Abstract
8p22–23-rs2254546 was firstly discovered to be associated with Kawasaki disease (KD) susceptibility by a genome-wide association study. However, only one Chinese replication study has been performed so far. To verify this association in another Chinese population, a hospital-based case-control study in Zhejiang province was conducted followed by an integrated meta-analysis, comprising five case-control studies of 1958 cases, 5615 controls and four transmission disequilibrium tests of 503 trios. In our case-control study, significant associations were observed between GG genotype or GG/GA genotypes of rs2254546 and increased KD risk (OR = 1.86, 95% CI = 1.01–3.41, P = 0.045; OR = 1.83, 95% CI = 1.01–3.33, P = 0.048), compared with AA genotype; however, no significant association was found in allelic model (OR = 1.20, 95% CI = 0.96–1.50, P = 0.117). The meta-analysis further revealed that the G allele was significantly associated with the increased KD risk without evidence of heterogeneity (OR = 1.55, 95% CI = 1.42–1.70, P < 0.001). In conclusion, rs2254546 polymorphism might significantly contribute to the risk of KD.
Kawasaki disease (KD; OMIM, 611775), also called Mucocutaneous Lymph Node Syndrome, is an acute, self-limited vasculitis that predominately affects infants and young children1,2,3. Although it was first described in Japan in 1967 by the Japanese pediatrician Tomisaku Kawasaki4, the etiology of KD is still not completely clear. The major clinical manifestations of KD include prolonged fever, bilateral non-purulent conjunctivitis, diffuse mucosal inflammation, polymorphous skin rashes, peripheral extremity changes, and cervical lymphadenopathy5. About 15–25% of untreated cases suffer coronary artery lesions, which is the most serious complication of KD, and makes KD the leading cause of acquired heart disease in children of developed countries6.
Genetic determinants have been suggested to contribute to KD susceptibility. Firstly, the risk of KD in siblings of affected children is ten times higher as compared with the general population7. Secondly, it is twice as high in children born to parents with a history of KD compared with the general population8. Thirdly, populations in Asian countries have higher incidence rates of KD than those in Western countries: Japan has the highest annual incidence rate9, followed by Korea10 and Taiwan11, regardless of their residences12. Accumulating clinical and epidemiological findings suggest that infectious agents trigger inflammatory responses, stirring up host immune dysregulation in part of genetically predisposed individuals13,14.
Genome-wide association studies (GWASs) have revolutionized the identification of genomic regions associated with complex diseases15, and so do with KD. Until now, a total of six published GWASs have identified multiple novel single nucleotide polymorphisms (SNPs) associated with KD or coronary artery lesions in Japanese, Europeans, Koreans, Taiwanese and so on16,17,18,19,20,21. Among these SNPs, rs2254546, located at an intergenic region between family with sequence similarity 167, member A (FAM167A, also known as C8orf13) and B lymphoid tyrosine kinase (BLK) at 8p22–23, was firstly discovered to be associated with KD by Onouchi et al.20, who firstly performed a GWAS in a large sample size and then validated this locus in two independent case-control panels of Japanese and transmission disequilibrium tests (TDTs) of other ancestries. Notably, the TDT based on family is considerably advantageous as less confounding caused by population admixture and is more important than case-control study in genetic association analysis22.
The results of KD research on Chinese are very important because of the high prevalence, but as far as we know, there is only one study which has replicated the association between rs2254546 and KD risk on a population from Sichuan province of Southwest China23. Therefore, we proposed to verify this association on another Chinese population from Zhejiang province of Southeast China. Subsequently, a meta-analysis, combining the current and previously published studies, and applying the method described by Kazeem and Farrall24, was performed to integrate the results from case-control studies and TDTs, and to provide more convincing evidence for this association.
Results
Results of our current case-control study
Population characteristics
A total of 428 cases and 478 controls participated in our current study. There were 263 (61.45%) males in cases and 294 (61.51%) males in controls, and no statistically significant difference was observed between these two groups in the distribution of gender (Pearson χ2 = 0.000, P = 0.986).
Association analysis
Among all the participants, 892 samples were genotyped successfully, so that the genotyping call rate of rs2254546 was 98.45%. The distribution of genotypes in controls was in agreement with Hardy-Weinberg equilibrium (HWE, P = 0.062) while no significant difference was observed in genotype frequencies between cases and controls (χ2 = 4.08, P = 0.130). As shown in Table 1, under the multivariate logistic regression model adjusted for gender, individuals with GG genotype showed a significant association with the increased risk of KD, compared with those carrying AA genotype (odds ratio (OR) = 1.86, 95% confidence interval (95%CI) = 1.01–3.41, P = 0.045). The same result was found under the dominant model (OR = 1.83, 95% CI = 1.01–3.33, P = 0.048). However, we failed to find the significant association in allelic model (OR = 1.20, 95% CI = 0.96–1.50, P = 0.117).
