ABSTRACT
As they mature, professional secretory cells like pancreatic acinar and gastric chief cells induce the transcription factor MIST1 (also known as BHLHA15) to substantially scale up production of large secretory granules in a process that involves expansion of apical cytoplasm and redistribution of lysosomes and mitochondria. How a scaling factor like MIST1 rearranges cellular architecture simply by regulating expression levels of its transcriptional targets is unknown. RAB26 is a MIST1 target whose role in MIST1-mediated secretory cell maturation is also unknown. Here, we confirm that RAB26 expression, unlike most Rabs which are ubiquitously expressed, is tissue specific and largely confined to MIST1-expressing secretory tissues. Surprisingly, functional studies showed that RAB26 predominantly associated with LAMP1/cathepsin D lysosomes and not directly with secretory granules. Moreover, increasing RAB26 expression – by inducing differentiation of zymogen-secreting cells or by direct transfection – caused lysosomes to coalesce in a central, perinuclear region. Lysosome clustering in turn caused redistribution of mitochondria into distinct subcellular neighborhoods. The data elucidate a novel function for RAB26 and suggest a mechanism for how cells could increase transcription of key effectors to reorganize subcellular compartments during differentiation.
KEY WORDS: MIST1, Zymogenic cell, LAMP1, Vesicle trafficking
INTRODUCTION
Rab proteins are the largest members of the Ras family of small protein GTPases with more than 70 members identified in humans (Schwartz et al., 2007). They are important regulators of intracellular membrane and vesicle positioning and trafficking. By cycling between GTP- and GDP-bound forms, they work like simple machines to define specific membrane-associated compartments within cells. Many Rabs – like RAB4, RAB5, RAB7 and RAB11 – are ubiquitously expressed (Gurkan et al., 2005; Jin et al., 2012; Zhang et al., 2007), performing essentially the same function in diverse cells. RAB7, for example, aids the maturation of early endosomes into late endosomes and ultimately coordinates transport to lysosomes (Bucci et al., 2000; Feng et al., 1995).
In contrast, expression of some Rabs is largely confined to cells that perform specific physiological functions. Among the best studied are the RAB27 and RAB3 families which are expressed highly in specialized secretory cells. RAB27A and RAB27B function in the transport and docking of mature vesicles destined for secretion (Fukuda, 2013) in melanocytes (Hume et al., 2001) and T lymphocytes (Ménasché et al., 2000). The RAB3 family includes RAB3A, RAB3B, RAB3C and RAB3D, the first three of which are predominantly neuronal, and are involved in neurotransmitter and neuroendocrine hormone release (Geppert et al., 1994; Johannes et al., 1994; Schlüter et al., 2004; Weber et al., 1994), whereas RAB3D is expressed in a variety of extraneuronal secretory cells, in particular exocrine serous cells in the pancreas, salivary glands and stomach (Chen et al., 2002; Millar et al., 2002; Ohnishi et al., 1997). Functionally, RAB3 isoforms all share a similar role in mature secretory vesicle docking (Fukuda, 2008).
The primary function of serous exocrine cells is to synthesize, store and secrete digestive enzyme precursors. MIST1 (also known as BHLHA15) is a basic helix-loop-helix (bHLH) transcription factor that acts as a scaling factor in these cells (Mills and Taghert, 2012). MIST1 expression is a feature of all serous exocrine cells (Lemercier et al., 1997; Pin et al., 2000). Recent work has begun to identify the molecular targets that it scales up to establish a high-capacity polarized secretory apparatus (Capoccia et al., 2013; Direnzo et al., 2012; Garside et al., 2010; Jia et al., 2008; Johnson et al., 2004; Pin et al., 2001; Rukstalis et al., 2003; Tian et al., 2010). For example, RAB3D has been identified as a target in both pancreatic and gastric exocrine cells (Johnson et al., 2004; Tian et al., 2010). We also identified RAB26, which is phylogenetically related to RAB3D (Fukuda, 2008; Pereira-Leal and Seabra, 2001) as a direct MIST1 target that is required for the establishment of large secretory granules (Tian et al., 2010). However, the mechanism of RAB26 function in secretory cells has not been characterized (Azouz et al., 2012).
Here, we present work showing that RAB26 is not like other members of its phylogenetic family because it does not directly associate with secretory vesicles, but rather with lysosomes. Accordingly, scaling up expression of RAB26 causes increasing centripetal coalescence of lysosomes to the perinuclear region in a manner dependent on GTP cycling. Furthermore, using electron microscopy, immunofluorescence microscopy and immunoblotting, we demonstrate that the lysosomal coalescence caused by increased RAB26 also results in redistribution of mitochondria into distinct cellular neighborhoods. Taken together, our data establish a role for RAB26 as a novel lysosome-associated Rab whose expression is transcriptionally induced specifically in secretory tissues to reorganize the cellular distribution of lysosomes and, indirectly, mitochondria.
RESULTS
RAB26 expression is induced in acinar secretory cells by MIST1
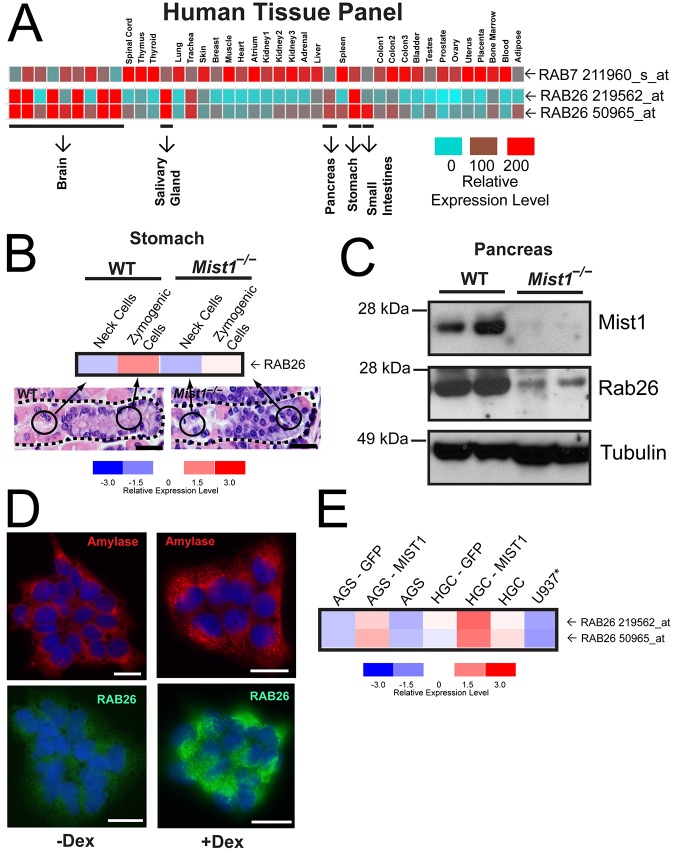
Previous reports have shown that RAB26 is expressed in secretory tissues (Nashida et al., 2006; Tian et al., 2010; Wagner et al., 1995; Yoshie et al., 2000). To further elucidate the role of RAB26, we assayed the expression pattern of this gene in a human tissue panel (Ge et al., 2005) (http://sbmdb.genome.rcast.u-tokyo.ac.jp/refexa/) (Fig. 1A). In contrast to a more ubiquitously expressed Rab (e.g. RAB7), RAB26 showed variable expression with high expression levels in exocrine secretory tissues and in parts of the brain. The two RAB26 probesets showed strong correlation among the tissues examined (Pearson's correlation coefficient of 0.9145) and mirrored the known tissue distribution of the secretory cell transcription factor MIST1 (Lemercier et al., 1997; Mills and Taghert, 2012; Pin et al., 2000). Moreover, within specific organs, RAB26 expression was confined to MIST1-expressing cell lineages like the serous exocrine, digestive-enzyme-secreting zymogenic (chief) cells of the stomach, but not their precursors, the mucus-secreting ‘neck cells’ (Capoccia et al., 2013; Geahlen et al., 2013; Fig. 1B). RAB26 expression was not scaled up in the absence of the transcription factor MIST1 in either gastric units from Mist1−/− mice (Fig. 1B) or in another tissue populated by digestive-enzyme secreting cells, the pancreas (Fig. 1C). We next decided to investigate RAB26 scalability in a cell culture system that would facilitate analysis of RAB26 expression level relative to its subcellular distribution and function. First, we analyzed the well-established secretory pancreatic cell line, AR42J, which expresses MIST1 (Jia et al., 2008) and can be differentiated with dexamethasone treatment to upregulate MIST1 target gene expression (Limi et al., 2012; Qiu et al., 2001) and increase amylase-containing secretory vesicles (Logsdon, 1986; Rinn et al., 2012) (Fig. 1D). In these cells, we found that upon differentiation, as in the stomach and pancreas in vivo, RAB26 expression was induced substantially (Fig. 1D). We confirmed this relationship between MIST1 and RAB26 in human gastric cell lines (Fig. 1E). Expression of RAB26 was endogenously low in both AGS and HGC-27 cells (and absent in non-epithelial U937 monocyte cells), but increased in a scalable manner upon MIST1 (but not GFP control) transfection (Fig. 1E; Pearson's coefficient of 0.9660, indicating strong correlation). These data confirm that RAB26 expression is MIST1-dependent and, based on our earlier studies showing direct MIST1 binding to the RAB26 promoter (Tian et al., 2010), we conclude that RAB26 is a direct transcriptional target whose expression is scaled up by MIST1.
Fig. 1.
Expression of RAB26 is cell- and tissue-dependent, and inducible by the transcription factor MIST1. (A) Expression of RAB7 and RAB26 in the REFEXA database of human tissues (http://sbmdb.genome.rcast.u-tokyo.ac.jp/refexa/). The highly RAB26 expressing secretory tissues are highlighted below. Gene expression is shown with a relative scale (0–200) with red, high, and blue, low expression. (B) Microarray analysis of RAB26 gene expression from isolated populations of gastric ZCs and their precursor neck cells from wild-type and Mist1−/− mice. Arrows indicate the location of isolated cell populations in representative H&E-stained gastric gland images. The gene expression for the microarray analyses are shown with a relative expression scale (−3.0 to 3.0) with red, high, and blue, low expression. (C) Western blot analysis of indicated proteins from two wild-type and two Mist1−/− mice. (D). Immunofluorescence of AR42J acinar cell differentiation upon treatment with dexamethasone (Dex); amylase secretory vesicles are red; endogenous RAB26 is green. (E) Gene expression analysis of RAB26 expression from AGS and HGC-27 gastric cell lines before and after transfection with either GFP or MIST1; a non-epithelial monocyte control cell line is also shown (U937). Scale bars: 20 µm.
RAB26 localizes specifically to LAMP1 lysosomal membrane-associated vesicles
To study the functional role of RAB26, we performed experiments in HGC-27 cells because (1) they express low-level endogenous RAB26, even without MIST1 transfection (Fig. 1E); (2) we have previously shown that co-transfection of MIST1 and a cargo of digestive enzyme induces a network of large secretory granules that would allow us to study the interaction between RAB26 and those vesicles (Tian et al., 2010); and (3) they are more easily transfected and larger than AGS or AR42J cells, facilitating detailed microscopy. We engineered a version of RAB26 (EGFP–RAB26) with a monomerized EGFP fused to its N-terminus to aid in subsequent localization and trafficking studies.
We had previously shown that interfering with RAB26 function inhibited MIST1-mediated granulogenesis (Tian et al., 2010) and hypothesized, based on the initial descriptive publications (Nashida et al., 2006; Wagner et al., 1995; Yoshie et al., 2000), that RAB26 would function somehow to traffic nascent or maturing secretory granules. To study RAB26–secretory-granule interactions, we induced a network of secretory granules by the transfecting secretory cargo RFP-tagged Pepsinogen C, in cells stably expressing MIST1, a system we have previously described (Tian et al., 2010). Using live-cell timelapse confocal microscopy, we observed, unexpectedly, that the smaller EGFP–RAB26 vesicles did not fuse, or move in concert, with the larger PGC–RFP-containing secretory granules (supplementary material Movie 1). In addition, RAB26 vesicles showed no overlap with immature secretory vesicles labeled with antibody against the prohormone convertase Furin (supplementary material Fig. S1A). Finally, EGFP–RAB26 did not interact directly with amylase secretory granules in AR42J cells (data not shown).
