axis, because transmission does change with age of infection. Actual values are in units of per susceptible host (per day)2.
The above patterns of cross-covariance reveal that, in C57 mice, there is predominately a positive genetic covariance between transmission and virulence across all infection ages. In other words, strains with high transmission rates tend also to induce high mortality rates, regardless of the infection ages of interest. As a result, once the evolutionary dynamics of virulence are also included, this dampens the extent to which transmission evolves when compared with the case where transmission is assumed to evolve independently (compare Fig. 2A/B with 4A/B). Notice that this is particularly apparent in the full dataset for transmission at infection age 25, where the positive genetic covariance between transmission and virulence is particularly strong (Figs. 3A and 4A).
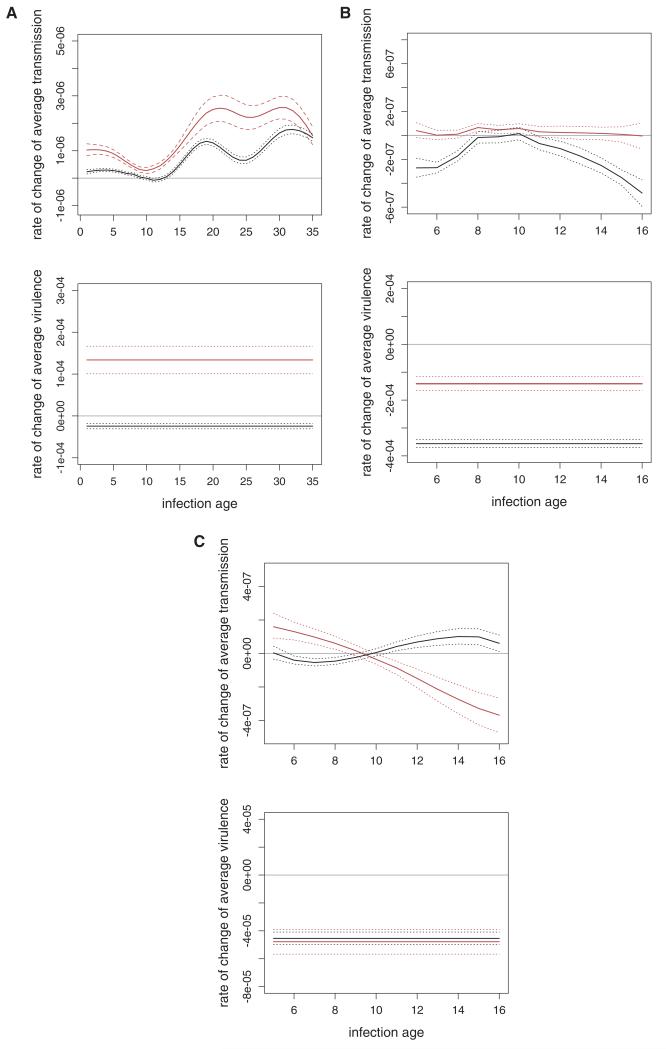
Figure 4. Evolutionary predictions for transmission rate (top graphs) and virulence (bottom graphs).
As before, (A) C57 mice, (B) C57 mice, truncated dataset, (C) MF1 mice. Red lines are predictions for epidemic conditions, black lines for endemic. Solid lines denote mean predictions and dotted lines denote confidence intervals. Actual values represent change per day in transmission rate or virulence at different infection ages. The scales in the top graphs are unchanged from Figure 2 to aid comparisons.