Table 1. Association between rs2254546 and KD risk in the current study.
| Genotype | Cases N (%) | Controls N (%) | OR | 95%CI | P† |
|---|---|---|---|---|---|
| AA | 17 (4.07) | 34 (7.17) | Ref. | Ref. | |
| AG | 138 (33.01) | 156 (32.91) | 1.77 | 0.95–3.31 | 0.073 |
| GG | 263 (62.92) | 284 (59.92) | 1.86 | 1.01–3.41 | 0.045 |
| G/A allele | 1.20 | 0.96–1.50 | 0.117 | ||
| Dominant model | 1.83 | 1.01–3.33 | 0.048 | ||
| Recessive model | 1.14 | 0.87–1.49 | 0.347 | ||
| Additive model | 1.19 | 0.95–1.48 | 0.123 |
†The P values were adjusted for gender.
The positive results were in bold.
Results of meta-analysis
Study characteristics
After comprehensive searching, two published articles met the inclusion criteria, one GWAS and one replication study respectively20,23. Thus, a total of eight previous reports plus our current study, comprising five case-control studies of 1958 cases, 5615 controls and four TDTs of 503 trios were finally included in this meta-analysis. Of the nine studies, six studies were conducted in Asians, one in Europeans, one in Hispanics and the other one in multi-ethnic population. Table 2 shows the characteristics of the included studies.
Table 2. The characteristics of included studies in meta-analysis.
| First author | Year | Country | Ethnicity | Study design | Study type | Genotyping method | Sample size |
|---|---|---|---|---|---|---|---|
| Onouchi | 2012 | Japan | Asian | CC | GWAS | Illumina Human Hap550v3 BeadChip | 428/3379 |
| Onouchi | 2012 | Japan | Asian | CC | Replication | Invader Assay | 470/378 |
| Onouchi | 2012 | Japan | Asian | CC | Replication | Invader Assay | 284/569 |
| Onouchi | 2012 | USA | European | TDT | Replication | Invader Assay | 248 |
| Onouchi | 2012 | USA | Asian | TDT | Replication | Invader Assay | 72 |
| Onouchi | 2012 | USA | Hispanic | TDT | Replication | Invader Assay | 94 |
| Onouchi | 2012 | USA | Mixed | TDT | Replication | Invader Assay | 89 |
| Yan | 2013 | China | Asian | CC | Replication | TaqMan SNP Genotyping Assays | 358/815 |
| Current study | 2013 | China | Asian | CC | Replication | TaqMan SNP Genotyping Assays | 418/474 |
Abbreviations: CC, case-control study; TDT, transmission/disequilibrium test.
Combined results of case-control studies and TDTs
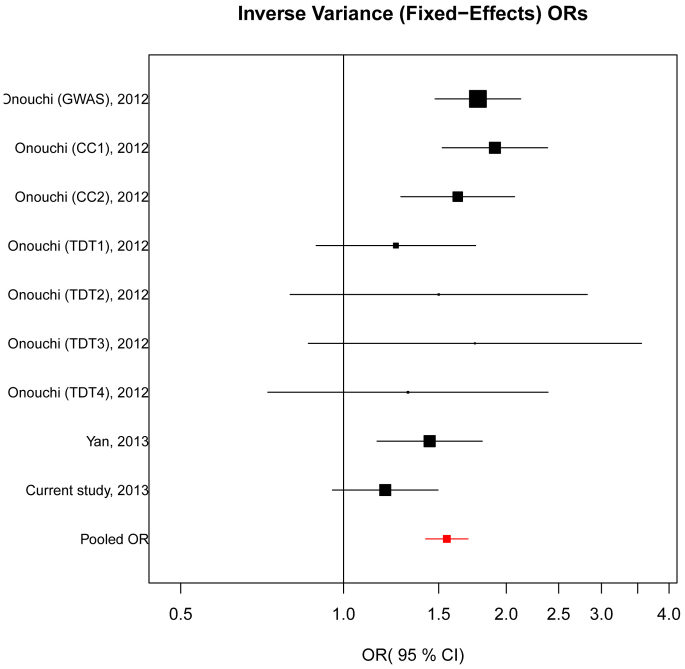
No significant evidence of heterogeneity was observed among the five case-control studies and four TDTs (Q = 13.00, Pheterogeneity = 0.112); thus, the fixed-effects model was employed to assess the pooled association between rs2254546 and KD susceptibility. In the overall meta-analysis, the G allele conferred an increased risk of KD compared with the A allele (OR = 1.55, 95% CI = 1.42–1.70, P < 0.001; Figure 1).
