Abstract
There has been a growing interest in understanding the ways in which bacteria interact with nano-structured surfaces. As a result, there is a need for innovative approaches to enable researchers to visualize the biological processes taking place, despite the fact that it is not possible to directly observe these processes. We present a novel approach for the three-dimensional visualization of bacterial interactions with nano-structured surfaces using the software package Autodesk Maya. Our approach comprises a semi-automated stage, where actual surface topographic parameters, obtained using an atomic force microscope, are imported into Maya via a custom Python script, followed by a ‘creative stage', where the bacterial cells and their interactions with the surfaces are visualized using available experimental data. The ‘Dynamics' and ‘nDynamics' capabilities of the Maya software allowed the construction and visualization of plausible interaction scenarios. This capability provides a practical aid to knowledge discovery, assists in the dissemination of research results, and provides an opportunity for an improved public understanding. We validated our approach by graphically depicting the interactions between the two bacteria being used for modeling purposes, Staphylococcus aureus and Pseudomonas aeruginosa, with different titanium substrate surfaces that are routinely used in the production of biomedical devices.
Titanium and titanium alloys1 are biomaterials that are widely used in biomedical applications2,3,4,5,6. They represent substrata upon which bacteria can attach, and successfully colonize, forming biofilms4,5,6. Bacterial colonization of biomaterial surfaces and their organization in complex microbial aggregates have received considerable attention over the past few decades6,7,8,9,10,11,12. As a result, intensive research has been focused on modifying the surfaces of these materials at the nano-scale10,11,12 and/or creating a nano-structured surface architecture, often by mimicking naturally occurring surfaces, in order to restrict and eliminate bacterial attachment13,14. Despite numerous efforts to create bacterial-resistant surfaces, there is still a lack of understanding of the initial processes taking place when bacterial cells first interact with nano-structured surfaces. It is difficult to obtain this understanding, mainly because of challenges associated with the small dimensions of the objects under investigation, and the difficulty in capturing images during continuous real-time processes.
Three-dimensional (3D) visualizations and computer-generated animations may, in part, fill this gap in understanding by providing researchers with the ability to effectively display surface topographical data, together with possible animated scenarios, to describe the bacterial cell—surface interactions that are taking place. 3D visualization has had a great impact in many fields, including nanotechnology and biomedical research, where objects under investigation can be visualized on a micron- and nano-metric scale15,16,17,18. The aim of this work was to develop and evaluate a practical approach for the 3D visualization of nano- and microscopic objects and their interactions with surfaces using the 3D animation software package Autodesk Maya (http://usa.autodesk.com/maya/). Visualization of the different topographic surface architectures of several metallic surfaces was developed using data files that were generated using Atomic Force Microscopy (AFM), and the bacterial cell-surface interaction scenarios developed using Maya's Dynamics capabilities. The resulting 3D visualizations allow an increased insight into the bacterial attachment processes that are taking place. The resulting animations are informative, and greatly enhance the ability to visualize the interaction, which assists in the dissemination of research to both scientific and public audiences, and perhaps provide additional motivation for future developments in the area of direct imaging of bacterial attachment onto nano-structured surfaces.
Results
Dynamic, three-dimensional visualizations of the interactions between two bacterial species (S. aureus and P. aeruginosa) and three different types of titanium surfaces that represent characteristic examples of micron-, nano- and sub-nano-metric-scale surface topographies25,26 have been developed. The 3D visualizations were constructed using Maya and based on an approach that consists of two stages, an initial semi-automated stage, followed by a subsequent ‘creative' stage. (i) During the first stage, the AFM data (in comma-separated, CSV, format) were imported into Maya using a custom developed Python script. A color map was applied to the surface, based on the minimum and maximum data values. Scaling of the height of the surface features was maintained in order to emphasize the topographic features. (ii) In the second, creative stage, Maya's geometric modeling and texturing capabilities were employed to create realistic bacterial images and animations. Dynamic properties were associated with the bacterial models, allowing them to interact with simple force fields. Importantly, using experimental data pertaining to the substrates ensured the scientific accuracy of the visualizations was retained. This was not possible, however, for the modeled bacteria. In the future, as improved analytic or numerical models of surface interactions become available, additional Maya Embedded Language (MEL) or Python scripts may be utilized to further increase the scientific accuracy of the resulting animations. The key features of the two steps of the visualization process are as follows.
