ABSTRACT
Contraction of the actomyosin ring (AMR) provides the centripetal force that drives cytokinesis. In budding yeast (Saccharomyces cerevisiae), assembly and contraction of the AMR is coordinated with membrane deposition and septum formation at the bud neck. A central player in this process is Iqg1, which promotes recruitment of actin to the myosin ring and links AMR assembly with that of septum-forming components. We observed early actin recruitment in response to inhibition of cyclin-dependent kinase 1 (Cdk1) activity, and we find that the Cdk1-dependent phosphorylation state of Iqg1 is a determining factor in the timing of bud neck localization of both Iqg1 and actin, with both proteins accumulating prematurely in cells expressing nonphosphorylatable Iqg1 mutants. We also identified the primary septum regulator Hof1 as a binding partner of Iqg1, providing a regulatory link between the septation and contractile pathways that cooperate to complete cytokinesis.
KEY WORDS: Iqg1, Cdk1, Actomyosin ring, Cytokinesis
INTRODUCTION
Cytokinesis requires the spatial and temporal regulation of multiple cellular and extracellular components at the site of cell division. In animal and fungal cells, a ring of actin filaments joined by myosin bundles forms along the plasma membrane (Mabuchi, 1994; Satterwhite and Pollard, 1992). As myosin motor activity contracts the ring and draws the membrane inwards, new plasma membrane and extracellular matrix components are deposited at the division site. The cell must coordinate these processes with each other and relative to other cell cycle events to ensure that cell division occurs only after completion of mitosis.
In the budding yeast, Saccharomyces cerevisiae, cytokinesis is carried out by structures and processes that are largely equivalent to those in animal cells (Wloka and Bi, 2012), including a contractile actomyosin ring (AMR) (Bi et al., 1998) and a cell wall deposition pathway that is analogous to mammalian extracellular matrix remodeling (Bi and Park, 2012). However, the budding yeast life cycle, with its ‘pre-furrowed’ division site, enables perturbation of either process without lethal effects (Lippincott and Li, 1998b; Rodriguez and Paterson, 1990; Schmidt et al., 2002), making S. cerevisiae a valuable model for study of the regulatory relationships among the proteins involved.
The yeast AMR is assembled sequentially throughout the cell cycle (Lippincott and Li, 1998b), beginning with the ring-shaped accumulation of septin proteins at the future bud site (Drubin and Nelson, 1996; Pringle et al., 1995). A ring of the type-II myosin heavy chain Myo1 forms at the nascent bud neck in a septin-dependent manner. Next, the septin-binding protein Bni5 (Lee et al., 2002) and the formins Bnr1 and Bni1 (Vallen et al., 2000) arrive at and depart from the bud neck in a distinct order, interacting with the Myo1 tail to guide Myo1 into its final position (Fang et al., 2010). The myosin light chain protein Mlc1 arrives in mitosis (Boyne et al., 2000) and facilitates recruitment of the essential protein Iqg1 (Shannon and Li, 2000). Iqg1 both stabilizes the Myo1 ring during mitosis (Fang et al., 2010) and is required for the recruitment of filamentous actin (F-actin), which completes AMR assembly and immediately precedes ring contraction (Epp and Chant, 1997; Lippincott and Li, 1998b; Shannon and Li, 1999).
AMR contraction is coupled with the onset of cell wall formation, which begins with the centripetal deposition of the chitinous primary septum by chitin synthase II (Chs2) (Sburlati and Cabib, 1986; Shaw et al., 1991). A secondary septum is then deposited along the primary septum and becomes the cell wall upon digestion of the primary septum. Like AMR assembly, primary septum deposition follows the sequential bud neck localization of several organizing proteins, most importantly Hof1, Cyk3 and Inn1 (Devrekanli et al., 2012; Kamei et al., 1998; Korinek et al., 2000; Lippincott and Li, 1998a; Nishihama et al., 2009; Sanchez-Diaz et al., 2008).
Although disruption of AMR contraction and primary septum deposition together is lethal (Vallen et al., 2000), neither process alone is essential for viability (Bi et al., 1998; Lippincott and Li, 1998b; Rodriguez and Paterson, 1990; Schmidt et al., 2002). However, each process is defective in the absence of the other, as myo1Δ cells deposit a misoriented primary septum (Fang et al., 2010; Rodriguez and Paterson, 1990) and chs2Δ cells produce unstable and asymmetric AMRs (Schmidt et al., 2002; VerPlank and Li, 2005). The two processes are at once physically distinct, mutually interdependent and partially redundant, a relationship that invites and facilitates more detailed molecular investigation. Genetic studies have illuminated the dependencies underlying sequential protein recruitment to the bud neck (Jendretzki et al., 2009; Meitinger et al., 2010; Roh et al., 2002), but information on the temporal regulation of these components remains limited.
The 173-kDa protein Iqg1 is one of the few cytokinesis components that is essential for viability (Epp and Chant, 1997). Iqg1 is homologous to the mammalian IQGAP proteins that bundle F-actin (Bashour et al., 1997; Fukata et al., 1997) and regulate actin-dependent processes including cytokinesis (Hart et al., 1996; White et al., 2012). Iqg1 is required for actin recruitment to the bud neck (Bi et al., 1998; Epp and Chant, 1997; Lippincott and Li, 1998b) and interacts genetically with primary septum regulators, such as Cyk3 (Korinek et al., 2000). Its large size and multi-domain architecture, together with its essential function, make Iqg1 a likely physical link between the AMR and primary septum machinery.
Cytokinesis occurs during a period defined by the inactivation of cyclin-dependent kinase 1 (Cdk1) and the dephosphorylation of Cdk1 protein substrates, brought on by mitotic cyclin degradation and Cdc14 phosphatase activation (Stegmeier and Amon, 2004). Dephosphorylation of Cdk1 substrates underlies many events of late mitosis and cytokinesis (Bloom et al., 2011; Chin et al., 2012; Khmelinskii et al., 2009; Mishima et al., 2004; Sullivan et al., 2008; Teh et al., 2009). Cdc14 localizes to the bud neck prior to the onset of cytokinesis and is known to promote primary septum deposition (Palani et al., 2012). Similarly, the mitotic exit network (MEN) that sustains Cdc14 activity regulates septin dynamics (Cid et al., 2001; Lippincott et al., 2001), actin localization (Sanchez-Diaz et al., 2012), primary septum function (Meitinger et al., 2010) and AMR contraction (Lippincott et al., 2001; Luca et al., 2001; Vallen et al., 2000).
Here, we addressed the mechanisms by which Cdk1 substrate dephosphorylation governs AMR assembly and contraction. We identified Cdk1-mediated phosphorylation of Iqg1 as a determinant of bud neck localization of Iqg1 itself and of proteins in the AMR assembly and primary septum deposition pathways, providing insight into the mechanisms by which Cdks control the timing and coordination of cytokinetic events.
RESULTS
Inhibition of Cdk1 is sufficient for pre-anaphase recruitment of F-actin to the bud neck
In several species, experimental inhibition of Cdk1 in mitotically arrested cells can induce premature cytokinetic events (Dischinger et al., 2008; Niiya et al., 2005; Sanchez-Diaz et al., 2012). In budding yeast, expression of the Cdk1 inhibitory protein Sic1 in mitotic cells promotes AMR assembly and partial contraction, as well as septum formation (Sanchez-Diaz et al., 2012). To explore these findings further, we used confocal fluorescence microscopy in combination with an analog-sensitive allele of CDK1 (cdk1-as) that is specifically inhibited by the purine analog 1-NM-PP1 (Bishop et al., 2000). Wild-type and cdk1-as cells were arrested in a metaphase-like state by treatment with nocodazole, and 1-NM-PP1 was added to the culture for 15 min. Cells were then fixed and stained with fluorophore-conjugated phalloidin to visualize F-actin structures. A ring of F-actin at the bud neck, indicating a prematurely assembled AMR, was clearly visible in only ∼7% of wild-type cells arrested in metaphase, irrespective of 1-NM-PP1 treatment. This number rose to 27% in mock-treated cdk1-as cells, and 73% when these cells were treated with 1-NM-PP1 (Fig. 1A,B). Thus, chemical inhibition of Cdk1 results in premature actin ring assembly. The partial phenotype observed in the absence of inhibitor is consistent with previous observations that uninhibited Cdk1-as is a moderately weakened kinase (Bishop et al., 2000).