Figure 1. Forest plot of the association between rs2254546 G allele and KD risk.
For each study, the estimate of OR and its 95% CI is plotted with a box and a horizontal line. Fixed-effects pooled OR = 1.55, 95% CI = 1.42–1.70, P < 0.001; Q = 13.00, Pheterogeneity = 0.112.
Stratified analysis
In consideration of our strategy of meta-analysis by integrating case-control studies and TDTs, the data were firstly stratified by study design. As shown in Table 3, significant heterogeneity was detected in case-control subgroup but not in TDT subgroup (Q = 10.84, Pheterogeneity = 0.028; Q = 0.83, Pheterogeneity = 0.843). Meanwhile, significant associations were found between the polymorphism and increased KD risk in both case-control and TDT subgroups (OR = 1.57, 95% CI = 1.34–1.85, P < 0.001; OR = 1.35, 95% CI = 1.05–1.74, P = 0.018), under random-effects model and fixed-effects model respectively.
Table 3. Meta-analysis of rs2254546 in association with KD risk stratified by study design.
| Case-control | TDT | OR (95%CI) | ||||
|---|---|---|---|---|---|---|
| Study ID | Cases G/A | Controls G/A | Transmitted G | Non-transmitted G | Case-control | TDT |
| Onouchi (GWAS), 2012 | 702/154 | 4866/1892 | - | - | 1.77 (1.48–2.13) | - |
| Onouchi (CC1), 2012 | 761/179 | 522/234 | - | - | 1.91 (1.52–2.38) | - |
| Onouchi (CC2), 2012 | 454/114 | 808/330 | - | - | 1.63 (1.28–2.07) | - |
| Onouchi (TDT1), 2012 | - | - | 75 | 60 | - | 1.25 (0.89–1.76) |
| Onouchi (TDT2), 2012 | - | - | 24 | 16 | - | 1.50 (0.80–2.82) |
| Onouchi (TDT3), 2012 | - | - | 21 | 12 | - | 1.75 (0.86–3.56) |
| Onouchi (TDT4), 2012 | - | - | 25 | 19 | - | 1.32 (0.72–2.39) |
| Yan, 2013 | 590/126 | 1246/384 | - | - | 1.44 (1.15–1.81) | - |
| Current study, 2013 | 664/172 | 724/224 | - | - | 1.19 (0.95–1.50) | - |
| Pooled | 3171/745 | 8166/3064 | 145 | 107 | 1.57 (1.34–1.85)† | 1.35 (1.05–1.74)‡ |
†Random-effects pooled OR, P < 0.001; Q = 10.84, Pheterogeneity = 0.028.
‡Fixed-effects pooled OR, P = 0.018; Q = 0.83, Pheterogeneity = 0.843.
We further stratified the data by ethnicity. Because there was less than three studies conducted in Europeans or Hispanics, we performed a meta-analysis in Asian population only. Similarly, we found a significant association between the G allele of rs2254546 and increased KD risk under random-effects model due to the heterogeneity (OR = 1.58, 95% CI = 1.44–1.74, P < 0.001, Q = 10.87, Pheterogeneity = 0.054; Table 4).
Table 4. Meta-analysis of rs2254546 in association with KD risk in Asian population.
| Case-control | TDT | ||||
|---|---|---|---|---|---|
| Study ID | Cases G/A | Controls G/A | Transmitted G | Non-transmitted G | OR (95%CI) |
| Onouchi (GWAS), 2012 | 702/154 | 4866/1892 | - | - | 1.77 (1.48–2.13) |
| Onouchi (CC1), 2012 | 761/179 | 522/234 | - | - | 1.91 (1.52–2.38) |
| Onouchi (CC2), 2012 | 454/114 | 808/330 | - | - | 1.63 (1.28–2.07) |
| Onouchi (TDT2), 2012 | - | - | 24 | 16 | 1.50 (0.80–2.82) |
| Yan, 2013 | 590/126 | 1246/384 | - | - | 1.44 (1.15–1.81) |
| Current study, 2013 | 664/172 | 724/224 | - | - | 1.19 (0.95–1.50) |
| Pooled | 3171/745 | 8166/3064 | 24 | 16 | 1.58 (1.44–1.74)† |
†Random-effects pooled OR, P < 0.001; Q = 10.87, Pheterogeneity = 0.054.
Sensitivity analysis
To evaluate the influence of each study on the overall estimate, sensitivity analysis was performed. A series of pooled ORs with 95% CIs produced similarly before and after eliminating each study at a time, suggesting that our results were robust (Table 5).