Surface visualization
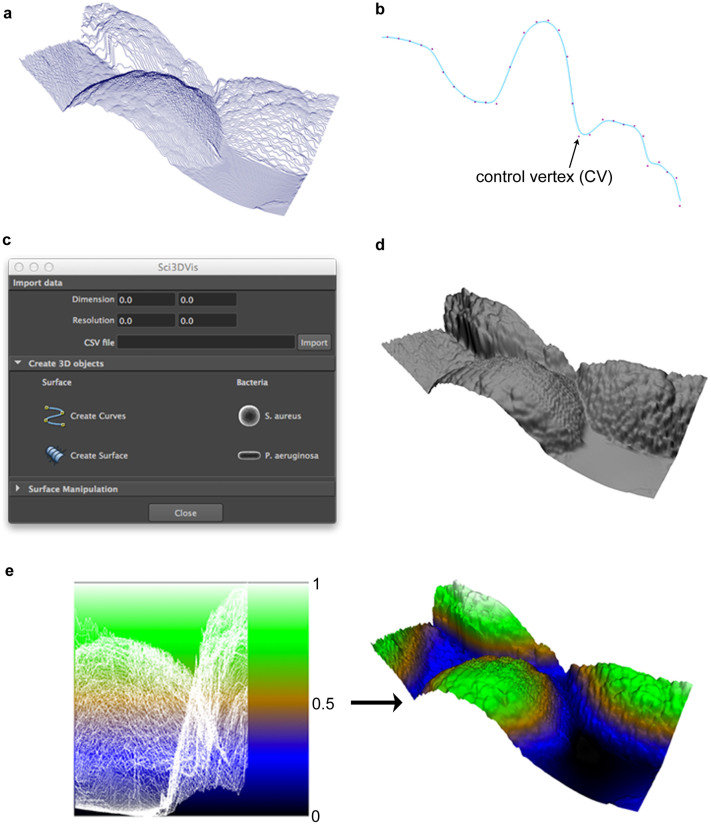
During the file import process, the Python script reads each row of the CSV data file, parsing it into a set of height values that are equally spaced along the row. A Maya curve object is then created, based on the height values for each line (Fig. 1a). Every curve consists of control vertices (CVs) to which values appropriate to each dimension (X, Y and Z axis) are assigned (Fig. 1b), along with an interpolation function; for simplicity, a B-spline is used. The values in each row file are mapped, in Maya's world coordinates, to the Y-axis values of the curve's CVs. Before the curves are created, the resolution and dimensions of the surface can be clarified and used to increment the corresponding X and Z values for each Y value. Finally, selecting all of the curves and choosing the “Create Surface” option from the script interface (Fig. 1c) allows the 3D surface to be constructed (Fig. 1d). The “Create Surface” command activates Maya's loft tool. The script also includes an option for creating the basic shapes of the bacteria, e.g., spherical S. aureus cells and rod-shaped P. aeruginosa cells, which can be scaled proportionally according the surface dimensions.
Figure 1. Visualization of titanium surfaces in the Maya software package.
The AFM data files, in CSV format, were imported and a set of equally spaced curves created based on the surface height values from the file (a). Each curve consists of control vertices (CVs) to which values can be applied for each dimension (X, Y and Z) (b). By selecting all the curves and the “Create Surface” command from the script interface (c), a polygonal surface can be created (d). A planar UV map is then assigned to the surface geometry, in order to apply the ramp texture that is used to obtain the color map values of the surface (e). Autodesk screen shots reprinted with the permission of Autodesk, Inc.
In order to apply the color maps, the UV mapping option was used. UV coordinates are a two-dimensional texture coordinate system for the 3D model, which allows for accurate placement of 2D texture on a 3D model. A planar mapping was assigned to the surfaces along the X-axis. Planar mapping is not always an adequate way to map more general 3D models, as the UV coordinates may overlap, however, since it satisfied the requirements for the height-based texturing of the surfaces it was considered suitable in this instance. For the surfaces, a ramp texture that contained layers of colors (Fig. 1e) was applied to a Lambert material. Maya provides a great deal of flexibility in defining color maps. In this study, a terrain map was used to emphasize features that are either above (green/white) or below (blue/black) the average surface height (brown), however alternative methods can be employed to achieve a similar result.