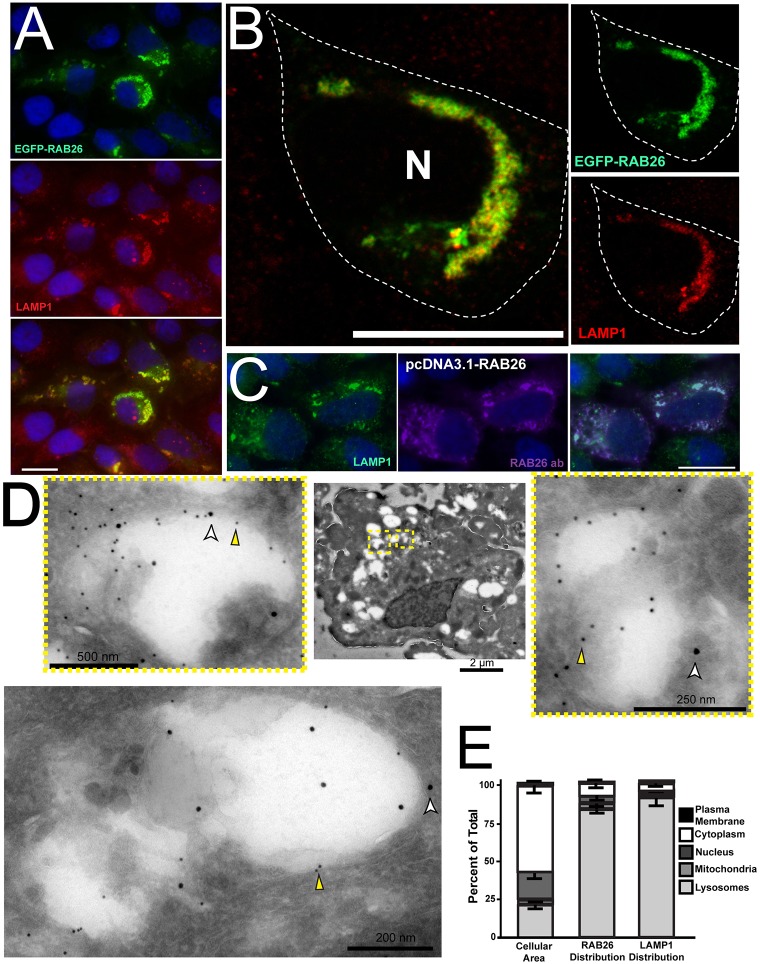
RAB26-associated vesicles similarly did not overlap substantially with markers of the following other organelles: the ER (calregulin), early endosomal (EEA1), Golgi (giantin), cis-Golgi (GM130), or trans-Golgi markers (TGN46) compartments (supplementary material Fig. S1B–F). The lack of RAB26 association with the Golgi was of interest, because recent studies have demonstrated that RAB26 might occupy a recycling endosomal (Chan et al., 2011) or giantin-positive Golgi compartment (Li et al., 2012). Because of the proximity, if not substantial direct overlap, of RAB26 vesicles with our trans-Golgi marker, we next examined membrane compartments that interact with the trans-Golgi. RAB26 showed slight overlap but were often also proximal to vesicles of the post Golgi sorting compartment, as marked by antibodies against CI-M6PR, AP1 and GGA2 (supplementary material Fig. S1G–J). The only other principal destination for microtubule-associated vesicles emerging from the Golgi – besides secretory vesicles – is lysosomes, so, by process of elimination, we reasoned RAB26 was likely to be associated with lysosomes. Accordingly, antibodies against the late endosome/lysosome marker LAMP1 substantially colabeled RAB26-associated vesicles (Fig. 2A,B). Confocal microscopy in RAB26-transfected HGC-27 cells showed near complete colocalization of RAB26 with LAMP1 (Fig. 2B). Furthermore, confocal live imaging of EGFP–RAB26 with LysoTracker revealed nearly identical movement of the two fluors (supplementary material Movie 2), indicating they both clearly defined the same dynamic vesicle population. In addition, transfection of a plasmid that was independently designed to encode endogenous, full-length and non-fluorescently tagged RAB26 revealed colocalization with anti-RAB26 and anti-LAMP1 antibodies (Fig. 2C). Finally, using immunoelectron microscopy, we found that RAB26-transfected cells showed significant labeling of large vesicular structures with both 12-nm anti-RAB26 and 18-nm anti-LAMP1 gold particles; 82.4% of total cellular RAB26 particles and 91.9% of LAMP1 associated with these structures, which sometimes contained identifiable cellular debris, consistent with lysosomes (Fig. 2D,E).
Fig. 2.
RAB26 localizes to LAMP1-positive lysosomal vesicles. (A) Immunofluorescence of HGC-27 cells transfected with EGFP–RAB26 (green) and co-immunostained with LAMP1 (red). (B) Confocal image of EGFP–RAB26-transfected (green) HGC-27 cells stained for LAMP1 (red). Insets (unmerged) show membranes positive for RAB26 (green) and LAMP1 (red). The cell border is outlined; N, nucleus. (C) HGC-27 cells transfected with RAB26 without a GFP tag (pcDNA3.1-RAB26) and co-stained with anti-LAMP1 (green) and anti-RAB26 (purple). (D) Immunoelectron micrographs of HGC-27 cells transfected with EGFP–RAB26. A representative low-magnification cell is shown with insets and additional panel highlighting large labeled vesicles labeled with antibody against RAB26 (12-nm gold particles, yellow arrowheads) and LAMP1 (18-nm gold particles, white arrowheads). (E) Graph with left bar showing the mean percentage total cellular area of lysosomes, mitochondria, nucleus, cytoplasm and plasma membrane in multiple quantified cells. Right bars show the fraction of total RAB26 or LAMP1 gold particles associated with each of those compartments. Scale bars: 20 µm unless indicated.
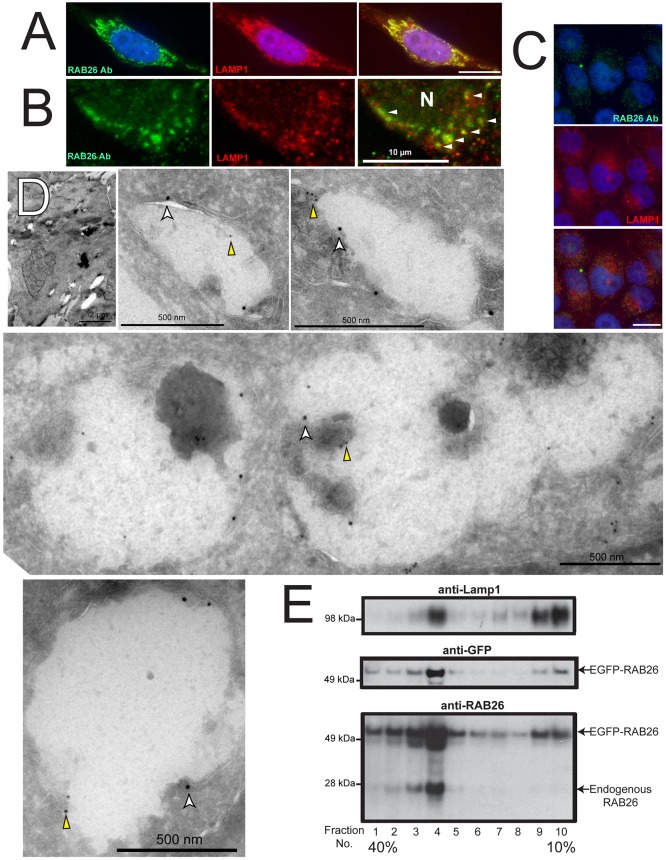
As a control for potential mislocalization caused by overexpression, we next surveyed endogenous RAB26 localization. Although expression of native RAB26 in HGC cells is relatively low (Fig. 1E; Fig. 3C), we found endogenous RAB26 associated with LAMP1 lysosomes in rare untransfected cells with higher levels (Fig. 3A,B). Immunoelectron microscopy showed that, although untransfected cells lacked the larger grouped LAMP1-associated vesicular complexes found in RAB26 overexpressing cells, the LAMP1-labeled vesicles that were present were also decorated with lower level RAB26 labeling (Fig. 3D). Density gradient cell fractionation further confirmed that both EGFP–RAB26 (detectable by antibodies to both GFP and RAB26) and endogenous RAB26 protein were enriched in the purified lysosomal fraction as defined by positive LAMP1 immunoreactivity (Fig. 3E). Finally, as an additional control, we designed and transfected an N-terminally EGFP-fused RAB3D plasmid, a close family member of RAB26 and also a MIST1 target (Johnson et al., 2004; Tian et al., 2010). RAB3D had a distinct cellular distribution to that of RAB26 with no lysosomal colocalization (supplementary material Fig. S2A).
Fig. 3.
Endogenous RAB26 is also lysosome-associated. (A) Immunofluorescence imaging of endogenous RAB26 staining (green) in a rare HGC-27 cell with moderate RAB26 levels, co-stained for LAMP1 (red). (B) Confocal immunomicroscopy of endogenous RAB26 (green) and LAMP1 (red) (N, nucleus). Arrowheads indicate colocalization of RAB26 and LAMP1 vesicles. (C) Lower magnification epifluorescence images of HGC-27 cells with more representative, low-level endogenous RAB26 (green) and LAMP1 (red). (D) Immunoelectron micrographs of control GFP-transfected HGC-27 cells. A representative cell is shown at low magnification with several panels showing electron lucent structures labeled with anti-LAMP1 antibody (18-nm gold particles, white arrowheads) and sparsely labeled with anti-RAB26 antibody (12-nm gold particles, yellow arrowheads). (E) Density gradient cell fractionation experiment with numbered density fractions analyzed for lysosomes with anti-LAMP1 antibody, EGFP-tagged RAB26 by both anti-GFP and anti-RAB26 antibodies (middle and lower panel), and endogenous RAB26 by anti-RAB26 immunoblotting (lower panel). Scale bars: 20 µm unless indicated.
Our findings were applicable beyond HGC-27 cells because similar lysosome–RAB26 colocalization occurred in another gastric cell line (supplementary material Fig. S2B), and epithelial cells from other tissues, including 5637 bladder cells (supplementary material Fig. S2C). Likewise, in pancreatic AR42J cells, we found that RAB26 vesicles formed a unique vesicle population separate from amylase-containing secretory vesicles (data not shown), and again overlapping nearly completely with LAMP1 (supplementary material Fig. S2D,E).
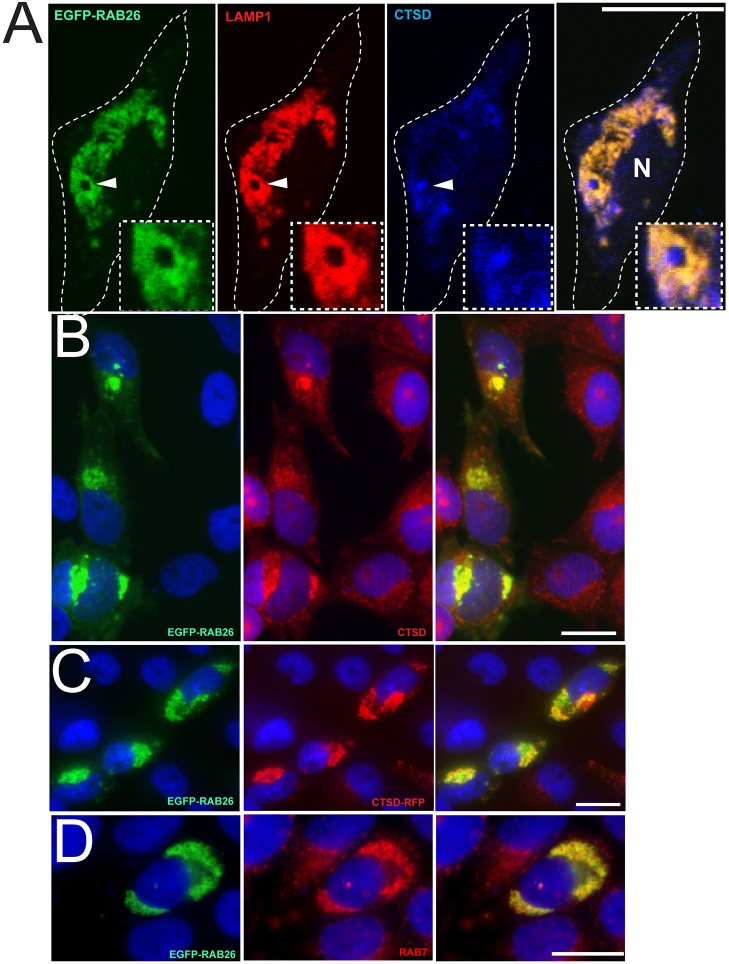
There was also significant overlap of RAB26 and LAMP1 with the lysosomal acid hydrolase cathepsin D (CTSD) (Fig. 4A–C). RAB26 and LAMP1 often defined the membrane portion of vesicles, whose interiors were filled with cathepsin D, as expected (Fig. 4A). Moreover, we designed an RFP-tagged cathepsin D protein (CTSD–RFP) that also showed significant colocalization with EGFP-RAB26 upon co-transfection (Fig. 4C), providing an independent non-antibody confirmation of the direct association of RAB26 with the lysosomal compartment. EGFP–RAB26 also overlapped with another known Rab protein, RAB7 (Fig. 4D), whose role in lysosomal trafficking has been well-established (Bucci et al., 2000). Combined, these experiments definitively show that RAB26 defines the lysosomal subcellular location in multiple cells.