Table 5. Sensitivity analysis of allelic model.
| Study omitted | Q | Pheterogeneity | OR (95%CI) | P |
|---|---|---|---|---|
| Onouchi (GWAS), 2012 | 10.32 | 0.171 | 1.49 (1.34–1.65) | <0.001 |
| Onouchi (CC1), 2012 | 9.17 | 0.241 | 1.49 (1.35–1.65) | <0.001 |
| Onouchi (CC2), 2012 | 12.83 | 0.076 | 1.51 (1.31–1.75) | <0.001 |
| Onouchi (TDT1), 2012 | 11.31 | 0.126 | 1.58 (1.44–1.73) | <0.001 |
| Onouchi (TDT2), 2012 | 12.98 | 0.072 | 1.53 (1.34–1.75) | <0.001 |
| Onouchi (TDT3), 2012 | 12.89 | 0.075 | 1.52 (1.34–1.74) | <0.001 |
| Onouchi (TDT4), 2012 | 12.69 | 0.080 | 1.54 (1.35–1.76) | <0.001 |
| Yan, 2013 | 12.51 | 0.085 | 1.54 (1.33–1.79) | <0.001 |
| Current study, 2013 | 6.76 | 0.454 | 1.63 (1.48–1.80) | <0.001 |
Cumulative meta-analysis
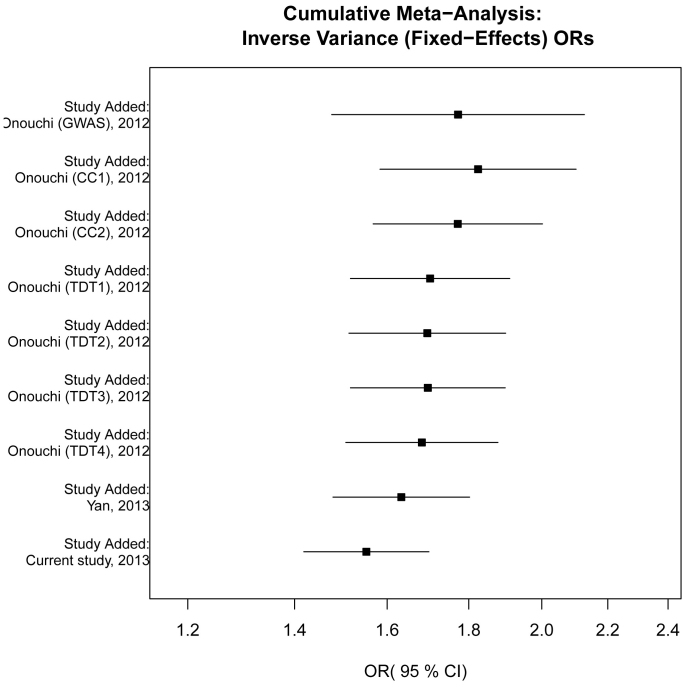
Cumulative meta-analysis was conducted via adding one study at a time in chronological order. Although the cumulative effects tended to be null significant in time sequence, they still kept exceeding the threshold 1.0. Meanwhile, the 95% CIs became increasingly narrower with the accumulation of more data, indicating a progressive improvement in the precision of the estimates (Figure 2).
Figure 2. Forest plot of cumulative meta-analysis between rs2254546 G allele and KD risk in chronological order.
The estimate of ORs and their 95% CIs are plotted with boxes and horizontal lines.
Publication bias
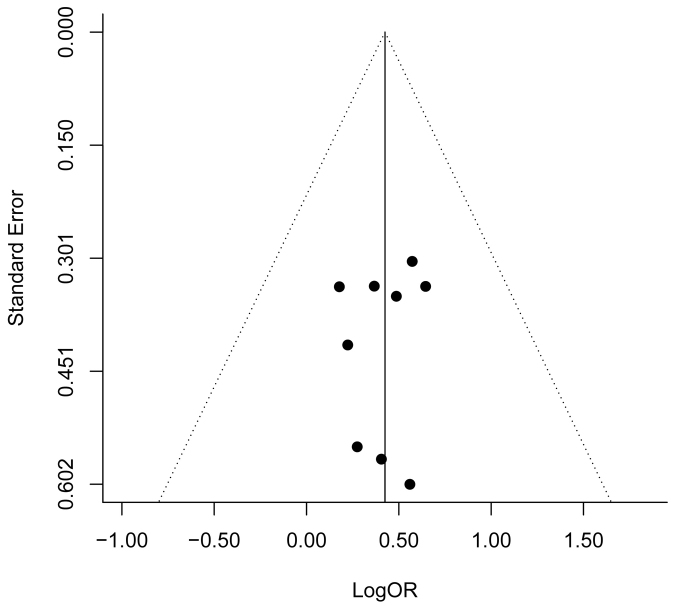
Publication bias was firstly assessed by visual inspection of the funnel plot. The dots were nearly symmetrically distributed, suggesting little evidence of publication bias (Figure 3). The result of regression test for funnel plot asymmetry further indicated no publication bias in this meta-analysis (P = 0.815).