Fig. 1.
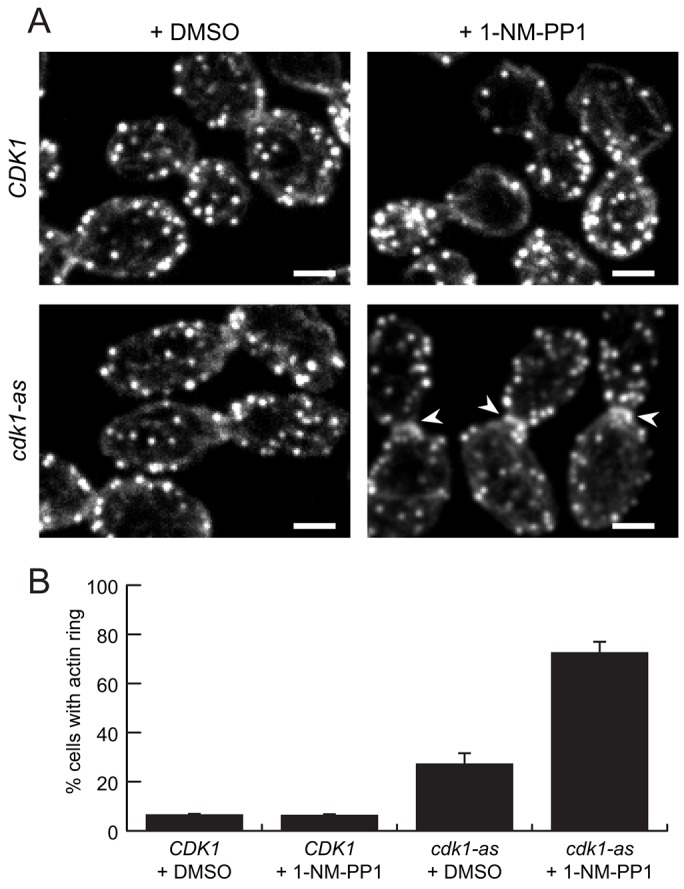
Actin ring assembly in response to Cdk1 inhibition. (A) Phalloidin-stained F-actin in nocodazole-arrested cells treated with 10 µM 1-NM-PP1 or DMSO for 15 min. Arrowheads: F-actin rings, confirmed by 3D analysis of ring shape (see Materials and Methods). Scale bars: 2 µm. (B) Actin ring frequency in nocodazole-arrested cells; results are means±s.e.m. for three experimental repeats.
Cells expressing nonphosphorylatable Iqg1 assemble a pre-anaphase actin ring
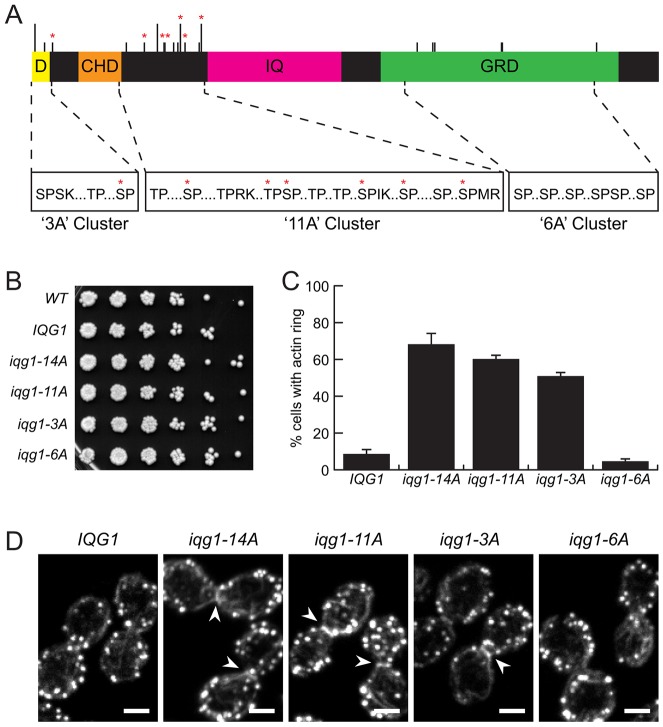
We set out to identify one or more Cdk1 substrates whose dephosphorylation was sufficient to reproduce the effects of Cdk1 inhibition on actin recruitment. Iqg1 was an appealing candidate, owing to its essential role in AMR assembly and because it contains 20 sites matching the minimal Cdk1 recognition motif (S/T-P), seven of which are known to be phosphorylated in mitotic cells and rapidly dephosphorylated after Cdk1 inhibition (Holt et al., 2009) (Fig. 2A). In the related yeast Candida albicans, the Iqg1 ortholog is phosphorylated in vitro by Cdk1, and mutation of Cdk1 consensus sites stabilizes Iqg1 and alters AMR assembly and disassembly (Li et al., 2008). Iqg1 contains multiple functional domains, including an anaphase-promoting complex/cyclosome (APC/C) recognition sequence (Ko et al., 2007; Tully et al., 2009), a calponin homology domain (CHD), which is required for actin recruitment and is homologous to known mammalian actin-bundling domains (Mateer et al., 2004), several IQ repeats that localize Iqg1 to the bud neck through an interaction with Mlc1 (Shannon and Li, 2000), and a degenerate GTPase-activating protein (GAP)-related domain (GRD) associated with AMR contraction (Shannon and Li, 1999). The 20 Cdk consensus sites occur in three clusters: three sites are near the N-terminal APC/C recognition sequence, 11 sites are between the CHD and the IQ-repeat region, and six sites are within the GRD near the C-terminus (Fig. 2A).
Fig. 2.
Iqg1 phosphorylation controls actin ring assembly. (A) Functional domains and Cdk1 consensus phosphorylation sites of Iqg1. D, APC/C recognition motif; CHD, calponin homology domain; IQ, eleven IQ repeats; GRD, GAP-related domain; short lines, minimal Cdk1 consensus motifs (S/T-P); tall lines, full Cdk1 consensus motifs (S/T-P-x-K/R); asterisks, sites identified as Cdk1-dependent phosphosites in vivo by mass spectrometry (Holt, et al., 2009). Site clusters are named for their associated alanine-mutant alleles. (B) iqg1 phosphomutants grow at wild-type rates. Serial dilutions of log-phase cultures grown on rich medium at 30°C. (C) The actin ring frequency in nocodazole-arrested cells is increased in iqg1 phosphomutants. iqg1-14A is mutated in both the ‘3A’ and ‘11A’ clusters. Results are means±s.e.m. for two experimental repeats. (D) Representative nocodazole-arrested cells stained with phalloidin. Arrowheads, F-actin rings. Scale bars: 2 µm.
We analyzed the functions of these clusters individually and in combination by introducing mutant alleles of IQG1 at its endogenous locus. Serine or threonine residues were replaced with alanine residues to generate the alleles iqg1-3A, iqg1-11A, iqg1-6A and iqg1-14A (iqg1-11A and iqg1-3A in combination) (Fig. 2A; supplementary material Tables S1, S2). All mutants proliferated at normal rates under nutrient-rich conditions (Fig. 2B). The mutants were arrested by nocodazole treatment and stained with phalloidin. After 2 h of nocodazole treatment, the iqg1-3A, -11A and -14A populations showed clear F-actin rings in a majority of metaphase-arrested cells, compared to only 9% for wild-type IQG1 (Fig. 2C,D), demonstrating an inhibitory role for Cdk1-dependent phosphorylation of Iqg1 in AMR assembly. The iqg1-14A actin ring frequency did not differ significantly from that observed in our earlier experiments with Cdk1-as inhibition, suggesting that dephosphorylation of Iqg1 is sufficient to explain the effect of Cdk1 inhibition on AMR assembly. iqg1-6A cells displayed no difference from wild-type in this and other experiments (Fig. 2C,D; data not shown), so the six C-terminal sites were excluded from further analysis.
