ABSTRACT
The phosphoinositide 3-kinase (PI3K) family is important to nearly all aspects of cell and tissue biology and central to human cancer, diabetes and aging. PI3Ks are spatially regulated and multifunctional, and together, act at nearly all membranes in the cell to regulate a wide range of signaling, membrane trafficking and metabolic processes. There is a broadening recognition of the importance of distinct roles for each of the three different PI3K classes (I, II and III), as well as for the different isoforms within each class. Ongoing issues include the need for a better understanding of the in vivo complexity of PI3K regulation and cellular functions. This Cell Science at a Glance article and the accompanying poster summarize the biochemical activities, cellular roles and functional requirements for the three classes of PI3Ks. In doing so, we aim to provide an overview of the parallels, the key differences and crucial interplays between the regulation and roles of the three PI3K classes.
KEY WORDS: Class I PI3K, Class II PI3K, Class III PI3K, PI3-kinases, PI3K, Phosphoinositide
Introduction
This Cell Science at a Glance article and the accompanying poster provide a broad overview of the current knowledge and emerging themes related to the important phosphoinositide 3-kinase (PI3K) family. Although widely appreciated for crucial roles in cell signaling or membrane trafficking, new research directions continue to reveal important complexities in specific PI3K class regulation and functions. Here, we will not elaborate on the details of specific pathways or processes that engage individual PI3K subfamily members in normal and human disease states, which have been reviewed in depth elsewhere. We instead will highlight the similarities and differences in the molecular-cellular requirements for each of three PI3K classes (I, II and III).
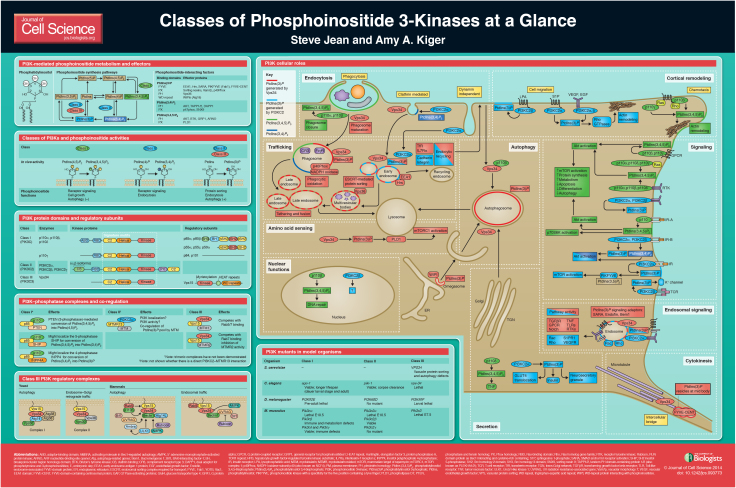
Class I, II and III PI3Ks synthesize three distinct phosphoinositides
Most PI3K functions are mediated by phosphoinositides, the low-abundance phosphorylated forms of phosphatidylinositol (PtdIns) (see poster) (Di Paolo and De Camilli, 2006; Sasaki et al., 2009). All three PI3K classes (I, II and III) phosphorylate the 3′-position hydroxyl of the D-myo-inositol head group to generate specific phosphoinositide forms (see poster) (Vanhaesebroeck et al., 2010a). In vitro, all classes can generate phosphatidylinositol 3-phosphate [PtdIns(3)P], class I and II can synthesize phosphatidylinositol (3,4)-bisphosphate [PtdIns(3,4)P2], and only class I can produce phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3]. An understanding of more-selective in vivo PI3K activities has been derived from approaches that combine the use of genetic mutants or pharmacological inhibitors with phosphoinositide analysis by chromatography, microscopy imaging of fluorescent biosensors and epistasis with specific phosphoinositide 3-phosphatases. In vivo, there is significant support for class I PI3K synthesis of PtdIns(3,4,5)P3 [and indirectly, PtdIns(3,4)P2], class III PI3K synthesis of PtdIns(3)P, and to a lesser extent, class II PI3K synthesis of PtdIns(3)P and PtdIns(3,4)P2 (see poster) (Backer, 2008; Vanhaesebroeck et al., 2010a; Falasca and Maffucci, 2012; Posor et al., 2013; Schink et al., 2013). Despite their overlapping selectivities, the different PI3Ks exhibit non-redundant functions in cells and animals, as discussed below. The PI3K family appears restricted to eukaryotes, and only the class III PI3K is conserved from yeast to human (Brown and Auger, 2011). In contrast, class I and II PI3Ks have evolved conserved multidomains, distinct adaptors and expanded targets that are relevant to multicellular life (Engelman et al., 2006).
PI3K phosphoinositide functions are mediated by effector recruitment
A common role for phosphoinositides is the recruitment of effector proteins through phosphoinositide-binding protein domains. The best-characterized 3-phosphoinositide-binding domains are FYVE and a subset of PX and PH domains (Lemmon, 2008) (see poster). The relatively low phosphoinositide-binding affinity of these domains in combination with 3-phosphoinositide phosphatase activities permits highly reversible effector localization and responses. In addition, ‘coincidence detection’ of both a phosphoinositide and another membrane-localized protein can further promote effector specificity (Di Paolo and De Camilli, 2006; Jean and Kiger, 2012). There is a broad range of known phosphoinositide-regulated effector functions. One category of effectors directs localized membrane remodeling events, such as membrane tubulation, fusion, fission or transport. This is seen with the endosomal sorting and tubulation performed by the PtdIns(3)P-binding PX-domain-containing sorting nexins (SNXs) (Cullen, 2008). A second category of effectors mediates membrane-localized signaling. Examples of this are signal transduction at the plasma membrane via the PtdIns(3,4,5)P3-binding PH-domain-containing proteins Akt and Grp1 (Hawkins et al., 2006). PI3K phosphoinositide activities can also serve other roles by locally modifying the biophysical properties of membranes, such as electrostatic interactions or a less well-understood function in regulating membrane curvature (McLaughlin and Murray, 2005; Lemmon, 2008).