Figure 3. Funnel plot of natural logarithm of OR against inverse standard error in each study.
Regression test for funnel plot asymmetry P = 0.815.
Discussion
In our current case-control study, individuals carrying GG genotype or GG/GA genotypes of rs2254546 showed significant associations with KD susceptibility, compared with AA genotype. Moreover, the following meta-analysis, integrating five case-control studies and four TDTs, provided better evidence of the significant relationship between the polymorphism and increased KD risk, which was further confirmed by cumulative analysis in chronological order. Since the discrepancy in genetic backgrounds has been implied to be one possible reason for the distinction of diseases or traits in different ethnicities25, and the prevalence of KD is overwhelmingly higher in Asians, it was deemed meaningful to assess the association of rs2254546 with KD risk in Asian ethnicity alone. Besides, sensitivity analysis indicated the stability of the results and little publication bias existed.
These findings are biologically plausible on some level. rs2254546 is located at an intergenic region between FAM167A and BLK at 8p22–23, a region harboring SNPs associated with several autoimmune diseases, such as systemic lupus erythematosus26,27, rheumatoid arthritis28 and systemic sclerosis29,30. FAM167A encodes a ubiquitously expressed gene but has not been functionally characterized30. BLK encodes B-lymphoid tyrosine kinase, a Scr family tyrosine kinase expressed primarily in the B-cell lineage, and transduces signal downstream following stimulation of B-cell receptors26,31. Little is known about the function of rs2254546 so far, and all that we know are the statistical associations of this polymorphism with KD20,23 or primary Sjogren's syndrome32. However, Onouchi et al.20 have demonstrated that two other SNPs, rs13277113 and rs2736340, which have been repeatedly associated with other autoimmune diseases, were in high linkage disequilibrium with rs2254546 in Japanese (D′ = 0.99, r2 = 0.87; D′ = 0.99, r2 = 0.91). It has been indicated that the risk A allele of rs13277113 was associated with reduced expression of BLK mRNA in transformed B-cell lines26. Meanwhile, Chang et al. have also provided evidence that the risk T allele of rs2736340 was associated with lower expression of BLK in peripheral blood B cells during the acute stage of KD, and decreased BLK expression in peripheral blood B cells may alter B cell function and predispose individuals to KD33. All of the above indicate the reasonability of our results. What's more, it is well known that most of the variants identified by GWASs cannot be per se causal but imply the probability of being in linkage with the “real” causal variants34, so we can speculate that rs2254546, like the two relevant SNPs, rs2736340 and rs13277113, is possibly the real causal variant, but more likely to be just in linkage equilibrium with the real causal variant.
The current study had its own special advantages. Firstly, we adopted a comprehensive analysis strategy (a case-control study and followed by a meta-analysis) to discern the association between rs2254546 and KD susceptibility; Secondly, in this meta-analysis, significant associations were detected in both case-control and TDT subgroups, which confirmed the statistical consistency of the two study designs. The study power was strongly enhanced by integrating these two study designs together due to the enlargement of sample size. Thirdly, the between-study heterogeneity in the overall meta-analysis was so negligible that these studies were considered compatible. Finally, the results of cumulative analysis, sensitivity analysis and publication bias further provided statistical evidence of the high quality of the meta-analysis.
Despite of the clear advantages we have mentioned above, several limitations should be figured out at the same time. Firstly, our own case-control study failed to replicate the significant association in allelic model, and the relatively small sample size and low statistical power might be taken into account. Secondly, the association between rs2254546 and KD risk is merely on statistical level, so the lack of functional experiments makes its role in KD pathogenesis still an enigma. Thirdly, KD is a complex disease caused by both genetic and environmental factors. However, the insufficient information about the environment limited the further evaluation of gene-environment interaction.
In summary, our case-control study combined with the following meta-analysis might provide convincing evidence for the significant association between rs2254546 polymorphism and KD susceptibility. Fine-mapping and sequencing studies, as well as functional studies are further warranted to identify and validate the potential causal variants.