Cells expressing nonphosphorylatable Iqg1 assemble F-actin rings prior to anaphase in an unperturbed cell cycle
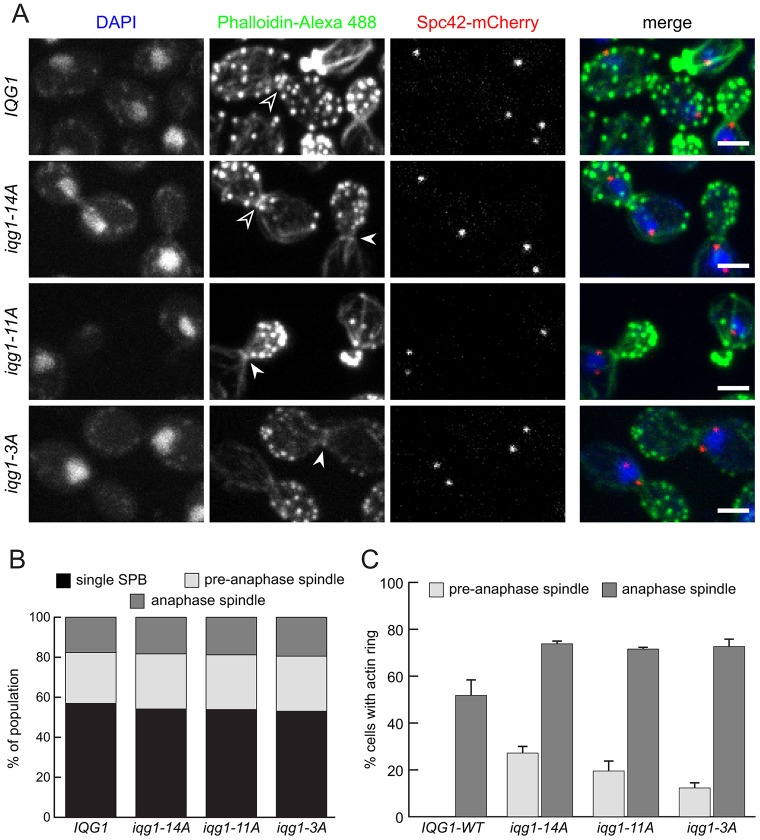
To assess the timing of AMR assembly in an unperturbed cell cycle, we observed the frequency of phalloidin-stained F-actin rings in an asynchronous, exponentially growing culture (Fig. 3A). The spindle pole body (SPB)-associated protein Spc42 was visualized by the red fluorescent protein mCherry, permitting the sorting of cells into three cell cycle stages based on the spacing between the two SPBs. The proportion of cells in each stage was not significantly altered in any mutant, suggesting no overall defect in cell cycle progression (Fig. 3B). Actin rings were not observed in G1 or S phase cells with unseparated SPBs (data not shown). In wild-type cells, rings were observed only in cells whose wide SPB spacing indicated an elongated anaphase spindle, and even then in only 52% of cells (Fig. 3A,C). By contrast, iqg1-14A populations displayed F-actin rings in 27% of cells with short pre-anaphase spindles and in 74% of cells with anaphase spindles (Fig. 3A,C). Long-spindled iqg1-14A cells without rings are likely cells fixed after ring contraction but before cell separation, rather than cells with late-forming AMRs, as suggested by the 100% ring frequency in iqg1-14A cells fixed in early anaphase with a single elongated DAPI mass (data not shown). iqg1-3A and -11A populations showed similar but slightly lower ring frequencies than the iqg1-14A combination. Thus, F-actin recruitment to the AMR occurs only after the initiation of spindle elongation in wild-type cells, but prior to anaphase in the Iqg1 phosphomutants.
Fig. 3.
Actin rings form prior to anaphase in iqg1 phosphomutants. (A) Three-color imaging of cells fixed during asynchronous exponential growth. White arrowheads, F-actin rings in pre-anaphase cells. Black arrowheads, F-actin rings in anaphase cells. Scale bars: 2 µm. (B) Normal overall cell cycle progression in iqg1 phosphomutants. Asynchronous cells were sorted based on DAPI and Spc42–mCherry data from the experiment shown in A. No spindle, cells with a single distinguishable SPB focus; pre-anaphase spindle, cells with a single round DAPI mass and two distinguishable SPBs spaced less than 2.5 µm apart; anaphase spindle, cells with an elongated or split DAPI mass and SPB spacing greater than 2.5 µm. (C) Actin ring frequency among cells within the ‘pre-anaphase’ and ‘anaphase’ spindle groups. No actin rings were observed in cells with a single SPB focus. Results are means±s.e.m. for two experimental repeats.
These observations demonstrate that the colocalization of actin and myosin is not sufficient for contraction. Stable AMRs incompetent for contraction have been described previously in the case of an iqg1 mutant lacking the GRD (Shannon and Li, 1999), but our observation of rings that assemble early but contract normally reveals that budding yeast regulate AMR assembly and AMR contraction separately.
Nonphosphorylatable Iqg1 arrives at the bud neck prematurely
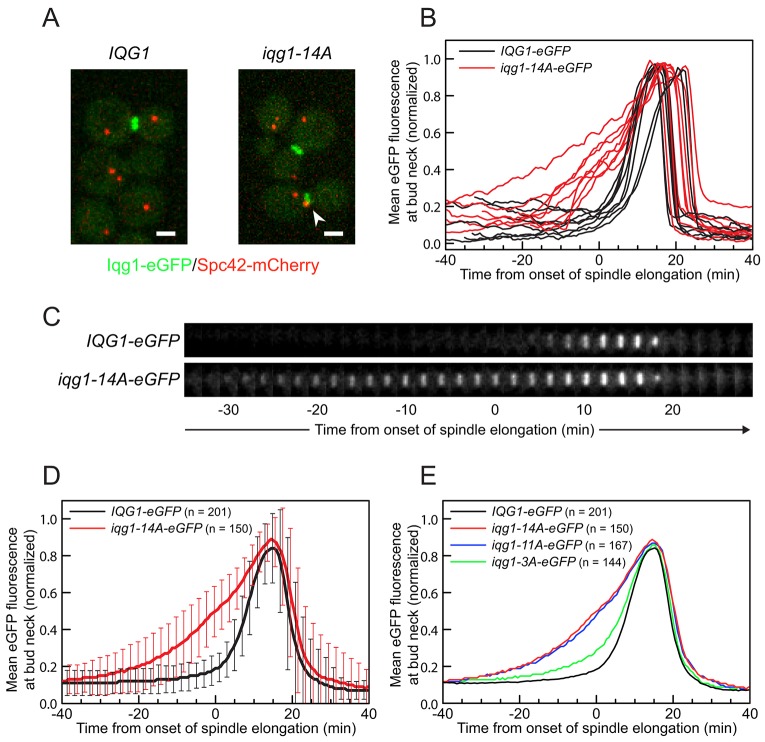
The premature recruitment of F-actin to the bud neck in IQG1 phosphomutants might be explained by an increase in binding affinity between Iqg1 and F-actin, or by premature localization of Iqg1 itself, or by a combination of both. To address these possibilities, we marked Iqg1 with enhanced green fluorescent protein (eGFP) and observed Iqg1–eGFP dynamics over the cell cycle in live cells by time-lapse microscopy, again using Spc42–mCherry as a marker for cell cycle progression (Fig. 4A).
Fig. 4.