PI3K protein domains and regulatory subunits
All PI3Ks possess a ‘PI3K signature motif’ that is composed of a C2 domain, which likely binds membranes, a helical domain and the catalytic kinase domain (see poster) (Vanhaesebroeck et al., 2010a). The classification of PI3Ks into the three different classes is based mainly on the presence of additional protein domains and their interactions with regulatory subunits. The nomenclature of the PI3K subunits is shown in Table 1.
Table 1. Nomenclature of PI3K subunits.
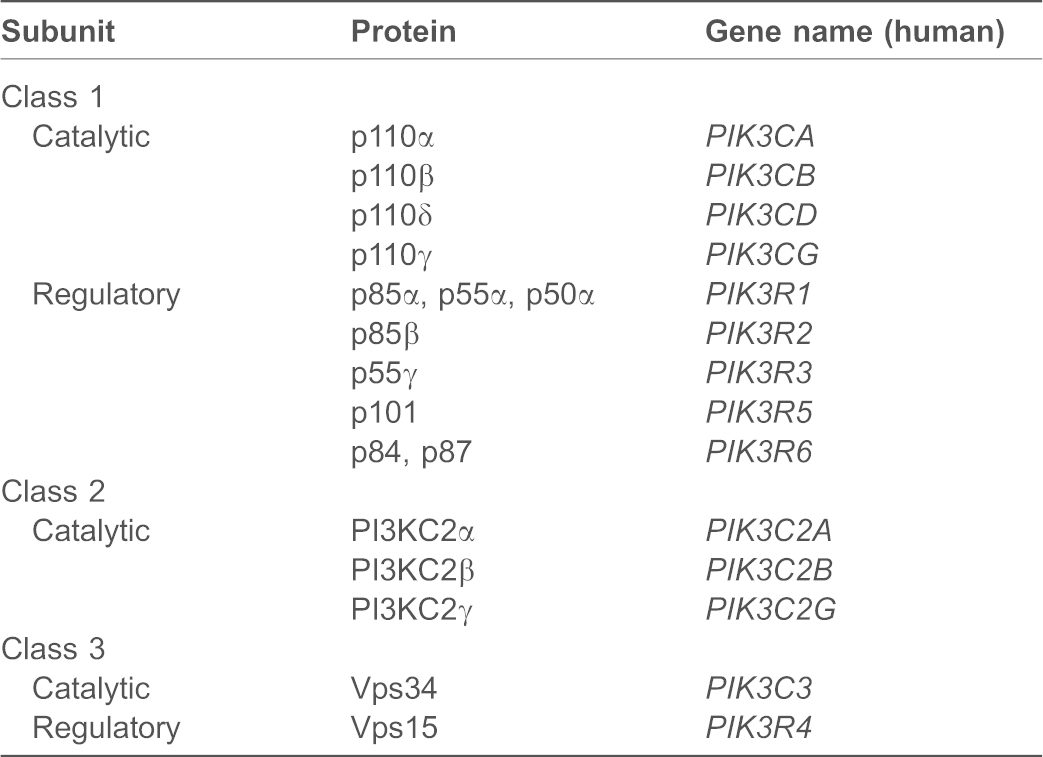
The class I PI3K subfamily comprises four members in vertebrates (see poster), only one member in worm and fly, and there are none in yeast (Hawkins et al., 2006; Brown and Auger, 2011). Class I PI3Ks function as heterodimers consisting of one of four catalytic p110 subunits (p110α, β, δ or γ) and a regulatory subunit [p85α (or its splice variants p55α and p50α), p85β, p55γ, p101 or p84]. (Vanhaesebroeck et al., 2010a; Vadas et al., 2011). There are two major classes of regulatory subunits, each represented by alternative isoforms. Alternative p85 regulatory subunits (p85, p55, p50), which each harbor two Src homology 2 domains (nSH2, cSH2) and an intervening p110-binding region (iSH2), constitutively interact with the p110α, p110β and p110δ catalytic subunits through an N-terminal adaptor-binding domain (ABD). Acting as an adaptor, p85 recruits the complex to phosphorylated tyrosine commonly downstream of activated receptor tyrosine kinases (RTKs). p85 also negatively regulates the kinase activity of p110α through a helical domain interaction, with important effects in cancer (Vadas et al., 2011). In contrast, p110γ does not have a clear p85-binding domain. Instead, p110γ heterodimers form with the regulatory subunits p101 or p84 that are devoid of SH2 domains and are almost exclusively activated by G protein-coupled receptors (GPCRs). Class I PI3Ks also harbor a Ras-binding domain (RBD) in the N-terminal extension, and p110α, p110δ and p110γ are each stimulus-dependent Ras effectors (Vanhaesebroeck et al., 2010a). In contrast, the p110β RBD interacts with Rab5 GTPase (Christoforidis et al., 1999) and Rho GTPase family members (Fritsch et al., 2013), and specifically, Rac potentiates p110β GPCR responses (Guillermet-Guibert et al., 2008; Dbouk et al., 2012; Fritsch et al., 2013).