Methods
Study participants
This study contained 428 children diagnosed with KD, all of which were unrelated ethnic Han Chinese and were consecutively recruited between April 2009 and September 2012 from Children's Hospital, Zhejiang University School of Medicine, China. The diagnosis of KD was based on the 5th revised edition of the guidelines established by the Kawasaki Disease Research Committee in Japan in 2002 (http://kawasaki-disease.org/diagnostic/). The controls consisted of 478 gender-matched unrelated healthy Han Chinese children without any evidence of infection, part of which have ever been included in our previous study35. All the controls were recruited from children who took the entrance physical examination for primary school in Children's Hospital, Zhejiang University School of Medicine.
This study was approved by the ethics committee of the Children's Hospital, Zhejiang University School of Medicine, and the methods were carried out in accordance with the approved guidelines. Informed consent was obtained from all participants or parents/caregivers of all the subjects who were studied.
Genotyping
Genomic DNA was extracted from 2 ml peripheral blood sample that was collected from each participant, using the RelaxGene Blood System DP319-02 (Tiangen, Beijing, China) according to the manufacturer's protocol. The concentration and the optical density of DNA was confirmed by NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Genotypes were determined by TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA) as described below, which was performed without knowledge of participants' disease status. The final reaction volume per well was 5 μL for a 384-well plate, including 20× working stock of SNP Genotyping Assay (0.25 μL), TaqMan Universal PCR Master Mix (2×), No AmpErase UNG (2.5 μL), DNase-free water (1.25 μL), and Template DNA (1 μL). The first two of which were customized by Applied Biosystems. The reaction was carried out with an initial step of 10 minutes at 95°C, which was the optimal temperature for AmpliTaq Gold Enzyme Activation; then followed by 50 cycles of 15 seconds at 92°C and 90 seconds at 60°C. After PCR amplification, allelic discrimination plate read and analysis were performed using Applied Biosystems 7900HT Fast Real-Time PCR System, followed the manufacturer's protocol.
For quality control, a 5% random samples were tested twice to assess the reproducibility, with a concordance rate of 100%. A total of 892 samples were genotyped successfully, so that the genotyping call rate of rs2254546 was 98.45%.
Statistical analysis
HWE was assessed by the goodness-of –fit χ2 test for genotypes in the control group. Pearson's χ2 test was applied to estimate the differences in gender and distribution of genotypes between cases and controls. After adjusting for gender, unconditional multivariate logistic regression was employed to estimate ORs and their 95% CIs for the effect of the SNP on KD susceptibility. To avoid the assumption of genetic models, dominant, recessive and additive model were all analyzed respectively. All above statistical analyses were performed in SPSS V13.0 and all P values were two-tailed with a significant level of 0.05.
Meta-analysis of rs2254546 in association with KD risk
To further confirm the association between rs2254546 and KD susceptibility, a meta-analysis combining published literatures and our current study was conducted. We searched various databases including PubMed, EMBASE, ISI Web of Science and Google Scholar up to October 2013 without language restriction, using the keywords ‘rs2254546 or FAM167A-BLK' combined with ‘Kawasaki disease'. We further checked five large Chinese biomedical bibliographic databases to expand the coverage of our searches36. References listed in the retrieved articles and reviews were also scanned for missing information. The included articles had to meet the following criteria: (1) either case-control study or TDT design; (2) availability of allelic frequency necessary for calculating ORs and their 95% CIs; (3) clear definition of KD, either based on the diagnostic criteria from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association5, or the diagnostic guidelines for Kawasaki disease (the 5th revised edition, http://kawasaki-disease.org/diagnostic/), published by the Kawasaki Disease Research Committee of Japan; (4) studies on humans. When multiple articles had overlapped study participants, the one with more complete design or larger sample size was finally selected.
For each of the eligible studies, the following information was extracted independently by two reviewers (W.W. and J.L.): first author's name, year of publication, country, ethnicity of study participants, study design, study type, genotyping method, sample size, frequencies of alleles in case and control group in case-control studies and numbers of transmitted and non-transmitted alleles from heterozygous parents to affected offspring in family-based studies. The Cochran's χ2-based Q statistic test was applied to assess between-study heterogeneity. As regard of the p-value cut-off, both the use of a significance level of 0.1 or 0.05 are commonly accepted in this type of test37,38. In this study, we chose 0.1 as the significance p-value cut-off for the heterogeneity. If P < 0.1, a random-effects model, using DerSimonian and Laird method39, was employed to compute the pooled ORs; otherwise, a fixed-effects model, using Mantel-Haenszel method40, was adopted. For the synthesis of case-control studies and TDTs, a method described by Kazeem and Farrall24 was used to obtain a combined estimate of the magnitude of the association. The “Catmap” software41 for this method was downloaded from the comprehensive R archive network http://www.r-project.org. On condition that the heterogeneity existed, a meta-regression analysis would be conducted to explore the possible sources of the heterogeneity across studies42. Stratified analysis was carried out subsequently, on the basis of the suspected sources of heterogeneity reported by the meta-regression analysis. Afterwards, sensitivity analysis was performed to assess the influence of each study on the overall estimate. Cumulative meta-analysis was also conducted by adding studies one at a time in chronological order. Funnel plot and regression test were utilized to assess publication bias. All statistical analyses and figures were performed in R software V2.13.2 and P values less than 0.05 were considered statistically significant for all tests except for Q test for heterogeneity.