Fig. 4.
RAB26 localizes with other lysosomal markers. (A) Confocal image of a HGC-27 cell transfected with EGFP–RAB26 (green) and immunostained for LAMP1 (red) and cathepsin D (blue). The cell border is outlined; N, nucleus. Insets show a higher magnification of a lysosomal vesicle (arrowhead) that contains membranous RAB26 and LAMP1 with interior cathepsin D. (B) Immunofluorescence staining HGC-27 cells transfected with EGFP–RAB26 (green) and co-stained for cathepsin D (red). (C) Epifluorescence microscopy of HGC-27 cells co-transfected with EGFP–RAB26 (green) and RFP-tagged cathepsin D (CTSD–RFP, red). (E) Immunofluorescence staining HGC-27 cells transfected with EGFP–RAB26 (green) and co-stained for RAB7 (red). Scale bars: 20 µm.
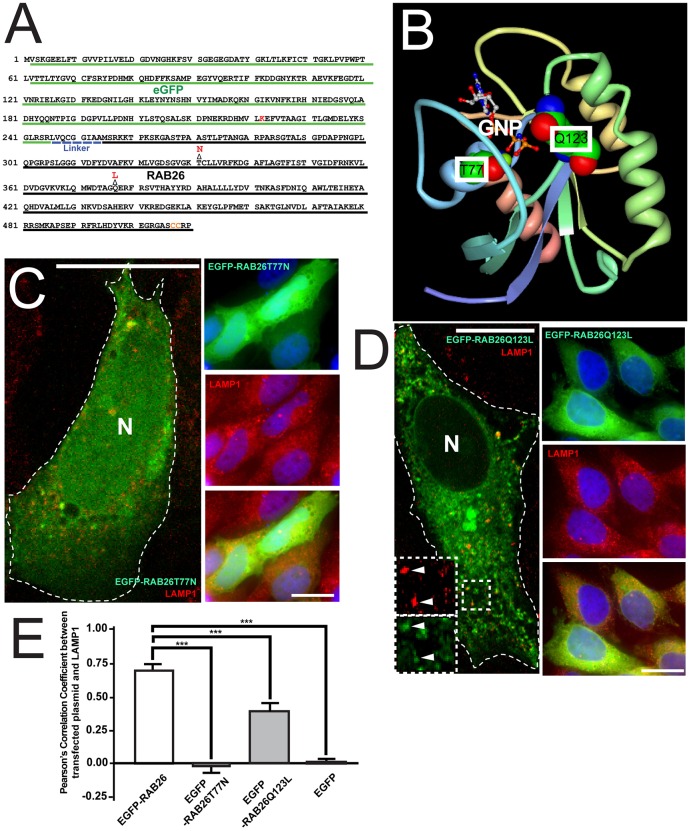
To confirm that RAB26 associates with lysosomes actively, that is, based on normal GTP–GDP cycling and trafficking, we next designed single-point mutant EGFP–RAB26 variants: one with a threonine to an asparagine mutation (EGFP–RAB26T77N) and another with a glutamine to leucine mutation (EGFP–RAB26Q123L) (Fig. 5A). Based on published RAB26 structural models [Protein Data Bank (PDB) accession code 2G6B] (Wang et al., 2006) and previous studies of Ras (Barbacid, 1987; Feig, 1999; Scheffzek et al., 1997) and orthologous Rabs (Brondyk et al., 1993; Burstein et al., 1992; Chen et al., 2002), EGFP–RAB26T77N would abolish Mg2+-coordinated GTP binding in the G2 box region, and EGFP–RAB26Q123L would disrupt GTP hydrolysis mediated by the G3 box region (Fig. 5B). When transfected, EGFP–RAB26T77N showed a diffuse cytoplasmic distribution (Fig. 5C), and EGFP–RAB26Q123L showed a more-dispersed vesicular distribution (Fig. 5D). Neither EGFP–RAB26T77N nor EGFP control localized with LAMP1, and EGFP–RAB26Q123L showed some correlation with lysosomes. Specifically, although most lysosomes still colocalized with EGFP–RAB26Q123L, some EGFP–RAB26Q123L signal overlapped with non-LAMP1-labeled structures (Fig. 5D,E; Pearson's coefficient of 0.384 compared to wild-type EGFP–RAB26 with substantial co-labeling and Pearson's coefficient of 0.702). We next analyzed RAB26–EGFP-transfected cells in the presence of a prenylation inhibitor and found wild-type RAB26 no longer trafficked to lysosomes, and when we blocked GTP hydrolysis with non-hydrolyzable GTP analogs, we found that RAB26–EGFP trafficked only partially to lysosomes, phenocopying the Q123L GTP-hydrolysis-deficient mutant (data not shown). To further investigate extra-lysosomal EGFP–RAB26Q123L vesicle localization, we co-stained with EEA1, giantin and the CI-M6PR (data not shown) revealing no substantial colocalization with Golgi, but partial overlap with the endosomal compartments. Thus, we conclude RAB26 requires prenylation and GTP binding to target to lysosomes, and dynamic GTP cycling to maintain localization on lysosomes and avoid trafficking to other membranes as lysosomes recycle (Sridhar et al., 2013).
Fig. 5.
Design and characterization of RAB26 point mutants. (A) Amino acid sequence of EGFP–RAB26 plasmid with EGFP sequence (green) and human RAB26 sequence (black) labeled. EGFP–RAB26T77N and EGFP–RAB26Q123L were constructed by the indicated point mutations (shown in red) in the RAB26 sequence, converting a threonine residue into an asparagine residue, and a glutamine residue into a leucine residue, respectively. (B) A ribbon diagram of the crystal structure of RAB26 generated by FirstGlance in Jmol (ver. 1.03) from http://bioinformatics.org/firstglance/fgij//fg.htm?mol = 2G6B (Wang et al., 2006)]. The atomic structure of GNP-bound RAB26 is shown in the diagram; the threonine (T77) and glutamine (Q123) that are within the GNP-binding pocket are labeled. (C) Confocal fluorescence microscopy of the distribution of EGFP–RAB26T77N (green) in transfected HGC-27 cells immunostained with anti-LAMP1 antibody (red). The cell border is outlined; N, nucleus. Right panels show epifluorescence microscopy of EGFP–RAB26T77N (green) co-stained for LAMP1 (red). (D) Confocal fluorescence microscopy of the localization of EGFP–RAB26Q123L (green) in transfected HGC-27 cells immunostained with anti-LAMP1 antibody (red). Insets indicate areas of overlap of EGFP–RAB26Q123L vesicles with lysosomes (arrowheads). The cell border is outlined; N, nucleus. Right panels show epifluorescence microscopy of EGFP–RAB26Q123L (green) co-stained for LAMP1 (red). (E) Pearson's correlation coefficient quantification of pixel-to-pixel colocalization of multiple cells transfected with EGFP–RAB26, EGFP–RAB26T77N, EGFP–RAB26Q123L and GFP with LAMP1 lysosomes. ***P<0.001, one-way ANOVA test with Dunnett's correction. Scale bars: 20 µm.
Increased RAB26 is sufficient to traffic lysosomes centripetally
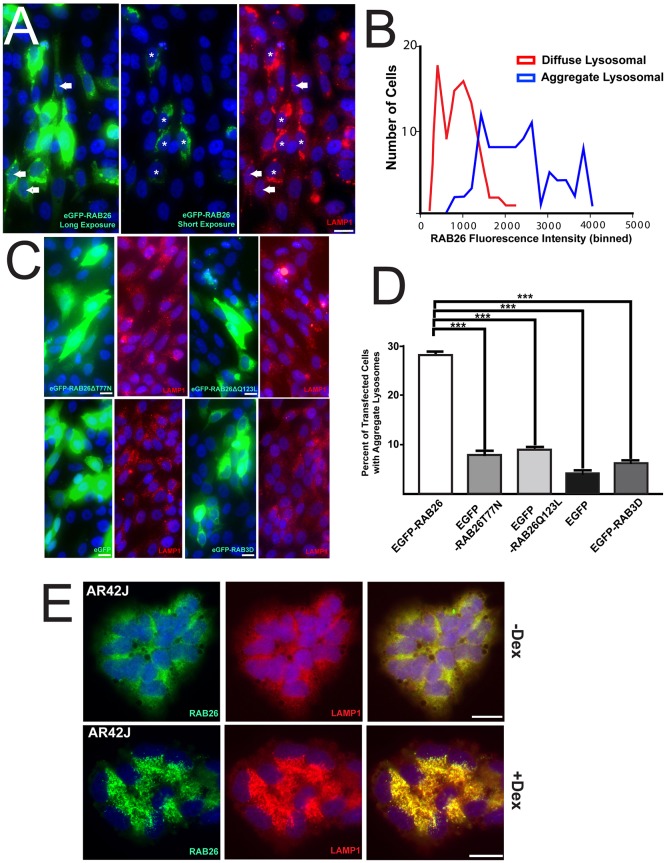
To determine the consequences of RAB26 association with lysosomes, we analyzed the effects of scaling up RAB26 expression, which occurs in tissues when differentiating secretory cells increase RAB26 levels in response to the transcription factor MIST1. To specifically analyze the effects of scaling up RAB26 without the effects of other MIST1 targets, we took advantage of the varying levels of expression induced by transient transfection in HGC-27 cells, whose endogenous RAB26 levels are usually low. We noticed that HGC-27 cells with the highest levels of RAB26, as determined by EGFP fluorescence intensity, showed lysosomal coalescence into a central, perinuclear region, whereas cells with lower levels showed more-diffuse peripheral lysosomes (Fig. 6A,B). Moreover, when we quantified this on a cell-by-cell basis, we found cells with coalesced lysosomes almost always showed higher levels of RAB26 [a mean fluorescence intensity (MFI)>1000 on a normalized 16-bit scale] than cells retaining diffuse lysosomes (MFI<1000; Fig. 6B). Transfection of the two RAB26 point mutants did not recapitulate this lysosome-repositioning phenomenon (Fig. 6C,D), indicating that the ability of RAB26 to bind and cycle GTP is necessary for this process. In addition, cells transfected with EGFP or EGFP–RAB3D also failed to show centripetal lysosomal movement (Fig. 6C,D), revealing that this process is independent of GFP overexpression and is RAB26 specific. Quantification showed that, of cells with detectable EGFP–RAB26, 30% displayed this central perinuclear lysosomal phenotype (Fig. 6D), whereas only ∼5–10% of EGFP-positive cells transfected with various control constructs (EGFP alone, EGFP–RAB26T77N, EGFP–RAB26Q123L and EGFP–RAB3D) showed this pattern of lysosome distribution. These other transfected cells showed no change in lysosome distribution no matter the levels of expression.
Fig. 6.