The class II PI3K (PI3KC2) subfamily has three members in vertebrates (see poster), only one member in worm and fly, and also none in yeast (Brown and Auger, 2011; Falasca and Maffucci, 2012). This class has additional domains in both N- and C-terminal extensions. There is no known obligatory regulatory subunit, but the class II enzymes interact with proteins that could serve adaptor functions. PI3KC2α and PI3KC2β contain an N-terminal clathrin-binding (CB) region, suggesting a link with clathrin-coated vesicles. The PI3KC2α N-terminal region appears to inhibit kinase activity, which can be released by clathrin binding (Gaidarov et al., 2001), and PI3KC2α has been implicated in clathrin-mediated endocytosis (Posor et al., 2013). The PI3KC2β N-terminus also binds the scaffold protein intersectin, which promotes increased PtdIns(3)P synthesis (Das et al., 2007). Unlike the PI3KC2α and -β isoforms, PI3KC2γ protein interactions have not been tested. We identified a scaffold for the 3-phosphoinositide phosphatase myotubularin (MTM) as a possible PI3KC2 adaptor, as discussed below (Jean et al., 2012). Class II PI3Ks also harbor a Ras-binding domain (RBD), although these signaling inputs are not well characterized. All three PI3KC2 subfamily members possess a unique C-terminal extension that carries a C2 domain and a PX domain that preferentially binds to phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] (Stahelin et al., 2006).
A single class III PI3K is conserved in all eukaryotes (see poster) (Backer, 2008). This enzyme was first identified as vacuolar protein sorting 34 (Vps34), the sole PI3K in yeast (Schu et al., 1993). Vps34 binds to the adaptor protein Vps15 which is N-terminally myristoylated and regulates the intracellular membrane localization of Vps34 and its activity (Backer, 2008). Vps15 also engages with other key membrane proteins, such as the Rab5 GTPase to coordinate Vps34 activity at endosomes (Christoforidis et al., 1999). Vps34 does not contain known protein domains outside of the ‘PI3K signature motif’, but it engages in a growing list of protein interactions that regulate distinct PtdIns(3)P pools (see below) (Simonsen and Tooze, 2009; Kim et al., 2013).
PI3K and phosphatase co-regulation
An emerging theme is the co-regulation of specific PI3Ks and PtdIns-phosphatases through shared adaptor protein interactions (see poster) (Jean and Kiger, 2012). Although first identified as a p110 regulatory subunit, p85 has since been shown to also bind, regulate share phenotypes with the antagonizing phosphatase and tensin homologue (PTEN) 3-phosphatase (Chagpar et al., 2010). In this way, p85 reversibly regulates the conversion of PtdIns(4,5)P2 to PtdIns(3,4,5)P3. p85 also interacts with the 5-phosphatase SHIP (Jackson et al., 1995) and with the 4-phosphatase INPP4 (Munday et al., 1999), which have possible significance in the successive endosomal conversion of class I PI3K PtdIns(3,4,5)P3 to PtdIns(3,4)P2 and PtdIns(3)P, respectively (Ivetac et al., 2005; Shin et al., 2005). Likewise, we identified a conserved physical interaction between class II PI3K and MTMR13/Sbf, an adaptor for 3-phosphoinositide phosphatase MTMR2/Mtm, and we also found that all these components functionally co-regulate a PtdIns(3)P pool in endosomal trafficking (Jean et al., 2012). Finally, the Vps15 adaptor for class III Vps34 was identified to exist in a trimeric complex with the 3-phosphatases MTM1 or MTMR2 that compete with Rab5 and Rab7 for Vps15 binding (Cao et al., 2007). These parallels suggest that shared kinase–phosphatase adaptor interactions provide a tight spatiotemporal control of distinct phosphoinositide pools and thus of their specific cellular functions.
PI3K cellular functions and pathways
The collective cellular and organismal functions for the PI3K family extend into all parts of the cell, cell-types and developmental stages (see poster). Each PI3K class has multiple cellular roles through the regulation of distinct phosphoinositide pools. The direct roles of PI3Ks can be categorized predominantly as acting in cell signaling (class I, II) or membrane trafficking (class II, III). Although not yet widely addressed, members of different PI3K classes can act at successive steps in shared pathways and processes (Dou et al., 2010; Lu et al., 2012). There are also emerging descriptions of PI3K localization and/or roles in the nucleus (Kumar et al., 2011). In addition, there are many indirect consequences of these diverse PI3K functions. Below is a brief overview of the cellular roles and pathways depicted on the poster and as reviewed elsewhere.
Class I PI3Ks
There is a rich literature on the functions for this founding class of the PI3K family. Numerous growth factor pathways are under the control of activated RTKs or GPCRs that recruit p85–p110 complexes to the plasma membrane, where upon relief of p85 inhibition, p110 converts PtdIns(4,5)P2 in to PtdIns(3,4,5)P3 to elicit signaling responses (Vanhaesebroeck et al., 2010a). Notably, PtdIns(3,4,5)P3 recruits the kinase Akt, which controls multiple pathways (including activation of mTORC1, FOXO and others), to regulate cell growth, proliferation, survival, metabolism and autophagy (Vanhaesebroeck et al., 2012). Localized class I PI3K activity also plays a role in cortical F-actin dynamics, which underlies chemotaxis and phagocytosis of large particles (Leverrier et al., 2003; Tamura et al., 2009; Hawkins et al., 2010; Flannagan et al., 2012; Weiger and Parent, 2012). For example, at the neutrophil leading edge, p110γ-induced PtdIns(3,4,5)P3 formation results in the recruitment of Rac GTPase, which promotes F-actin polymerization, lamellipodia formation and cell migration (Yoo et al., 2010). Consistent with a central role for PtdIns(3,4,5)P3 in all of these cellular processes, the 3-phosphatase PTEN downregulates PtdIns(3,4,5)P3 and these class I PI3K-activated pathways (Song et al., 2012).