Author Contributions
Conceived and designed the study strategy: X.M. and F.G. Designed the experiment: W.W. and J.L. Recruited the participants and collected their information and blood samples: J.L. and X.L. Performed the experiments: J.L., X.L. and N.S. Analysis the data: W.W., J.L. and Y.Q. Collected and checked the information of eligible studies included in meta-analysis: W.W. and J.L. Statistical analyses: W.W., N.S., Y.W. and L.Z. Reference collection and data management: J.D., J.K. and Q.Z. Wrote the manuscript: W.W. and J.L. Prepared the tables and figures: W.W. and R.Z. All authors reviewed the manuscript.
Acknowledgments
This work is supported by grants from The National Natural Science Foundation of China (81270177), Ministry of Health Research Foundation of China (201339378), The Health Bureau of Zhejiang Province (2009A124, 2009CA072) and Population and family planning commission of Zhejiang province (JSW2013-A15).
References
- Burns J. C. & Glodé M. P. Kawasaki syndrome. The Lancet 364, 533–544 (2004). [DOI] [PubMed] [Google Scholar]
- McCrindle B. W. Kawasaki disease: A childhood disease with important consequences into adulthood. Circulation 120, 6–8 (2009). [DOI] [PubMed] [Google Scholar]
- Rowley A. H. Kawasaki disease: Novel insights into etiology and genetic susceptibility. Annu. Rev. Med. 62, 69–77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T. Acute febrile mucocutaneous syndrome with lymphoid involvement with specific desquamation of the fingers and toes in children. Arerugi 16, 178–222 (1967). [PubMed] [Google Scholar]
- Newburger J. W. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: A statement for health professionals from the committee on rheumatic fever, endocarditis, and Kawasaki disease, council on cardiovascular disease in the young, American Heart Association. Pediatrics 114, 1708–1733 (2004). [DOI] [PubMed] [Google Scholar]
- Kato H. et al. Long-term consequences of Kawasaki disease. A 10- to 21-year follow-up study of 594 patients. Circulation 94, 1379–1385 (1996). [DOI] [PubMed] [Google Scholar]
- Fujita Y. et al. Kawasaki disease in families. Pediatrics 84, 666–669 (1989). [PubMed] [Google Scholar]
- Uehara R., Yashiro M., Nakamura Y. & Yanagawa H. Kawasaki disease in parents and children. Acta Paediatr. 92, 694–697 (2003). [DOI] [PubMed] [Google Scholar]
- Nakamura Y. et al. Epidemiologic features of Kawasaki disease in Japan: Results of the 2009–2010 nationwide survey. J. Epidemiol. 22, 216–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. W. et al. Epidemiological features of Kawasaki disease in Korea, 2006–2008. Pediatr. Int. 53, 36–39 (2011). [DOI] [PubMed] [Google Scholar]
- Lue H. C. et al. Epidemiological features of Kawasaki disease in Taiwan, 1976–2007: Results of five nationwide questionnaire hospital surveys. Pediatr. Neonatol. Published Online First: 10 Oct 2013. http://dx.doi.org/10.1016/j.pedneo.2013.07.010 (2013). [DOI] [PubMed] [Google Scholar]
- Holman R. C. et al. Kawasaki syndrome in Hawaii. Pediatr. Infect. Dis. J. 24, 429–433 (2005). [DOI] [PubMed] [Google Scholar]
- Lin C. Y., Lin C. C., Hwang B. & Chiang B. Serial changes of serum interleukin-6, interleukin-8, and tumor necrosis factor alpha among patients with Kawasaki disease. J. Pediatr. 121, 924–926 (1992). [DOI] [PubMed] [Google Scholar]
- Matsubara T., Furukawa S. & Yabuta K. Serum levels of tumor necrosis factor, interleukin 2 receptor, and interferon-gamma in Kawasaki disease involved coronary-artery lesions. Clin. Immunol. Immunopathol. 56, 29–36 (1990). [DOI] [PubMed] [Google Scholar]
- Manolio T. A. Bringing genome-wide association findings into clinical use. Nat. Rev. Genet. 14, 549–558 (2013). [DOI] [PubMed] [Google Scholar]
- Burgner D. et al. A genome-wide association study identifies novel and functionally related susceptibility loci for Kawasaki disease. PLoS Genet. 5, e1000319 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J. et al. A genome-wide association analysis reveals 1p31 and 2p13.3 as susceptibility loci for Kawasaki disease. Hum. Genet. 129, 487–495 (2011). [DOI] [PubMed] [Google Scholar]