In the truncated dataset for C57 mice, the same general conclusions apply but the effect of including the evolutionary dynamics of virulence is even more pronounced. For example, in the endemic case, we predict that transmission will evolve to lower levels at most infection ages because of the trade-off between transmission and virulence. The reason for this outcome is the relatively small genetic variance in transmission across infection ages 6–16 in the truncated dataset, as compared with the genetic variance at ages 25+ in the full dataset (Fig. 2). In this case, the effect of direct selection for increased transmission cannot compensate for the effect of indirect selection that arises from selection for lower virulence. On the other hand, in the epidemic case for the truncated dataset, the strength of selection for increased transmission is larger owing to the higher abundance of susceptible hosts. As a result, even though there is again relatively little genetic variance in transmission rate in this case, the larger strength of direct selection is better able to counterbalance the effect of indirect selection on virulence. Indeed the two almost exactly cancel, resulting in essentially no evolutionary response in transmission rate at any infection age (Fig. 4B). These predictions can be qualitatively altered, for example, by changing the way that within-host parasite densities are mapped to transmission rates. Increasing this linear coefficient by one order of magnitude increases the genetic variation in transmission rate, though the qualitative patterns of auto- and cross-covariance remain unchanged. With this mapping, selection for increasing transmission can overcome the effect of indirect selection on virulence in the epidemic case, leading to the evolution of higher transmission rates (Appendix B, Figure B2). In contrast, the evolutionary predictions for transmission rates for the other two datasets are not qualitatively altered, since these are also constrained by negative covariances between transmission at different ages which also increase with this different mapping (Appendix B, Figure B2).
For the dataset in MF1 mice, the evolutionary predictions for transmission rate are qualitatively similar to the patterns we found in the one trait case, i.e., increasing transmission early at the cost of reduced transmission later in the epidemic case, and vice versa for the endemic case (Fig. 4C). The biggest quantitative difference in predictions that occurs when we allow virulence to evolve as well is that the response to selection for higher transmission at late ages in the endemic case is now weaker. This is due to the fact that the trade-off between transmission and virulence is strongest for later infection ages (Fig. 3C), meaning that any increase in transmission rate at these ages comes with a high associated cost of also increasing virulence. In the epidemic case, the predicted response to selection for higher transmission rates early is not constrained by this trade-off, since the cross-covariance between transmission at early ages and virulence is actually negative, i.e., the strains that have the highest transmission at early infection ages are least virulent. So, selection will favor these strains, resulting in average transmission rates increasing at early ages, at the cost of decreasing transmission rates at later ages, and with the correlated effect of decreasing virulence.
Discussion
We have shown that the function-valued trait approach for studying disease life-history evolution (Day et al. 2011) can be applied to real data. Unlike traditional approaches for studying disease evolution that nest within-host models into between-host epidemiological frameworks, this approach characterizes the within-host dynamics through covariance functions measured at the level of the host. Although this approach circumvents much of the need for a mechanistic understanding of within-host processes, it still requires a considerable amount of high-quality infection data. Specifically, it requires long-term data collection that covers as much of the normal course of infection as is logistically possible (because evolutionary predictions were altered with the truncated time series), and it requires data from replicated infections with a number of different parasite genotypes to capture as much of the natural genetic variation as possible.
We have used two comprehensive within-host infection datasets (Bell et al. 2006; Reece et al. 2008) to demonstrate the function-valued trait approach. However, there are still some shortcomings with these data; in particular, definitions of disease life-history traits were constrained by available measures. Ideally, for malaria, one would have actual measures of transmission to mosquitoes as a function of infection age (or within-host gametocyte densities as a reasonable proxy for transmission potential) as well as infection-age dependent mortality rates. Further, our results demonstrate that evolutionary predictions can depend on how these host-level measures are mapped to disease life-history traits, even when only the magnitudes of the variance–covariance functions change with different mappings and not the qualitative patterns (Appendix B, Figure B2).
Despite not having sufficiently complete datasets to make broad conclusions about malaria evolution, we have still uncovered some biologically interesting findings about disease life history evolution more generally. First, our results show that the genetic covariance functions necessary for making evolutionary predictions vary even within the same host–parasite system (compare Fig. 2B,C and Fig. 3B,C). The qualitatively different patterns of covariance in transmission and virulence apparent in the two datasets could be due to a difference in the hosts used in each experiment (e.g., different strains, different sex, likely different initial sizes and ages, etc.) or it could be that the empirically estimated covariance functions are not very repeatable across experiments. Unfortunately, with these data, we simply cannot discern between these two possibilities. However, the differences we observed, presumably a small subset of those possible, are important: differences between the two studies produced qualitatively different patterns of within-host dynamics that affect evolution (see also Reece et al. 2009). This work therefore sets the stage for asking which host, parasite, and environmental factors alter evolutionary trajectories of disease life history. Of particular interest would be precisely replicated datasets from experiments that manipulate a range of factors. For example, quantifying the relative importance of host heterogeneity in systems where host genetics can be controlled will be important for determining the ability of this modeling framework to make accurate predictions for wild systems. Once there is a clearer idea of the factors that generate differences in genetic covariances and, therefore, of the stability and repeatability of the genetic covariances involved, it will be easier to interpret analogous experimental data. Experiments that test the effects of interventions on dynamics, for example, with vaccinated and unvaccinated hosts, or other manipulations that mimic the natural variation in hosts that parasites might be exposed to in real infections, for example, immunocompromised versus immunocompetent, and anemic versus “healthy,” offer important opportunities to study disease life-history evolution.
