Abstract
PhiC31 integrase-mediated gene delivery has been extensively used in gene therapy and animal transgenesis. However, random integration events are observed in phiC31-mediated integration in different types of mammalian cells; as a result, the efficiencies of pseudo attP site integration and evaluation of site-specific integration are compromised. To improve this system, we used an attB-TK fusion gene as a negative selection marker, thereby eliminating random integration during phiC31-mediated transfection. We also excised the selection system and plasmid bacterial backbone by using two other site-specific recombinases, Cre and Dre. Thus, we generated clean transgenic bovine fetal fibroblast cells free of selectable marker and plasmid bacterial backbone. These clean cells were used as donor nuclei for somatic cell nuclear transfer (SCNT), indicating a similar developmental competence of SCNT embryos to that of non-transgenic cells. Therefore, the present gene delivery system facilitated the development of gene therapy and agricultural biotechnology.
PhiC31 integrase is a DNA recombinase derived from Streptomyces phage φC311. This enzyme can mediate recombination between two sequences, namely, attB and attP2,3. PhiC31 integrase does not require perfect attP site conservation to achieve recombination between attP and attB sites4. These imperfect attP sites or pseudo attP sites resemble a wild-type attP site5 and are present in transcriptionally active areas of a genome6. PhiC31 integrase recognizes relatively short but moderately specific sequences in mammalian genomes7. Thus, a phiC31 integrase system exhibits several characteristics, such as site-specificity and unidirectional recombination, and allow the successful use of integrase in various fields of study, including gene delivery in vitro5 or in vivo8, gene therapy9, and production of transgenic animals10.
Thyagarajan et al.5 demonstrated that phiC31 integrase can mediate attB-containing plasmid integration in an unmodified mammalian genome with an increased efficiency compared with random integration. However, unwanted random integration remains involved in phiC31-mediated integration at various background levels in different types of mammalian cells or embryonic stem cell lines5,11,12,13; as a result, the efficiency of pseudo-attP site integration and evaluation of site-specific integration are compromised. Although a high ratio between phiC31-integrase-expressing plasmid and attB-containing plasmid is extensively used to decrease random background integration14,15, high amounts of phiC31 integrase plasmid may cause an unwanted insertion of an integrase-expressing plasmid that can induce chromosome arrangement16,17. To reduce the cytotoxicity and genotoxicity induced by persistent integrase expression, we can replace integrase-expressing plasmids with integrase mRNA18,19,20, cell-permeable TAT-phiC31 protein21, or inducible phiC31 integrase versions22; however, this replacement cannot essentially prevent random integration. In addition, phiC31 system is less efficient in some cell types, such as haematopoietic T cells23, because phiC31 integrase interacts with certain cellular proteins, such as DAXX24. Such an interaction may also lead to random integration in specific cell types during phiC31-mediated transfection.
We presented an approach that used a negative selection strategy to exclude random integration during phiC31-mediated transfection. We also excised a selection system and plasmid bacterial backbone by using two other recombinases, namely, Cre and Dre.