Development of the 3D model for bacterial cells
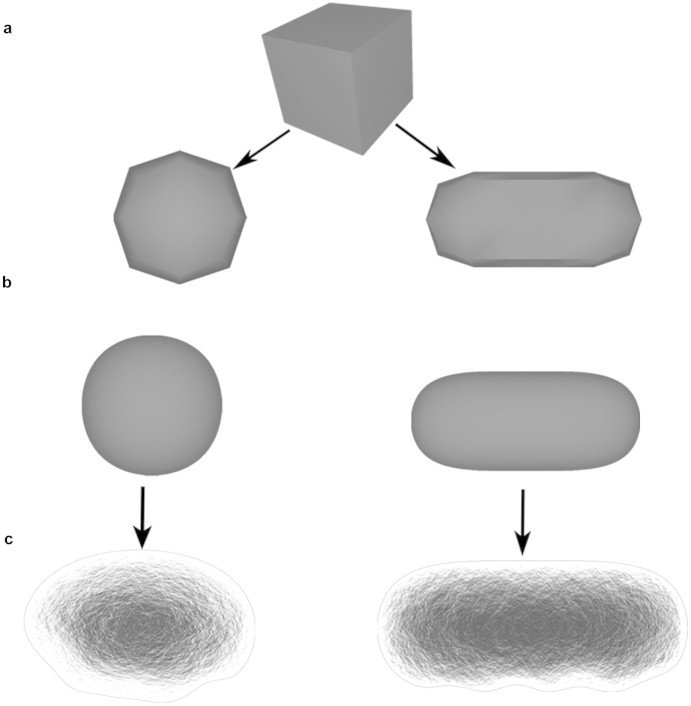
The models of the bacterial cells, S. aureus (spherical) and P. aeruginosa (cylindrical shape with round caps) were created as polygonal models using Maya's standard modeling tools (Fig. 2a and 2b). A 2D bump texture was assigned to allow a more detailed representation of the bacterial shape, whilst maintaining the underlying 3D model of the cells at lower resolution (Fig. 2c). This allows the work to be simplified and avoids the generation of complex geometrical structures until they are rendered in the final stages of the visualization process.
Figure 2. 3D models of bacterial cells.
The round shaped bacteria S. aureus and rod shaped P. aeruginosa cells were created using Maya's modeling tools. A cube is used as a base mesh for the development of the models (a). The ‘smooth' and ‘extrude' tool were used for the further modeling of the bacterial cell shapes (b). A 2D bump texture was then assigned in order to create bacteria with a more realistic appearance (c) and at the same time to preserve a lower quality mesh. Autodesk screen shots reprinted with the permission of Autodesk, Inc.
Dynamic interactions in the visualization process
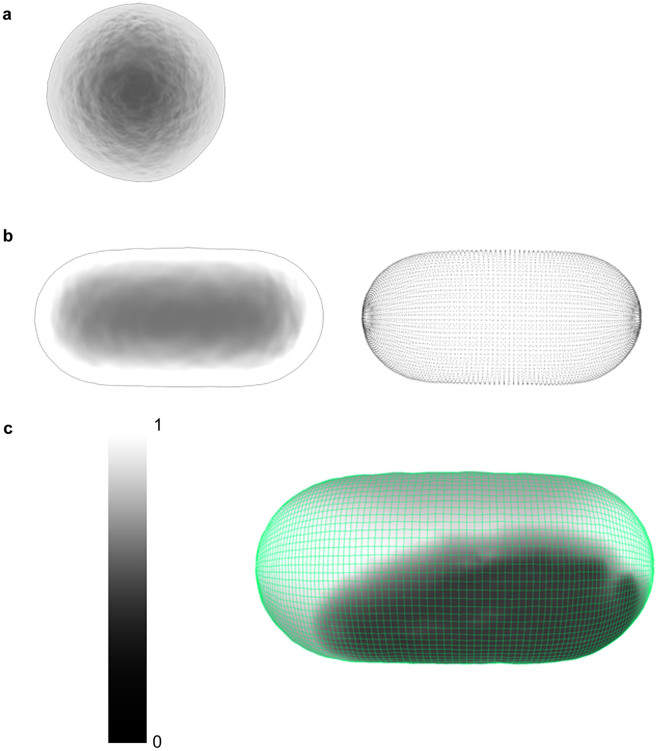
Maya's Dynamics function supports the construction of active or passive, rigid or soft bodies, and particle and fluid effects, while simulating their interactions within different force fields such as gravity. Passive bodies in Maya's environment are bodies that can interact with other objects, but these bodies are not influenced by the interaction. For example, when a sphere (which is, in this instance, an active body) is bouncing on a plane, the plane will be passive and it will not move when the sphere contacts the surface of the plane. When a soft body is assigned to a 3D geometry, Maya creates a corresponding particle object. The combination of the particle and the geometry of the object define the soft body. When a force field affects the particle system, the vertices of the 3D object will move in response to the changes in the particle position. This affords a level of flexibility to the 3D geometry that would be very difficult to model on a frame-by-frame basis. The amount of flexibility is controlled by adjusting the ‘weighting' assigned to the particle. This can range from 0 (flexible) to 1 (rigid) (Fig. 3c). Maya's ‘nDynamics' feature is an advanced dynamic simulation available in the menu function. It is based on the Maya Nucleus system. The Nucleus system consists of a series of nDynamic objects. These objects include particle systems (nParticles), objects created from a polygonal mesh from a connected network of particles that comprise the final dynamic mesh that represents the surface of the bacterial cells (nCloth), passive collision objects, dynamic constraints and the Nucleus solver. The Nucleus solver allows the simulation of collisions, which dictate the extent of some of the dynamic forces. Here, the nCloth function was used to create dynamic objects from 3D bacterial models. The attribute values of the nCloth function (e.g., ‘Collision', ‘Dynamic properties', ‘Pressure' and ‘Time') were adjusted until the desired bacterial body behavior was achieved.