Second, our results provide new perspectives on virulence evolution theory. A main premise of this theory is that diseases will evolve intermediate levels of virulence because of a trade-off between this trait and transmission (e.g., Anderson and May 1982; Ewald 1983). Although discussions of this hypothesis are not always cast in genetic terms, such trade-offs amount to patterns of genetic covariance that constrain evolution. The predominately positive covariance between virulence and transmission across most infection ages documented here therefore supports this hypothesis, and it also matches previous conclusions from similar data that were analyzed differently (Mackinnon and Read 1999). At the same time, however, the age-specific patterns of covariance presented here reveal important, and previously unappreciated, subtleties to the trade-off hypothesis. Data to support this trade-off hypothesis have been limited (reviewed in Lipsitch and Moxon 1997; Alizon et al. 2009). Alizon et al. (2009) discuss the many likely challenges to empirically demonstrating that such a trade-off exists, but one possibility not discussed is that any such trade-off may be obscured by the constraints acting on a single trait alone. For example, our analyses clearly demonstrate that even when a genetic virulence-transmission trade-off exists (i.e., a positive covariance between the two traits), selection can increase transmission over some infection ages without simultaneously increasing virulence (e.g., the patterns in Figure 4a and c). This could be interpreted as undermining the trade-off hypothesis, however we have shown that it is actually the result of trade-offs between transmission rate across infection ages (i.e., negative covariances between transmission at different ages). When there are trade-offs within transmission across ages, as may be expected, for example, if high parasite densities and high transmission early induce immune response that later clear parasites, then this alone can constrain evolution toward ever higher transmission rates. The resulting patterns of evolutionary change in transmission rates across infection ages will depend on the epidemiological dynamics, because these determine the strength of selection at different infection ages.
These same “within-trait” constraints on evolution could also occur for virulence or recovery (although we do not currently have the data to address this question). For example, some parasites may display intermediate levels of virulence, not because there is a trade-off between virulence and transmission, but because there are trade-offs within virulence alone, and therefore there are no strains that have zero virulence at all infection ages. It is likely that in natural infections, trade-offs exist both within and between traits. As a result, it is perhaps not surprising that simple (i.e., nonfunction-valued) trade-offs are sometimes difficult to find—the nature of the trade-off can be much more subtle and complex once infection age is accounted for and infections are treated as the dynamic phenomena that they are.
The approach we demonstrate here is similar to analyses of function-valued traits in quantitative genetics (e.g., Kirkpatrick and Heckman 1989; Kirkpatrick et al. 1990; 1994; Pletcher and Geyer 1999). A considerable amount of work in that field has focused on developing methods for describing estimated covariance matrices with functions (reviewed in Jaffrézic and Pletcher 2000; Kingsolver et al. 2001; Meyer and Kirkpatrick 2005). Using G functions, rather than estimated G matrices, circumvents problems that arise when traits are measured at irregular intervals (Pletcher and Geyer 1999) and also takes into account any measurement error in the data (Kirkpatrick and Heckman 1989). Further, the G functions themselves can be used directly to identify the types of evolutionary change for which there is (and is not) substantial genetic variation available (Kirkpatrick and Heckman 1989; Kirkpatrick et al. 1990). In contrast, we make explicit predictions for evolutionary change under different selection regimes, as determined by epidemiological dynamics. Our approach accounts for measurement error at the level of the data: by fitting spline functions to the original data (i.e., the traits themselves), we are able to simulate artificial datasets by incorporating the known error structure of these measured data (Mideo et al. 2008b; Miller et al. 2010). This also allows us to generate data for those infection ages where experimental measurements are missing. Each set of simulated data generates a particular set of parasite genotype average traits, a particular set of G matrices, and a particular set of evolutionary predictions. By repeating this process many times we are able to estimate the variation in the G matrices (not shown) and the evolutionary predictions (shown as confidence bands in Figures 2 and 4) that arises due to error in the original experimental measurements. Given the discrete life cycle of malaria parasites, it makes sense to think of infections (and parasite traits) in discrete time steps, however, there is no reason why future applications could not use standard quantitative genetic approaches to fit functions to measured G matrices, and then use these functions to generate evolutionary predictions with the equations we develop in the companion paper (Day et al. 2011).