Scaling of RAB26 expression leads to perinuclear localization of lysosomes. (A) Fluorescence microscopy of HGC-27 cells transfected with EGFP–RAB26 (green) and stained for anti-LAMP1 (red). A long and short exposure is shown to highlight the differences in RAB26 levels with the cells expressing highest (*) and more moderate (solid white arrows) levels of RAB26 indicated. (B) Histogram of EGFP–RAB26-transfected cells immunostained for LAMP1 with either diffuse (red line) or clustered (blue line) LAMP1 lysosomal distribution plotted against the binned fluorescence intensity of EGFP–RAB26. (C) Epifluorescence microscopy of HGC-27 cells transfected with EGFP–RAB26T77N, EGFP–RAB26Q123L, EGFP–RAB3D and EGFP expression plasmids (green) co-stained with anti-LAMP1 (red). (D) The percentage of HGC-27 cells transfected with EGFP–RAB26, EGFP–RAB26T77N, EGFP–RAB26Q123L, EGFP–RAB3D and EGFP showing aggregated lysosomes from multiple cell fields with coalesced perinuclear LAMP1 structures quantified by cell count and plotted. ***P<0.001, one-way ANOVA test with Dunnett's correction. (E) Immunofluorescence microscopy of untreated (–Dex) or dexamethasone treated (+Dex) AR42J cells stained for endogenous RAB26 (green) and LAMP1 (red). Scale bars: 20 µm.
In addition, we examined the AR42J pancreatic acinar cell system, in which we are able to induce differentiation, increase MIST1 expression and scale up RAB26 from moderate to high levels. Increasing RAB26 (marked by anti-RAB26) correlated with striking centripetal coalescence of LAMP1 vesicles to the perinuclear region (Fig. 6E). In other words, endogenous RAB26 scaled up by cellular differentiation phenocopied transfection-mediated RAB26 scaling.
Localization of other vesicular structures including endosomes, the Golgi and late endosomes (data not shown and supplementary material Fig. S3A,B) were unaffected by transfection of EGFP–RAB26 or any of its mutant forms. Transfection of RAB26 and RAB26 mutants had no significant effect on: (1) overall levels of LAMP1 protein in cell cultures; (2) levels of cathepsin D; (3) lysosome-mediated processing of cathepsin D to its smaller active form; and (4) mTOR/S6K signaling, which depends in part on lysosomal function (supplementary material Fig. S4A). In summary, RAB26 specifically localizes with lysosomes in a GTP/GDP-dependent manner, and, as RAB26 levels are scaled up, it reorients only lysosomes and not other vesicular compartments. Although global lysosomal function does not appear to be affected by RAB26, more experiments would be needed to analyze how RAB26 might mediate the many additional aspects of lysosome trafficking and function.
RAB26 coordinates reorganization of mitochondria
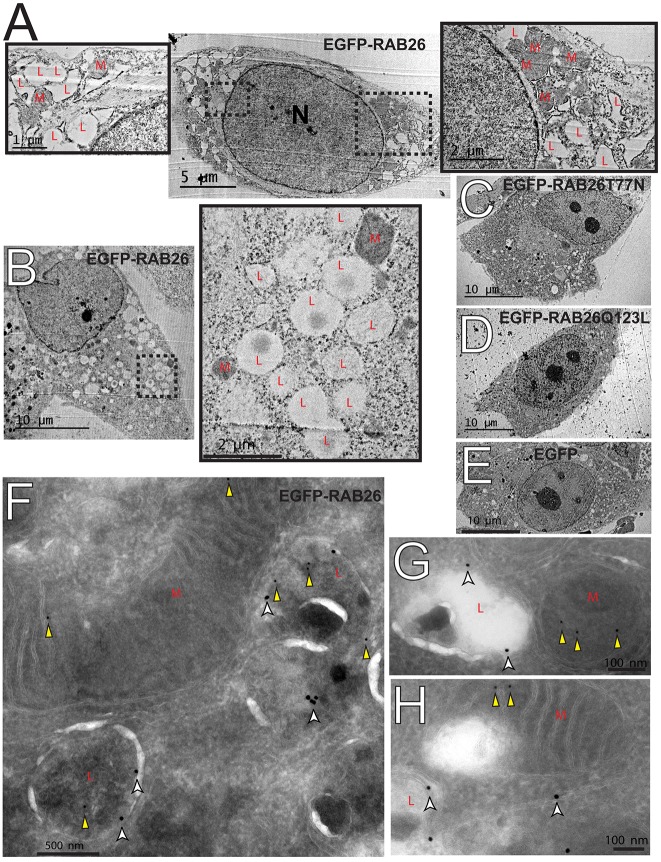
To begin to address the potential cellular effects of scaling up RAB26, we returned to ultrastructural analyses of transfected cells. Supporting our immunofluorescence and immunoelectron microscopy data, we found large clusters of electron-lucent vesicles, morphologically consistent with lysosomes (Fig. 2D), only in the RAB26-transfected cells (Fig. 7A–E). In our control cells, or in cells transfected with EGFP–RAB26T77N, EGFP–RAB26Q123L or EGFP, lysosomes were smaller and dispersed throughout the cytoplasm (Fig. 7C–E). We noticed that the lysosome clusters in the EGFP–RAB26-expressing cells largely excluded mitochondria (Fig. 7A,B). Thus, instead of the diffuse mitochondrial network seen in cells transfected with various control constructs (Fig. 7C–E), the mitochondria in RAB26-transfected were clustered into distinct cellular neighborhoods. In fact, upon returning to our initial immunoelectron micrographs, we found RAB26 occasionally labeling mitochondrial membranes (3.17% of total labeling) adjacent to LAMP1-labeled lysosomes. Thus, RAB26 might also directly or indirectly affect cellular mitochondrial organization.
Fig. 7.
RAB26 overexpressing cells show ultrastructural rearrangements. (A,B) Transmission electron micrographs of EGFP–RAB26-expressing HGC-27 cells with clustered electron-lucent lysosome vesicles (‘L’) highlighted in the insets with electron-dense mitochondrial clusters indicated (‘M’). N, nucleus. Transmission electron micrographs of HGC-27 cells transfected with (C) EGFP–RAB26T77N, (D) EGFP–RAB26Q123L and (E) EGFP. (F–H) Panels of immunoelectron microscopy of EGFP–RAB26-transfected HGC-27 cells labeled with LAMP1 (18-nm gold particles, white arrowheads) and RAB26 (12-nm gold particles, yellow arrowheads) with mitochondria (‘M’) and lysosomes (‘L’) highlighted.
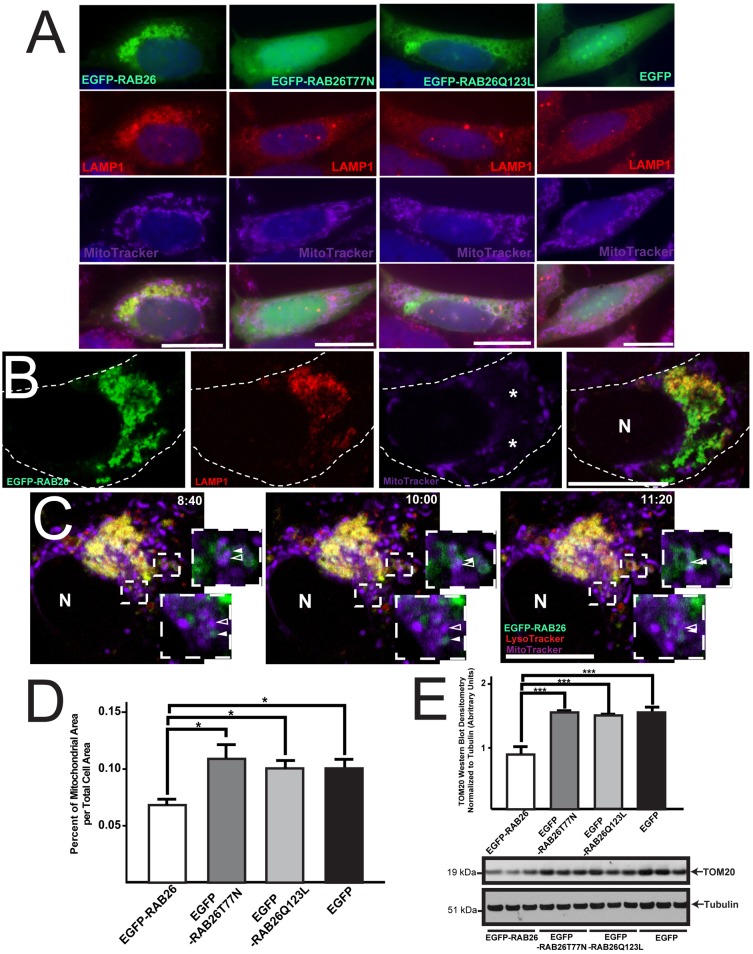
Accordingly, in cells expressing abundant EGFP–RAB26, we found that mitochondria, like lysosomes, were also clustered but into distinct subcellular regions that were specifically free of lysosomes (Fig. 8A,B). Control cells showed mitochondria organized in the expected diffuse, lattice-like network throughout the cell (Fig. 8A). Using live-cell confocal microscopy, we were able to observe mitochondrial interaction on the borders of the EGFP–RAB26 and LAMP1 lysosomal clusters (supplementary material Movie 2) including temporary areas of overlap with apparent mitochondrial–lysosomal fusion or membrane exchange, as highlighted in frames from supplementary material Movie 2 (Fig. 8C). When we quantified the cell area of mitochondria as a percentage of the total cell area in EGFP–RAB26-transfected cells, we found it to be significantly reduced relative to controls (Fig. 8D). In fact, when whole cellular populations were analyzed for mitochondrial density by western blotting for the mitochondrial outer membrane marker TOM20, we found a significant reduction (n = 3 independent transfections per construct) only when functional RAB26 was transfected, indicating trimming of mitochondria across the whole population (Fig. 8E). However, when mitochondrial function was analyzed using dyes sensitive to mitochondrial membrane potential [CMXRos (Poot et al., 1996)] and superoxide damage production [MitoSOX Red (Mukhopadhyay et al., 2007)], we found that although mitochondria were reduced in cells overexpressing RAB26, those that remained were largely intact and functional, as was the case for all controls analyzed (supplementary material Fig. S4B,C). Taken together, our data show that RAB26 repositions lysosomes and reorganizes mitochondria.
Fig. 8.
RAB26-induced lysosomal clustering leads to mitochondrial reorganization. (A) Fluorescence microscopy of HGC-27 cells transfected with EGFP–RAB26, EGFP–RAB26T77N, EGFP–RAB26Q123L and EGFP (green), co-stained for lysosomes (anti-LAMP1 antibody, red) and mitochondria (MitoTracker, purple). (B) Confocal section of EGFP–RAB26-expressing cell (green) stained for lysosomes (anti-LAMP1 antibody, red) and mitochondria (MitoTracker, purple). Perinuclear mitochondria free zones are indicated (*). The cell border is outlined; N, nucleus. (C) Live confocal timelapse microscopy of EGFP–RAB26 (green), LysoTracker-labeled lysosomes (red) and MitoTracker-labeled mitochondria (purple) vesicle dynamics. Insets highlight RAB26 (green, filled white arrowhead) and mitochondria (purple, open white arrowhead) interactions (time stamps of images from supplementary material Movie 2 are shown. N, nucleus. (D) Quantification of mitochondrial area over total cell area measured from multiple HGC-27 cells transfected with EGFP–RAB26, EGFP–RAB26T77N, EGFP–RAB26Q123L and EGFP. *P<0.05, one-way ANOVA test with Dunnett's correction. (E) Densitometry quantification of western blots (triplicate experiments) of mitochondrial membrane density using TOM20 and α/β tubulin markers for HGC-27 cells transfected with EGFP–RAB26, EGFP–RAB26T77N, EGFP–RAB26Q123L and EGFP. ***P<0.001, one-way ANOVA test with Dunnett's correction. Scale bars: 20 µm.
