ABSTRACT
The Hedgehog (Hh) family of secreted proteins governs many key developmental processes by regulating Ci/Gli transcription factors at multiple levels including nuclear–cytoplasmic shuttling. Here, we investigate the mechanism underlying the regulation of Ci/Gli subcellular localization by identifying and characterizing a novel nuclear localization sequence (NLS) in the N-terminal conserved domain of Ci/Gli that matches the PY-NLS consensus. We demonstrate that the PY-NLS functions in parallel with a previously identified bipartite NLS to promote nuclear localization and activity of full-length Ci. We find that Transportin (Trn), the Drosophila homolog of Kapβ2, is responsible for PY-NLS-mediated nuclear localization of Ci. Furthermore, we show that the tumor suppressor and conserved Hh pathway component Suppressor of fused (Sufu) opposes Trn-mediated Ci nuclear import by masking its PY-NLS. Finally, we provide evidence that Gli proteins also contain a functional PY-NLS and that mammal Sufu uses a similar mechanism to regulate nuclear translocation of Gli. Our study not only provides a mechanistic insight into how Sufu regulates Hh signaling and the subcellular localization of Ci/Gli, but also reveals a role for Trn/Kapβ2 in developmental regulation.
KEY WORDS: Hedgehog, Sufu, Ci, Gli, Transportin, PY-NLS
INTRODUCTION
The hedgehog (Hh) signaling pathway is a major and evolutionarily conserved signaling cascade that is essential for both embryonic development and adult tissue homeostasis (Jiang and Hui, 2008; Varjosalo and Taipale, 2008; Ingham et al., 2011; Briscoe and Thérond, 2013). Not surprisingly, deregulation of the pathway activity has been implicated in numerous human diseases, including birth defects and cancer (Taipale and Beachy, 2001; Nieuwenhuis and Hui, 2005; Jiang and Hui, 2008). In the canonical Hh pathway, Hh exerts its biological influence through a signaling cascade that culminates in controlling the balance between the activator and repressor forms of Ci (fly) and Gli (mammalian) transcription factors (Jiang and Hui, 2008; Robbins et al., 2012; Briscoe and Thérond, 2013). Ci/Gli activity is kept in check by multiple layers of regulation, including phosphorylation-mediated proteolytic processing, nuclear–cytoplasmic shuttling and ubiquitin-mediated degradation (Jiang, 2006; Jiang and Hui, 2008; Wilson and Chuang, 2010; Chen and Jiang, 2013).
In the absence of Hh, full-length Ci is phosphorylated by multiple kinases including protein kinase A (PKA), glycogen synthase kinase 3β (GSK3β) and casein kinase 1 (CK1), which recruits the SCFSlimb E3 ubiquitin ligase that targets Ci for ubiquitin/proteasome-mediated proteolysis to generate a truncated repressor form (CiR) (Jiang and Struhl, 1998; Wang et al., 1999; Jia et al., 2002; Price and Kalderon, 2002; Jia et al., 2005; Smelkinson et al., 2007; Zhang et al., 2013b). The kinesin-like protein Costal2 (Cos2) and the Ser/Thr kinase Fused (Fu) form a complex with Ci to prevent its nuclear localization (Chen et al., 1999; Wang et al., 2000; Wang and Holmgren, 2000; Wang and Jiang, 2004). In addition, Cos2 recruits PKA, GSK3β and CK1 to the complex to promote phosphorylation and proteolytic processing of Ci, whereas Hh signaling inhibits these processes and promotes nuclear translocation of Ci, at least in part, by dissociating the Ci–Cos2–kinase complexes (Zhang et al., 2005; Ruel et al., 2007; Shi et al., 2011). The full-length Ci forms a separate complex with the pathway inhibitor Suppressor of fused (Sufu) (Monnier et al., 1998; Lum et al., 2003), which keeps Ci in an inactive form by impeding its nuclear translocation, as well as by inhibiting its activity in the nucleus (Ohlmeyer and Kalderon, 1998; Méthot and Basler, 2000; Wang et al., 2000; Croker et al., 2006). In mammals, Sufu is a major negative regulator of the Hh signaling activity (Svärd et al., 2006; Varjosalo et al., 2006). Germline mutations in human Sufu predispose to childhood medulloblastoma and meningioma (Taylor et al., 2002; Aavikko et al., 2012). Sufu regulates proteolytic processing, nuclear localization and transcriptional activity of Gli proteins (Ding et al., 1999; Barnfield et al., 2005; Chen et al., 2009; Kise et al., 2009; Humke et al., 2010; Tukachinsky et al., 2010; Wang et al., 2010). Despite the conserved function of Sufu in the regulation of nuclear localization of Ci/Gli, the underlying mechanism remains poorly understood.
Ci and Gli proteins exhibit limited sequence similarities outside their Zn-finger DNA-binding domains; however, their N-terminal regions contain a 49 amino acid conserved domain called the N-terminal regulatory element (NR), which binds Sufu and is required for Sufu to retain Ci in the cytoplasm (Croker et al., 2006). Unexpectedly, we found here that the NR domain functions as a nuclear localization signal (NLS) that is masked by Sufu binding. Sequence analysis and mutagenesis study revealed that the NR domain harbors a PY family of NLS that is recognized by Transportin (Trn), the Drosophila homolog of Kapβ2 (also known as transportin-1 or TNPO1) (Chook and Süel, 2011). We demonstrate that the PY-NLS acts in parallel with a previously identified bipartite NLS to regulate Ci nuclear localization and activity, and that Sufu impedes Ci nuclear translocation by blocking binding of Trn to the NR domain. Furthermore, we provide evidence that Sufu regulates nuclear localization of Gli through a similar mechanism.
RESULTS
Identification of a PY-NLS in the N-terminal region of Ci
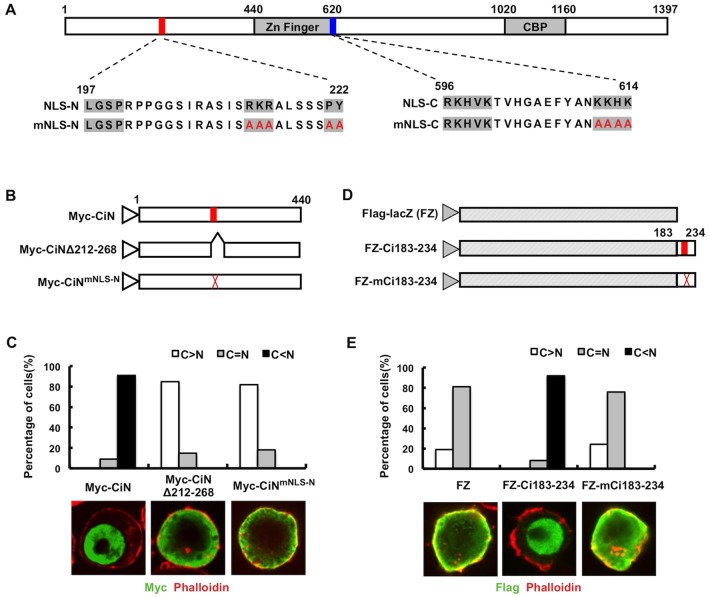
Ci contains a canonical bipartite NLS with two basic clusters (aa 596–600 and aa 611–614) lying at the end of its Zn-finger domain (Fig. 1A) (Wang and Holmgren, 1999). Therefore, we were surprised to find that a Myc-tagged N-terminal fragment of Ci containing amino acids 1–440 (Myc–CiN) is almost exclusively localized in the nucleus (Fig. 1B,C). The size of the Myc–CiN protein, which is estimated to be ∼60 kDa, exceeds the passive diffusion restriction (∼40 kDa) of nuclear pore complexes (Paine et al., 1975), suggesting that the CiN contains a NLS that mediates its active nuclear import.
Fig. 1.
The N-terminal region of Ci contains a PY-NLS. (A) Structure of Ci with gray boxes indicating the Zn finger DNA-binding domain and the CBP binding domain. The red and blue bars represent the newly identified PY-NLS (NLS-N) and previously identified bipartite NLS (NLS-C), respectively. The amino acid sequences for the wild-type and mutant NLSs are shown underneath the diagram. (B) Myc-tagged wild type CiN (Myc–CiN) and its variants with PY-NLS deleted (Myc–CiNΔ212-268) or mutated (Myc–CiNmNLS-N). (C) Quantification of nuclear or cytoplasmic localization of Myc–CiN and its variants. A total of 100 cells were randomly picked and categorized based on the differential nuclear or cytoplasmic distribution of Myc signal for each construct. The y-axis indicates the percentage of cells in each category. C>N, cells contain higher levels of cytoplasmic signal than nuclear signal; C = N, cells contain equal levels of cytoplasmic and nuclear signal; C<N, cells contain higher levels of nuclear signal than cytoplasmic signal. Representative images of S2 cells transfected with Myc–CiN, Myc–CiNΔ212-268 or Myc–CiNmNLS-N and immunostained with an anti-Myc antibody (green) and Phalloidin (red), a dye that highlights cell membrane are shown below. Mutation of the PY-NLS blocked the nuclear translocation of CiN. (D) Diagrams of FLAG-tagged LacZ (FLAG–LacZ or FZ) and LacZ Ci fusion proteins that contain the wild type (FZ–Ci183-234) or mutated (FZ–mCi183-234) PY-NLS sequence. (E) Quantification of nuclear and cytoplasmic localization of FZ and its variants. Representative images of S2 cells transfected with FZ, FZ-Ci183-234 or FZ-mCi183-234 and immunostained with an anti-FLAG antibody (green) and Phalloidin (red) are shown below. Fusion of the wild-type but not the mutated PY-NLS to LacZ promoted its nuclear localization.