The four class I catalytic isoforms share overlapping but distinct functions. The p110γ and p110δ isoforms are mainly restricted to functions in immune cells where they are expressed, whereas p110α and p110β are ubiquitous, but also exhibit isoform-specific cell-type- and context-dependent requirements. Most class I PI3K functions are related to their catalytic properties; however, there is growing evidence for kinase-independent scaffolding roles for p110γ and p110β (Patrucco et al., 2004; Hirsch et al., 2009; Dou et al., 2010; Rauch et al., 2011; Dou et al., 2013). Less well understood are the roles for the numerous regulatory adaptor isoforms in the regulation of class I p110 functions. Given the importance of class I PI3K function in cellular homeostasis and regulation, mutations in class I PI3Ks or in p85 adaptors are associated with a multitude of human diseases ranging from diabetes to cancer. In addition, PIK3CA-activating mutations have recently been linked to congenital lipomatous overgrowth with vascular, epidermal and skeletal anomalies, or CLOVES syndrome (Kurek et al., 2012). We refer readers to reviews that discuss the involvement of class I PI3K in human disease (Vicinanza et al., 2008; Kok et al., 2009; Vanhaesebroeck et al., 2010b; Wong et al., 2010).
Class II PI3Ks
Class II PI3Ks have received less research attention to date, so it is more difficult to generalize on their functions. One emerging theme is the requirement for class II PI3K functions at the cell cortex, as seen by its disparate roles in cell migration, cortical remodeling, glucose transport, insulin signaling, channel regulation, endocytosis and exocytosis (Mazza and Maffucci, 2011; Falasca and Maffucci, 2012). Class II PI3K functions have been mostly (although controversially) attributed to PtdIns(3)P, which has roles in either cell signaling or intracellular membrane trafficking. Growing evidence suggests that these two mechanisms could be interrelated at the level of ‘endosomal signaling’ (Yoshioka et al., 2012; Biswas et al., 2013), or endosome platforms for the formation of specific signal transduction complexes and responses (Platta and Stenmark, 2011). With regard to cell signaling roles, PI3KC2-induced PtdIns(3)P appears to mediate immune cell K+ channel activity (in the case of PI3KC2β), growth factor receptor responses, and activation of Rho GTPases in cell contraction and migration (in the cases of PI3KC2α and -β) (Bridges et al., 2012; Falasca and Maffucci, 2012; Yoshioka et al., 2012), whereas PI3KC2α-mediated PtdIns(3,4)P2 activity has been associated with insulin-induced Akt stimulation (Leibiger et al., 2010). With regard to membrane trafficking roles, PI3KC2α-induced PtdIns(3)P activity appears to direct endosomal trafficking in endocytic recycling (Krag et al., 2010; Jean et al., 2012; Yoshioka et al., 2012), phagosome maturation (Lu et al., 2012), late steps in exocytosis (Mazza and Maffucci, 2011; Falasca and Maffucci, 2012) and autophagy (Behrends et al., 2010; Devereaux et al., 2013), whereas PtdIns(3,4)P2 activity has been recently implicated with a central role in clathrin-mediated endocytosis (Posor et al., 2013).
The identification of class II PI3K effectors and pathways will be important for a broader understanding of their functions. Although not yet causally linked to human disease, PI3KC2 polymorphisms and studies in cell lines suggest that it might be potentially involved in diabetes (PI3KC2α and -γ) and cancer (PI3KC2α and -β) (Falasca and Maffucci, 2012). Recent mouse knockout results also suggest a possible role for human PI3KC2α in blood vessel formation and integrity (Yoshioka et al., 2012). Interestingly, phenotypes that result from an inactivation of specific MTM 3-phosphoinositide phosphatases are suppressed by a co-knockdown of class II PI3K in flies and worms (Lu et al., 2012). This suggests that class II PI3K inhibition might be a therapeutic avenue for treatment of MTM-associated myopathy and neuropathy disorders in humans (Vicinanza et al., 2008).
Class III PI3K
Vps34 predominantly regulates membrane trafficking, with central roles in endosomal protein sorting, endosome–lysosome maturation, autophagosome formation, autophagy flux and cytokinesis (Backer, 2008; Simonsen and Tooze, 2009; Nezis et al., 2010; Raiborg et al., 2013). Vps34 is found in an increasing variety of protein regulatory complexes that specify the synthesis of PtdIns(3)P pools at distinct intracellular membranes (see poster) (Backer, 2008). The three core Vps34 regulatory complex components include the Vps34 catalytic subunit, the Vps15 membrane adaptor (see above), and Vps30 (known as Beclin 1 in mammals, also called Atg6). In yeast, the complex I [Vps34, Vps15, Vps30, Atg14 and Atg38 (Araki et al., 2013)] is implicated in autophagy, whereas complex II (Vps34, Vps15, Vps30 and Vps38) is involved in membrane trafficking for vacuolar protein sorting, although not all Vps34 endosomal functions seem to require Vps30/Beclin 1. Other associated proteins – the extent and identity of which continues to emerge – specify Vps34 localization, activity and membrane accessibility (Backer, 2008; Simonsen and Tooze, 2009; Funderburk et al., 2010; Kim et al., 2013; Russell et al., 2013).