DISCUSSION
Here, we find, in multiple cell types, that RAB26 associates with and traffics lysosomes, which is surprising both because lysosome-associated Rabs are rare and because RAB26 was initially characterized in the rat pancreas as being associated with secretory vesicles (Wagner et al., 1995). However, these earlier results were generated using antibody staining in fixed tissue sections and cell fractionation density gradient enrichment of granules (Nashida et al., 2006; Yoshie et al., 2000). In fact, secretory granules and lysosomes are not easily distinguishable by density gradient properties (Pasquali et al., 1999), and the mature secretory granule markers, syntaxin 6 and γ-adaptin, used as markers of secretory granules in previous analyses of RAB26, are also involved in Golgi sorting to lysosomes and late endosomes (Ghosh et al., 2003; Robinson, 1990). Those markers have been found to be specifically removed from mature secretory granules (Klumperman et al., 1998). Moreover, in a large screen of mast cell Rabs that regulate secretion, transfected RAB26, which mast cells do not normally express, was also found to be perinuclear and associated with a vesicular compartment that was expected to be LAMP1 positive (Azouz et al., 2012).
Recent studies have found that RAB26 is associated with RAB11 recycling endosomes (Chan et al., 2011) in Drosophila neurons, and a giantin-positive Golgi sorting compartment (Li et al., 2012) when transfected into HEK293 cells. These results would seem to contradict those presented here, although we have focused here exclusively on exocrine secretory cells, trying to mimic the effects of scaling RAB26 as occurs in differentiating cells in tissue. Thus, RAB26 might traffic differently depending on cell type; however, there are other possible explanations for our seemingly differing findings. There is overlap between Golgi, endosome and lysosome markers because lysosomes mature through sorting and trafficking (Ghosh et al., 2003; Griffiths et al., 1990; Luzio et al., 2000; Moore et al., 2004; Peden et al., 2004; Saftig and Klumperman, 2009). RAB7, the most well-studied lysosomal Rab protein (Bucci et al., 2000), has also been implicated in endosomal trafficking (Feng et al., 1995). In fact, its close relative, RAB7B, has been shown to colocalize with both Golgi and lysosomal markers (Progida et al., 2010). The transient nature of these dynamic and constantly communicating compartments (Saraste and Goud, 2007) complicate all subcellular localization studies. Here, we have used multiple secretory cell types and multiple modalities to examine both endogenous and transfected RAB26, which we argue reinforces our conclusions that RAB26 is predominantly lysosome-associated in dedicated secretory cells.
That lysosomes cycle between peripheral and central regions has been well documented, with various motor proteins (Hollenbeck and Swanson, 1990; Lin and Collins, 1992), and cytoskeletal filaments (Cordonnier et al., 2001; Matteoni and Kreis, 1987) implicated in this traffic. Much work has also elucidated the role of the Rab family in lysosomal movement and function. Specifically, RAB4, RAB5, RAB9 and RAB11 have been shown to coordinate endosomal traffic into and out of maturing lysosomes (Saftig and Klumperman, 2009). However, only RAB7 has been well established as a key cellular regulator of late endosome to lysosome trafficking, and overexpression of RAB7 causes central lysosome movement, whereas RAB7 T22N and N125I point mutants leave lysosomes dispersed (Bucci et al., 2000). There is some evidence that two other ubiquitously expressed Rabs, RAB34 (Wang and Hong, 2002) and RAB36 (Chen et al., 2010), also function to redistribute lysosomes centrally (Colucci et al., 2005). Interestingly, these Rabs share a common effector, RAB7-interacting lysosomal protein (RILP), which is crucial for inducing dynein-mediated movement (Cantalupo et al., 2001; Jordens et al., 2001; Wang and Hong, 2005). RAB26 might also mediate lysosomal rearrangement through RILP interactions.
Mitochondria are constantly pruned and recycled in post-mitotic cells. Under conditions of oxidative stress, the normal steady-state mitochondrial recycling (Lipsky and Pedersen, 1981; Terman et al., 2010) is scaled up to maintain physiological function and forestall apoptosis in a process that depends on lysosome-mediated pruning (Green and Kroemer, 2004). Mitochondria are delivered to lysosomes via many routes (Ashrafi and Schwarz, 2013). For example, damaged mitochondria, as tracked by their membrane marker TOM20, can be delivered directly to lysosomes (Soubannier et al., 2012).
Our findings indicate that scaling up RAB26 might enhance the ongoing baseline direct trafficking events between lysosomes and mitochondria. Little is known about lysosome–mitochondria interactions in tissues that are not under constant oxidative damage. In exocrine tissues, mitochondria play a crucial role in secretagogue-mediated Ca2+ signaling (Tinel et al., 1999), which depends on their subcellular positioning (Park et al., 2001). Increasing RAB26 might be an inherent mechanism for secretory cells to reorganize mitochondria and to focus them in areas where they can facilitate Ca2+ delivery to secretory granules. We observed only a partial reduction in mitochondrial numbers, no matter how high expression of RAB26 was in a given cell; thus, RAB26 might prune or cluster, rather than destroy, mitochondria.
Our studies have confirmed that RAB26 is not ubiquitous but, rather, is specifically expressed in only certain cell lineages. Thus, we expect that it would be sufficient but not required for lysosome-mediated mitochondrial reorganization because other more universal mechanisms must be present in all cells. In fact, our two point mutants do not act as dominant negatives in affecting basal lysosomal movement or preventing mitochondrial degradation initiated by oxidative damage (data not shown). In addition, siRNA-mediated knockdown of endogenous RAB26 similarly had no effect on lysosomes or mitochondria (data not shown). RAB3D, another direct MIST1 target and secretory-cell-specific Rab protein shows a similar ability to ‘scale-up’ vesicle dynamics; when overexpressed in acinar cells, RAB3D enhances regulated secretion (Ohnishi et al., 1997). However, when Rab3d is deleted regulated secretion is unaffected (Riedel et al., 2002). Thus, these exocrine-cell-specific Rabs represent a subset of ‘scale-able’ cellular effectors that enhance cellular dynamics.
In digestive-enzyme-secreting tissues, such as the pancreas and stomach, RAB26 expression is almost wholly dependent on the transcription factor MIST1 (Tian et al., 2010). In the absence of MIST1 and, thus RAB26, exocrine secretory cells display aberrant secretory vesicle organization, maturation and secretion (Direnzo et al., 2012; Johnson et al., 2004; Luo et al., 2005; Pin et al., 2001; Ramsey et al., 2007; Tian et al., 2010). Interestingly, Mist1−/− exocrine cells also exhibit aberrant lysosomal trafficking (Capoccia et al., 2013; Direnzo et al., 2012; Pin et al., 2001). Furthermore, in cells expressing wild-type MIST1 mitochondria are organized into distinct subcellular neighborhoods (similar to what we see with increased RAB26), whereas in Mist1−/− cells, mitochondria are dispersed throughout the cell, resulting in abnormal Ca2+ flux (Luo et al., 2005). From our previous study, no other direct MIST1 targets seem to have clear roles in lysosome movement, mitochondria reorganization or Rab protein function (e.g. no guanine-nucleotide-exchange factors, GTPase-activating proteins or other identified Rab effectors are known MIST1 targets) (Tian et al., 2010). Thus, RAB26 function might be an important and crucial unrecognized aspect in MIST1-regulated cell secretory function.
Loss of MIST1 results in increased susceptibility to pancreatitis (Alahari et al., 2011; Kowalik et al., 2007; Zhu et al., 2004), and, in wild-type mice, pancreatitis leads to silencing of Mist1 (Shi et al., 2013; Shi et al., 2009). Acute pancreatitis is caused by inappropriate interaction between lysosomal enzymes and zymogen constituents of secretory granules (Gaiser et al., 2011; Halangk et al., 2000; Saluja et al., 1997), which leads to mitochondrial disorganization (Mareninova et al., 2006; Odinokova et al., 2009). Thus, one mechanism for how MIST1 might suppress acute pancreatitis is by trafficking of lysosomes and mitochondria through RAB26. Future experiments will have to address whether loss of RAB26 in MIST1-expressing cells causes disrupted lysosome/mitochondrial-mediated damage, although deletion of tissue-specific Rabs in vivo has not been that informative [e.g. Rab3d-null mice have only a mild phenotype (Riedel et al., 2002)], perhaps because they are sufficient for scaling up specific aspects of secretion but not absolutely required.
Our data clearly show that coalescence of lysosomes and mitochondria is caused by expression of higher levels of RAB26. The changes occur whether endogenous RAB26 expression is increased by MIST1 during differentiation or if it is directly transfected to higher levels. Thus, our studies exemplify how a transcription factor like MIST1 can scale up a specific target gene during maturation to redistribute organelles in a way that enhances the specific physiological function the cell must perform in the adult organism. In other words, we provide mechanistic evidence for how transcription factors like MIST1 can act as scaling factors to control cell architecture simply by increasing expression of specific genes that encode cellular effectors (Mills and Taghert, 2012). It will be interesting to determine how the scaling of RAB26 by MIST1 might work in more detail, and whether RAB26 acts as a rheostat (i.e. the more RAB26 expression increases, the more centralized lysosomal trafficking there is) or more like a binary switch (i.e. low levels do not change lysosomal distribution, high levels cause lysosomes to coalesce centrally).
In summary, we propose a mechanism whereby a transcriptionally controlled Rab protein can regulate crucial physiologically relevant cell structural adaptations. RAB26, through its ability to reposition lysosomes, can, in turn, effect mitochondrial reorganization. Mitochondria and lysosomes are crucial for the formation, maintenance and secretion of zymogen granules. Therefore, we propose that RAB26 is a novel, lysosome-associated, tissue- and cell-type-specific small protein GTPase that can be ‘scaled-up’ by the transcription factor MIST1 to enhance the secretory function of exocrine cells.
MATERIALS AND METHODS
GeneChip analysis
Human tissue expression data was obtained from the LSBM RefExA database (http://sbmdb.genome.rcast.u-tokyo.ac.jp/refexa/) (Ge et al., 2005). Human cell line expression data was prepared as previously described (Tian et al., 2010). H&E staining and mouse gene chip arrays were generated as previously described (Capoccia et al., 2013). Mice were maintained in a specific pathogen-free barrier facility. All experiments involving animals were performed according to protocols approved by the Washington University School of Medicine Animal Studies Committee. Heat maps of RAB26 expression levels were generated by dChip analysis (Zhong et al., 2003). Correlation between RAB26 probesets was calculated using GraphPad Prism to generate Pearson's correlation coefficients based on raw read values.
Cell lines and transfection
HGC-27 and AGS cells were maintained as previously described (Tian et al., 2010). 5637 cells (gift from Indira Mysorekar, Washington University) were maintained at 37°C in 5% CO2 in RPMI 1640 supplemented with 10% fetal bovine serum, 0.9% glutamine, 0.4% HEPES, 1% sodium pyruvate, 2.5% glucose, and 100 ng/ml each of penicillin and streptomycin. AR42J cells (ATCC, Manassas, VA) were maintained in F12K medium supplemented with 20% fetal bovine serum, 1.0% glutamine, and 100 ng/ml each of penicillin and streptomycin. Dexamethasone (100 nM, Sigma-Aldrich, St. Louis, MO) was added to cells for 48 h to induce secretory differentiation. For transient transfection, AGS or HGC-27 cells were transfected via electroporation using Nucleofector II (Lonza, Basel, Switzerland), program B-023, and cell line transfection solution V. For each electroporation, 3 µg PGC–RFP, 5 µg EGFP–RAB26, 5 µg EGFP–RAB26T77N, 5 µg EGFP–RAB26Q123L, 5 µg EGFP–RAB3D, 3 µg CTSD–RFP (see below for details), or 3 µg pmaxGFP (Lonza) plasmid was used. In some experiments and other cell lines, we also used TransIT-2020 and TransIT-LT1 Transfection Reagent (Mirus, Madison, WI) according to manufacturer's protocol. For visualization, cells were replated on Lab-Tek Chamber Slide 4-well Permanox slides (Thermo Fisher Scientific, Rochester, NY) and routinely analyzed 24–72 h post transfection. MIST1–EGFP expressing stable cell lines were established as previously described (Tian et al., 2010).