- Tsai F. J. et al. Identification of novel susceptibility loci for Kawasaki disease in a Han Chinese population by a genome-wide association study. PLoS One 6, e16853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor C. C. et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat. Genet. 43, 1241–1246 (2011). [DOI] [PubMed] [Google Scholar]
- Onouchi Y. et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat. Genet. 44, 517–521 (2012). [DOI] [PubMed] [Google Scholar]
- Lee Y. C. et al. Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis. Nat. Genet. 44, 522–525 (2012). [DOI] [PubMed] [Google Scholar]
- Lander E. S. & Schork N. J. Genetic dissection of complex traits. Science 265, 2037–2048 (1994). [DOI] [PubMed] [Google Scholar]
- Yan Y. et al. Combined analysis of genome-wide-linked susceptibility loci to Kawasaki disease in Han Chinese. Hum. Genet. 132, 669–680 (2013). [DOI] [PubMed] [Google Scholar]
- Kazeem G. R. & Farrall M. Integrating case-control and TDT studies. Ann. Hum. Genet. 69, 329–335 (2005). [DOI] [PubMed] [Google Scholar]
- Yeung R. S. Kawasaki disease: Update on pathogenesis. Curr. Opin. Rheumatol. 22, 551–560 (2010). [DOI] [PubMed] [Google Scholar]
- Hom G. et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 358, 900–909 (2008). [DOI] [PubMed] [Google Scholar]
- Yang W. et al. Population differences in SLE susceptibility genes: STAT4 and BLK, but not PXK, are associated with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 10, 219–226 (2009). [DOI] [PubMed] [Google Scholar]
- Ito I. et al. Replication of association between FAM167A(C8orf13)-BLK region and rheumatoid arthritis in a Japanese population. Ann. Rheum. Dis. 69, 936–937 (2010). [DOI] [PubMed] [Google Scholar]
- Gourh P. et al. Association of the C8orf13-BLK region with systemic sclerosis in North-American and European populations. J. Autoimmun. 34, 155–162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito I. et al. Association of the FAM167A-BLK region with systemic sclerosis. Arthritis Rheum. 62, 890–895 (2010). [DOI] [PubMed] [Google Scholar]
- Dymecki S. M., Niederhuber J. E. & Desiderio S. V. Specific expression of a tyrosine kinase gene, BLK, in B lymphoid cells. Science 247, 332–336 (1990). [DOI] [PubMed] [Google Scholar]
- Sun F. et al. Association studies of TNFSF4, TNFAIP3 and FAM167A-BLK polymorphisms with primary Sjogren's syndrome in Han Chinese. J. Hum. Genet. 58, 475–479 (2013). [DOI] [PubMed] [Google Scholar]
- Chang C. J. et al. Replication and meta-analysis of GWAS identified susceptibility loci in Kawasaki disease confirm the importance of B lymphoid tyrosine kinase (BLK) in disease susceptibility. PLoS One 8, e72037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Brown M. A., McCarthy M. I. & Yang J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Evaluation of intravenous immunoglobulin resistance and coronary artery lesions in relation to th1/th2 cytokine profiles in patients with Kawasaki disease. Arthritis Rheum. 65, 805–814 (2013). [DOI] [PubMed] [Google Scholar]
- Xia J., Wright J. & Adams C. E. Five large Chinese biomedical bibliographic databases: Accessibility and coverage. Health Info. Libr. J. 25, 55–61 (2008). [DOI] [PubMed] [Google Scholar]
- Li H., Hao X., Zhang W., Wei Q. & Chen K. The hOGG1 Ser326Cys polymorphism and lung cancer risk: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 17, 1739–1745 (2008). [DOI] [PubMed] [Google Scholar]
- Lu C. et al. CCND1 G870A polymorphism contributes to breast cancer susceptibility: a meta-analysis. Breast Cancer Res. Treat. 116, 571–575 (2009). [DOI] [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control. Clin. Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748 (1959). [PubMed] [Google Scholar]
- Nicodemus K. K. Catmap: Case-control and TDT meta-analysis package. BMC Bioinformatics 9, 130 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S. G. & Sharp S. J. Explaining heterogeneity in meta-analysis: A comparison of methods. Stat. Med. 18, 2693–2708 (1999). [DOI] [PubMed] [Google Scholar]