Careful sequence analysis uncovered a putative PY-NLS-like sequence (aa 197–222) that matches the consensus sequence: basic/hydrophobic motif-X(7∼12)-R/K/H-X(2∼5)-PY/L (Lee et al., 2006). This motif is located within the NR domain, which is highly conserved among Ci/Gli proteins (Fig. 1A) (Croker et al., 2006). Strikingly, either deletion or mutation of the C-terminal core (RKRALSSPY) of the PY-NLS motif (Myc–CiNΔ212-268 or Myc–CiNmNLS-N) prevented the nuclear localization of Myc–CiN, indicating that this PY motif is a bona fide NLS that is responsible for the nuclear localization of Myc–CiN (Fig. 1A–C).
To determine whether the PY-NLS motif is sufficient to mediate active nuclear import of a heterologous protein, we fused aa 183–234 containing either the wild-type or mutated PY motif to the C-terminus of FLAG–LacZ (FZ–Ci183-234 and FZ–mCi183-234; Fig. 1D) and expressed the fusion proteins in S2 cells. Whereas FLAG–LacZ (FZ) was distributed in both cytoplasm and nucleus, FZ–Ci183-234 exhibited exclusive nuclear staining (Fig. 1E). By contrast, FZ–mCi183-234, which contains the mutated PY-NLS motif, remained largely in the cytoplasm (Fig. 1E). Taken together, we conclude that the N-terminal region of Ci contains a functional PY-NLS. We named the N-terminal PY-NLS as NLS-N and the canonical bipartite NLS as NLS-C, respectively.
NLS-N acts in parallel with NLS-C to promote Ci nuclear localization and activity
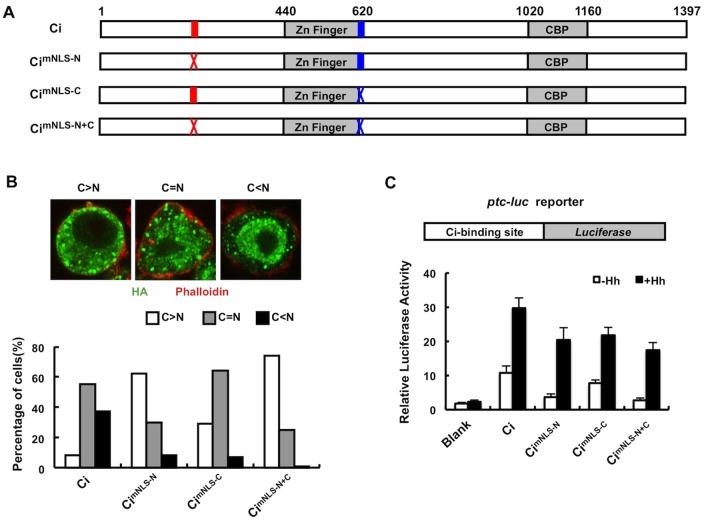
To determine whether full-length Ci is regulated by NLS-N, NLS-N was mutated either alone or in conjunction with NLS-C in the context of an HA-tagged full-length Ci (Fig. 2A), and the resulting Ci variants and wild-type Ci were expressed in S2 cells by transient transfection. Transfected cells were treated with the nuclear export inhibitor LMB to allow detection of nuclear Ci (Chen et al., 1999; Wang and Holmgren, 2000; Wang and Jiang, 2004). Owing to variation in the subcellular localization of Ci among different cells, we grouped cells into three categories: (1) more Ci in the cytoplasm than in the nucleus (C >N); (2) Ci evenly distributed in the cytoplasm and nucleus (C = N); (3) more Ci in the nucleus than in the cytoplasm (C<N) (Fig. 2B). After 2 hours of treatment with LMB, ∼37% of S2 cells transfected with wild-type Ci exhibited higher levels of Ci in the nucleus than in the cytoplasm (Fig. 2B). Mutation of NLS-N (HA–CimNLS-N) or NLS-C (HA–CimNLS-C) individually reduced the C<N population to less than 10%, whereas double mutation (HA–CimNLS-N+C) nearly eliminated the C<N population. The majority of cells expressing HA–CimNLS-N+C (>60%) exhibited higher levels of Ci in the cytoplasm than in the nucleus and a small fraction exhibited equal distribution of Ci in the cytoplasm and nucleus (Fig. 2B). The residual nuclear localization of CimNLS-N+C might be due to an incomplete inactivation of two nuclear localization signals or there could be a third, yet to be identified, NLS in Ci. Alternatively, CimNLS-N+C could enter the nucleus through an interacting protein. Nevertheless, the data indicated that NLS-N and NLS-C act in parallel to promote efficient nuclear import of Ci.
Fig. 2.
NLS-N acts in conjunction with NLS-C to regulate Ci nuclear localization and activity. (A) HA–Ci and its variants with NLS-N, NLS-C or both mutated. Red and blue bars represent wild-type NLS-N and NLS-C, respectively, whereas red and blue crosses represent mutated NLS-N and NLS-C, respectively. (B) S2 cells transfected with HA–Ci, HA–CimNLS-N, HA–CimNLS-C or HA–CimNLS-N+C were treated with 10 ng/ml LMB for 2 hours prior to immunostaining with an anti-HA antibody (green) and Phalloidin (red). Transfected cells show different patterns of nuclear and cytoplasmic distribution of tagged Ci or mutants. Quantification of nuclear and cytoplasmic localization of HA–Ci and its variants is shown below. (C) ptc-luc reporter structure and ptc-luc reporter assay in S2 cells expressing Ci or its variants in the absence or presence of Hh-conditioned medium. The y-axis represents normalized ptc-luc activity. Values are means ± s.d.
To determine the effect of mutating NLS-N/NLS-C on Ci activity, we carried out a ptc-luc reporter assay in S2 cells in the absence or presence of Hh-conditioned medium as described previously (Chen et al., 1999; Zhang et al., 2009). Overexpression of wild-type Ci activated the ptc-luc reporter gene, which was further enhanced by Hh (Fig. 2C) (Chen et al., 1999). Both HA–CimNLS-N and HA–CimNLS-C exhibited reduced activity whereas HA–CimNLS-N+C showed the lowest activity in the absence as well as presence of Hh (Fig. 2C), which is consistent with their defect in nuclear import.