Results
Suicide effect of different fusion TK proteins
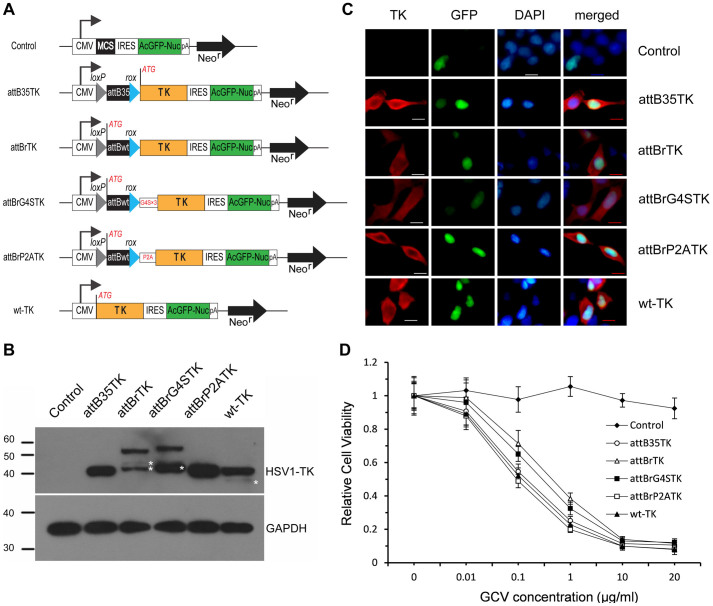
HEK293 cells were transiently transfected with different TK constructs (Figure 1A). Total cellular protein was then subjected to western blot analysis. In Figure 1B, a polyclonal antibody to HSV-1 TK detected an immunoreactive protein with a molecular weight of 40 kDa in the cells transfected with wt-TK (lane 6), which expressed a wild-type TK protein. Both attBrTK- (lane 3) and attBrG4STK-transfected (lane 4) cells contained proteins with a molecular weight of 55 kDa. These antibodies also detected lower molecular-weight proteins (indicated by stars in Figure 1B) that were probably the proteolytic breakdown products of fusion proteins. The result also showed that the wild-type TK protein was possibly a degradation target (lane 6). HEK293 cells transfected with attB35TK (lane 2) or attBrP2ATK (lane5) showed an immunoreactive protein of exactly the same size as the wild-type TK protein (lane 6). As expected, no TK gene product was detected in the cells transfected with the empty control vector (lane 1). Immunostaining results further confirmed that different TK fusion proteins were expressed in mammalian cells. In addition, no differences were observed in the subcellular localization of different fusion TK proteins, which were mainly localized in the cytosolic compartment, similar to the wild-type TK (Figure 1C).
Figure 1. Suicide effect of different fusion TK proteins.
(A) Schematic of different fusion TK constructs. CMV: Cytomegalovirus immediate early promoter; loxP and rox: recognition targets of Cre and Dre recombinases; attB35 and attBwt: minimal fuctional size and full-length of wild-type attB site; G4S × 3: (Gly4Ser)3 flexible linker; P2A: self-cleaving 2A peptide derived from porcine teschovirus-1; ATG: initiation codon flanked by a Kozak consensus sequence; control: empty vector pIRES2-AcGFP1-Nuc. (B) Western blot analysis of HEK293 cells transfected with different TK constructs. The blot was probed with polyclonal antibodies to HSV-1 TK or GAPDH. The stars denote probable proteolytic breakdown products. Full-length blots are presented in Supplementary Figure S8. (C) Immunofluorescence staining of HEK293 cells at 48 h post-transfection. The nuclei were stained with DAPI (blue); TK proteins (red) were stained with an anti-TK antibody and visualized with Cy3-labeled secondary antibodies. Scale bars = 10 μm. (D) Relative viability of HEK293 cells transfected with different TK constructs. The transfected cells selected by FACS were exposed to different concentrations of the nucleoside analog GCV for 4 d. Cytotoxicity was assessed by the WST-1 assay. “Control” represented the cells transfected with the empty vector pIRES2-AcGFP1-Nuc.
A cytotoxicity assay was performed on HEK293 cells to investigate the suicide effect of different fusion TK proteins. The results showed that all of the TK constructs except the control transfected HEK293 cells were sensitive to gancyclovir (GCV) treatment at high concentrations (Figure 1D). By contrast, the vector control cell line transfected with pIRES2-AcGFP1-Nuc was highly resistant to 10 μg/mL of GCV and exhibited 2.76% cell death. At 0.1 μg/mL of GCV, 51.1 ± 4.0% cells were dead in the attBrP2ATK-transfected group compared with 28.5 ± 7.7% (P = 0.001) and 34.8 ± 4.5% (P = 0.032) cell death rates in attBrTK- and attBrG4STK-transfected cells after a treatment period of 4 d. This result indicated that attBrP2ATK-transfected cells were more sensitive to GCV than the other two attB-fused TK-construct-transfected cells at relatively low GCV concentrations. By comparison, attBrP2ATK-transfected cells showed similar viability to attB35TK- and wt-TK-transfected cells, in which a wild-type TK protein was expressed in both groups. Thus, we chose attBrP2ATK as a representative TK construct fused with full-length attB for the subsequent studies.