Figure 3. Dynamic objects.
The S. aureus cells were created as rigid dynamic bodies (a), meaning that the cell does not deform when interacting with the substrate surface. The membrane flexibility and fluctuation of P. aeruginosa cells was achieved by assigning a soft dynamic body to the 3D polygonal mesh (b). When a soft body is created, a particle system (b, right) is assigned to every vertex of the polygonal mesh (b, left). The changes made to the particle positions by a force field, or another object, affects the corresponding vertex of the 3D model, resulting in a flexible object. This flexibility can be control by adjusting the ‘weighting' assigned to the particles, ranging from 0 (flexible) to 1 (rigid) (c). For the P. aeruginosa cells, the value of 0.75 was assigned in order to approximate the membrane fluctuations that would take place on the bacterial surface, however this number is arbitrarily assigned.
Practical applications of the proposed approach
The first two movies presented as Supplementary Video S1 and Supplementary Video S2 available online were based on the AFM surface topographical data obtained from two different titanium samples: as-received titanium and that of a 12 nm titanium thin film on a glass substrate (see the Methods section for details of the surface preparation). The titanium surfaces were created as Maya passive bodies, while the bacterial cell models were created as active bodies, allowing a more realistic representation of the bacterial motion using a gravity field. The bacterial models were positioned above the surfaces, and the interactions between the bacteria and the titanium surfaces were created using Maya's Dynamic functions. The dynamic representation of the cell membranes was created by assigning the Dynamic objects to the 3D polygonal model of the bacterial shape. S. aureus cells were created as Dynamic rigid bodies (Fig. 3a) while P. aeruginosa cells were created as Dynamic soft bodies (Fig. 3b), with the goal of recreating a realistic approximation of the membrane fluctuations experienced by the less rigid P. aeruginosa cell walls. Previous studies have shown that both species of bacteria interact differently with the two surface types25,26,27,28. The reasons for the difference in interaction have been reported elsewhere27. In order to highlight the variation in surface architecture, animations containing both the actual dimensions and an exaggerated scaling were developed. A scaling of 25 units along the axis perpendicular to the surface (the Y-axis in Maya world coordinates) was found to be suitable, but the selection of the scaling value to achieve this result was found to be somewhat arbitrary. In order to avoid non-proportional scaling of the bacterial shapes, the spherical and rod-shaped bacterial cells were animated as transparent objects. The ability to have full control over the attributes of individual elements within a scene is a feature common to many animation and modeling packages, however this functionality is not routinely included in domain-specific scientific visualization software. The annotated screenshots of the Videos S1 and S2 can be found in the Supplementary Figures S1 and S2, available online.