Immunofluorescence and cell imaging
For immunofluorescence analysis, cultured cells were transfected and stained as described previously (Tian et al., 2010). Fluorescence microscopy and imaging were performed using a Zeiss Axiovert 200 microscope with 20× (Plan-Apochromat, 0.8 NA), 40× (Plan-Neofluar 0.85 NA) and 63× objectives (Plan-Apochromat, 1.4 NA) with Axiocam MRM camera and AxioVision software. Additional, confocal microscopy and imaging were performed using a Zeiss LSM510 microscope with 40× (EC Plan-Neofluar, 0.75 NA) and 63× (Plan-Apochromat, 1.4 NA) objectives using LSM510 software. Live-cell imaging was performed using an Olympus FluoView FV1000 microscope equipped with a 60× objective (PLAPON 60XO, 1.42 NA) maintained at 37°C in 5% CO2, with images captured using the FV10-ASW software or using a Zeiss LSM510 confocal microscope with 63× objectives (Plan-Apochromat, 1.4 NA) maintained at 37°C in 5% CO2 with images captured using the Zeiss LSM510 software. Contrast (maximal, minimal and midtone) adjustment and fluorescent channel overlay and pseudocoloring were performed with Photoshop (Adobe Systems, San Jose, CA). All adjustments were performed on the entire image equally.
The primary antibodies used for cell immunostaining were: rabbit anti-amylase (1∶100, Calbiochem, CA), rabbit anti-RAB26 (1∶100, ProteinTech Group, Chicago, IL), rabbit anti-giantin (1∶1000, Covance), rabbit anti-EEA1 (1∶250, Abcam), rabbit anti-furin (1∶500, Thermo Scientific), goat anti-calregulin (1∶100, Santa Cruz Biotechnology), rabbit anti-GM130 (1∶250, BD Biosciences, San Jose, CA), sheep anti-TGN46 (1∶250, Serotec, Oxford, UK), rabbit anti-CI-M6PR (1∶500), mouse anti-AP1 (1∶500), rabbit anti-CTSD (1∶150), mouse anti-GGA2 (1∶500) (gifts from Stuart Kornfeld, Washington University), mouse anti-LAMP1 (1∶40, Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-LAMP1 (1∶50, clone H4A3, developed by J. Thomas August and James E. K. Hildreth and obtained from the Developmental Studies Hybridoma Bank under the auspices of the NICHD maintained by The University of Iowa), rabbit anti-RAB7 (1∶150, Cell Signaling Technology, Danvers, MA). Secondary antibodies used were Alexa-Fluor-488-, -594- and -647-conjugated donkey anti-goat, anti-rabbit, anti-sheep and anti-mouse Ig antibodies (1∶500; Invitrogen, Carlsbad, CA). LysoTracker and MitoTracker (Invitrogen) were used at 75 nM and 250 nM, respectively, according to manufacturer's instructions. MitoTracker CMXRos and MitoSOX Red (Invitrogen) were used at 100 nM and 2 µM, respectively, according to manufacturer's instructions.
Immunofluorescence quantification
For pixel-to-pixel overlap of EGFP–RAB26, EGFP–RAB26T77N, EGFP–RAB26Q123L, and GFP with LAMP1, cells were transfected and stained for LAMP1 as described and confocal images were taken from at least 10 cells per condition. Thresholded Pearson's correlation coefficients were calculated using Volocity image analysis software (PerkinElmer, Waltham, MA).
For lysosome distribution quantification, cells were transfected as described above with EGFP–RAB26, EGFP–RAB26T77N, EGFP–RAB26Q123L, EGFP–RAB3D or GFP plasmids. Post transfection, lysosomes were stained with a LAMP1 antibody as described. Blinded to treatment conditions, 16-bit images captured in Zeiss Axiovision software were analyzed with ImageJ software as follows. Central lysosomes (bright concentrated staining around perinuclear region), diffuse lysosomes (disperse cytoplasmic staining), transfected cells and total cells, determined by total nuclei counted using the technique developed by Selinummi et al (Selinummi et al., 2005), were scored. 65 low-power fields comprising three independent experiments were examined totaling ∼10,000 cells counted per condition.
For enhanced green fluorescent protein (EGFP)–RAB26 fluorescence intensity quantification, cells were transfected with EGFP–RAB26 and stained for LAMP1. 16-bit images captured in Zeiss Axiovision software were analyzed with ImageJ software as follows. We determined mean green cytoplasmic fluorescence intensities in each region after subtraction from the median background (green in an area with no cells). These same cells were then scored individually for their lysosome distribution (as defined above). A total of 180 cells were scored across three transfection experiments.
For mitochondrial area quantification, cells were transfected and stained as previously described. 16-bit images captured in Axiovision software were analyzed with ImageJ software as follows. The area selection tool was used to outline mitochondria, and the area was measured using the ‘Analyze area’ measurement tool. Multiple cells were scored across three transfection experiments.
Plasmid preparation
PGC–RFP, EGFP–RAB26 and EGFP–RAB26T77N mammalian expression plasmids were constructed as previously described (Tian et al., 2010). To generate EGFP–RAB26Q123L, EGFP–RAB26 was mutagenized by site-directed mutagenesis to convert the glutamine residue at amino acid position 123 to a leucine, and a new XhoI restriction site was introduced. The pcDNA3.1-RAB26 plasmid was constructed using the same approach as EGFP–RAB26, using backbone plasmid pcDNA3.1/V5-His-TOPO (Invitrogen) with the first amino acid of the His-tag modified to a stop codon. The monomeric EGFP–RAB3D expression plasmid was constructed with the coding region of human RAB3D cDNA (IMAGE identification number 3861912; Open Biosystems) added in-frame to the C-terminus of EGFP, replacing the RAB26-coding region in EGFP–RAB26 by ribocloning (Barnes, 1994; Barnes, 2006). PCR was performed on the vector region by using primers DNA3as (5′-GGCAATTCCACCACACTGGACTAGu-3′) and pcD3LKs (5′-GGGCAATTCTGCAGATATCCAGCAc-3′) on EGFP-RAB26 DNA. The RAB3D target was PCR amplified using DNA3s (5′-ACTAGTCCAGTGTGGTGGAATTGCc-3′) and hRAB3D-DNA3b (5′-GGGTGTCTCCAGCTGATGCCATGGCGGCAATTCCACCACACTGGACTAGTG-3′) with pcD3LKas (5′-GTGCTGGATATCTGCAGAATTGCCc-3′) and hRAB3D-pcD3LKs (5′-GATGCTCCAGCCCCCCAGCCCTCCTCTTGTAGCTGCTAGGGCAATTCTGCAGATATCCAGCAC-3′). The pcDNA3.1 CTSD–RFP fusion construct was generated with the human CTSD region of pcDNA3.1-hCTSD-myc (a gift from Stuart Kornfeld) added in-frame to the N-terminus of RFP, replacing the PGC coding region in PGC–RFP by ribocloning. PCR was performed on the vector region by using primers DNA3as and RFPs (5′-GCCTCCTCCGAGGACGTCAu-3′). The hCTSD target was PCR-amplified using DNA3s and hCTSD-DNA3b (5′-GCTGGAGGGCTGCATGGCGGCAATTCCACCACACTGGACTAGT-3′) with RFPas (5′-ATGACGTCCTCGGAGGAGGc-3′) and hCTSD-RFPs (5′-CGAGGCTGCCCGCCTCGCCTCCTCCGAGGACGTCAT-3′). All construct coding regions were verified to be correct by DNA sequencing.
Density gradient ultracentrifugation lysosomal enrichment
Lysosomes were isolated with the Pierce Lysosomal Enrichment Kit of Tissue and Cultured Cells (Rockford, IL) according to manufacturer's protocol. Briefly, cells were transfected as described, pelleted, and homogenized using a Dounce tissue grinder. Nuclei were removed and then lysate was ultracentrifuged on prepared Optiprep density gradients using Beckman Coulter Optima L-100 XP ultracentrifuge (Fullerton, CA). Fractions were removed and subjected to western blot analysis.
Western blot analysis
Mouse tissue was flash frozen in liquid nitrogen and homogenized in RIPA buffer using a PowerGen700 (Fischer Scientific, Pittsburg, PA). Cells were transfected as described and lysed in RIPA buffer. Proteins were quantified by DC protein assay (Bio-Rad) and then separated on NuPAGE Bis-Tris gels (Invitrogen), transferred onto Immobilon polyvinylidene difluoride (Millipore, Bedford, MA) or Amersham Hybond ECL nitrocellulose (GE Healthcare, Buckinghamshire, UK) membranes, and detected by Immobilon chemiluminescence (Millipore). Primary antibodies used were mouse anti-LAMP1 (1∶1000, Developmental Studies Hybridoma Bank), rabbit anti-GFP (1∶1500, Santa Cruz Biotechnology), rabbit anti-RAB26 (1∶800, ProteinTech Group Chicago, IL), rabbit anti-MIST1 [1∶500 (Pin et al., 2000)], mouse anti-TOM20 (1∶1000, BD Biosciences), rabbit anti-cathepsin D (1∶2000, a gift from Stuart Kornfeld, Washington University), rabbit anti-mTOR (1∶1000, Cell Signaling Technology), rabbit anti-phospho-p70S6 kinase (1∶1000, Cell Signaling Technology), rabbit anti-p70 S6 kinase (1∶1000, Cell Signaling Technology), and rabbit anti-α- and β-tubulin (α/β tubulin) (1∶1500, Cell Signaling Technology). Secondary antibodies were horseradish-peroxidase-conjugated donkey anti-rabbit and anti-mouse Ig (Santa Cruz Biotechnology, Santa Cruz, CA).
Quantifications of immunoblots were performed by scanning 16-bit images into ImageJ. Band intensities for TOM20 and α/β tubulin were selected and calculated by using the ‘Analyze mean gray value’ measurement tool. Standardized values were calculated determining the ratio of TOM20 signal to α/β tubulin signal.
Protein structure analysis
RAB26 protein structure was obtained from the Protein Data Bank, accession code 2G6B (Wang et al., 2006). The image was generated using Protein Workshop (Moreland et al., 2005) with surfaces (Xu and Zhang, 2009).
Electron microscopy
For transmission electron microscopy studies, cells were transfected as previously described and plated on Lab-Tek Chamber Slide 4-well Permanox slides (Thermo Fisher Scientific). After being rinsed in PBS, they were fixed in modified Karnovsky's fixative (2.5% glutaraldehyde, 2% paraformaldehyde in 0.1 mmol/liter cacodylate buffer). TEM thin sections were cut directly from cell cultures embedded on the original Permanox substrate.
For immunoelectron microscopy studies, HGC-27 cells transfected with EGFP–RAB26 or EGFP control were trypsinized, pelleted, and fixed on ice for 1 h in PBS, 4% paraformaldehyde and 0.05% glutaraldehyde. Subsequent sample processing was performed as previously described (Beatty, 2008). After incubation with primary antibodies against RAB26 (1∶50, ProteinTech Group) and LAMP1 (1∶25, Developmental Studies Hybridoma Bank) sections were then washed and probed with anti-rabbit conjugated to 12-nm and anti-mouse conjugated to 18-nm colloidal gold (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Quantification of the labeling specificity of RAB26 and LAMP1 was performed by calculating cellular areas of lysosomes, mitochondria, nucleus, cytoplasm, and plasma membrane using the ImageJ ‘Area selection’ tool and ‘Analyze area’ measurement tool. Areas were compared to counted 12-nm RAB26 and 18-nm LAMP1 labeling within 100 nm of the same cellular areas. Multiple cells from five fields of view were counted.
Graphing and Statistics
All graphs and statistics were made with GraphPad Prism and then visualized using Adobe Illustrator. Statistical analysis was, in the case of simple control-versus-experimental condition comparison, by Student's t-test. Otherwise, significances were determined by one-way analysis of variance (ANOVA) test with Dunnett's multiple-comparison correction.
Supplementary Material
Acknowledgments
The authors wish to thank the following people at Washington University: Edward Oates and Xavier Jirau for plasmid preparation assistance, Wandy Beatty and Karen Green for EM sample preparation and expertise, Saso Cemerski, Wandy Beatty and Dennis Oakley for confocal microscopy help, Indira Mysorekar for use of 5637 cells, and David Sibley for use of the ultracentrifuge. In addition, we would like to thank (also from Washington University) Marielle Boonen, Eline van Meel, and Stuart Kornfeld for sharing valuable antibodies. We acknowledge the Advanced Imaging and Tissue Analysis Core of the Washington University Digestive Disease Core Center (DDRCC). Thanks also go to Benjamin Moore and Benjamin Cappocia for thoughtful review of the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
R.U.J. performed all experiments and drafted the manuscript. J.C.M. edited the manuscript and provided funding. R.U.J. and J.C.M. conceptualized, designed and interpreted the experiments.