NLS-N regulates nuclear localization and activity of Ci in vivo
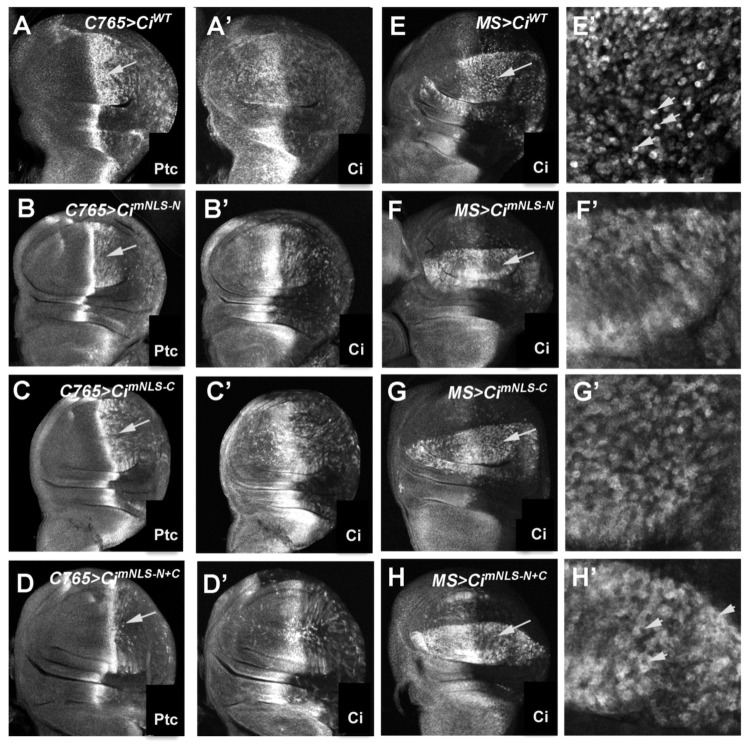
To further determine the role of the newly identified NLS in nuclear localization and activity of Ci in vivo, we turned to the Drosophila wing imaginal disc, which is one of the best systems to study Hh signaling in vivo (Jiang and Hui, 2008). In Drosophila wing imaginal discs, Hh is expressed exclusively in the posterior (P) compartment cells, whereas Ci is only expressed in the anterior (A) compartment cells. Owing to the limited amounts of Hh moving into the A-compartment, only A-compartment cells near the A/P boundary are exposed to Hh and express Hh target genes, such as ptc. The P-compartment cells do not express any Hh target genes even when they are exposed to Hh because they do not express endogenous Ci; however, when exogenously expressed Ci is provided, it allows P-compartment cells to ectopically activate Hh target genes (Fig. 3A) (Wang et al., 1999). To compare the activity of Ci variants containing mutated NLSs with that of wild-type Ci, UAS transgenes expressing UAS-HA-CiWT, UAS-HA-CimNLS-N, UAS-HA-CimNLS-C, UAS-HA-CimNLS-N+C were introduced into the 75B1 locus using the phiC31 integration system to ensure similar levels of transgene expression (Bischof et al., 2007). UAS transgenes were expressed in wing discs using a weak Gal4 driver, C765, or a strong Gal4 driver, MS1096 (MS for short). Compared with wild-type Ci (C765 >CiWT), C765 >CimNLS-N and C765 >CimNLS-C induced ectopic ptc expression in P-compartment cells at lower levels (arrows in Fig. 3B,C, compare with Fig. 3A). Mutation of both NLS-N and NLS-C (C765 >CimNLS-N+C) diminished the ectopic expression of ptc (arrow in Fig. 3D), consistent with a requirement of both NLS-N and NLS-C for optimal Ci activity.
Fig. 3.
The PY-NLS motif is required for efficient nuclear translocation of Ci and optimal Ci activity in vivo. (A–D′) Wing discs expressing UAS-HA-CiWT (A,A′), UAS-HA-CimNLS-N (B,B′), UAS-HA-CimNLS-C (C,C′) or UAS-HA-CimNLS-N+C (D,D′) with the C765 Gal4 driver were immunostained to show the expression of Ptc (A–D) and Ci (A′–D′). Arrows indicate the posterior compartments. (E–H′) Low (E–H) and high (E′–H′) magnification views of wing discs expressing UAS-HA-CiWT (E,E′), UAS-HA-CimNLS-N (F,F′), UAS-HA-Ci-mNLS-C (G,G′) or UAS-HA-CimNLS-N+C (H,H′) with the MS1096 Gal4 driver were treated with LMB 50 ng/ml for 2 hours prior to immunostaining with an anti-Ci antibody and a nuclear dye (not shown). Arrows in E–H indicate the posterior compartments that are enlarged in panels E′–H′. Arrowheads in E′ and H′ highlight individual cells exhibiting predominant nuclear localization of CiWT and predominant cytoplasmic localization of CimNLS-N+C, respectively.
In wing discs treated with LMB, CiWT accumulated predominantly in the nuclei of P-compartment cells (Fig. 3E,E′) whereas CimNLS-N and CimNLS-C exhibited both nuclear and cytoplasmic staining (Fig. 3F–G′). A previous study also indicated that mutation of the NLS-C partially affected nuclear localization of Ci in vivo (Sisson et al., 2006). By contrast, CimNLS-N+C accumulated predominantly in the cytoplasm (Fig. 3H,H′), suggesting that NLS-N and NLS-C exhibit partially redundant functions, but both are required for effective Ci nuclear import, which is essential for maximal Hh pathway activation.
Trn is responsible for NLS-N-mediated Ci nuclear import
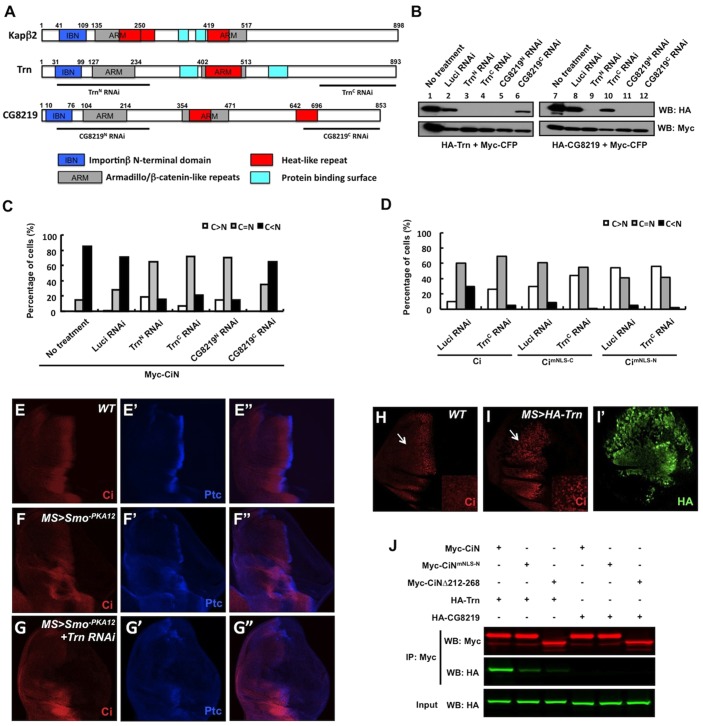
The NLS-mediated nuclear import of substrate requires its being recognized by distinct members of the Kapβ transport receptor superfamily (Chook and Süel, 2011). In contrast to the classic NLS, which contains either a single or two clusters of basic residues recognized by the Kapα–Kapβ1 heterodimer (Kap60p–Kap95p in yeast) (Conti and Izaurralde, 2001), the PY-NLS, which comprises a large linear signal with diverse sequences, is specifically recognized by Kapβ2 (Kap104p in yeast) (Lee et al., 2006). There are two homologs of Kapβ2 in Drosophila, Trn (74% identity) and CG8219 (58% identity) (Fig. 4A). To determine whether they regulate Ci localization through the NLS-N, we tested whether they are required for the nuclear localization of Myc–CiN. To perturb the activity of Trn or CG8219, we performed RNAi to knockdown endogenous Trn or CG8219 in S2 cells transfected with Myc–CiN. RNAi efficiency and specificity were verified in S2 cells transfected with HA-tagged Trn or CG8219 because antibodies against endogenous Trn or CG8129 are unavailable. The N-terminal regions of Trn and CG8219 are highly conserved and contain the putative protein binding surfaces and two ARM domains, whereas the C-terminal regions of these two proteins are more divergent (Fig. 4A). Consistently, TrnN RNAi and CG8219N RNAi, which target an N-terminal region of Trn and CG8219, respectively, knocked down both proteins, whereas TrnC RNAi and CG8219C RNAi, which target a C-terminal region of Trn and CG8219, respectively, specifically knocked down the corresponding genes (Fig. 4A,B). Compared with Luc RNAi control, which did not significantly affect the nuclear localization of Myc–CiN, knockdown of Trn by TrnC RNAi dramatically decreased the nuclear localization of Myc–CiN (Fig. 4C). By contrast, CG8219C RNAi, which specifically targets CG8219, did not cause any obvious change in the subcellular localization of Myc–CiN (Fig. 4C). Furthermore, knockdown of both Trn and CG8219 by either TrnN RNAi or CG8219N RNAi had an effect similar to that seen with knockdown of Trn alone (Fig. 4C). Taken together, these results indicate that Trn, but not CG8219, is essential for promoting the nuclear import of Myc–CiN.
Fig. 4.
Trn interacts with CiN and is required for NLS-N-mediated Ci nuclear localization. (A) Schematics of human Kapβ2, Trn and Drosophila protein encoded by CG8219 with various domains demarcated by colored boxes. Note the overlap between ARM and Heat-like repeats. Lines underneath each protein indicate the regions targeted by the corresponding RNAi constructs. (B) Western blot analysis to confirm the targeting efficiency and specificity for the indicated RNAi constructs. (C) Knockdown of Trn but not CG8219 affects CiN nuclear localization. S2 cells pre-treated without or with the indicated dsRNA were transfected with Myc–CiN expression construct and immunostained with an anti-Myc antibody and Phalloidin. For each treatment, 100 cells were randomly picked and categorized based on the differential nuclear and cytoplasmic distribution of Myc signal. (D) Knockdown of Trn affects nuclear localization of full-length Ci in a manner dependent on the intact NLS-N. S2 cells pre-treated with control or TrnC dsRNA were transfected with HA–Ci, HA–CimNLS-C or HA–CimNLS-N. After treatment with LMB, transfected cells were immunostained to visualize subcellular localization of Ci. (E–G″) Late third instar wild-type wing disc (E,E″) or wing discs expressing UAS-Smo-PKA12 either alone (F,F″) or together with UAS-Trn-RNAi (G,G″) with MS1096 were immunostained to show the expression of Ci (red) and Ptc (blue). Trn RNAi enhanced the ptc expression defect caused by overexpression of Smo-PKA12. (H–I′) Low and high magnification (inset) views of a wild-type wing disc (H) and a wing disc expressing HA–Trn with MS1096 (I,I′) treated with 50 ng/ml LMB for 2 hours prior to immunostaining with anti-Ci (red) and anti-HA (green) antibodies. Arrows indicate the anterior compartments. In wild-type disc, LMB treatment leads to Ci nuclear accumulation only in anterior cells abutting the A/P boundary (H). Overexpression of Trn promotes Ci nuclear localization in A-compartment cells distant from the A–P boundary (I,I′). (J) Trn but not CG8219 binds CiN in a manner dependent on the PY-NLS. Purified Myc-tagged CiN or its variants were incubated with immunopurified HA–Trn or HA–CG8219. The protein complexes were immunoprecipitated with an anti-Myc antibody, followed by western blot analysis.