Approaches that account for the complex functional relationships between traits are rapidly developing (e.g., Blows and Hoffmann 2005; Walsh and Blows 2009; Agrawal and Stinchcombe 2009). Such comprehensive approaches are important, given the long-standing recognition that evolutionary trajectories of particular traits can be altered when functionally related traits are ignored (Lande and Arnold 1983). Our results reiterate this point: evolutionary predictions for transmission can be altered when we allow virulence to evolve and account for the genetic relationships between these two traits. Standard evolutionary quantitative genetic studies may be confronted with large numbers of correlated life-history traits contributing to survival and fecundity, and a main goal is determining which of these are the most important (e.g., Hine and Blows 2006). Studies of disease life history evolution may be slightly less complicated because only three traits—transmission, virulence, and recovery—contribute to a disease’s “surivival” and “fecundity”. Disease systems for which these traits are directly measurable offer exciting possibilities for studying the joint evolution of all three traits.
Supplementary Material
Table 1. Estimates of virulence of different parasite genotypes in different host backgrounds.
For the C57 mice, the estimates in parentheses represent virulences for the truncated dataset.
ACKNOWLEDGEMENTS
We thank M. Morrissey, D. Nussey and A. Wilson for discussion; two anonymous reviewers for helpful suggestions on the manuscript; and the Wellcome Trust (NM, SR, AB, AR), an NSERC Discovery Grant (WN), an NSERC Steacie Fellowship and the Canada Research Chairs Program (TD) for funding.
LITERATURE CITED
- Agrawal AF, Stinchcombe JR. How much do genetic covariances alter the rate of adaptation? Proc. R. Soc. B. 2009;276:1183–1191. doi: 10.1098/rspb.2008.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizon S, van Baalen M. Emergence of a convex trade off between transmission and virulence. Am. Nat. 2005;165:E155–E167. doi: 10.1086/430053. [DOI] [PubMed] [Google Scholar]
- Alizon S, van Baalen M. Multiple infections, immune dynamics, and the evolution of virulence. Am. Nat. 2008;172:E150–E168. doi: 10.1086/590958. [DOI] [PubMed] [Google Scholar]
- Alizon S, Hurford A, Mideo N, van Baalen M. Virulence evolution and the trade off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 2009;22:245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- André JB, Ferdy JB, Godelle B. Within-host parasite dynamics, emerging trade off, and evolution of virulence with immune system. Evolution. 2003;57:1489–1497. doi: 10.1111/j.0014-3820.2003.tb00357.x. [DOI] [PubMed] [Google Scholar]
- André JB, Gandon S. Vaccination, within-host dynamics, and virulence evolution. Evolution. 2006;60:13–23. [PubMed] [Google Scholar]
- Antolin MF. Unpacking β: Within-host dynamics and the evolutionary ecology of pathogen transmission. Annu. Rev. Ecol. Evol. Syst. 2008;39:415–437. [Google Scholar]
- van Baalen M, Sabelis MW. The dynamics of multiple infection and the evolution of virulence. Am. Nat. 1995;146:881–910. [Google Scholar]
- Barnes KI, White NJ. Population biology and antimalarial resistance: the transmission of antimalarial drug resistance in plasmodium falciparum. Acta. Trop. 2005;94:230–240. doi: 10.1016/j.actatropica.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Bell AS, de Roode JC, Sim D, Read AF. Within-host competition in genetically diverse malaria infections: Parasite virulence and competitive success. Evolution. 2006;60:1358–1371. [PubMed] [Google Scholar]
- Blows MW, Hoffmann AA. A reassessment of genetic limits to evolutionary change. Ecology. 2005;86:1371–1384. [Google Scholar]
- Bremermann HJ, Pickering J. A game-theoretical model of parasite virulence. J. Theor. Biol. 1983;100:411–426. doi: 10.1016/0022-5193(83)90438-1. [DOI] [PubMed] [Google Scholar]
- Collins WE, Jeffery GM. A retrospective examination of mosquito infection on humans infected with Plasmodium falciparum. Am. J. Trop. Med. Hyg. 2003;68:366–371. [PubMed] [Google Scholar]