Iqg1 phosphomutants accumulate at bud neck prematurely. (A) Live-cell images of cells expressing C-terminal fusions of eGFP and mCherry at the endogenous loci of IQG1 and SPC42, respectively. Arrowhead, pre-anaphase Iqg1p–eGFP accumulation at the bud neck in the iqg1 phosphomutant. Scale bars: 2 µm. (B) Quantification of green fluorescence signal in the bud neck region over time for eight representative cells from each genotype, normalized and smoothed. (C) Time series of Iqg1–eGFP dynamics at the bud neck in typical wild-type and iqg1 phosphomutant cells, at 2-min intervals. (D) Population average of normalized mean eGFP fluorescence at each time point for the indicated number of cells, pooling data from two experiments (±s.d.). (E) Population average of normalized mean eGFP fluorescence at each time point for all four iqg1 alleles tested.
The iqg1 phosphomutants matched wild-type cells in measures of overall cell cycle duration and time between major SPB dynamics milestones (data not shown). However, the behavior of Iqg1–eGFP in relation to those milestones was altered dramatically. A ring-shaped fluorescence signal became visible 4–8 min after the onset of spindle elongation in a typical IQG1-eGFP cell but 15–20 min prior to elongation onset in a typical iqg1-14A-eGFP cell (Fig. 4A,C).
To obtain a more quantitative understanding, we measured the mean fluorescence per pixel within the bud neck region at each time point. The results confirmed early Iqg1–eGFP accumulation in the phosphomutant relative to onset of spindle elongation (Fig. 4B,D). Results for iqg1-11A did not differ significantly from those for iqg1-14A, whereas iqg1-3A cells showed a similar but less severe phenotype (Fig. 4E), as in our studies of actin localization (Fig. 3C). The timing of peak Iqg1–eGFP concentration was not affected, suggesting that the full recruitment of Iqg1 depends on additional factors. Considering the actin results presented above, this suggests that sub-peak levels of dephosphorylated Iqg1 at the bud neck are sufficient for F-actin recruitment. Iqg1 ring contraction and degradation proceeded with wild-type timing in all strains (data not shown).
Iqg1 interacts in vivo with Mlc1 and the primary septum deposition regulator Hof1
On the basis of the premature localization of Iqg1 phosphomutants to the bud neck, we hypothesized that Cdk1 phosphorylation weakens the binding affinity between Iqg1 and one or more proteins that recruit Iqg1 to the bud neck. Previous studies have identified Mlc1 as a binding partner of Iqg1 whose localization profile matches that of the hypothesized recruiter (Boyne et al., 2000; Shannon and Li, 2000). To test the possibility that Iqg1 phosphorylation inhibits interactions with its binding partners, we analyzed Iqg1-associated proteins in immunoprecipitates of Iqg1–eGFP from cell lysates. Cells were arrested by nocodazole treatment to ensure high Iqg1 levels and high bud neck concentrations in the phosphomutant. Sypro-Ruby staining (Fig. 5) revealed specific binding of Iqg1–eGFP and a number of other proteins, including a 16-kDa protein that seemed to have near 1∶1 stoichiometry to the Iqg1–eGFP bait. Mass spectrometric analysis identified this protein as Mlc1 (supplementary material Table S3A). We did not observe reproducible differences in Mlc1 band intensity between the wild-type and iqg1-14A immunoprecipitates, suggesting that phosphorylation does not interfere with this interaction. We note, however, that microscopic analysis of insoluble lysate fractions revealed many largely intact Iqg1–eGFP rings, so the interactions analyzed in this experiment might be limited to soluble Iqg1–Mlc1 complexes that are not incorporated into the AMR, where the phosphomutant phenotypes were observed.
Fig. 5.
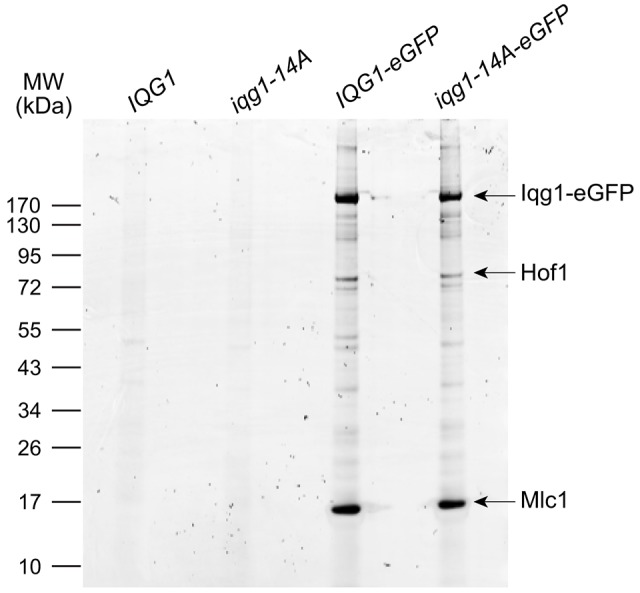
Iqg1 associates with Mlc1 and Hof1 in vivo. Sypro-Ruby stain of Iqg1–GFP immunoprecipitates from lysates of nocodazole-arrested cells. Hof1 and Mlc1 were identified by mass spectrometry (supplementary material Table S3). There were no reproducible differences between the IQG1-eGFP and iqg1-14A-eGFP preparations.
Mass spectrometry identified the second-most prominent Iqg1-associated protein as the F-BAR protein Hof1 (supplementary material Table S3B), a bud neck protein that regulates primary septum deposition through interactions with Inn1 and Chs2 (Nishihama et al., 2009; Oh et al., 2013). Iqg1 interaction with Inn1 has been demonstrated previously (Sanchez-Diaz et al., 2008), and Iqg1 interacts genetically with another Inn1-regulating protein, Cyk3 (Korinek et al., 2000). Our evidence for a physical interaction between Iqg1 and Hof1 provides further support for the notion that Iqg1 contributes to the regulation of primary septum deposition.
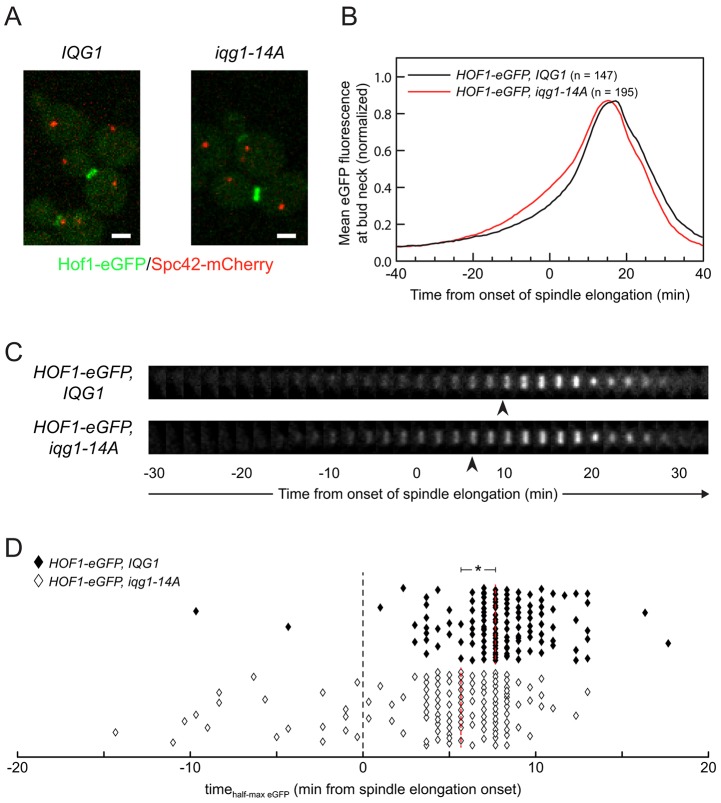
Hof1 localizes prematurely at the bud neck in cells expressing nonphosphorylatable Iqg1
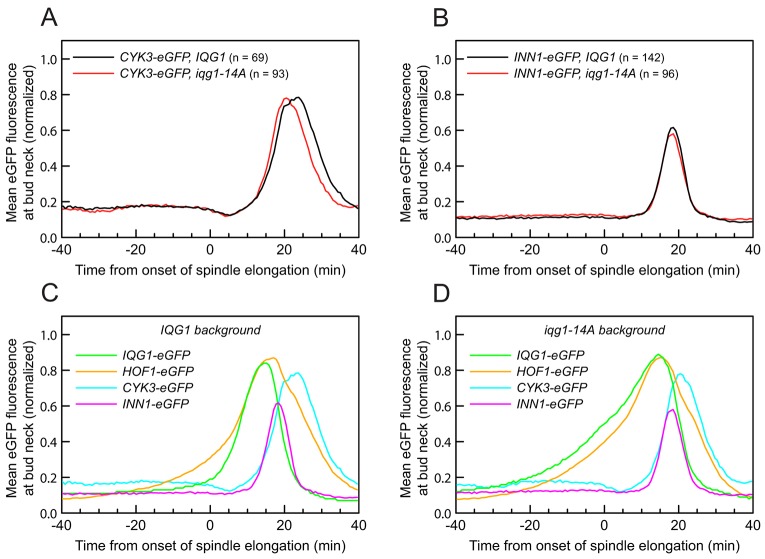
The interaction between Iqg1 and Hof1 led us to hypothesize that Iqg1 phosphorylation influences Hof1 localization and thereby links the AMR and septum deposition pathways. To explore this possibility, we analyzed Hof1–eGFP dynamics in relation to spindle elongation in IQG1 or iqg1-14A cells (Fig. 6A). The timing of the onset and the peak of bud-neck-localized Hof1–eGFP signal were not significantly affected, but the average rate of accumulation was increased slightly in the iqg1-14A background (Fig. 6B,C). The distribution of accumulation rates among individual cells was analyzed by marking the time at which each bud neck exceeded the half-maximal eGFP fluorescence: 14% of the counted iqg1-14A cells accumulated more than half of their bud neck Hof1 before the onset of spindle elongation, compared to less than 2% of IQG1 cells (Fig. 6D). iqg1-11A yielded a nearly identical distribution to iqg1-14A, whereas iqg1-3A again displayed an intermediate phenotype (10% of cells) (data not shown). We conclude that phosphorylation of Iqg1 helps control the timing of recruitment of Hof1 to the bud neck, although it is not likely to be the primary determinant of that timing.