We also examined the effect of Trn inactivation on the subcellular localization of full-length Ci and its variants lacking either NLS-C (CimNLS-C) or NLS-N (CimNLS-N). Trn RNAi increased the cytoplasmic localization of both Ci and CimNLS-C, but had little if any effect on the subcellular localization of CimNLS-N (Fig. 4D). We noticed that the effect of Trn RNAi on Ci subcellular localization was less dramatic than mutation of NLS-N, probably because of incomplete inactivation of Trn by RNAi. Hence, Trn is responsible for NLS-N-mediated nuclear localization of Ci.
Trn modulates Hh pathway activity in vivo
Because Trn regulates a large number of nuclear proteins that harbor the PY-NLS (Lee et al., 2006), complete loss of Trn function is likely to cause a pleiotropic effect. In addition, the presence of multiple NLSs for regulating Ci nuclear import predicts that a substantial amount of Ci would enter the nucleus in the absence of Trn, meaning that inactivation of Trn might not have a dramatic effect on Hh pathway activity. To overcome these problems, we examined whether inactivation of Trn could modulate Hh signaling activity in a genetic sensitized background in which the pathway activity was downregulated by expressing a dominant-negative form of Smo (MS>Smo−PKA12) (Jia et al., 2004; Chen et al., 2010). In wing discs expressing MS>Smo−PKA12, ptc expression was reduced (Fig. 4F,F″ compare with 4E,E″). We found that Trn RNAi could further decrease ptc expression in discs expressing MS>Smo−PKA12 (Fig. 4G,G″), supporting the notion that Trn positively regulates Hh signaling activity.
To determine whether Trn regulates nuclear translocation of Ci in vivo, we also generated UAS-HA-Trn transgenic flies. Treating wild-type wing discs with LMB induced nuclear localization of Ci in A-compartment cells near the A/P boundary, but failed to do so in A-compartment cells distant from the A/P boundary (arrow in Fig. 4H). Interestingly, overexpressing Trn using MS1096 (MS>HA-Trn) resulted in nuclear localization of Ci in anterior cells distant from the A/P border (arrow in Fig. 4I), suggesting that excessive Trn can promote nuclear localization of Ci in vivo.
Trn physically interacts with Ci through NLS-N
To further corroborate that Trn regulates Ci nuclear import through NLS-N, we examined whether Trn binds Ci through NLS-N using purified epitope-tagged proteins derived from S2 cells. Immunopurified Myc–CiN, Myc–CiNΔ212-268 or Myc–CiNmNLS-N proteins were incubated with purified HA–Trn or HA–CG8219 and the protein complexes were immunoprecipitated using an anti-Myc antibody, resolved by SDS-PAGE followed by western blot analysis with an anti-HA antibody. As shown in Fig. 4J, HA–Trn coimmunoprecipitated with Myc–CiN. Mutation of the NLS-N either by deletion (Myc–CiNΔ212-268) or amino acid substitution (Myc–CiNmNLS-N) diminished the binding of HA–Trn, suggesting that Trn interacts with Ci through NLS-N. Consistent with our finding that CG8219 is not involved in the regulation of CiN, HA–CG8219 was not pulled down by either wild-type or mutant forms of Myc–CiN (Fig. 4J). Taken together, these results suggest that Trn binds to NLS-N to promote nuclear import of Ci.
Sufu impedes nuclear localization of Ci by blocking binding of Trn to NLS-N
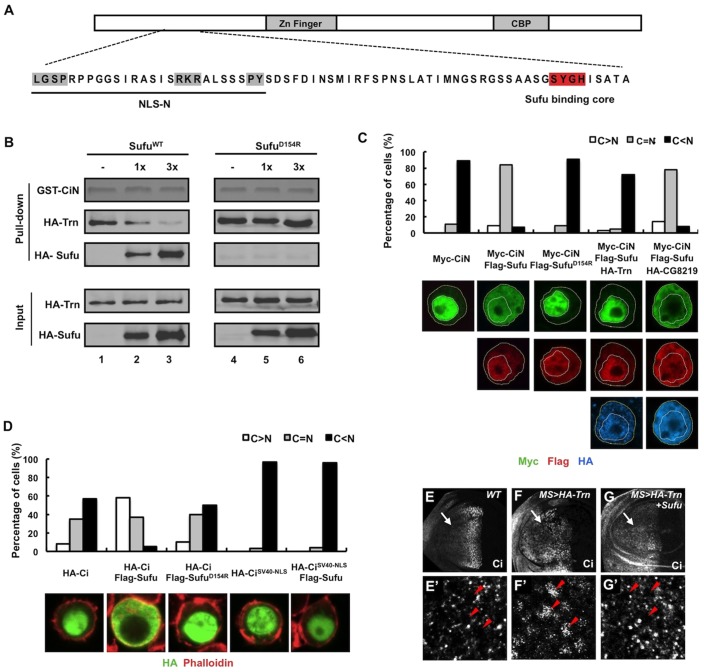
Previous studies demonstrated that Sufu binds to an N-terminal domain in Ci/Gli proteins via a core sequence SYGH (Dunaeva et al., 2003), which is in close proximity to NLS-N (Fig. 5A). This raises an interesting possibility that binding of Sufu to CiN could mask the NLS-N and prevent Trn from binding to Ci, which could be an underlying mechanism by which Sufu impedes the nuclear localization of Ci.
Fig. 5.
Sufu inhibits Ci nuclear translocation by blocking the interaction of Trn and Ci. (A) Structure of Ci with sequence of the NR region indicated underneath. The PY-NLS motif is highlighted in gray and the Sufu-binding core in red. (B) Binding of Sufu to CiN precludes Trn binding. Equal amounts of GST–CiN bound to Glutathione beads were incubated with a fixed amount of purified HA–Trn without or with increasing amounts of purified HA–SufuWT or HA–SufuD154R. Pull-down and input proteins were analyzed by western blot. GST–CiN was visualized by Coomassie Blue staining. (C) Quantification of subcellular localization of CiN. 100 cells were randomly picked for each transfection and categorized based on the differential nuclear/cytoplasmic distributions of Myc signal. Representative images of S2 cells transfected with the indicated constructs and immunostained with anti-Myc (green), anti-FLAG (red) and anti-HA (blue) antibodies are shown below. Yellow and white dashed lines outline cellular and nuclear membrane, respectively. (D) Quantification of subcellular localization of Ci and CiSV40-NLS. 100 cells were randomly picked for each transfection and categorized based on the differential nuclear and cytoplasmic distributions of HA signal. Representative images of S2 cells transfected with HA–Ci or HA–CiSV40-NLS either alone or with FLAG–Sufu or FLAG–SufuD154R. The transfected cells were treated with 10 ng/ml LMB for 2 hours before immunostaining with an anti-HA antibody (green) and Phalloidin (red). (E–G′) Low (E–G) and high (E′–G′) magnification views of a wild-type wing disc (E,E′), a wing disc expressing HA–Trn (F,F′), and a wing disc coexpressing HA–Trn and Sufu (G,G′) with MS1096 treated with 50 ng/ml LMB for 2 hours prior to immunostaining with an anti-Ci (red) antibody. Arrows indicate the anterior compartments; arrowheads indicate nuclei. Coexpression of Sufu blocked Trn-induced nuclear localization of Ci in A-compartment cells away from the A/P boundary.