On early endosomes, Rab5 GTPase activates specific Vps34 complexes for endosomal maturation (see poster). The synthesis of endosomal PtdIns(3)P leads to the recruitment of effectors, such as endosomal sorting complex required for transport (ESCRT) components that are involved in sorting of protein cargo, and the homotypic fusion and protein sorting (HOPS) complex that mediates endosome fusion and trafficking to lysosomes (Raiborg et al., 2013). In a similar fashion, Rab5 and Vps34 are required for phagosome maturation (Flannagan et al., 2012). Distinct Vps34 complexes that contain the autophagy core factor Atg14 have been associated with autophagy. PtdIns(3)P synthesis at autophagosome precursor membranes – potentially at omegasomes that arise at the endoplasmic reticulum (ER) and on nascent autophagosomes – recruits the protein WD-repeat protein interacting with phosphoinositides 1 (WIPI1; Atg18 in yeast) and elicits a hierarchical cascade that directs autophagosome formation (Simonsen and Tooze, 2009; Jaber et al., 2012; Schink et al., 2013). A growing list of regulators controls the activity of the Vps34 complex in autophagy (see poster), including the direct and multi-faceted roles for AMPK that promote Vps34 activity in response to nutrient stress (Kim et al., 2013). Signaling functions are also becoming more broadly established for Vps34, with roles in yeast pheromone signaling (Backer, 2008), regulation of various developmental receptor pathways (von Zastrow and Sorkin, 2007; Platta and Stenmark, 2011; Wada and Sun-Wada, 2013) and amino acid sensing in mTORC1 activation (Yoon et al., 2011; Zoncu et al., 2011; Jaber et al., 2012). In cytokinesis, Vps34 activity at the midbody leads to the synthesis of a PtdIns(3)P pool that recruits FYVE-CENT and associated proteins that regulate the role of ESCRT-III in abscission (Nezis et al., 2010; Schink et al., 2013).
So far, no human diseases are associated with mutations in Vps34, although genetic linkage studies found a correlation between Vps34 promoter variants and schizophrenia (Backer, 2008). In addition, low levels of PtdIns(3)P in brain have been found in humans affected with Alzheimer's disease (Morel et al., 2013), and co-expression of Vps15 and Vps34 could suppress aspects of Danon autophagic vacuolar myopathy (AVM) disease in human patient muscle cells (Nemazanyy et al., 2013).
PI3K studies in model organisms
PI3K mutants isolated in model organisms have been instrumental in describing cellular roles for PI3Ks (Vanhaesebroeck et al., 2012) (see poster). Roles for class I PI3K in insulin-signaling-meditated regulation of cell size, growth and lifespan were first shown in Drosophila and Caenorhabditis elegans (Oldham and Hafen, 2003; Kenyon, 2005), the class II PI3K subfamily was first identified and studied in flies (MacDougall et al., 1995), and class III PI3K functions in trafficking were identified through its mutant phenotypes in yeast (Schu et al., 1993; Raiborg et al., 2013). Nearly all of the PI3Ks and regulatory subunits have now been targeted by gene deletions in mouse, with extensive phenotypic variation depending on the expression pattern and genetic redundancy of the targeted isoform (Vanhaesebroeck et al., 2005; Falasca and Maffucci, 2012; Jaber et al., 2012; Yoshioka et al., 2012). Although most PI3K isoforms are required for mouse viability, tissue-specific knockout and knock-in strategies are now commonly used to facilitate the in vivo characterization of PI3K function. Care must be taken in interpreting PI3K isoform knockouts, owing to both the possible compensation by the non-targeted isoforms in animals with multigene families and the potential for the involvement of kinase-dependent and -independent functions. The use of compound inhibitors has advanced our understanding of class I and class III PI3K functions (Vanhaesebroeck et al., 2012). The development of isoform-specific class I and class III PI3K inhibitors are becoming valuable tools both for use in research and as the focus of clinical trials for use as chemotherapeutics (Wu et al., 2009; Wong et al., 2010). However, class II PI3Ks are generally less sensitive to the first-generation pan-inhibitors, and no specific class II PI3K compounds have been described to date.
Perspectives
A vast amount of knowledge on PI3Ks has been acquired since their discovery nearly 30 years ago (Vanhaesebroeck et al., 2012). Class I PI3K isoforms represent important druggable targets for multiple human diseases, and key lessons such as on feedback loops and crosstalk within and between PI3K and other signaling pathways have been uncovered in pursuit of class I-related therapies (Carracedo and Pandolfi, 2008; Castellano and Downward, 2011). The central role for class III PI3K in autophagy, along with the increasing relevance of autophagy to human disease, has sparked basic and applied research aimed at better understanding the regulation and function of the Vps34 complex. There are still many basic functions that remain to be better illuminated: a deeper understanding of the structure, generalized roles and isolation of selective compounds for PI3KC2; composition and in vivo regulation of PI3K protein complex assemblies; and the direct or indirect effects of phosphoinositide phosphatases on PI3K functions. The expanding knowledge on PI3Ks clearly continues to raise new questions with central relevance to both basic biology and human health.
Supplementary Material
Acknowledgments
We apologize to our colleagues for not citing important contributions owing to the lack of space.
Footnotes
Competing interests
The authors declare no competing interests.