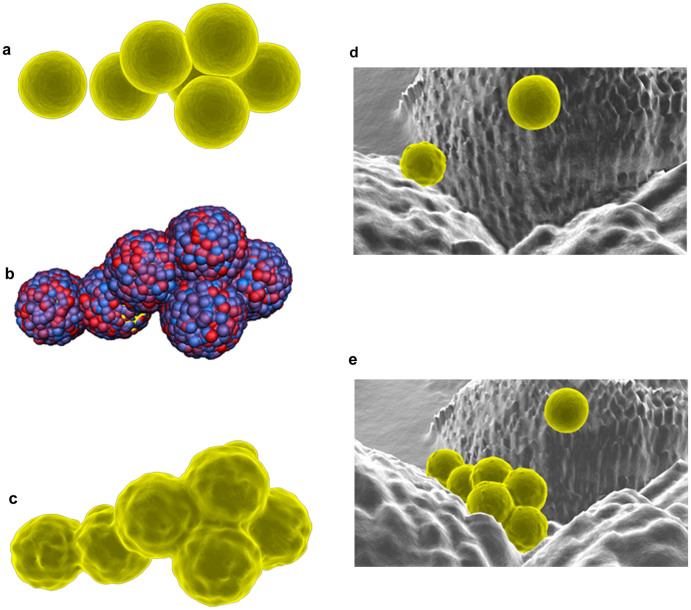
The third movie sequence (see Supplementary Video S3, available online) involved the interaction of S. aureus bacterial cells with a titanium surface that has been modified with laser treatment so that it mimics the surface architecture of a Lotus leaf. The titanium surfaces were created as passive collision objects. As is the case with passive bodies that are manipulated using the Dynamic menu, these objects can interact with other nDynamic objects without being affected by that interaction. The S. aureus cell-like 3D polygonal meshes were made as nCloth objects, allowing a better representation of the deformation of the shape of a spherical cell as it came into contact with the substrate surface. After adjusting the Nucleus's attributes until the required interaction processes were realized (e.g., by increasing the number of substeps in Maya's nDynamic simulations, causing a greater number of calculations to be made per frame), multiple nCaches were saved for each nDynamic object. The nCaching option allowed the nDynamic calculations to be stored and played back when needed without the nDynamic objects being active. The nCache file can also be used to scale the overall duration of the simulation within the Maya timeline, as well as selecting the time at which the simulation should start (using the Trax Editor). An additional simulation of this process was created using Maya's nParticle system, which was used to recreate the process of secretion of the extracellular polymetric substances (EPS) by the cells attaching to the surface (Fig. 4a–c). The EPS were recreated by assigning an nParticles system to each bacterial model. An emitter, a node that generates the particles, was set to emit the particles from the object mesh. After adjusting some of the attributes of the nParticle (e.g., the shape attributes such as Particle Size, Collisions, Output Mesh, Dynamic Properties), the final simulation was saved using the nCaching option. Finally, these particles were converted into a polygonal geometry for later rendering into the final animation sequence.
Figure 4. Bacteria on the Lotus-like titanium surface.
The bacteria polygonal modes were created as ‘nCloth' objects, resulting in more advanced, deformable objects rather just simple rigid objects (a). The exo-polymeric substances (EPS) were created using an ‘nParticle' system (b), which was converted to a polygonal mesh (c). The animation of the interaction between the S. aureus cells with the Lotus-like surface was developed with the ‘nDynamic' tools and objects; images presented here represent the first minute (d) and the 30th minute (e) of the bacterial interaction. Autodesk screen shots reprinted with the permission of Autodesk, Inc.
The final adjustment of the nCloth attributes resulted in representing the distinguishing dynamic motion of the cells, together with their moving membrane behavior. An example of the functionality that was available using the nCloth simulation functionality can be seen in the process where the bacteria cells slide over the surface, which is a function of the membrane flexibility and the extent to which the membrane sticks to the substrate surface, resulting in the final attachment and grouping of cells (Fig. 4d and 4e). This animation was based on the interpretation of bacterial cell interactions with Lotus leaf-like titanium surfaces. It reflects the typical number of bacterial cells that are able to attach onto the surfaces during the first thirty minutes of interaction, as has been reported elsewhere26. The annotated screenshots of Video S3 can be found in the Supplementary Figure S3, available online.
Discussion
The proposed approach comprises two main stages: the first is a semi-automated stage, where the data pertaining to the actual surface topography are loaded into Maya. This is followed by a ‘creative stage', where the interaction between the bacterial cells and surface is modeled using Maya's Dynamic systems. The key difference between these two stages is that with the former, the surfaces are visualized using a set of topographical parameters that are obtained directly using atomic force microscopy. The development of the bacterial cell models and their interactions with the surfaces (‘creative stage'), however, was part of a design process that took into account the size of the cells, their geometry and scaled proportionally to match the size of the surface models. The nDynamic interactions were then developed, and based upon a careful analysis of the scanning electron microscopy (SEM) and confocal scanning electron microscopy (CLSM) micrographs. Despite the fact that the micrographs were not directly processed using Maya, as was the case for the surface topographical data obtained from the AFM scans, the visual information contained in the SEM and optical images was utilized for the development of the dynamic interaction that was imagined to have taken place between the cells and the substrate surface. This included the time frame of the attachment and/or the number and pattern of the cells that attached to the surface over a given period of time.