To determine whether Sufu inhibits Trn binding to CiN, we carried out in vitro binding experiments using a GST pull-down assay. GST–CiN fusion proteins were incubated with purified HA–Trn in the absence or presence of increasing amounts of purified HA–Sufu. Protein complexes were pulled down using glutathione beads, separated by SDS-PAGE followed by western blot to visualize bound proteins. As shown in Fig. 5B, HA–Trn was readily pulled down by GST-CiN in the absence of Sufu (lane 1); however, increasing amounts of Sufu resulted in a gradual decrease in the amount of HA–Trn bound to GST–CiN and a concomitant increase in the number of complexes of FLAG–Sufu and GST–CiN (Fig. 5B, lanes 2–3), suggesting that binding of Sufu to CiN precluded Trn binding. To further test this notion, we carried out competition experiments using the mutant form of Sufu with D154 replaced with R (SufuD154R). In a crystal structure of the hSufu–Gli1 complex, D159 of hSufu (corresponding to D154 of Drosophila Sufu) is located in the binding pocket that contacts the SYGH motif of Gli1 (Zhang et al., 2013a). Indeed, SufuD154R failed to bind CiN (Fig. 5B). Importantly, we found that SufuD154R no longer inhibited binding of Trn to CiN (Fig. 5B, lanes 4–6). Taken together, these results suggest that binding of Sufu to the SYGH motif precludes the recognition of NLS-N by Trn.
To further explore the antagonistic relationship between Trn and Sufu in the regulation of Ci nuclear import and determine the underlying mechanism, we turned to an in vitro cell-based system. As shown in Fig. 5C, Myc–CiN was predominantly localized in the nucleus when transfected into S2 cells. Co-transfection of FLAG–Sufu retained a large fraction of Myc–CiN in the cytoplasm. Addition of HA–Trn but not HA–CG8219 in the transfection restored the predominant nuclear localization of Myc–CiN. Consistent with its inability to block Trn binding to CiN, SufuD154R failed to impede the nuclear localization of CiN (Fig. 5C). In addition, wild-type Sufu but not SufuD154R retained full-length Ci in the cytoplasm (Fig. 5D). In additional support of the notion that Sufu regulates Ci subcellular localization by masking its NLSs, we found that Sufu failed to impede the nuclear import of a Ci variant (CiSV40-NLS) (Wang and Jiang, 2004), which contains an exposed SV40-derived NLS inserted at its N-terminus that is no longer masked by binding of Sufu to Ci (Fig. 5D).
To examine the competitive relationship between Trn and Sufu in regulating Ci nuclear translocation in vivo, we coexpressed Sufu with Trn using MS1096 and found that excessive Sufu suppressed Trn-induced ectopic Ci nuclear localization in A-compartment cells away from the A/P boundary (Fig. 5E–G′). Hence, Sufu opposes the function of Trn in vivo.
A conserved PY-NLS function in Gli proteins
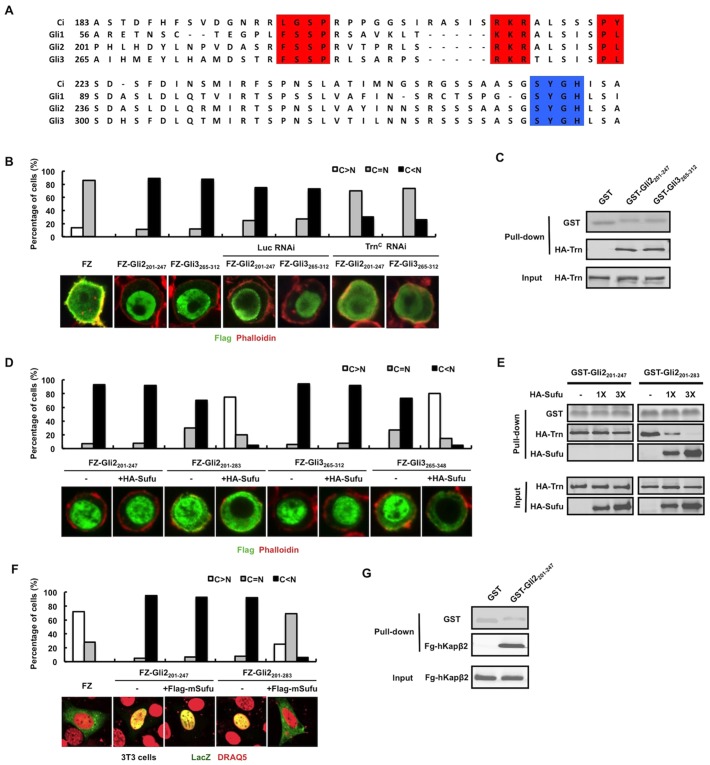
Sequence alignment revealed that Gli proteins also contain a putative PY-NLS motif, although in this case the PY core is replaced by a PL core that is also found in a subfamily of PY-NLS containing proteins (Fig. 6A) (Lee et al., 2006). Because Gli2 and Gli3 are the primary mediators of vertebrate Hh signaling and there is evidence that Hh induces nuclear translocation of Gli2 and Gli3 (Kim et al., 2009; Humke et al., 2010), we focused our functional analysis on the putative PY-NLS motif present in Gli2 and Gli3. N-terminal fragments of Gli2 and Gli3 containing the putative PY-NLS sequences were fused to the C-terminus of FLAG–LacZ (FZ) to generate FZ–Gli2201-247 and FZ–Gli3265-312, or to GST to generate GST–Gli2201-247 and GST–Gli3265-312. As shown in Fig. 6B, FZ–Gli2201-247 and FZ–Gli3265-312 were localized almost exclusively in the nucleus, whereas FZ is largely in the cytoplasm, suggesting that the N-terminal fragments of Gli2 and Gli3 can promote nuclear localization of a heterologous protein. Both GST–Gli2201-247 and GST–Gli3265-312 pulled down HA–Trn (Fig. 6C). Furthermore, Trn RNAi diminished the positive effect of the Gli2 and Gli3 fragments on the nuclear translocation of FZ chimeric proteins (Fig. 6B), suggesting these N-terminal fragments contain a functional PY-NLS. To determine whether Sufu regulates nuclear translocation of Gli by blocking Trn, we fused longer Gli2 and Gli3 fragments that contained both the PY-NLS and the Sufu binding motif SYGH to FZ to generate FZ–Gli2201-283 and FZ–Gli3265-348. We found that coexpression of Sufu sequestered FZ–Gli2201-283 and FZ–Gli3265-348 in the cytoplasm but did not affect the nuclear localization of FZ–Gli2201-247 and FZ–Gli3265-312 (Fig. 6D). In the GST pull-down assay, we found that increasing amounts of HA–Sufu diminished the amounts of HA–Trn pulled down by GST-Gli2201-283 but did not affect the association between GST–Gli2201-247 and HA–Trn (Fig. 6E). These results support the notion that binding of Sufu to the Gli2 and Gli3 SYGH core prevents Trn/Kapβ2 from binding to the adjacent PY-NLS motif and thus inhibits PY-NLS-mediated nuclear translocation of Gli proteins.
Fig. 6.
NLS-N is conserved in Gli proteins and regulated by Sufu. (A) Sequence alignment of the N-terminal conserved region of Ci/Gli (human Gli1, mouse Gli2 and human Gli3) with the PY-NLS motif highlighted in red and the Sufu binding core highlighted in blue. (B) Quantification of nuclear and cytoplasmic localization of FZ and its variants. Representative images of S2 cells transfected with FLAG–LacZ (FZ), FZ–Gli2201-247 and FZ–Gli3265-312 without or with control or Trnc dsRNA treatment and immunostained with anti-FLAG antibody (green) and Phalloidin (red) are shown below. (C) GST pull-down assay to show that HA–Trn is associated with GST–Gli2201-247 and GST–Gli3265-312. Input and bound HA–Trn proteins were visualized by western blot, whereas GST fusion proteins were detected by Coomassie Blue staining. (D) Quantification of nuclear and cytoplasmic localization of FZ and its variants. Representative images of S2 cells transfected with the indicated FZ–Gli chimera either alone or together with Sufu and immunostained with an anti-FLAG antibody (green) and Phalloidin (red) are shown below. Nuclear translocation of FZ–Gli2201-283 or FZ–Gli3265-348 but not FZ–Gli2201-247 and FZ–Gli3265-312 was blocked by Sufu coexpression. (E) GST pull-down assay to show that increasing amount of HA–Sufu blocked the binding of HA–Trn to GST–Gli2201-283 but not to GST–Gli3201-247. (F) Quantification of nuclear and cytoplasmic localization of FZ and its variants. Representative images of NIH3T3 cells transfected with the indicated FZ–Gli2 chimera either alone or together with mouse Sufu (mSufu) and immunostained with an anti-LacZ antibody (green) and DRAQ5 nuclear dye (pseudo red) are shown below. (G) GST pull-down assay to show that HA–hKapβ2 is associated with GST–Gli2201-247. Input and bound HA–hKapβ2 proteins were visualized by western blot whereas GST fusion proteins were detected by Coomassie Blue staining.