Funding
Support for A.A.K. was provided from the National Institutes of Health; and for S.J. from Fonds de la recherche en santé du Québec and the American Heart Association. Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster is available for downloading in the online version of this article at jcs.biologists.org. Individual poster panels are available as JPEG files at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.093773/-/DC1
References
- Araki Y., Ku W-C., Akioka M., May A. I., Hayashi Y., Arisaka F., Ishihama Y., Ohsumi Y. (2013). Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J. Cell Biol. 203, 299–313 10.1083/jcb.201304123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J. M. (2008). The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 410, 1–17 10.1042/BJ20071427 [DOI] [PubMed] [Google Scholar]
- Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010). Network organization of the human autophagy system. Nature 466, 68–76 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K., Yoshioka K., Asanuma K., Okamoto Y., Takuwa N., Sasaki T., Takuwa Y. (2013). Essential role of class II phosphatidylinositol-3-kinase-C2α in sphingosine 1-phosphate receptor-1-mediated signaling and migration in endothelial cells. J. Biol. Chem. 288, 2325–2339 10.1074/jbc.M112.409656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D., Ma J. T., Park S., Inoki K., Weisman L. S., Saltiel A. R. (2012). Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol. Biol. Cell 23, 2955–2962 10.1091/mbc.E11-12-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R., Auger K. R. (2011). Phylogenomics of phosphoinositide lipid kinases: perspectives on the evolution of second messenger signaling and drug discovery. BMC Evol. Biol. 11, 4 10.1186/1471-2148-11-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C., Laporte J., Backer J. M., Wandinger-Ness A., Stein M-P. (2007). Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic 8, 1052–1067 10.1111/j.1600-0854.2007.00586.x [DOI] [PubMed] [Google Scholar]
- Carracedo A., Pandolfi P. P. (2008). The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene 27, 5527–5541 10.1038/onc.2008.247 [DOI] [PubMed] [Google Scholar]
- Castellano E., Downward J. (2011). RAS interaction with PI3K: more than just another effector pathway. Genes Cancer 2, 261–274 10.1177/1947601911408079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagpar R. B., Links P. H., Pastor M. C., Furber L. A., Hawrysh A. D., Chamberlain M. D., Anderson D. H. (2010). Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 107, 5471–5476 10.1073/pnas.0908899107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S. C., Waterfield M. D., Backer J. M., Zerial M. (1999). Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1, 249–252 10.1038/12075 [DOI] [PubMed] [Google Scholar]
- Cullen P. J. (2008). Endosomal sorting and signalling: an emerging role for sorting nexins. Nat. Rev. Mol. Cell Biol. 9, 574–582 10.1038/nrm2427 [DOI] [PubMed] [Google Scholar]
- Das M., Scappini E., Martin N. P., Wong K. A., Dunn S., Chen Y. J., Miller S. L., Domin J., O'Bryan J. P. (2007). Regulation of neuron survival through an intersectin-phosphoinositide 3′-kinase C2beta-AKT pathway. Mol. Cell. Biol. 27, 7906–7917 10.1128/MCB.01369-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dbouk H. A., Vadas O., Shymanets A., Burke J. E., Salamon R. S., Khalil B. D., Barrett M. O., Waldo G. L., Surve C., Hsueh C. et al. (2012). G protein-coupled receptor-mediated activation of p110β by Gβγ is required for cellular transformation and invasiveness. Sci. Signal. 5, ra89 10.1126/scisignal.2003264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereaux K., Dall'armi C., Alcazar-Roman A., Ogasawara Y., Zhou X., Wang F., Yamamoto A., De Camilli P., Di Paolo G. (2013). Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS ONE 8, e76405 10.1371/journal.pone.0076405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Dou Z., Chattopadhyay M., Pan J. A., Guerriero J. L., Jiang Y. P., Ballou L. M., Yue Z., Lin R. Z., Zong W. X. (2010). The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J. Cell Biol. 191, 827–843 10.1083/jcb.201006056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z., Pan J. A., Dbouk H. A., Ballou L. M., DeLeon J. L., Fan Y., Chen J. S., Liang Z., Li G., Backer J. M. et al. (2013). Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol. Cell 50, 29–42 10.1016/j.molcel.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman J. A., Luo J., Cantley L. C. (2006). The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7, 606–619 10.1038/nrg1879 [DOI] [PubMed] [Google Scholar]
- Falasca M., Maffucci T. (2012). Regulation and cellular functions of class II phosphoinositide 3-kinases. Biochem. J. 443, 587–601 10.1042/BJ20120008 [DOI] [PubMed] [Google Scholar]
- Flannagan R. S., Jaumouillé V., Grinstein S. (2012). The cell biology of phagocytosis. Annu. Rev. Pathol. 7, 61–98 10.1146/annurev-pathol-011811-132445 [DOI] [PubMed] [Google Scholar]
- Fritsch R., de Krijger I., Fritsch K., George R., Reason B., Kumar M. S., Diefenbacher M., Stamp G., Downward J. (2013). RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell 153, 1050–1063 10.1016/j.cell.2013.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk S. F., Wang Q. J., Yue Z. (2010). The Beclin 1-VPS34 complex – at the crossroads of autophagy and beyond. Trends Cell Biol. 20, 355–362 10.1016/j.tcb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidarov I., Smith M. E., Domin J., Keen J. H. (2001). The class II phosphoinositide 3-kinase C2alpha is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol. Cell 7, 443–449 10.1016/S1097-2765(01)00191-5 [DOI] [PubMed] [Google Scholar]
- Guillermet-Guibert J., Bjorklof K., Salpekar A., Gonella C., Ramadani F., Bilancio A., Meek S., Smith A. J. H., Okkenhaug K., Vanhaesebroeck B. (2008). The p110beta isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110gamma. Proc. Natl. Acad. Sci. USA 105, 8292–8297 10.1073/pnas.0707761105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins P. T., Anderson K. E., Davidson K., Stephens L. R. (2006). Signalling through Class I PI3Ks in mammalian cells. Biochem. Soc. Trans. 34, 647–662 10.1042/BST0340647 [DOI] [PubMed] [Google Scholar]