An additional distinction between the stages is their degree of influence into the eventual visualization of the processes taking place when a cell comes in contact with the substrate surface. For example, in the initial semi-automated stage, most of the procedures require no human intervention. The creative stage, however, requires a degree of subjective interpretation in order to visualize the process of bacterial cell – surface interactions. As demonstrated in this study, utilizing 3D animation software packages such as Maya for the creation of scientific animations provide the opportunity to visualize the data and to propose data-informed research hypotheses. For example, the Avizo Standard software (http://www.vsg3d.com/avizo/standard) has been used to display the topographical surface features obtained from AFM analysis data21. Whilst this software can be used to describe the qualitative and quantitative characteristics of surfaces, Avizo does not currently provide the necessary functionality to allow the animation or simulation of bacterial interactions with surfaces.
In the context of 3D visualization in Maya, the simulation of a scientific process should not be mistaken with an actual scientific simulation. If actual dynamic data were available, it would be possible to accurately represent the actual forces that exist between the bacteria and the nano-structured surfaces. Instead, the Dynamics functionality of Maya is a useful alternative for the recreation of natural motions and collisions between objects in order to capture the essence of an interaction. Simulations in Maya originate from processes of interaction between objects that are affected by some forces, or each other, and therefore the calculations represent an interpretation of the changes taking place in the shape of an object. Simulations performed using Maya's Dynamics and nDynamics functionalities do not currently support the option to directly import an object's actual dynamic attributes. However, as has been shown in this study using the AFM data, it is possible to create scripts that could allow the importation of additional experimental, analytical or numerical data as new nodes into the software and to use these to create what is hoped to be more accurate and realistic dynamic interactions between the bacteria and the substrate surfaces. Developing and sharing scripts that can import scientific data directly into applications such as Maya could make these software packages more independent, easier to use, and more cost effective as an addition to existing domain-specific scientific visualization packages19,20,22,23,24. The visualization of this type of research data can be employed for different purposes, and can be used in the preparation of research publications and in the preparation of more graphic and interesting research presentations. The approach used in this study has the potential to open new avenues for experimental data analysis, including the possible further development and improvement of the tools currently available for the visualization of bacterial cell shapes and dynamic interactions within the Maya software.
Methods
Software
Autodesk Maya is one of the most commonly-used software packages [along with, for example, 3Ds Max (e.g., http://usa.autodesk.com/3ds-max/), Blender (http://www.blender.org/), Houdini (http://www.sidefx.com/), Lightwave (https://www.lightwave3d.com/), CINEMA4D (http://www.maxon.net/products/cinema-4d-prime/), etc.] for developing high-quality 3D animations with special effects. All of these packages were designed to take into account the needs of the entertainment industry, and are not designed to work using scientific data formats.
Maya's applicability for visualizing scientific research data has been previously recognized29,30,31. Maya was designed as a database for storing graphical information, which is deposited in objects called ‘nodes'. The nodes have properties (or attributes) that store information that pertains to their changeable characteristics, and data can flow between nodes. Maya's graphical user interface (GUI) consists of over 900 commands, allowing users to create, modify and manipulate these nodes. Behind every GUI-accessible command is a script written in the Maya Embedded Language (MEL). MEL supports the customization of existing commands or the development of new commands to perform specific tasks that are not already part of Maya's default menu set. Maya also provides application programming interfaces (APIs) for C++ and the Python programming language (http://www.python.org).
Titanium surfaces
The surfaces of three different titanium substrates, containing sub-nanometric, nanometric and micro-/nano-structured surface topographies were examined in this study. In order to create accurate 3D models, AFM surfaces topography data files were used. A scanning probe microscope (Solver P7LS, NT-MDT) was used to obtain images of the surface morphology and to quantitatively measure and analyze the surface roughness of these metallic surfaces on the nanometer scale, as described elsewhere25. All of the roughness data presented are the average obtained from four separate scans of 10 μm × 10 μm areas. Five parameters were used for the characterization of surfaces: average roughness (Ra), root mean square (RMS) roughness (Rq), maximum height (Rmax), skewness (Rskw) and kurtosis (Rkur)14,28,32.
Titanium surfaces with sub-nanometer surface roughness were obtained via the deposition of 12 nm thickness titanium films on silicon wafers (henceforth referred to as 12 nm films). These films were fabricated using a Kurt J Lesker CMS - 18 magnetron sputtering thin film deposition system as previously described27. This approach allowed the controlled atomic deposition of titanium onto the substrates for the purposes of producing metallic thin films with sub-nanoscopic and nanoscopic surface roughness33. The surfaces of the 12 nm films were found to be remarkably smooth on the sub-nanometer scale, i.e., Ra of 0.20 nm and Rq of 0.24 nm on the 10 μm × 10 μm scanning areas34. The superhydrophobic titanium surfaces (with a water contact angle of θW = 166°) were fabricated using a femtosecond laser ablation technique. The resulting surfaces contained two tier micro- and nanoscale quasi-periodic self-organized structures that mimicked the surface of the Nelumbo nucifera lotus leaf13. Surface topography data were derived from AFM data analysis.