To determine whether the Gli PY-NLS can function in the mammalian system, we examined the subcellular localization of FZ–Gli2 chimeric proteins in NIH3T3 cells. Similar to the observation in S2 cells, we found that both FZ–Gli2201-247 and FZ–Gli2201-283 were exclusively localized in the nucleus, and that coexpression of mouse Sufu (mSufu) retains only FZ–Gli2201-283 but not FZ–Gli2201-247 in the cytoplasm (Fig. 6F). Finally, we performed a GST pull-down assay and found that GST–Gli2201-247 interacts with the human Kapβ2 protein (Fig. 6G).
DISCUSSION
Signal-induced nuclear translocation of Ci/Gli transcription factors represents a crucial step in Hh signal transduction (Chen et al., 1999; Wang et al., 2000; Wang and Holmgren, 2000; Humke et al., 2010). The subcellular localization of Ci/Gli is controlled by both nuclear import and export; however, Hh signaling appears to promote nuclear translocation of Ci/Gli by regulating import rather than export (Chen et al., 1999; Wang et al., 2000; Wang and Holmgren, 2000). Thus, understanding how nuclear import of Ci/Gli is regulated represents an important unsolved question (Briscoe and Thérond, 2013). A key to this problem is to identify all the nuclear localization signals in Ci/Gli and the corresponding Kapβ family members responsible for their nuclear import. Here, we identified a novel NLS located in the conserved N-terminal region of Ci/Gli and showed that this NLS acts in conjunction with the previously identified bipartite NLS to effectively promote Ci nuclear import and activity. Furthermore, we demonstrated that this new NLS, which we named NLS-N, is subject to regulation by Sufu.
Compared with the classic mono- or bi-partite NLS, PY-NLS sequences are very divergent and difficult to predict (Chook and Süel, 2011). Indeed, previous sequence analysis failed to identify Ci/Gli proteins as putative Kapβ2 substrates (Lee et al., 2006). According to four general criteria to verify a putative sequence as a functional NLS (Damelin et al., 2002), we provide several lines of evidence suggesting that the N-terminal conserved region of Ci/Gli harbors a bona fide PY-NLS. (1) They can mediate nuclear import of a heterologous protein in a manner depending on Trn/Kapβ2. (2) They physically interact with Trn/Kapβ2. (3) Inactivation of Trn affects nuclear localization of Ci. (4) The NLS-N in Ci acts independently of the NLS-C to regulate optimal nuclear import and activity of full-length Ci in both S2 cells and wing imaginal discs.
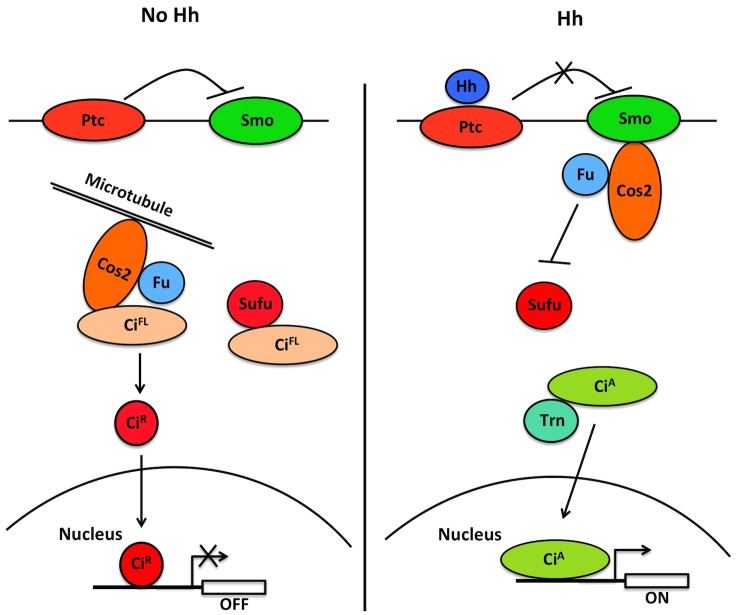
Our study also provides a mechanistic insight into how Sufu regulates Ci/Gli nuclear–cytoplasmic shuttling (Fig. 7). In the absence of Hh, Ci forms a trimeric complex with Cos2 and Fu as well as a dimeric complex with Sufu, although low levels of a tetrameric complex containing Ci, Cos2, Fu and Sufu might also exist (Stegman et al., 2000; Lum et al., 2003; Wang and Jiang, 2004). Dimeric Sufu–Gli complexes have also been observed (Tukachinsky et al., 2010). Our previous study revealed that Cos2 retains Ci in the cytoplasm through microtubule tethering as well as masking of the NLS (Wang and Jiang, 2004), but how Sufu impedes nuclear localization of Ci remained unknown. The close proximity between the PY-NLS motif and the Sufu-binding core in Ci/Gli raised an interesting possibility that binding of Sufu to the Ci/Gli N-terminal region masks the NLS-N. Indeed, our in vitro binding assay using purified proteins demonstrates that binding of Sufu to CiN inhibits Trn binding. We also provide evidence that Sufu and Trn regulate Ci nuclear import antagonistically. Furthermore, inhibition of Trn binding and NLS-N-mediated nuclear localization depends on the interaction between Sufu and the SYGH motif because amino acid substitution of an essential residue (D154) in Sufu that contacts the SYGH motif renders the mutant Sufu incapable of doing so. Binding of Sufu to the SYGH motif could prevent Trn from binding to NLS-N through steric hindrance. Alternatively, the NLS-N sequence could also be part of the Sufu-binding domain and binding of Sufu to CiN might directly compete with Trn for binding to NLS-N. Consistent with the latter possibility, we found that amino acid substitutions in the PY-NLS motif compromised Sufu binding (data not shown). Because SufuD154R also failed to sequester full-length Ci, whose nuclear import is mediated by both NLS-N and NLS-C, interaction between Sufu and the N-terminal region of Ci might also be required for masking the NLS-C. In addition, our unpublished data revealed that Sufu binds full-length Ci more stably than CiN, suggesting that Sufu could simultaneously interact with CiN and a C-terminal domain similar to the dual binding to Gli1 (Merchant et al., 2004), and this dual interaction might contribute to the masking of both NLS-N and NLS-C.
Fig. 7.
A model for the regulation of Ci subcellular localization by Trn and Sufu in Hh signaling. In the absence of Hh, full-length Ci (CiFL) forms complexes with Sufu and Cos2/Fu, which retain CiFL in the cytoplasm by NLS masking and microtubule tethering. In addition, CiFL is converted into a truncated repressor form (CiR) through ubiquitin/proteasome-mediated proteolysis. In the presence of Hh, Ci dissociates from Cos2 and Sufu, and Trn binds the exposed PY-NLS of Ci to promote its nuclear translocation.
How does Hh signaling promote the nuclear import of Ci/Gli? In response to Hh, Ci dissociates from Cos2 (Ruel et al., 2007; Shi et al., 2011). A reduction in interaction between Sufu and the N-terminal region of Ci has also been observed by FRET analysis in response to Hh pathway activation (Shi et al., 2011), which might expose the PY-NLS and allow Trn to bind Ci and promote its nuclear translocation. Similarly, Shh can dissociate Gli proteins from Sufu (Humke et al., 2010; Tukachinsky et al., 2010), which could expose their NLSs. A remaining question is how Hh signaling regulates the binding between Sufu and Ci/Gli. In Drosophila, Hh induces phosphorylation of Sufu but phosphorylation-deficient forms of Sufu behave similar to the wild-type Sufu with respect to regulation of Ci (Zhou and Kalderon, 2011). It is interesting to note that the NR domain of Ci/Gli contains many conserved Ser/Thr residues (Croker et al., 2006), raising the possibility that phosphorylation of NR domain modulates its interaction with Sufu or Trn/Kapβ2. Indeed, activation of the Hh pathway can induce phosphorylation and nuclear translocation of Gli3 (Humke et al., 2010). Therefore, it would be interesting to determine whether Hh stimulates phosphorylation of the NR domain to regulate the association of Ci/Gli with Sufu or Trn/Kapβ2 in the future.