Funding
This work was supported the National Institutes of Health [grant numbers R01 DK-079798, DK094989, 2P30 DK052574 to J.C.M.]; and the American Cancer Society [grant number DDC-115769 to D.M.C.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.138776/-/DC1
References
- Alahari S., Mehmood R., Johnson C. L., Pin C. L. (2011). The absence of MIST1 leads to increased ethanol sensitivity and decreased activity of the unfolded protein response in mouse pancreatic acinar cells. PLoS ONE 6, e28863 10.1371/journal.pone.0028863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G., Schwarz T. L. (2013). The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42 10.1038/cdd.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azouz N. P., Matsui T., Fukuda M., Sagi-Eisenberg R. (2012). Decoding the regulation of mast cell exocytosis by networks of Rab GTPases. J. Immunol. 189, 2169–2180 10.4049/jimmunol.1200542 [DOI] [PubMed] [Google Scholar]
- Barbacid M. (1987). ras genes. Annu. Rev. Biochem. 56, 779–827 10.1146/annurev.bi.56.070187.004023 [DOI] [PubMed] [Google Scholar]
- Barnes W. M. (1994). PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc. Natl. Acad. Sci. USA 91, 2216–2220 10.1073/pnas.91.6.2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes W. M. (2006). Ribocloning: DNA cloning and gene construction using PCR primers terminated with a ribonucleotide. Cold Spring Harb. Protoc. 2006, pdb.prot4142. [DOI] [PubMed] [Google Scholar]
- Beatty W. L. (2008). Late endocytic multivesicular bodies intersect the chlamydial inclusion in the absence of CD63. Infect. Immun. 76, 2872–2881 10.1128/IAI.00129-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondyk W. H., McKiernan C. J., Burstein E. S., Macara I. G. (1993). Mutants of Rab3A analogous to oncogenic Ras mutants. Sensitivity to Rab3A-GTPase activating protein and Rab3A-guanine nucleotide releasing factor. J. Biol. Chem. 268, 9410–9415 [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. (2000). Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11, 467–480 10.1091/mbc.11.2.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein E. S., Brondyk W. H., Macara I. G. (1992). Amino acid residues in the Ras-like GTPase Rab3A that specify sensitivity to factors that regulate the GTP/GDP cycling of Rab3A. J. Biol. Chem. 267, 22715–22718 [PubMed] [Google Scholar]
- Cantalupo G., Alifano P., Roberti V., Bruni C. B., Bucci C. (2001). Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20, 683–693 10.1093/emboj/20.4.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoccia B. J., Jin R. U., Kong Y. Y., Peek R. M., Jr, Fassan M., Rugge M., Mills J. C. (2013). The ubiquitin ligase Mindbomb 1 coordinates gastrointestinal secretory cell maturation. J. Clin. Invest. 123, 1475–1491 10.1172/JCI65703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. C., Scoggin S., Wang D., Cherry S., Dembo T., Greenberg B., Jin E. J., Kuey C., Lopez A., Mehta S. Q. et al. (2011). Systematic discovery of Rab GTPases with synaptic functions in Drosophila. Curr. Biol. 21, 1704–1715 10.1016/j.cub.2011.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Edwards J. A., Logsdon C. D., Ernst S. A., Williams J. A. (2002). Dominant negative Rab3D inhibits amylase release from mouse pancreatic acini. J. Biol. Chem. 277, 18002–18009 10.1074/jbc.M201248200 [DOI] [PubMed] [Google Scholar]
- Chen L., Hu J., Yun Y., Wang T. (2010). Rab36 regulates the spatial distribution of late endosomes and lysosomes through a similar mechanism to Rab34. Mol. Membr. Biol. 27, 23–30 10.3109/09687680903417470 [DOI] [PubMed] [Google Scholar]
- Colucci A. M., Campana M. C., Bellopede M., Bucci C. (2005). The Rab-interacting lysosomal protein, a Rab7 and Rab34 effector, is capable of self-interaction. Biochem. Biophys. Res. Commun. 334, 128–133 10.1016/j.bbrc.2005.06.067 [DOI] [PubMed] [Google Scholar]
- Cordonnier M. N., Dauzonne D., Louvard D., Coudrier E. (2001). Actin filaments and myosin I alpha cooperate with microtubules for the movement of lysosomes. Mol. Biol. Cell 12, 4013–4029 10.1091/mbc.12.12.4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Direnzo D., Hess D. A., Damsz B., Hallett J. E., Marshall B., Goswami C., Liu Y., Deering T., Macdonald R. J., Konieczny S. F. (2012). Induced Mist1 expression promotes remodeling of mouse pancreatic acinar cells. Gastroenterology 143, 469–480 10.1053/j.gastro.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L. A. (1999). Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1, E25–E27 10.1038/10018 [DOI] [PubMed] [Google Scholar]
- Feng Y., Press B., Wandinger-Ness A. (1995). Rab 7: an important regulator of late endocytic membrane traffic. J. Cell Biol. 131, 1435–1452 10.1083/jcb.131.6.1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. (2008). Regulation of secretory vesicle traffic by Rab small GTPases. Cell. Mol. Life Sci. 65, 2801–2813 10.1007/s00018-008-8351-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. (2013). Rab27 effectors, pleiotropic regulators in secretory pathways. Traffic 14, 949–963 10.1111/tra.12083 [DOI] [PubMed] [Google Scholar]
- Gaiser S., Daniluk J., Liu Y., Tsou L., Chu J., Lee W., Longnecker D. S., Logsdon C. D., Ji B. (2011). Intracellular activation of trypsinogen in transgenic mice induces acute but not chronic pancreatitis. Gut 60, 1379–1388 10.1136/gut.2010.226175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garside V. C., Kowalik A. S., Johnson C. L., DiRenzo D., Konieczny S. F., Pin C. L. (2010). MIST1 regulates the pancreatic acinar cell expression of Atp2c2, the gene encoding secretory pathway calcium ATPase 2. Exp. Cell Res. 316, 2859–2870 10.1016/j.yexcr.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Yamamoto S., Tsutsumi S., Midorikawa Y., Ihara S., Wang S. M., Aburatani H. (2005). Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 86, 127–141 10.1016/j.ygeno.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Geahlen J. H., Lapid C., Thorell K., Nikolskiy I., Huh W. J., Oates E. L., Lennerz J. K., Tian X., Weis V. G., Khurana S. S. et al. (2013). Evolution of the human gastrokine locus and confounding factors regarding the pseudogenicity of GKN3. Physiol. Genomics 45, 667–683 10.1152/physiolgenomics.00169.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geppert M., Bolshakov V. Y., Siegelbaum S. A., Takei K., De Camilli P., Hammer R. E., Südhof T. C. (1994). The role of Rab3A in neurotransmitter release. Nature 369, 493–497 10.1038/369493a0 [DOI] [PubMed] [Google Scholar]
- Ghosh P., Dahms N. M., Kornfeld S. (2003). Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 4, 202–213 10.1038/nrm1050 [DOI] [PubMed] [Google Scholar]
- Green D. R., Kroemer G. (2004). The pathophysiology of mitochondrial cell death. Science 305, 626–629 10.1126/science.1099320 [DOI] [PubMed] [Google Scholar]
- Griffiths G., Matteoni R., Back R., Hoflack B. (1990). Characterization of the cation-independent mannose 6-phosphate receptor-enriched prelysosomal compartment in NRK cells. J. Cell Sci. 95, 441–461 [DOI] [PubMed] [Google Scholar]
- Gurkan C., Lapp H., Alory C., Su A. I., Hogenesch J. B., Balch W. E. (2005). Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol. Biol. Cell 16, 3847–3864 10.1091/mbc.E05-01-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halangk W., Lerch M. M., Brandt-Nedelev B., Roth W., Ruthenbuerger M., Reinheckel T., Domschke W., Lippert H., Peters C., Deussing J. (2000). Role of cathepsin B in intracellular trypsinogen activation and the onset of acute pancreatitis. J. Clin. Invest. 106, 773–781 10.1172/JCI9411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck P. J., Swanson J. A. (1990). Radial extension of macrophage tubular lysosomes supported by kinesin. Nature 346, 864–866 10.1038/346864a0 [DOI] [PubMed] [Google Scholar]
- Hume A. N., Collinson L. M., Rapak A., Gomes A. Q., Hopkins C. R., Seabra M. C. (2001). Rab27a regulates the peripheral distribution of melanosomes in melanocytes. J. Cell Biol. 152, 795–808 10.1083/jcb.152.4.795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Sun Y., Konieczny S. F. (2008). Mist1 regulates pancreatic acinar cell proliferation through p21 CIP1/WAF1. Gastroenterology 135, 1687–1697 10.1053/j.gastro.2008.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin E. J., Chan C. C., Agi E., Cherry S., Hanacik E., Buszczak M., Hiesinger P. R. (2012). Similarities of Drosophila rab GTPases based on expression profiling: completion and analysis of the rab-Gal4 kit. PLoS ONE 7, e40912 10.1371/journal.pone.0040912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L., Lledo P. M., Roa M., Vincent J. D., Henry J. P., Darchen F. (1994). The GTPase Rab3a negatively controls calcium-dependent exocytosis in neuroendocrine cells. EMBO J. 13, 2029–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. L., Kowalik A. S., Rajakumar N., Pin C. L. (2004). Mist1 is necessary for the establishment of granule organization in serous exocrine cells of the gastrointestinal tract. Mech. Dev. 121, 261–272 10.1016/j.mod.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., Neefjes J. (2001). The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680–1685 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Klumperman J., Kuliawat R., Griffith J. M., Geuze H. J., Arvan P. (1998). Mannose 6-phosphate receptors are sorted from immature secretory granules via adaptor protein AP-1, clathrin, and syntaxin 6-positive vesicles. J. Cell Biol. 141, 359–371 10.1083/jcb.141.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik A. S., Johnson C. L., Chadi S. A., Weston J. Y., Fazio E. N., Pin C. L. (2007). Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein-induced pancreatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1123–G1132 10.1152/ajpgi.00512.2006 [DOI] [PubMed] [Google Scholar]
- Lemercier C., To R. Q., Swanson B. J., Lyons G. E., Konieczny S. F. (1997). Mist1: a novel basic helix-loop-helix transcription factor exhibits a developmentally regulated expression pattern. Dev. Biol. 182, 101–113 10.1006/dbio.1996.8454 [DOI] [PubMed] [Google Scholar]
- Li C., Fan Y., Lan T. H., Lambert N. A., Wu G. (2012). Rab26 modulates the cell surface transport of α2-adrenergic receptors from the Golgi. J. Biol. Chem. 287, 42784–42794 10.1074/jbc.M112.410936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limi S., Ojakian G., Raffaniello R. (2012). Rab3D regulates amylase levels, not agonist-induced amylase release, in AR42J cells. Cell. Mol. Biol. Lett. 17, 258–273 10.2478/s11658-012-0008-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. X., Collins C. A. (1992). Immunolocalization of cytoplasmic dynein to lysosomes in cultured cells. J. Cell Sci. 101, 125–137 [DOI] [PubMed] [Google Scholar]
- Lipsky N. G., Pedersen P. L. (1981). Mitochondrial turnover in animal cells. Half-lives of mitochondria and mitochondrial subfractions of rat liver based on [14C]bicarbonate incorporation. J. Biol. Chem. 256, 8652–8657 [PubMed] [Google Scholar]
- Logsdon C. D. (1986). Glucocorticoids increase cholecystokinin receptors and amylase secretion in pancreatic acinar AR42J cells. J. Biol. Chem. 261, 2096–2101 [PubMed] [Google Scholar]
- Luo X., Shin D. M., Wang X., Konieczny S. F., Muallem S. (2005). Aberrant localization of intracellular organelles, Ca2+ signaling, and exocytosis in Mist1 null mice. J. Biol. Chem. 280, 12668–12675 10.1074/jbc.M411973200 [DOI] [PubMed] [Google Scholar]
- Luzio J. P., Rous B. A., Bright N. A., Pryor P. R., Mullock B. M., Piper R. C. (2000). Lysosome-endosome fusion and lysosome biogenesis. J. Cell Sci. 113, 1515–1524 [DOI] [PubMed] [Google Scholar]