AFM data import
The raw data obtained using AFM were converted to a text file format using the free, open-source application Gwyddion (http://gwyddion.net/). The text files were then re-formatted as comma-separated value (CSV) files. The surface topography values from the CSV files were then imported into the Maya software package as 3D objects, using a custom Python script. Python, an open-source dynamic programming language, became part of Maya's API in version 8.5. The script was written in Maya's Script Editor, which simplifies the creation of interface components, such as a user window where the functions can be accessed from within the application menu. The surface models were visualized using a polygonal geometry consisting of 3D points (vertices) connected with lines (edges).
Rendering and post-production
The final animations were rendered using the mental ray plug-in (http://www.mentalimages.com/products/mental-ray) as a sequence of images in the TARGA format. To enable the greatest flexibility in future applications of the animations, full-HD 1080p (1920 × 1080 pixels) resolution and the production quality preset were selected. Post-production was completed using Adobe Premier CS5.5, where additional information was added to the movie sequences, such as titles and text overlays of color map values. The movies were then exported in an MPEG format.
Author Contributions
Conceived the project: E.P.I. Designed the experiments: C.J.F., E.P.I. Performed the experiments: V.B. Analyzed the data: C.J.F., E.P.I., V.B., R.J.C. Wrote the paper: V.B., C.J.F., E.P.I., R.J.C.
Supplementary Material
NEW Supplementary Information File
Acknowledgments
This study was supported in part by the Australian Research Council (ARC). Autodesk, AutoCAD, DWG, the DWG logo, and Inventor are registered trademarks or trademarks of Autodesk, Inc., and/or its subsidiaries and/or affiliates in the USA and other countries.
References
- Liu X., Chu P. K. & Ding C. Surface modification of titanium, titanium alloys, and related materials for biomedical applications. Mater. Sci. Eng. R. 47, 49–121 (2004). [Google Scholar]
- Crawford R. J. et al. Surface topographical factors influencing bacterial attachment. Adv. Colloid Interface Sci 179–182, 142–149 (2012). [DOI] [PubMed] [Google Scholar]
- Kim S. & Park C. B. Bio-inspired synthesis of minerals for energy, environment, and medicinal applications. Adv. Functional Mater. 23, 10–25 (2013). [Google Scholar]
- Krishna Alla R. et al. Surface roughness of implants: A review. Trends Biomater. Artif. Organs. 25, 112–118 (2011). [Google Scholar]
- Norowski Jr P. A. & Bumgardner J. D. Biomaterial and antibiotic strategies for peri-implantitis. J. Biomed. Mater. Res. - Part B Appl. Biomater. 88, 530–543 (2009). [DOI] [PubMed] [Google Scholar]
- Valiev R. Z. et al. Nanostructured titanium for biomedical applications. Adv. Eng. Mater. 10, B15–B17+702 (2008). [Google Scholar]
- Bjursten L. M. et al. Titanium dioxide nanotubes enhance bone bonding in vivo. J. Biomed. Mater. Res. - Part A. 92, 1218–1224 (2010). [DOI] [PubMed] [Google Scholar]
- Dreaden E. C. & El-Sayed M. A. Detecting and destroying cancer cells in more than one way with noble metals and different confinement properties on the nanoscale. Acc. Chem. Res. 45, 1854–1865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi S. et al. The significance of crystallographic texture of titanium alloy substrates on pre-osteoblast responses. Biomater. 27, 3532–3539 (2006). [DOI] [PubMed] [Google Scholar]
- Lamolle S. F. et al. Titanium implant surface modification by cathodic reduction in hydrofluoric acid: Surface characterization and in vivo performance. J. Biomed. Mater. Res. - Part A. 88, 581–588 (2009). [DOI] [PubMed] [Google Scholar]
- Okawa S. & Watanabe K. Chemical mechanical polishing of titanium with colloidal silica containing hydrogen peroxide - Mirror polishing and surface properties. Dent. Mater. J. 28, 68–74 (2009). [DOI] [PubMed] [Google Scholar]
- Subramani K. Titanium surface modification techniques for implant fabrication - From microscale to the nanoscale. J. Biomim. Biomater. Tissue Eng. 5, 39–56 (2010). [Google Scholar]
- Fadeeva E. et al. Bacterial retention on superhydrophobic titanium surfaces fabricated by femtosecond laser ablation. Langmuir. 27, 3012–3019 (2011). [DOI] [PubMed] [Google Scholar]
- Webb H. K. et al. Nature inspired structured surfaces for biomedical applications. Curr. Med. Chem. 18, 3367–3375 (2011). [DOI] [PubMed] [Google Scholar]
- Hortolà P. Generating 3D and 3D-like animations of strongly uneven surface microareas of bloodstains from small series of partially out-of-focus digital SEM micrographs. Micron. 41, 1–6 (2010). [DOI] [PubMed] [Google Scholar]
- Ostadi H. et al. 3D visualization and characterization of nano structured materials in Nanotechnology (IEEE-NANO), 2011 11th IEEE Conference on. (2011).