MATERIALS AND METHODS
Transgenes
UAS-Trn-RNAi (VDRC #105181) was obtained from VDRC Stock Center. To generate UAS-Myc-CiN construct, a fragment encoding Ci N-terminal fragment (aa 1–440) was amplified by PCR and inserted in-frame between BglII and Asp718 sites of pUAST-Myc vector (Brand and Perrimon, 1993; Tong and Jiang, 2007). Using PCR-based mutagenesis, internal deletion and point mutations were further introduced to generate UAS-Myc-CiNΔ212-268 and UAS-Myc-CiNmNLS-N. To generate UAS-FLAG-LacZ-Ci/Gli-NLS chimeric constructs, a NotI-Asp718 fragment encoding full-length LacZ amplified from pcDNA3.1/V5-LacZ (Invitrogen) and Asp718-XbaI fragments encoding regions including the putative PY-NLSs of Ci/Gli proteins (aa 183–234 of Ci; aa 201–247 or 201–283 of mouse Gli2, and aa 265–312 and 265–348 of human Gli3) were fused and inserted between NotI and XbaI sites of the pUAST-FLAG vector. Point mutations were further introduced into UAS-FZ-Ci183-234 to generate UAS-FZ-mCi183-234 for Ci. FZ-Gli2201-247 and FZ-Gli2201-283 were subcloned between EcoRI and XbaI sites in pcDNA3.1+ for transfection in NIH3T3 cells. FLAG–hKapβ2 was a gift from Dr Yuh Min Chook's Lab. To generate UAS-HA-Ci, an EcoRI-Asp718 fragment encoding three copies of HA tag and an Asp718-XbaI fragment encoding Ci were fused and subcloned between EcoRI and XbaI sites of pUAST vector. Point mutations were then introduced by PCR-based mutagenesis to generate Ci variants including UAS-HA-CimNLS-N, UAS-HA-CimNLS-C, and UAS-HA-CimNLS-N+C. To generate UAS-HA-Trn and UAS-HA-CG8219, the coding regions of Trn and CG8219 were amplified by PCR and inserted in-frame between BglII and Asp718 sites of the pUAST-HA vector. To generate GST–Ci/Gli fusions, DNA fragments encoding the corresponding Ci/Gli sequences were subcloned between BamHI and NotI sites of pGEX4T3 vector. UAS-FLAG-Sufu and UAS-Myc-CFP have been described previously (Zhang et al., 2006; Zhang et al., 2009). C765 and MS1096 Gal4 drivers have been described previously (Chen et al., 2010). For generating flies with transgenes inserted at the 75B1 attP locus, the coding regions for HA-tagged Ci variants were subcloned between EcoRI and XbaI sites of a modified pUAST vector with an attB sequence inserted upstream of the UAS-binding sites (Liu et al., 2007).
Luciferase reporter assay
For luciferase reporter assays, S2 cells were cultured in 12-well plates and for each well transfected with 1 μg ptc-luc reporter construct, 50 ng RL-PolIII Renilla construct (an internal control), together with 0.5 μg Ci or its variant constructs. After incubation for 24 hours, transfected cells were split into control or Hh-conditioned media for an additional 24 hours. The luciferase reporter assays were performed using the Dual-Luciferase reporter assay system (Promega). Measurements for each sample were performed in triplicate using FLUOstar OPTIMA (BMG Labtech).
Cell culture, transfection, immunostaining, immunoprecipitation and western blot analysis
Drosophila S2 and S2R+ cells were cultured in Drosophila SFM (Invitrogen) with 10% fetal bovine serum (FBS), 100 U/ml of penicillin, and 100 mg/ml of streptomycin at 24°C. Transfections were carried out by Calcium Phosphate Transfection Kit (Specialty Media) according to the manufacturer's instructions. Immunoprecipitation and western blot analysis were carried out using standard protocols as previously described (Zhang et al., 2005). For immunostaining, S2 cells transfected with indicated constructs and treated without or with 10 ng/ml LMB for 2 hours were harvested and washed with PBS, fixed with 4% formaldehyde at room temperature for 20 minutes, and incubated with primary antibodies for 2 hours. Then, cells were washed three times with PBS followed by secondary antibody staining for 1.5 hours. NIH3T3 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), L-glutamine, 1 mM sodium pyruvate and penicillin. Transfection was carried out using a GenJet Plus in vitro DNA transfection kit (SignaGen). For immunofluorescence analysis, cells were seeded on LAB-TEK chamber slides, transfected with indicated constructs and stained using standard protocols. Antibodies and dyes used for this study were mouse anti-FLAG (M2, Sigma), rabbit anti-FLAG (Thermo), mouse anti-HA (F7, Santa Cruz), rat anti-HA (3F10), mouse anti-Myc (9E10, Santa Cruz), rabbit anti-LacZ (Affinity Bioreagents), Phalloidin (Life Technology) and DRAQ5 (Cell Signaling Technology).
RNAi in Drosophila S2 cells
DNA templates corresponding to Trn 100–800 bp, Trn 2100–2682 bp, CG8219 100–800 bp, and CG8219 2100–2562 bp were generated by PCR and used to make dsRNA targeting the N-terminus or C-terminus of Trn and CG8219, respectively. dsRNA targeting the firefly luciferase coding sequence was used as a control. All the dsRNAs were generated by MEGAscript High-Yield Transcription Kit (Ambion, AM1334). For the RNAi knockdown experiments, S2 cells were first cultured in serum-free medium containing the indicated dsRNA for 12 hours at 24°C. After adding FBS to a final concentration of 10%, dsRNA-treated cells were cultured overnight before transfection with the indicated constructs. After additional culturing for 2 days, cells were collected for immunostaining or western blot analysis.
Protein purification and pull-down assay
HA-tagged wild-type or D154R mutant Sufu, HA–Trn, and HA–CG8219 were expressed in S2R+ cells and purified with anti-HA agarose (Pierce) according to the manufacturer's protocol. The bound proteins were eluted with HA peptides (Pierce) at 1 mg/ml concentration. GST–Ci/Gli fusion proteins were produced in E. coli and purified with Glutathione–agarose beads (GE Healthcare). GST fusion proteins bound to beads were washed three times with ice-cold PBS containing 1% NP-40 and incubated with purified HA–Trn or HA–CG8219 at 4°C for 1.5 hours. For the Sufu competition assay, increasing amount of purified HA–SufuWT or D154R mutant was applied before the incubation. After washing three times with cell lysis buffer, the beads were boiled in 1× SDS loading buffer, followed by western blot analysis.
Acknowledgments
We thank Dr Qing Zhang for discussions, Drs Yuh Min Chook, Bing Li and Jiang Wu, DSHB, VDRC and Bloomington Stock Centers for reagents and Shuangxi Li for technical assistance.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
Q.S. and J.J. designed the experiments. Q.S. and Y.H. conducted the experiments. J.J. wrote the manuscript.
Funding
This work is supported by grants from the National Institutes of Health [grant numbers GM061269, GM067045]; National Science Foundation of China [grant number 31328017]; and Welch foundation [grant number I-1603]. J.J. is a Eugene McDermott Endowed Scholar in Biomedical Science at UTSW. Deposited in PMC for release after 12 months.
Reference
- Aavikko M., Li S. P., Saarinen S., Alhopuro P., Kaasinen E., Morgunova E., Li Y., Vesanen K., Smith M. J., Evans D. G. et al. (2012). Loss of SUFU function in familial multiple meningioma. Am. J. Hum. Genet. 91, 520–526 10.1016/j.ajhg.2012.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnfield P. C., Zhang X., Thanabalasingham V., Yoshida M., Hui C. C. (2005). Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation 73, 397–405 10.1111/j.1432-0436.2005.00042.x [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312–3317 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- Briscoe J., Thérond P. P. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Chen Y., Jiang J. (2013). Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 23, 186–200 10.1038/cr.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., von Kessler D. P., Park W., Wang B., Ma Y., Beachy P. A. (1999). Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell 98, 305–316 10.1016/S0092-8674(00)81960-1 [DOI] [PubMed] [Google Scholar]
- Chen M. H., Wilson C. W., Li Y. J., Law K. K., Lu C. S., Gacayan R., Zhang X., Hui C. C., Chuang P. T. (2009). Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 23, 1910–1928 10.1101/gad.1794109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Li S., Tong C., Zhao Y., Wang B., Liu Y., Jia J., Jiang J. (2010). G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes Dev. 24, 2054–2067 10.1101/gad.1948710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook Y. M., Süel K. E. (2011). Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta 1813, 1593–1606 10.1016/j.bbamcr.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E., Izaurralde E. (2001). Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 13, 310–319 10.1016/S0955-0674(00)00213-1 [DOI] [PubMed] [Google Scholar]
- Croker J. A., Ziegenhorn S. L., Holmgren R. A. (2006). Regulation of the Drosophila transcription factor, Cubitus interruptus, by two conserved domains. Dev. Biol. 291, 368–381 10.1016/j.ydbio.2005.12.020 [DOI] [PubMed] [Google Scholar]
- Damelin M., Silver P. A., Corbett A. H. (2002). Nuclear protein transport. Methods Enzymol. 351, 587–607 10.1016/S0076-6879(02)51870-X [DOI] [PubMed] [Google Scholar]
- Ding Q., Fukami S., Meng X., Nishizaki Y., Zhang X., Sasaki H., Dlugosz A., Nakafuku M., Hui C. (1999). Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr. Biol. 9, 1119–1122 10.1016/S0960-9822(99)80482-5 [DOI] [PubMed] [Google Scholar]