Fig. 6.
Iqg1 phosphorylation affects Hof1 bud neck accumulation. (A) Live-cell images of cells expressing C-terminal fusions of eGFP and mCherry at the endogenous loci of HOF1 and SPC42, respectively. Scale bars: 2 µm. (B) Population average of normalized mean eGFP fluorescence at each time point, pooling data from two experiments. (C) Time series of Hof1–eGFP dynamics at the bud neck in typical wild-type and iqg1-14A cells, at 2-min intervals. Arrowheads, time at which the mean eGFP signal exceeds the half-maximal value. (D) Distribution of the time points at which a cell exceeded the half-maximal Hof1p–eGFP bud neck accumulation. 133 representative cells are depicted for each population. *P<0.0004, 2-min difference in median values (dual ranksum and resampling tests).
Late primary septum regulator dynamics are not affected by Iqg1 phosphorylation
Current models (Devrekanli et al., 2012; Nishihama et al., 2009) suggest that Hof1 promotes primary septum formation by cooperating with Cyk3 to position Inn1 to activate the chitin synthase Chs2. Given that Iqg1 phosphomutants display altered Hof1 accumulation dynamics, we also analyzed the dynamics of Cyk3 and Inn1 in these mutants, employing C-terminal eGFP tags. No significant differences were observed for either protein (Fig. 7A,B). Comparisons of the timing of protein recruitment to the ring revealed that in wild-type and iqg1-14A cells, Cyk3 and Inn1 both accumulated at the bud neck after both Iqg1 and Hof1 reached peak concentrations. The bud neck levels of Inn1, like those of Iqg1 (Fig. 4C) (Ko et al., 2007; Tully et al., 2009), declined in parallel with AMR contraction, whereas Hof1 and particularly Cyk3 disappeared from the bud neck several minutes after the completion of AMR contraction (Fig. 7C,D; see also Fig. 6C).
Fig. 7.
Accumulation profile of multiple protein components during cytokinesis. (A,B) Iqg1 phosphorylation does not significantly affect the bud neck accumulation of Cyk3 or Inn1. Cyk3–eGFP or Inn1–eGFP was analyzed by the same methods used for other eGFP-tagged proteins. Population averages of normalized mean Cyk3–eGFP or Inn1–eGFP fluorescence at each time point. (C) Overlay of population averages for all analyzed proteins in the wild-type IQG1 background. (D) Overlay of population averages for all analyzed proteins in the iqg1-14A background. Note that the decline in Iqg1 levels at the bud neck coincides with AMR contraction (see Fig. 4C) (Ko et al., 2007; Tully et al., 2009), whereas the decline in Hof1 levels occurs after the completion of contraction (see Fig. 6C). The decline in Cyk3 at the bud neck also occurs long after the completion of contraction.
DISCUSSION
Cdk1 regulates protein assembly at the bud neck through Iqg1
Our results demonstrate that the phosphorylation of Iqg1 by Cdk1 is a major determinant of the timing of AMR assembly in preparation for cytokinesis. Previous studies have shown that Iqg1 is extensively phosphorylated at multiple Cdk1 consensus sites in cells arrested in mid-mitosis by expression of a nondegradable mutant form of the cyclin Clb2, and then rapidly dephosphorylated at these sites upon chemical inhibition of Cdk1 (Holt et al., 2009). It therefore seems likely that phosphorylation of Iqg1 by Cdk1 prior to anaphase inhibits its association with the bud neck and thereby inhibits actin recruitment; dephosphorylation in late anaphase then promotes AMR assembly.
We employed quantitative live-cell microscopy to obtain a temporally precise portrait of the ordered assembly of Iqg1 and other proteins at the site of cell division, using the onset of spindle elongation as a fixed reference point. These methods allowed us to measure the timing and rates of recruitment of different proteins in terms of population averages and variance among individual cells. Our analyses of Iqg1 levels suggest that preventing Cdk1-dependent phosphorylation accelerates the onset of the early phase of Iqg1 recruitment but does not influence the time at which peak Iqg1 levels are achieved. We therefore suspect that an additional regulatory trigger governs Iqg1 recruitment to peak levels.
Previous studies of Iqg1 in C. albicans (Li et al., 2008) yielded results that are both similar to and distinct from our results in S. cerevisiae, suggesting that the two species share some but not all features of Iqg1 regulation. In C. albicans, mutation of Cdk1-dependent phosphorylation sites in Iqg1 results in premature actin localization at the bud neck in nocodazole-arrested cells, and also stabilizes the Iqg1 protein and causes defects in AMR disassembly after contraction, leading to cytokinesis defects. Perhaps phosphorylation in C. albicans somehow promotes degradation by an APC/C-dependent mechanism like that identified previously in S. cerevisiae (Ko et al., 2007; Tully et al., 2009). However, our quantitative microscopy revealed no evidence for an effect of phosphomutants on overall levels of Iqg1 at the bud neck or in measurements taken throughout the cytosol (data not shown), and we did not see any impact of the mutations on the dynamics of AMR contraction or disassembly, and no significant effect on cytokinesis. This suggests that, in contrast to the C. albicans case, the Iqg1 phosphomutant is ubiquitylated and destroyed normally in S. cerevisiae, and that the function of Iqg1 phosphorylation in this species is related exclusively to the timing of AMR assembly, inhibiting both Iqg1 and actin accumulation prior to anaphase.
Iqg1 coordinates AMR assembly with primary septum deposition through mutual physical interactions
The processes of AMR contraction and primary septum deposition are partially redundant and mutually interdependent (Bi, 2001; Vallen et al., 2000). Iqg1 is essential for both processes. This study demonstrates that the expression of nonphosphorylatable Iqg1 is sufficient to accelerate the assembly of both the AMR and primary septum-depositing complexes, identifying Iqg1 as a physical link and as a shared point of regulation. Our results help explain the tight temporal coordination between the two processes and support the conception of a unified and autonomous ‘septation apparatus’ proposed in a previous study of ectopic contractile complexes (Roh et al., 2002).