- Hawkins P. T., Stephens L. R., Suire S., Wilson M. (2010). PI3K signaling in neutrophils. Curr. Top. Microbiol. Immunol. 346, 183–202 10.1007/82_2010_40 [DOI] [PubMed] [Google Scholar]
- Hirsch E., Braccini L., Ciraolo E., Morello F., Perino A. (2009). Twice upon a time: PI3K's secret double life exposed. Trends Biochem. Sci. 34, 244–248 10.1016/j.tibs.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Ivetac I., Munday A. D., Kisseleva M. V., Zhang X. M., Luff S., Tiganis T., Whisstock J. C., Rowe T., Majerus P. W., Mitchell C. A. (2005). The type Ialpha inositol polyphosphate 4-phosphatase generates and terminates phosphoinositide 3-kinase signals on endosomes and the plasma membrane. Mol. Biol. Cell 16, 2218–2233 10.1091/mbc.E04-09-0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber N., Dou Z., Chen J. S., Catanzaro J., Jiang Y. P., Ballou L. M., Selinger E., Ouyang X., Lin R. Z., Zhang J. et al. (2012). Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. USA 109, 2003–2008 10.1073/pnas.1112848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P., Schoenwaelder S. M., Matzaris M., Brown S., Mitchell C. A. (1995). Phosphatidylinositol 3,4,5-trisphosphate is a substrate for the 75 kDa inositol polyphosphate 5-phosphatase and a novel 5-phosphatase which forms a complex with the p85/p110 form of phosphoinositide 3-kinase. EMBO J. 14, 4490–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean S., Kiger A. A. (2012). Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat. Rev. Mol. Cell Biol. 13, 463–470 10.1038/nrm3379 [DOI] [PubMed] [Google Scholar]
- Jean S., Cox S., Schmidt E. J., Robinson F. L., Kiger A. (2012). Sbf/MTMR13 coordinates PI(3)P and Rab21 regulation in endocytic control of cellular remodeling. Mol. Biol. Cell 23, 2723–2740 10.1091/mbc.E12-05-0375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. (2005). The plasticity of aging: insights from long-lived mutants. Cell 120, 449–460 10.1016/j.cell.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Kim J., Kim Y. C., Fang C., Russell R. C., Kim J. H., Fan W., Liu R., Zhong Q., Guan K. L. (2013). Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303 10.1016/j.cell.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok K., Geering B., Vanhaesebroeck B. (2009). Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem. Sci. 34, 115–127 10.1016/j.tibs.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Krag C., Malmberg E. K., Salcini A. E. (2010). PI3KC2α, a class II PI3K, is required for dynamin-independent internalization pathways. J. Cell Sci. 123, 4240–4250 10.1242/jcs.071712 [DOI] [PubMed] [Google Scholar]
- Kumar A., Redondo-Muñoz J., Perez-García V., Cortes I., Chagoyen M., Carrera A. C. (2011). Nuclear but not cytosolic phosphoinositide 3-kinase beta has an essential function in cell survival. Mol. Cell. Biol. 31, 2122–2133 10.1128/MCB.01313-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek K. C., Luks V. L., Ayturk U. M., Alomari A. I., Fishman S. J., Spencer S. A., Mulliken J. B., Bowen M. E., Yamamoto G. L., Kozakewich H. P. et al. (2012). Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am. J. Hum. Genet. 90, 1108–1115 10.1016/j.ajhg.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibiger B., Moede T., Uhles S., Barker C. J., Creveaux M., Domin J., Berggren P. O., Leibiger I. B. (2010). Insulin-feedback via PI3K-C2alpha activated PKBalpha/Akt1 is required for glucose-stimulated insulin secretion. FASEB J. 24, 1824–1837 10.1096/fj.09-148072 [DOI] [PubMed] [Google Scholar]
- Lemmon M. A. (2008). Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- Leverrier Y., Okkenhaug K., Sawyer C., Bilancio A., Vanhaesebroeck B., Ridley A. J. (2003). Class I phosphoinositide 3-kinase p110beta is required for apoptotic cell and Fcgamma receptor-mediated phagocytosis by macrophages. J. Biol. Chem. 278, 38437–38442 10.1074/jbc.M306649200 [DOI] [PubMed] [Google Scholar]
- Lu N., Shen Q., Mahoney T. R., Neukomm L. J., Wang Y., Zhou Z. (2012). Two PI 3-kinases and one PI 3-phosphatase together establish the cyclic waves of phagosomal PtdIns(3)P critical for the degradation of apoptotic cells. PLoS Biol. 10, e1001245 10.1371/journal.pbio.1001245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall L. K., Domin J., Waterfield M. D. (1995). A family of phosphoinositide 3-kinases in Drosophila identifies a new mediator of signal transduction. Curr. Biol. 5, 1404–1415 10.1016/S0960-9822(95)00278-8 [DOI] [PubMed] [Google Scholar]
- Mazza S., Maffucci T. (2011). Class II phosphoinositide 3-kinase C2alpha: what we learned so far. Int. J. Biochem. Mol. Biol. 2, 168–182 [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S., Murray D. (2005). Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 10.1038/nature04398 [DOI] [PubMed] [Google Scholar]
- Morel E., Chamoun Z., Lasiecka Z. M., Chan R. B., Williamson R. L., Vetanovetz C., Dall'Armi C., Simoes S., Point Du Jour K. S., McCabe B. D. et al. (2013). Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat. Commun. 4, 2250 10.1038/ncomms3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday A. D., Norris F. A., Caldwell K. K., Brown S., Majerus P. W., Mitchell C. A. (1999). The inositol polyphosphate 4-phosphatase forms a complex with phosphatidylinositol 3-kinase in human platelet cytosol. Proc. Natl. Acad. Sci. USA 96, 3640–3645 10.1073/pnas.96.7.3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemazanyy I., Blaauw B., Paolini C., Caillaud C., Protasi F., Mueller A., Proikas-Cezanne T., Russell R. C., Guan K. L., Nishino I. et al. (2013). Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol. Med. 5, 870–890 10.1002/emmm.201202057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis I. P., Sagona A. P., Schink K. O., Stenmark H. (2010). Divide and ProsPer: the emerging role of PtdIns3P in cytokinesis. Trends Cell Biol. 20, 642–649 10.1016/j.tcb.2010.08.010 [DOI] [PubMed] [Google Scholar]