- Mareninova O. A., Sung K. F., Hong P., Lugea A., Pandol S. J., Gukovsky I., Gukovskaya A. S. (2006). Cell death in pancreatitis: caspases protect from necrotizing pancreatitis. J. Biol. Chem. 281, 3370–3381 10.1074/jbc.M511276200 [DOI] [PubMed] [Google Scholar]
- Matteoni R., Kreis T. E. (1987). Translocation and clustering of endosomes and lysosomes depends on microtubules. J. Cell Biol. 105, 1253–1265 10.1083/jcb.105.3.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménasché G., Pastural E., Feldmann J., Certain S., Ersoy F., Dupuis S., Wulffraat N., Bianchi D., Fischer A., Le Deist F. et al. (2000). Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat. Genet. 25, 173–176 10.1038/76024 [DOI] [PubMed] [Google Scholar]
- Millar A. L., Pavios N. J., Xu J., Zheng M. H. (2002). Rab3D: a regulator of exocytosis in non-neuronal cells. Histol. Histopathol. 17, 929–936 [DOI] [PubMed] [Google Scholar]
- Mills J. C., Taghert P. H. (2012). Scaling factors: transcription factors regulating subcellular domains. Bioessays 34, 10–16 10.1002/bies.201100089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. H., Millman E. E., Alpizar-Foster E., Dai W., Knoll B. J. (2004). Rab11 regulates the recycling and lysosome targeting of beta2-adrenergic receptors. J. Cell Sci. 117, 3107–3117 10.1242/jcs.01168 [DOI] [PubMed] [Google Scholar]
- Moreland J. L., Gramada A., Buzko O. V., Zhang Q., Bourne P. E. (2005). The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics 6, 21 10.1186/1471-2105-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P., Rajesh M., Yoshihiro K., Haskó G., Pacher P. (2007). Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem. Biophys. Res. Commun. 358, 203–208 10.1016/j.bbrc.2007.04.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashida T., Imai A., Shimomura H. (2006). Relation of Rab26 to the amylase release from rat parotid acinar cells. Arch. Oral Biol. 51, 89–95 10.1016/j.archoralbio.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Odinokova I. V., Sung K. F., Mareninova O. A., Hermann K., Evtodienko Y., Andreyev A., Gukovsky I., Gukovskaya A. S. (2009). Mechanisms regulating cytochrome c release in pancreatic mitochondria. Gut 58, 431–442 10.1136/gut.2007.147207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi H., Samuelson L. C., Yule D. I., Ernst S. A., Williams J. A. (1997). Overexpression of Rab3D enhances regulated amylase secretion from pancreatic acini of transgenic mice. J. Clin. Invest. 100, 3044–3052 10.1172/JCI119859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. K., Ashby M. C., Erdemli G., Petersen O. H., Tepikin A. V. (2001). Perinuclear, perigranular and sub-plasmalemmal mitochondria have distinct functions in the regulation of cellular calcium transport. EMBO J. 20, 1863–1874 10.1093/emboj/20.8.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali C., Fialka I., Huber L. A. (1999). Subcellular fractionation, electromigration analysis and mapping of organelles. J. Chromatogr. B Biomed. Sci. Appl. 722, 89–102 10.1016/S0378-4347(98)00314-4 [DOI] [PubMed] [Google Scholar]
- Peden A. A., Oorschot V., Hesser B. A., Austin C. D., Scheller R. H., Klumperman J. (2004). Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 164, 1065–1076 10.1083/jcb.200311064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal J. B., Seabra M. C. (2001). Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313, 889–901 10.1006/jmbi.2001.5072 [DOI] [PubMed] [Google Scholar]
- Pin C. L., Bonvissuto A. C., Konieczny S. F. (2000). Mist1 expression is a common link among serous exocrine cells exhibiting regulated exocytosis. Anat. Rec. 259, 157–167 [DOI] [PubMed] [Google Scholar]
- Pin C. L., Rukstalis J. M., Johnson C., Konieczny S. F. (2001). The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J. Cell Biol. 155, 519–530 10.1083/jcb.200105060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M., Zhang Y. Z., Krämer J. A., Wells K. S., Jones L. J., Hanzel D. K., Lugade A. G., Singer V. L., Haugland R. P. (1996). Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J. Histochem. Cytochem. 44, 1363–1372 10.1177/44.12.8985128 [DOI] [PubMed] [Google Scholar]
- Progida C., Cogli L., Piro F., De Luca A., Bakke O., Bucci C. (2010). Rab7b controls trafficking from endosomes to the TGN. J. Cell Sci. 123, 1480–1491 10.1242/jcs.051474 [DOI] [PubMed] [Google Scholar]
- Qiu X., Valentijn J. A., Jamieson J. D. (2001). Carboxyl-methylation of Rab3D in the rat pancreatic acinar tumor cell line AR42J. Biochem. Biophys. Res. Commun. 285, 708–714 10.1006/bbrc.2001.5224 [DOI] [PubMed] [Google Scholar]
- Ramsey V. G., Doherty J. M., Chen C. C., Stappenbeck T. S., Konieczny S. F., Mills J. C. (2007). The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development 134, 211–222 10.1242/dev.02700 [DOI] [PubMed] [Google Scholar]
- Riedel D., Antonin W., Fernandez-Chacon R., Alvarez de Toledo G., Jo T., Geppert M., Valentijn J. A., Valentijn K., Jamieson J. D., Südhof T. C. et al. (2002). Rab3D is not required for exocrine exocytosis but for maintenance of normally sized secretory granules. Mol. Cell. Biol. 22, 6487–6497 10.1128/MCB.22.18.6487-6497.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn C., Aroso M., Prüssing J., Islinger M., Schrader M. (2012). Modulating zymogen granule formation in pancreatic AR42J cells. Exp. Cell Res. 318, 1855–1866 10.1016/j.yexcr.2012.05.025 [DOI] [PubMed] [Google Scholar]
- Robinson M. S. (1990). Cloning and expression of gamma-adaptin, a component of clathrin-coated vesicles associated with the Golgi apparatus. J. Cell Biol. 111, 2319–2326 10.1083/jcb.111.6.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukstalis J. M., Kowalik A., Zhu L., Lidington D., Pin C. L., Konieczny S. F. (2003). Exocrine specific expression of Connexin32 is dependent on the basic helix-loop-helix transcription factor Mist1. J. Cell Sci. 116, 3315–3325 10.1242/jcs.00631 [DOI] [PubMed] [Google Scholar]
- Saftig P., Klumperman J. (2009). Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- Saluja A. K., Donovan E. A., Yamanaka K., Yamaguchi Y., Hofbauer B., Steer M. L. (1997). Cerulein-induced in vitro activation of trypsinogen in rat pancreatic acini is mediated by cathepsin B. Gastroenterology 113, 304–310 10.1016/S0016-5085(97)70108-2 [DOI] [PubMed] [Google Scholar]
- Saraste J., Goud B. (2007). Functional symmetry of endomembranes. Mol. Biol. Cell 18, 1430–1436 10.1091/mbc.E06-10-0933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997). The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 10.1126/science.277.5324.333 [DOI] [PubMed] [Google Scholar]
- Schlüter O. M., Schmitz F., Jahn R., Rosenmund C., Südhof T. C. (2004). A complete genetic analysis of neuronal Rab3 function. J. Neurosci. 24, 6629–6637 10.1523/JNEUROSCI.1610-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. L., Cao C., Pylypenko O., Rak A., Wandinger-Ness A. (2007). Rab GTPases at a glance. J. Cell Sci. 120, 3905–3910 10.1242/jcs.015909 [DOI] [PubMed] [Google Scholar]
- Selinummi J., Seppälä J., Yli-Harja O., Puhakka J. A. (2005). Software for quantification of labeled bacteria from digital microscope images by automated image analysis. Biotechniques 39, 859–863 10.2144/000112018 [DOI] [PubMed] [Google Scholar]
- Shi G., Zhu L., Sun Y., Bettencourt R., Damsz B., Hruban R. H., Konieczny S. F. (2009). Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology 136, 1368–1378 10.1053/j.gastro.2008.12.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G., DiRenzo D., Qu C., Barney D., Miley D., Konieczny S. F. (2013). Maintenance of acinar cell organization is critical to preventing Kras-induced acinar-ductal metaplasia. Oncogene 32, 1950–1958 10.1038/onc.2012.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V., McLelland G. L., Zunino R., Braschi E., Rippstein P., Fon E. A., McBride H. M. (2012). A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 22, 135–141 10.1016/j.cub.2011.11.057 [DOI] [PubMed] [Google Scholar]
- Sridhar S., Patel B., Aphkhazava D., Macian F., Santambrogio L., Shields D., Cuervo A. M. (2013). The lipid kinase PI4KIIIβ preserves lysosomal identity. EMBO J. 32, 324–339 10.1038/emboj.2012.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman A., Kurz T., Navratil M., Arriaga E. A., Brunk U. T. (2010). Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxid. Redox Signal. 12, 503–535 10.1089/ars.2009.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Jin R. U., Bredemeyer A. J., Oates E. J., Błazewska K. M., McKenna C. E., Mills J. C. (2010). RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol. Cell. Biol. 30, 1269–1284 10.1128/MCB.01328-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel H., Cancela J. M., Mogami H., Gerasimenko J. V., Gerasimenko O. V., Tepikin A. V., Petersen O. H. (1999). Active mitochondria surrounding the pancreatic acinar granule region prevent spreading of inositol trisphosphate-evoked local cytosolic Ca(2+) signals. EMBO J. 18, 4999–5008 10.1093/emboj/18.18.4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. C., Strowski M. Z., Göke B., Williams J. A. (1995). Molecular cloning of a new member of the Rab protein family, Rab 26, from rat pancreas. Biochem. Biophys. Res. Commun. 207, 950–956 10.1006/bbrc.1995.1277 [DOI] [PubMed] [Google Scholar]
- Wang T., Hong W. (2002). Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol. Biol. Cell 13, 4317–4332 10.1091/mbc.E02-05-0280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Hong W. (2005). Assay and functional properties of Rab34 interaction with RILP in lysosome morphogenesis. Methods Enzymol. 403, 675–687 10.1016/S0076-6879(05)03058-2 [DOI] [PubMed] [Google Scholar]
- Wang J., Tempel W., Shen Y., Shen L., Yaniw D., Arrowsmith C., Edwards A., Sundstrom M., Weigelt J., Bochkarev A. et al. (2006). Crystal Structure of Human RAB26 in Complex with a GTP Analogue. Structural Genomics Consortium (SGC) [Google Scholar]
- Weber E., Berta G., Tousson A., St John P., Green M. W., Gopalokrishnan U., Jilling T., Sorscher E. J., Elton T. S., Abrahamson D. R. et al. (1994). Expression and polarized targeting of a rab3 isoform in epithelial cells. J. Cell Biol. 125, 583–594 10.1083/jcb.125.3.583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Zhang Y. (2009). Generating triangulated macromolecular surfaces by Euclidean Distance Transform. PLoS ONE 4, e8140 10.1371/journal.pone.0008140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshie S., Imai A., Nashida T., Shimomura H. (2000). Expression, characterization, and localization of Rab26, a low molecular weight GTP-binding protein, in the rat parotid gland. Histochem. Cell Biol. 113, 259–263 [DOI] [PubMed] [Google Scholar]
- Zhang J., Schulze K. L., Hiesinger P. R., Suyama K., Wang S., Fish M., Acar M., Hoskins R. A., Bellen H. J., Scott M. P. (2007). Thirty-one flavors of Drosophila rab proteins. Genetics 176, 1307–1322 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Li C., Wong W. H. (2003). ChipInfo: Software for extracting gene annotation and gene ontology information for microarray analysis. Nucleic Acids Res. 31, 3483–3486 10.1093/nar/gkg598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Tran T., Rukstalis J. M., Sun P., Damsz B., Konieczny S. F. (2004). Inhibition of Mist1 homodimer formation induces pancreatic acinar-to-ductal metaplasia. Mol. Cell. Biol. 24, 2673–2681 10.1128/MCB.24.7.2673-2681.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.