- Long F., Zhou J. & Peng H. Visualization and analysis of 3D microscopic images. PLoS Comput. Biol. 8, e1002519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros L. C. S. et al. Visualizing the 3D architecture of multiple erythrocytes infected with plasmodium at nanoscale by focused ion beam-scanning electron microscopy. PLoS ONE. 7, e33445 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samak D., Fischer A. & Rittel D. 3D Reconstruction and Visualization of Microstructure Surfaces from 2D Images. CIRP Ann-Manuf. Techn. 56, 149–152 (2007). [Google Scholar]
- Sander B. & Golas M. M. Visualization of bionanostructures using transmission electron microscopical techniques. Micros. Res. Tech. 74, 642–663 (2011). [DOI] [PubMed] [Google Scholar]
- Binnig G., Quate C. F. & Gerber C. Atomic force microscope. Phys. Rev. Lett. 56, 930–933 (1986). [DOI] [PubMed] [Google Scholar]
- Bolliger M. J., Buck U., Thali M. J. & Bolliger S. A. Reconstruction and 3D visualisation based on objective real 3D based documentation. Forensic Sci. Med. Pathol. 8, 208–217 (2011). [DOI] [PubMed] [Google Scholar]
- Czech J., Dittrich M. & Stiles J. R. Rapid creation, Monte Carlo simulation, and visualization of realistic 3D cell models. Methods Mol. Biol. 500, 237–287 (2009). [DOI] [PubMed] [Google Scholar]
- Li W. H., Guo W. & Feng G. H. Modeling and simulation of broad-leaf plants. J. Jilin University. 38, 1146–1150 (2008). [Google Scholar]
- Truong V. K. et al. The influence of nano-scale surface roughness on bacterial adhesion to ultrafine-grained titanium. Biomater. 31, 3674–3683 (2010). [DOI] [PubMed] [Google Scholar]
- Truong V. K. et al. Air-directed attachment of coccoid bacteria to the surface of superhydrophobic lotus-like titanium. Biofouling. 28, 539–550 (2012). [DOI] [PubMed] [Google Scholar]
- Ivanova E. P. et al. Impact of nanoscale roughness of titanium thin film surfaces on bacterial retention. Langmuir. 26, 1973–1982 (2010). [DOI] [PubMed] [Google Scholar]
- Ivanova E. P. et al. Natural bactericidal surfaces: Mechanical rupture of Pseudomonas aeruginosa cells by cicada wings. Small. 8, 2489–2494 (2012). [DOI] [PubMed] [Google Scholar]
- Fung L. et al. Three-dimensional study of pectoralis major muscle and tendon architecture. Clin. Anat. 22, 500–508 (2009). [DOI] [PubMed] [Google Scholar]
- Perry J. L. & Kuehn D. P. Three-dimensional computer reconstruction of the levator veli palatini muscle in situ using magnetic resonance imaging. Cleft Palate Craniofac J. 44, 421–3 (2007). [DOI] [PubMed] [Google Scholar]
- Wu F. T. et al. Computational representation of the aponeuroses as NURBS surfaces in 3D musculoskeletal models. Comput Methods Programs Biomed. 88, 112–22 (2007). [DOI] [PubMed] [Google Scholar]
- Webb H. K. et al. Physico-mechanical characterisation of cells using atomic force microscopy - Current research and methodologies. J. Microbiol. Methods. 86, 131–139 (2011). [DOI] [PubMed] [Google Scholar]
- Wang J. Y., Ghantasala M. K. & McLean R. J. Bias sputtering effect on ultra-thin SmCo 5 films exhibiting large perpendicular coercivity. Thin Solid Films. 517, 656–660 (2008). [Google Scholar]
- Ivanova E. P. et al. Differential attraction and repulsion of Staphylococcus aureus and Pseudomonas aeruginosa on molecularly smooth titanium films. Sci. Rep. 1, 165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NEW Supplementary Information File