- Coombs D, Gilchrist MA, Ball CL. Evaluating the importance of within- and between-host selection pressures on the evolution of chronic pathogens. Theor. Popul. Biol. 2007;72:576–591. doi: 10.1016/j.tpb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Day T. Parasite transmission modes and the evolution of virulence. Evolution. 2001;55:2389–2400. doi: 10.1111/j.0014-3820.2001.tb00754.x. [DOI] [PubMed] [Google Scholar]
- Day T. On the evolution of virulence and the relationship between various measures of mortality. Proc. R. Soc. B. 2002;269:1317–1323. doi: 10.1098/rspb.2002.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T. Virulence evolution and the timing of disease life-history events. Trends. Ecol. Evol. 2003;18:113–118. [Google Scholar]
- Day T, Alizon S, Mideo N. Bridging scales in the evolution of infectious disease life histories: Theory. 2011 doi: 10.1111/j.1558-5646.2011.01394.x. (in press) [DOI] [PubMed] [Google Scholar]
- Ewald PW. Host-parasite relations, vectors, and the evolution of disease severity. Ann. Rev. Ecol. Syst. 1983;14:465–485. [Google Scholar]
- Frank S. Models of parasite virulence. Quart. Rev. Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- Ganusov V, Bergstrom C, Antia R. Within-host population dynamics and the evolution of microparasites in a heterogeneous host population. Evolution. 2002;56:213–223. doi: 10.1111/j.0014-3820.2002.tb01332.x. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, Coombs D. Evolution of virulence: Interdependence, constraints, and selection using nested models. Theor. Popul. Biol. 2006;69:145–153. doi: 10.1016/j.tpb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, Sasaki A. Modeling host–parasite coevolution: a nested approach based on mechanistic models. J. Theor. Biol. 2002;218:289–308. doi: 10.1006/jtbi.2002.3076. [DOI] [PubMed] [Google Scholar]
- Grant AJ, Restif O, McKinley TJ, Sheppard M, Maskell D, Mastroeni P. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol. 2008;6:e74. doi: 10.1371/journal.pbio.0060074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine E, Blows MW. Determining the effective dimensionality of the genetic variance-covariance matrix. Genetics. 2006;173:1135–1144. doi: 10.1534/genetics.105.054627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrézic F, Pletcher SD. Statistical models for estimating the genetic basis of repeated measures and other function-valued traits. Genetics. 2000;156:913–922. doi: 10.1093/genetics/156.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsolver JG, Gomulkiewicz R, Carter PA. Variation, selection and evolution of function-valued traits. Genetica. 2001;112-113:87–104. [PubMed] [Google Scholar]
- Kirkpatrick M, Heckman N. A quantitative genetic model for growth, shape, reaction norms, and other infinite-dimensional characters. J. Math. Biol. 1989;27:429–450. doi: 10.1007/BF00290638. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Hill WG, Thompson R. Estimating the covariance structure of traits during growth and ageing, illustrated with lactation in dairy cattle. Genet. Res. 1994;64:57–69. doi: 10.1017/s0016672300032559. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Lofsvold D, Bulmer M. Analysis of the inheritance, selection and evolution of growth trajectories. Genetics. 1990;124:979–993. doi: 10.1093/genetics/124.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Quantitative genetic analysis of multivariate evolution, applied to brain: body allometry. Evolution. 1979;33:402–416. doi: 10.1111/j.1558-5646.1979.tb04694.x. [DOI] [PubMed] [Google Scholar]
- Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Levin S, Grenfell B, Hastings A, Perelson A. Mathematical and computational challenges in population biology and ecosystem science. Science. 1997;275:334–343. doi: 10.1126/science.275.5298.334. [DOI] [PubMed] [Google Scholar]