Because Hof1 normally arrives at the bud neck slightly earlier than Iqg1 (Fig. 7C), it is surprising that the phosphorylation of Iqg1 can influence the timing of Hof1 recruitment. We suspect that Hof1 recruitment depends on two regulatory mechanisms, such that some Iqg1-independent mechanism normally promotes early localization before the arrival of Iqg1 further stimulates Hof1 concentrations to peak levels.
Although iqg1 phosphomutants begin assembling Iqg1 and actin at the bud neck prematurely, they reach full Iqg1 concentrations and undergo AMR contraction with wild-type timing, suggesting that there is an additional regulatory trigger that is needed for completion of cytokinesis. It is possible that dephosphorylation of additional Cdk1 substrates would serve this role. Cyk3 provides an appealing candidate. Overexpression of Cyk3 can rescue defects in both AMR and primary septum pathways, and induces premature Inn1 localization (Jendretzki et al., 2009). Cyk3 is also a Cdk1 substrate in vivo (Holt et al., 2009). Given that we found that Cyk3 and Inn1 localize normally in iqg1-14A cells (Fig. 7A,B), an intriguing possibility is that Cdk1 phosphorylation of Iqg1 delays early steps of septation apparatus assembly, whereas Cdk1 phosphorylation of Cyk3 holds off later steps. Analysis of cyk3 and iqg1 phosphomutants in combination would shed light on this possibility.
Cdk1 regulates Iqg1 via phosphorylation at distinct and distant sites
Our studies raise the question of how phosphorylation affects Iqg1 at the molecular level. Because the effect of phosphorylation on AMR assembly is inhibitory, the simplest possibility is that phosphorylation disrupts an interaction between Iqg1 and one or more other proteins. Alternatively, the inhibition of Iqg1 bud neck localization might be carried out by the promotion of binding to a third-party protein that competes with the bud neck for Iqg1 binding. Iqg1 might also autoinhibit its own recruitment if phosphorylation causes the protein to fold so as to obscure a binding site. Such models are appealing because none of the phosphosites in question fall within the IQ repeat region known to be responsible for Iqg1 localization.
The persistence of the phenotype in both the iqg1-3A and iqg1-11A mutants (albeit to different degrees) supports the possibility that Iqg1 must be phosphorylated at two distant regions to fully inhibit AMR assembly. This might be explained by Iqg1 binding to its recruiter(s) via multiple surfaces, with either surface sufficient for recruitment, or by a folding structure wherein the two regions form a single binding surface. Another possibility is a kinase-priming mechanism, wherein phosphorylation of one site promotes phosphorylation of other sites to produce a downstream effect. This mode of regulation might be common in Cdk1 regulatory motifs (Kõivomägi et al., 2013), but has thus far not been demonstrated in a case where the two phosphorylated residues are so far apart or separated by a functional domain. More detailed analysis of the specific Iqg1 phosphosites that are responsible for these phenotypes will be the first step towards determining the mechanism of inhibition.
The stoichiometric relationship observed between Iqg1 and Mlc1 in our co-immunoprecipitation analysis suggests that Iqg1 might arrive at the bud neck as part of a soluble complex, expanding the possibilities for the mechanistic impact of phosphorylation. Further analysis of Iqg1-binding partners and the timing of their interactions should be combined with structural studies and the investigation of other phosphoproteins, such as Cyk3, to more fully understand the regulatory circuits that enable the robust and timely coordination of multiple complex processes during the final stage of cell reproduction.
MATERIALS AND METHODS
Yeast procedures
Yeast strains were made in the W303 strain background and are listed in supplementary material Table S1. Cultures were grown at 30°C in YEP plus 2% D-glucose, except where noted. To achieve mitotic arrest, 15 µg/ml nocodazole was added to a mid-log-phase culture for 2 h. For Cdk1 inhibition, 10 µM 1-NM-PP1 was added for an additional 15 min following nocodazole treatment.
Plasmid construction and mutagenesis
Plasmids are listed in supplementary material Table S2. To create pSGN092, PCR-amplified full-length IQG1, including all 5′ and 3′ untranslated regions, was subcloned into the pRS306 integrating plasmid. Synthetic fragments of iqg1 alanine mutants (Invitrogen LifeTech gene synthesis) were subcloned into the pSGN092 backbone to generate plasmids containing full-length iqg1 phosphomutant alleles.
For endogenous genomic replacement of IQG1, PCR from these plasmids was used to amplify the 5′ half of IQG1 along with a URA3 selection cassette and a homology region to promote insertion at the IQG1 locus. Transformation of wild-type yeast with these PCR products and sequential selection for and then against the URA3 marker left the genomic locus unchanged, aside from the alanine mutations and two introduced restriction sites that do not affect amino acid sequence. A wild-type control strain underwent parallel transformations using wild-type plasmid (pSGN092) as a PCR template. The C-terminal mutant, iqg1-6A, was made separately by integration of the full pSGN094 plasmid, followed by a similar selection and counterselection process. In all cases, mutagenesis was verified by PCR and sequencing of the full IQG1 locus.
To construct C-terminal molecular fusions, we used PCR from pYM28 (eGFP, HIS3 selection) or pSGN099 (mCherry, kanMX selection), with primers to target genomic insertion in place of the stop codon of the gene. Transformants were verified by PCR and by fluorescence microscopy.
Visualization of F-actin
Cells were fixed by mixing 1.35 ml culture (OD600 = 0.4–0.6) and 150 µl of 37% formaldehyde at room temperature for 10 min. All subsequent reagents were in 0.1 M KH2PO4/K2HPO4 buffer at pH 7.0, and all centrifugation steps were 5 min at 1000 g at room temperature. Cells were washed by centrifugation, resuspended in 3.7% formaldehyde, and mixed for 16 h at 4°C. Cells were washed, treated with 10 mM ethanolamine to quench formaldehyde, briefly sonicated to separate cell clusters, permeabilized with 0.2% Triton X-100 for 15 min, and incubated for 45 min in 60 µl of 0.2% Triton X-100 plus 3 U/ml of Alexa-Fluor-488-conjugated phalloidin (Invitrogen Molecular Probes), agitating periodically to keep cells in suspension. Cells were mounted on concanavalin-A-coated coverslips in Vectashield mounting medium (Vector Laboratories) containing 1 µg/ml DAPI, and imaged within hours by spinning disk confocal microscopy in the Nikon Imaging Center at UCSF.
Microscopy
For phalloidin experiments, samples were illuminated by 491 nm and 561 nm lasers and images were captured as stacks of 19 Z-planes spaced 0.4 µm apart, each 512×512 pixels (10.6 px/µm), using a 100× objective under the control of µManager software (Edelstein et al., 2010) (Microscope, Nikon Ti, Spinning Disk Confocal, Yokogawa CSU-22; camera, Photometrics Evolve EMCCD). Cells were analyzed by comparison of a Z-projected (by maximum value) single image with the slice-by-slice three-dimensional (3D) data for each field of view. Only bud necks that showed both a clear horizontal band across the bud neck in the projected image and a curved pattern of staining that traced the plasma membrane in the 3D data were scored as F-actin rings. In nocodazole experiments, cells that lacked a single DAPI mass and symmetrical actin membrane patch distribution (10–20% of total) were not counted as arrested cells.
For eGFP experiments, mid-log-phase cells in synthetic complete medium plus 2% D-glucose (SD) were harvested by centrifugation and sealed between a coverslip and a 20 mm×20 mm pad of recently cast 1% agarose in SD. Microscopy was performed as above, except using a 60× objective and ten Z-planes spaced 0.6 µm apart every 40 s for 2 h over five nearly adjacent viewfields, each 512×512 pixels (6.4 px/µm).