- Oldham S., Hafen E. (2003). Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13, 79–85 10.1016/S0962-8924(02)00042-9 [DOI] [PubMed] [Google Scholar]
- Patrucco E., Notte A., Barberis L., Selvetella G., Maffei A., Brancaccio M., Marengo S., Russo G., Azzolino O., Rybalkin S. D. et al. (2004). PI3Kgamma modulates the cardiac response to chronic pressure overload by distinct kinase-dependent and -independent effects. Cell 118, 375–387 10.1016/j.cell.2004.07.017 [DOI] [PubMed] [Google Scholar]
- Platta H. W., Stenmark H. (2011). Endocytosis and signaling. Curr. Opin. Cell Biol. 23, 393–403 10.1016/j.ceb.2011.03.008 [DOI] [PubMed] [Google Scholar]
- Posor Y., Eichhorn-Gruenig M., Puchkov D., Schöneberg J., Ullrich A., Lampe A., Müller R., Zarbakhsh S., Gulluni F., Hirsch E. et al. (2013). Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature 499, 233–237 10.1038/nature12360 [DOI] [PubMed] [Google Scholar]
- Raiborg C., Schink K. O., Stenmark H. (2013). Class III phosphatidylinositol 3-kinase and its catalytic product PtdIns3P in regulation of endocytic membrane traffic. FEBS J. 280, 2730–2742 10.1111/febs.12116 [DOI] [PubMed] [Google Scholar]
- Rauch J., Volinsky N., Romano D., Kolch W. (2011). The secret life of kinases: functions beyond catalysis. Cell Commun. Signal. 9, 23 10.1186/1478-811X-9-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., Guan K. L. (2013). ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Takasuga S., Sasaki J., Kofuji S., Eguchi S., Yamazaki M., Suzuki A. (2009). Mammalian phosphoinositide kinases and phosphatases. Prog. Lipid Res. 48, 307–343 10.1016/j.plipres.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Schink K. O., Raiborg C., Stenmark H. (2013). Phosphatidylinositol 3-phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. Bioessays 35, 900–912 [DOI] [PubMed] [Google Scholar]
- Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., Emr S. D. (1993). Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88–91 10.1126/science.8385367 [DOI] [PubMed] [Google Scholar]
- Shin H. W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M. R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A. et al. (2005). An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170, 607–618 10.1083/jcb.200505128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Tooze S. A. (2009). Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 186, 773–782 10.1083/jcb.200907014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. S., Salmena L., Pandolfi P. P. (2012). The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13, 283–296 [DOI] [PubMed] [Google Scholar]
- Stahelin R. V., Karathanassis D., Bruzik K. S., Waterfield M. D., Bravo J., Williams R. L., Cho W. (2006). Structural and membrane binding analysis of the Phox homology domain of phosphoinositide 3-kinase-C2alpha. J. Biol. Chem. 281, 39396–39406 10.1074/jbc.M607079200 [DOI] [PubMed] [Google Scholar]
- Tamura N., Hazeki K., Okazaki N., Kametani Y., Murakami H., Takaba Y., Ishikawa Y., Nigorikawa K., Hazeki O. (2009). Specific role of phosphoinositide 3-kinase p110alpha in the regulation of phagocytosis and pinocytosis in macrophages. Biochem. J. 423, 99–108 10.1042/BJ20090687 [DOI] [PubMed] [Google Scholar]
- Vadas O., Burke J. E., Zhang X., Berndt A., Williams R. L. (2011). Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci. Signal. 4, re2 10.1126/scisignal.2002165 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Ali K., Bilancio A., Geering B., Foukas L. C. (2005). Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem. Sci. 30, 194–204 10.1016/j.tibs.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. (2010a). The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11, 329–341 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Vogt P. K., Rommel C. (2010b). PI3K: from the bench to the clinic and back. Curr. Top. Microbiol. Immunol. 347, 1–19 10.1007/82_2010_65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Stephens L., Hawkins P. (2012). PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol. 13, 195–203 10.1038/nrm3290 [DOI] [PubMed] [Google Scholar]
- Vicinanza M., D'Angelo G., Di Campli A., De Matteis M. A. (2008). Function and dysfunction of the PI system in membrane trafficking. EMBO J. 27, 2457–2470 10.1038/emboj.2008.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zastrow M., Sorkin A. (2007). Signaling on the endocytic pathway. Curr. Opin. Cell Biol. 19, 436–445 10.1016/j.ceb.2007.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y., Sun-Wada G. H. (2013). Positive and negative regulation of developmental signaling by the endocytic pathway. Curr. Opin. Genet. Dev. 23, 391–398 10.1016/j.gde.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Weiger M. C., Parent C. A. (2012). Phosphoinositides in chemotaxis. Subcell. Biochem. 59, 217–254 10.1007/978-94-007-3015-1_7 [DOI] [PubMed] [Google Scholar]
- Wong K-K., Engelman J. A., Cantley L. C. (2010). Targeting the PI3K signaling pathway in cancer. Curr. Opin. Genet. Dev. 20, 87–90 10.1016/j.gde.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Liu T., Hu Y. (2009). PI3K inhibitors for cancer therapy: what has been achieved so far? Curr. Med. Chem. 16, 916–930 10.2174/092986709787581905 [DOI] [PubMed] [Google Scholar]
- Yoo S. K., Deng Q., Cavnar P. J., Wu Y. I., Hahn K. M., Huttenlocher A. (2010). Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev. Cell 18, 226–236 10.1016/j.devcel.2009.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M. S., Du G., Backer J. M., Frohman M. A., Chen J. (2011). Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J. Cell Biol. 195, 435–447 10.1083/jcb.201107033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K., Yoshida K., Cui H., Wakayama T., Takuwa N., Okamoto Y., Du W., Qi X., Asanuma K., Sugihara K. et al. (2012). Endothelial PI3K-C2α, a class II PI3K, has an essential role in angiogenesis and vascular barrier function. Nat. Med. 18, 1560–1569 10.1038/nm.2928 [DOI] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., Sabatini D. M. (2011). mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.