- Lipsitch M, Moxon E. Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol. 1997;5:31–37. doi: 10.1016/S0966-842X(97)81772-6. [DOI] [PubMed] [Google Scholar]
- Mackinnon MJ, Read AF. Genetic relationships between parasite virulence and transmission in the rodent malaria Plasmodium chabaudi. Evolution. 1999;53:689–703. doi: 10.1111/j.1558-5646.1999.tb05364.x. [DOI] [PubMed] [Google Scholar]
- Meyer K, Kirkpatrick M. Up hill, down dale: quantitative genetics of curvaceous traits. Phil. Trans. R. Soc. B. 2005;360:1443–1455. doi: 10.1098/rstb.2005.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mideo N, Alizon S, Day T. Linking within- and between-host dynamics in the evolutionary epidemiology of infectious diseases. Trends. Ecol. Evol. 2008a;23:511–517. doi: 10.1016/j.tree.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Mideo N, Barclay VC, Chan BHK, Savill NJ, Read AF, Day T. Understanding and predicting strain-specific patterns of pathogenesis in the rodent malaria Plasmodium chabaudi. Am. Nat. 2008b;172:E214–E238. doi: 10.1086/591684. [DOI] [PubMed] [Google Scholar]
- Mideo N, Day T, Read AF. Modelling malaria pathogenesis. Cell. Microbiol. 2008c;10:1947–1955. doi: 10.1111/j.1462-5822.2008.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Råberg L, Read AF, Savill NJ. Quantitative analysis of immune response and erythropoiesis during rodent malarial infection. PLoS Comput. Biol. 2010;6:e1000946. doi: 10.1371/journal.pcbi.1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul REL, Bonnet S, Boudin C, Tchuinkam T, Robert V. Aggregation in malaria parasites places limits on mosquito infection rates. Infect. Genet. Evol. 2007;7:577–586. doi: 10.1016/j.meegid.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Pletcher SD, Geyer CJ. The genetic analysis of age-dependent traits: modeling the character process. Genetics. 1999;153:825–835. doi: 10.1093/genetics/153.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece S, Ramiro RS, Nussey DH. Plastic parasites: sophisticated strategies for survival and reproduction? Evol. Appl. 2009;2:11–23. doi: 10.1111/j.1752-4571.2008.00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece SE, Drew DR, Gardner A. Sex ratio adjustment and kin discrimination in malaria parasites. Nature. 2008;453:609–615. doi: 10.1038/nature06954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V, Read AF, Essong J, Tchuinkam T, Mulder B, Verhave JP, Carnevale P. Effect of gametocyte sex ratio on infectivity of Plasmodium falciparum to Anopheles gambiae. Trans. R. Soc. Trop. Med. Hyg. 1996;90:621–624. doi: 10.1016/s0035-9203(96)90408-3. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Iwasa Y. Optimal-growth schedule of pathogens within a host: switching between lytic and latent cycles. Theor. Popul. Biol. 1991;39:201–239. doi: 10.1016/0040-5809(91)90036-f. [DOI] [PubMed] [Google Scholar]
- Sinden RE, Dawes EJ, Alavi Y, Waldock J, Finney O, Mendoza J, Butcher GA, Andrews L, Hill A, Gilbert SC, et al. Progression of Plasmodium berghei through Anopheles stephensi is density-dependent. PLoS Pathog. 2007;3:e195. doi: 10.1371/journal.ppat.0030195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, McKenzie FE, Snow RW, Hay SI. Revisiting the basic reproductive number for malaria and its implications for malaria control. PLoS Biol. 2007;5:e42. doi: 10.1371/journal.pbio.0050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh B, Blows MW. Abundant genetic variation + strong selection = multivariate genetic constraints: a geometric view of adaptation. Annu. Rev. Ecol. Syst. 2009;40:41–59. [Google Scholar]
- Wargo AR, de Roode JC, Huijben S, Drew DR, Read AF. Transmission stage investment of malaria parasites in response to in-host competition. Proc. R. Soc. B. 2007;274:2629–2638. doi: 10.1098/rspb.2007.0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S. Partially specified ecological models. Ecol. Monogr. 2001;71:1–25. [Google Scholar]
- Wood S. Generalized Additive Models: An Introduction with R. Chapman & Hall/CRC; Boca Raton, USA: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.