- Dunaeva M., Michelson P., Kogerman P., Toftgard R. (2003). Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 278, 5116–5122 10.1074/jbc.M209492200 [DOI] [PubMed] [Google Scholar]
- Humke E. W., Dorn K. V., Milenkovic L., Scott M. P., Rohatgi R. (2010). The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24, 670–682 10.1101/gad.1902910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W., Nakano Y., Seger C. (2011). Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406 10.1038/nrg2984 [DOI] [PubMed] [Google Scholar]
- Jia J., Amanai K., Wang G., Tang J., Wang B., Jiang J. (2002). Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature 416, 548–552 10.1038/nature733 [DOI] [PubMed] [Google Scholar]
- Jia J., Tong C., Wang B., Luo L., Jiang J. (2004). Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature 432, 1045–1050 10.1038/nature03179 [DOI] [PubMed] [Google Scholar]
- Jia J., Zhang L., Zhang Q., Tong C., Wang B., Hou F., Amanai K., Jiang J. (2005). Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev. Cell 9, 819–830 10.1016/j.devcel.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Jiang J. (2006). Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle 5, 2457–2463 10.4161/cc.5.21.3406 [DOI] [PubMed] [Google Scholar]
- Jiang J., Hui C. C. (2008). Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812 10.1016/j.devcel.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Struhl G. (1998). Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391, 493–496 10.1038/35154 [DOI] [PubMed] [Google Scholar]
- Kim J., Kato M., Beachy P. A. (2009). Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA 106, 21666–21671 10.1073/pnas.0912180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kise Y., Morinaka A., Teglund S., Miki H. (2009). Sufu recruits GSK3beta for efficient processing of Gli3. Biochem. Biophys. Res. Commun. 387, 569–574 10.1016/j.bbrc.2009.07.087 [DOI] [PubMed] [Google Scholar]
- Lee B. J., Cansizoglu A. E., Süel K. E., Louis T. H., Zhang Z., Chook Y. M. (2006). Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543–558 10.1016/j.cell.2006.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Cao X., Jiang J., Jia J. (2007). Fused-Costal2 protein complex regulates Hedgehog-induced Smo phosphorylation and cell-surface accumulation. Genes Dev. 21, 1949–1963 10.1101/gad.1557407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L., Zhang C., Oh S., Mann R. K., von Kessler D. P., Taipale J., Weis-Garcia F., Gong R., Wang B., Beachy P. A. (2003). Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol. Cell 12, 1261–1274 10.1016/S1097-2765(03)00426-X [DOI] [PubMed] [Google Scholar]
- Merchant M., Vajdos F. F., Ultsch M., Maun H. R., Wendt U., Cannon J., Desmarais W., Lazarus R. A., de Vos A. M., de Sauvage F. J. (2004). Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol. Cell. Biol. 24, 8627–8641 10.1128/MCB.24.19.8627-8641.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méthot N., Basler K. (2000). Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development 127, 4001–4010 [DOI] [PubMed] [Google Scholar]
- Monnier V., Dussillol F., Alves G., Lamour-Isnard C., Plessis A. (1998). Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway.. Curr. Biol. 8, 583–S2 10.1016/S0960-9822(98)70227-1 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis E., Hui C. C. (2005). Hedgehog signaling and congenital malformations. Clin. Genet. 67, 193–208 10.1111/j.1399-0004.2004.00360.x [DOI] [PubMed] [Google Scholar]
- Ohlmeyer J. T., Kalderon D. (1998). Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396, 749–753 10.1038/25533 [DOI] [PubMed] [Google Scholar]
- Paine P. L., Moore L. C., Horowitz S. B. (1975). Nuclear envelope permeability. Nature 254, 109–114 10.1038/254109a0 [DOI] [PubMed] [Google Scholar]
- Price M. A., Kalderon D. (2002). Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell 108, 823–835 10.1016/S0092-8674(02)00664-5 [DOI] [PubMed] [Google Scholar]
- Robbins D. J., Fei D. L., Riobo N. A. (2012). The Hedgehog signal transduction network. Sci. Signal. 5, re6 10.1126/scisignal.2002906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel L., Gallet A., Raisin S., Truchi A., Staccini-Lavenant L., Cervantes A., Thérond P. P. (2007). Phosphorylation of the atypical kinesin Costal2 by the kinase Fused induces the partial disassembly of the Smoothened-Fused-Costal2-Cubitus interruptus complex in Hedgehog signalling. Development 134, 3677–3689 10.1242/dev.011577 [DOI] [PubMed] [Google Scholar]
- Shi Q., Li S., Jia J., Jiang J. (2011). The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development 138, 4219–4231 10.1242/dev.067959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson B. E., Ziegenhorn S. L., Holmgren R. A. (2006). Regulation of Ci and Su(fu) nuclear import in Drosophila. Dev. Biol. 294, 258–270 10.1016/j.ydbio.2006.02.050 [DOI] [PubMed] [Google Scholar]
- Smelkinson M. G., Zhou Q., Kalderon D. (2007). Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev. Cell 13, 481–495 10.1016/j.devcel.2007.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegman M. A., Vallance J. E., Elangovan G., Sosinski J., Cheng Y., Robbins D. J. (2000). Identification of a tetrameric hedgehog signaling complex. J. Biol. Chem. 275, 21809–21812 10.1074/jbc.C000043200 [DOI] [PubMed] [Google Scholar]
- Svärd J., Heby Henricson K., Persson-Lek M., Rozell B., Lauth M., Bergström A., Ericson J., Toftgård R., Teglund S. (2006). Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell 10, 187–197 10.1016/j.devcel.2005.12.013 [DOI] [PubMed] [Google Scholar]
- Taipale J., Beachy P. A. (2001). The Hedgehog and Wnt signalling pathways in cancer. Nature 411, 349–354 10.1038/35077219 [DOI] [PubMed] [Google Scholar]
- Taylor M. D., Liu L., Raffel C., Hui C. C., Mainprize T. G., Zhang X., Agatep R., Chiappa S., Gao L., Lowrance A. et al. (2002). Mutations in SUFU predispose to medulloblastoma. Nat. Genet. 31, 306–310 10.1038/ng916 [DOI] [PubMed] [Google Scholar]
- Tong C., Jiang J. (2007). Using immunoprecipitation to study protein-protein interactions in the Hedgehog-signaling pathway. Methods Mol. Biol. 397, 215–229 10.1007/978-1-59745-516-9_15 [DOI] [PubMed] [Google Scholar]
- Tukachinsky H., Lopez L. V., Salic A. (2010). A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 191, 415–428 10.1083/jcb.201004108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M., Taipale J. (2008). Hedgehog: functions and mechanisms. Genes Dev. 22, 2454–2472 10.1101/gad.1693608 [DOI] [PubMed] [Google Scholar]
- Varjosalo M., Li S. P., Taipale J. (2006). Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev. Cell 10, 177–186 10.1016/j.devcel.2005.12.014 [DOI] [PubMed] [Google Scholar]
- Wang Q. T., Holmgren R. A. (1999). The subcellular localization and activity of Drosophila cubitus interruptus are regulated at multiple levels. Development 126, 5097–5106 [DOI] [PubMed] [Google Scholar]
- Wang Q. T., Holmgren R. A. (2000). Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development 127, 3131–3139 [DOI] [PubMed] [Google Scholar]
- Wang G., Jiang J. (2004). Multiple Cos2/Ci interactions regulate Ci subcellular localization through microtubule dependent and independent mechanisms. Dev. Biol. 268, 493–505 10.1016/j.ydbio.2004.01.008 [DOI] [PubMed] [Google Scholar]
- Wang G., Wang B., Jiang J. (1999). Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 13, 2828–2837 10.1101/gad.13.21.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Amanai K., Wang B., Jiang J. (2000). Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 14, 2893–2905 10.1101/gad.843900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Pan Y., Wang B. (2010). Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development 137, 2001–2009 10.1242/dev.052126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. W., Chuang P. T. (2010). Mechanism and evolution of cytosolic Hedgehog signal transduction. Development 137, 2079–2094 10.1242/dev.045021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhao Y., Tong C., Wang G., Wang B., Jia J., Jiang J. (2005). Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev. Cell 8, 267–278 10.1016/j.devcel.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Zhang L., Wang B., Ou C. Y., Chien C. T., Jiang J. (2006). A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev. Cell 10, 719–729 10.1016/j.devcel.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Shi Q., Chen Y., Yue T., Li S., Wang B., Jiang J. (2009). Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 106, 21191–21196 10.1073/pnas.0912008106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Fu L., Qi X., Zhang Z., Xia Y., Jia J., Jiang J., Zhao Y., Wu G. (2013a). Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci. Nat. Commun. 4, 2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lv X., Yin W. C., Zhang X., Feng J., Wu W., Hui C. C., Zhang L., Zhao Y. (2013b). Ter94 ATPase complex targets k11-linked ubiquitinated ci to proteasomes for partial degradation. Dev. Cell 25, 636–644 10.1016/j.devcel.2013.05.006 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Kalderon D. (2011). Hedgehog activates fused through phosphorylation to elicit a full spectrum of pathway responses. Dev. Cell 20, 802–814 10.1016/j.devcel.2011.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]