For quantitative analysis of eGFP-tagged proteins in live cells, average-value Z-projections were stack-registered to compensate for drift in the field of view, and a single time-projected image was created by taking the maximum-value signal across all 180 time points for each pixel. Regions of interest (bud necks) were defined based on this time-projected image, and the average eGFP signal intensity within each region was measured for each time point in the average Z-projected image. Data for each bud neck was normalized vertically such that mean fluorescence value ranged from 0 to 1 over the time course. The time point corresponding to onset of spindle elongation was defined as the time when SPBs moved apart from each other and continued to move apart in subsequent time points.
For Hof1–eGFP data, time points corresponding to half-maximal fluorescence were determined by smoothing normalized data of each cell (reporting fluorescence value at each time point as the rolling average of the nearest four time points), then identifying the first time point reported as greater than 0.5.
Immunopurification of Iqg1–eGFP
1.5 l of nocodazole-arrested cultures were chilled on ice for 20 min, harvested by centrifugation, washed with chilled IP buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA), dripped directly into liquid nitrogen, and stored at −80°C. 9 g pellets were ground into fine powder by cryo-grinding (Retsch MM301 Ball Mill, 3× 3 min, 30 Hz). The powder was resuspended in 12 ml IP buffer plus protease and phosphatase inhibitors and lysis completed by 4× 30 s blending with a tissue homogenizer (Polytron PT 1200 E). The soluble fraction was isolated following 1 h of ultracentrifugation (Beckman 70.1 Ti rotor, 50,000 rpm at 4°C), and incubated for 4 h at 4°C with 35 µl of sepharose beads slurry coated with monoclonal GFP-Trap antibody (Chromotek, Allele Biotech) for 4 h at 4°C. Beads were collected by centrifugation, washed three times, resuspended in 40 µl SDS-PAGE buffer (20 mM Tris-HCl pH 6.8, 2% SDS, 4% glycerol, 10 mM β-mercaptoethanol) and boiled for 60 s. 15 µl of supernatant was analyzed on a 4–12% gradient SDS-PAGE gel and stained with Sypro-Ruby (Invitrogen) or GelCode Blue Stain (Thermo Scientific).
Mass spectrometry
Protein bands or lane fragments were in-gel digested according to a published protocol (http://msf.ucsf.edu/ingel.html). Briefly, SDS and chemicals used for staining/destaining were removed with 20 mM ammonium bicarbonate buffer in 50% acetonitrile/water. Disulfide bonds were reduced with DTT, and free sulfhydryls were alkylated with iodoacetamide. Reagent excess was removed with the above buffer, and proteins were incubated with side-chain-protected porcine trypsin (Promega, Madison, WI) overnight at 37°C. The resulting peptides were extracted with 5% formic acid in 50% acetonitrile/water and fractionated by reversed-phase chromatography on a C18 column (75 µm×150 mm) with gradient elution (starting with 2% B up to 35% B in 35 min; solvent A, 0.1% formic acid in water; solvent B, 0.1% formic acid in acetonitrile; flow rate: 600 nl/min) using a nanoACQUITY uHPLC system (Waters, Milford, MA) directly linked to a linear ion trap – Orbitrap hybrid tandem mass spectrometer (LTQ-Orbitrap XL, Thermo Fisher Scientific, San Jose, CA). The six most abundant multiply-charged ions of each mass spectrometry survey were automatically selected for CID analysis. The precursor masses were measured in the Orbitrap, and CID data were acquired in the linear ion trap. Dynamic exclusion was enabled. In-house software (PAVA) was used for peak list generation. Database searching was conducted using Protein Prospector 5.10.10, against the SwissProt database, downloaded March 21, 2013. Saccharomyces, Homo sapiens, Bos taurus, Sus scrofa proteins and the GFP sequence were searched (35417/535248 entries searched).
Supplementary Material
Acknowledgments
We thank Kurt Thorn and the UCSF Nikon Imaging Center for valuable assistance with quantitative microscopy, Dan Lu (UCSF) for automation of analytical processes, and Heather Eshleman, Nick Lyons, and Juliet Girard (UCSF) for comments on the manuscript. Mass spectrometry analyses were performed in the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (Director, A.L. Burlingame), with support from the National Institutes of Health [grant number 8P41-GM103481].
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
S.G.N. performed all experiments and wrote the manuscript, with guidance from D.O.M.
Funding
This work was supported by funding from the National Institute of General Medical Sciences [grant number R01-GM069901]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.144097/-/DC1
References
- Bashour A. M., Fullerton A. T., Hart M. J., Bloom G. S. (1997). IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links microfilaments. J. Cell Biol. 137, 1555–1566 10.1083/jcb.137.7.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. (2001). Cytokinesis in budding yeast: the relationship between actomyosin ring function and septum formation. Cell Struct. Funct. 26, 529–537 10.1247/csf.26.529 [DOI] [PubMed] [Google Scholar]
- Bi E., Park H-O. (2012). Cell polarization and cytokinesis in budding yeast. Genetics 191, 347–387 10.1534/genetics.111.132886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D. J., Salmon E. D., McMillan J. N., Yeh E., Pringle J. R. (1998). Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142, 1301–1312 10.1083/jcb.142.5.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., Blethrow J., Shimizu E., Tsien J. Z., Schultz P. G., Rose M. D. et al. (2000). A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401 10.1038/35030148 [DOI] [PubMed] [Google Scholar]
- Bloom J., Cristea I. M., Procko A. L., Lubkov V., Chait B. T., Snyder M., Cross F. R. (2011). Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J. Biol. Chem. 286, 5434–5445 10.1074/jbc.M110.205054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne J. R., Yosuf H. M., Bieganowski P., Brenner C., Price C. (2000). Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J. Cell Sci. 113, 4533–4543 [DOI] [PubMed] [Google Scholar]
- Chin C. F., Bennett A. M., Ma W. K., Hall M. C., Yeong F. M. (2012). Dependence of Chs2 ER export on dephosphorylation by cytoplasmic Cdc14 ensures that septum formation follows mitosis. Mol. Biol. Cell 23, 45–58 10.1091/mbc.E11-05-0434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid V. J., Adamiková L., Sánchez M., Molina M., Nombela C. (2001). Cell cycle control of septin ring dynamics in the budding yeast. Microbiology 147, 1437–1450 [DOI] [PubMed] [Google Scholar]
- Devrekanli A., Foltman M., Roncero C., Sanchez-Diaz A., Labib K. (2012). Inn1 and Cyk3 regulate chitin synthase during cytokinesis in budding yeasts. J. Cell Sci. 125, 5453–5466 10.1242/jcs.109157 [DOI] [PubMed] [Google Scholar]
- Dischinger S., Krapp A., Xie L., Paulson J. R., Simanis V. (2008). Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network. J. Cell Sci. 121, 843–853 10.1242/jcs.021584 [DOI] [PubMed] [Google Scholar]
- Drubin D. G., Nelson W. J. (1996). Origins of cell polarity. Cell 84, 335–344 10.1016/S0092-8674(00)81278-7 [DOI] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. (2010). Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. Chapter 14, Unit14.20 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp J. A., Chant J. (1997). An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr. Biol. 7, 921–929 10.1016/S0960-9822(06)00411-8 [DOI] [PubMed] [Google Scholar]
- Fang X., Luo J., Nishihama R., Wloka C., Dravis C., Travaglia M., Iwase M., Vallen E. A., Bi E. (2010). Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J. Cell Biol. 191, 1333–1350 10.1083/jcb.201005134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M., Kuroda S., Fujii K., Nakamura T., Shoji I., Matsuura Y., Okawa K., Iwamatsu A., Kikuchi A., Kaibuchi K. (1997). Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J. Biol. Chem. 272, 29579–29583 10.1074/jbc.272.47.29579 [DOI] [PubMed] [Google Scholar]
- Hart M. J., Callow M. G., Souza B., Polakis P. (1996). IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 15, 2997–3005 [PMC free article] [PubMed] [Google Scholar]
- Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009). Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendretzki A., Ciklic I., Rodicio R., Schmitz H-P., Heinisch J. J. (2009). Cyk3 acts in actomyosin ring independent cytokinesis by recruiting Inn1 to the yeast bud neck. Mol. Genet. Genomics 282, 437–451 10.1007/s00438-009-0476-0 [DOI] [PubMed] [Google Scholar]
- Kamei T., Tanaka K., Hihara T., Umikawa M., Imamura H., Kikyo M., Ozaki K., Takai Y. (1998). Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 273, 28341–28345 10.1074/jbc.273.43.28341 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Roostalu J., Roque H., Antony C., Schiebel E. (2009). Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell 17, 244–256 10.1016/j.devcel.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Ko N., Nishihama R., Tully G. H., Ostapenko D., Solomon M. J., Morgan D. O., Pringle J. R. (2007). Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol. Biol. Cell 18, 5139–5153 10.1091/mbc.E07-05-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek W. S., Bi E., Epp J. A., Wang L., Ho J., Chant J. (2000). Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr. Biol. 10, 947–950 10.1016/S0960-9822(00)00626-6 [DOI] [PubMed] [Google Scholar]
- Kõivomägi M., Örd M., Iofik A., Valk E., Venta R., Faustova I., Kivi R., Balog E. R., Rubin S. M., Loog M. (2013). Multisite phosphorylation networks as signal processors for Cdk1. Nat. Struct. Mol. Biol. 20, 1415–1424 10.1038/nsmb.2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. R., Song S., Ro H. S., Park C. J., Lippincott J., Li R., Pringle J. R., De Virgilio C., Longtine M. S., Lee K. S. (2002). Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 6906–6920 10.1128/MCB.22.19.6906-6920.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. R., Wang Y. M., Wang Y. (2008). The IQGAP Iqg1 is a regulatory target of CDK for cytokinesis in Candida albicans. EMBO J. 27, 2998–3010 10.1038/emboj.2008.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. (1998a). Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143, 1947–1960 10.1083/jcb.143.7.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. (1998b). Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140, 355–366 10.1083/jcb.140.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Shannon K. B., Shou W., Deshaies R. J., Li R. (2001). The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J. Cell Sci. 114, 1379–1386 [DOI] [PubMed] [Google Scholar]
- Luca F. C., Mody M., Kurischko C., Roof D. M., Giddings T. H., Winey M. (2001). Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol. Cell. Biol. 21, 6972–6983 10.1128/MCB.21.20.6972-6983.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I. (1994). Cleavage furrow: timing of emergence of contractile ring actin filaments and establishment of the contractile ring by filament bundling in sea urchin eggs. J. Cell Sci. 107, 1853–1862 [DOI] [PubMed] [Google Scholar]
- Mateer S. C., Morris L. E., Cromer D. A., Benseñor L. B., Bloom G. S. (2004). Actin filament binding by a monomeric IQGAP1 fragment with a single calponin homology domain. Cell Motil. Cytoskeleton 58, 231–241 10.1002/cm.20013 [DOI] [PubMed] [Google Scholar]
- Meitinger F., Petrova B., Lombardi I. M., Bertazzi D. T., Hub B., Zentgraf H., Pereira G. (2010). Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J. Cell Sci. 123, 1851–1861 10.1242/jcs.063891 [DOI] [PubMed] [Google Scholar]
- Mishima M., Pavicic V., Grüneberg U., Nigg E. A., Glotzer M. (2004). Cell cycle regulation of central spindle assembly. Nature 430, 908–913 10.1038/nature02767 [DOI] [PubMed] [Google Scholar]
- Niiya F., Xie X., Lee K. S., Inoue H., Miki T. (2005). Inhibition of cyclin-dependent kinase 1 induces cytokinesis without chromosome segregation in an ECT2 and MgcRacGAP-dependent manner. J. Biol. Chem. 280, 36502–36509 10.1074/jbc.M508007200 [DOI] [PubMed] [Google Scholar]
- Nishihama R., Schreiter J. H., Onishi M., Vallen E. A., Hanna J., Moravcevic K., Lippincott M. F., Han H., Lemmon M. A., Pringle J. R. et al. (2009). Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J. Cell Biol. 185, 995–1012 10.1083/jcb.200903125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y., Schreiter J., Nishihama R., Wloka C., Bi E. (2013). Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol. Biol. Cell 24, 1305–1320 10.1091/mbc.E12-11-0804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palani S., Meitinger F., Boehm M. E., Lehmann W. D., Pereira G. (2012). Cdc14-dependent dephosphorylation of Inn1 contributes to Inn1-Cyk3 complex formation. J. Cell Sci. 125, 3091–3096 10.1242/jcs.106021 [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Bi E., Harkins H. A., Zahner J. E., De Virgilio C., Chant J., Corrado K., Fares H. (1995). Establishment of cell polarity in yeast. Cold Spring Harb. Symp. Quant. Biol. 60, 729–744 10.1101/SQB.1995.060.01.079 [DOI] [PubMed] [Google Scholar]
- Rodriguez J. R., Paterson B. M. (1990). Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil. Cytoskeleton 17, 301–308 10.1002/cm.970170405 [DOI] [PubMed] [Google Scholar]
- Roh D-H., Bowers B., Schmidt M., Cabib E. (2002). The septation apparatus, an autonomous system in budding yeast. Mol. Biol. Cell 13, 2747–2759 10.1091/mbc.E02-03-0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Diaz A., Marchesi V., Murray S., Jones R., Pereira G., Edmondson R., Allen T., Labib K. (2008). Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat. Cell Biol. 10, 395–406 10.1038/ncb1701 [DOI] [PubMed] [Google Scholar]
- Sanchez-Diaz A., Nkosi P. J., Murray S., Labib K. (2012). The Mitotic Exit Network and Cdc14 phosphatase initiate cytokinesis by counteracting CDK phosphorylations and blocking polarised growth. EMBO J. 31, 3620–3634 10.1038/emboj.2012.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterwhite L. L., Pollard T. D. (1992). Cytokinesis. Curr. Opin. Cell Biol. 4, 43–52 10.1016/0955-0674(92)90057-J [DOI] [PubMed] [Google Scholar]
- Sburlati A., Cabib E. (1986). Chitin synthetase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae. J. Biol. Chem. 261, 15147–15152 [PubMed] [Google Scholar]
- Schmidt M., Bowers B., Varma A., Roh D-H., Cabib E. (2002). In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115, 293–302 [DOI] [PubMed] [Google Scholar]
- Shannon K. B., Li R. (1999). The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol. Biol. Cell 10, 283–296 10.1091/mbc.10.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K. B., Li R. (2000). A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr. Biol. 10, 727–730 10.1016/S0960-9822(00)00539-X [DOI] [PubMed] [Google Scholar]
- Shaw J. A., Mol P. C., Bowers B., Silverman S. J., Valdivieso M. H., Durán A., Cabib E. (1991). The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114, 111–123 10.1083/jcb.114.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. (2004). Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38, 203–232 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Sullivan M., Holt L., Morgan D. O. (2008). Cyclin-specific control of ribosomal DNA segregation. Mol. Cell. Biol. 28, 5328–5336 10.1128/MCB.00235-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh E. M., Chai C. C., Yeong F. M. (2009). Retention of Chs2p in the ER requires N-terminal CDK1-phosphorylation sites. Cell Cycle 8, 2965–2976 10.4161/cc.8.18.9542 [DOI] [PubMed] [Google Scholar]
- Tully G. H., Nishihama R., Pringle J. R., Morgan D. O. (2009). The anaphase-promoting complex promotes actomyosin-ring disassembly during cytokinesis in yeast. Mol. Biol. Cell 20, 1201–1212 10.1091/mbc.E08-08-0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E. A., Caviston J., Bi E. (2000). Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11, 593–611 10.1091/mbc.11.2.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L., Li R. (2005). Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol. Biol. Cell 16, 2529–2543 10.1091/mbc.E04-12-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. D., Erdemir H. H., Sacks D. B. (2012). IQGAP1 and its binding proteins control diverse biological functions. Cell. Signal. 24, 826–834 10.1016/j.cellsig.2011.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C., Bi E. (2012). Mechanisms of cytokinesis in budding yeast. Cytoskeleton 69, 710–726 10.1002/cm.21046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.