Site-specific genomic integration in HEK293 cells mediated by phiC31 integrase
Different TK constructs were electroporated separately into HEK293 cells in the presence of functional phiC31 integrase mRNA or inactive mutant integrase mRNA to determine the effect of phiC31 integrase on site-specific integration. These integrase mRNAs were produced by in vitro transcription as described previously20. At 12 d after electroporation, individual cell colonies were obtained by G418 screening or G418/GCV dual selection. Table 1 shows the number of stably transfected HEK293 colonies derived from different TK constructs in the presence of functional integrase or mutant inactive integrase. We performed G418 screening and applied a functional phiC31 integrase. Our results showed that the full-length attB-fused TK construct (attBrP2ATK) exhibited the greatest number of integrants, followed by attB35TK. Moreover, wt-TK construct showed the lowest colony-forming ability. The attB-containing TK constructs showed substantially increased colony number in the presence of functional integrase compared with mutant inactive integrase; by comparison, wt-TK constructs formed a similar number of cell clones when these cells were co-transfected with either functional integrase or inactive integrase. In addition, 16.6%, 21.0%, and 94.9% of attBrP2ATK-, attB35TK-, and wt-TK-derived G418-resistant clones were GFP positive, respectively. GFP-positive cell clones may indicate that a random integration event occurred. Few surviving clones were also GFP positive when G418/GCV dual selection was performed. The attB site recombination test was also performed to determine whether or not the loss of fluorescent signal is caused by site-specific integration. The results showed that the specific band of non-recombined attB was not detected in the pooled genomic DNA of GFP-negative cell colonies screened by G418/GCV dual selection (Supplementary Fig. S1A).
Table 1. Colony number of HEK293 cells transfected with different TK constructs.
| GFP+/G418r | GCV | − | − | + | + |
|---|---|---|---|---|---|
| phiC31 | − | + | − | + | |
| attBrP2ATK | 59/63 | 48/289 | 0 | 0/187 | |
| attB35TK | 61/68 | 51/243 | 0 | 0/156 | |
| wt-TK | 67/72 | 74/78 | 0/2 | 0 |
A two-step nested PCR was performed on genomic DNA to detect 19q13.31 pseudo-site integration and investigate whether or not phiC31-integrase mediates the site-specific integration of these TK constructs6. Only the cells co-transfected with attB-containing donor vector and functional phiC31 integrase showed an appropriately sized PCR product (Supplementary Fig. S1B). This result indicated that a unique transgene was inserted at the intended genomic site. Among 12 analyzed pools derived from the attBrP2ATK construct, 10 pools contained insertions in both orientations in this particular site (Table 2). In the two other pools, insertions were detected in the forward orientation. The insertion at 19q13.31 pseudo-attP site in one orientation was detected in 8 of 12 attB35TK-derived pools; 6 pools contained at least one insertion in the opposite orientation. Considering the different colony numbers in the selected pools derived from donor plasmids containing full-length and reduced attB sites, we could not infer whether or not a full-length attB prefers integrating in the 19q13.31 site compared with the reduced attB. Both full-length and 35 bp attB sites were effective in our system; however, full-length attB showed a slightly higher colony-forming ability than the reduced attB.
Table 2. Detection of phiC31-mediated insertions at pseudo attP site 19q13.31.
| Number of pools which contain at least one orientation insertion into 19q13.31 site | |||
|---|---|---|---|
| Donor vector | attB length | Forward orientation | Reverse orientation |
| attBrP2ATK | 285 bp | 12/12 | 10/12 |
| attB35TK | 35 bp | 8/12 | 6/12 |
Site-specific recombinase-based integration and excision in primary isolated bovine fetal fibroblasts
The CMV promoter was replaced with a CAGGS promoter to maintain the high expression levels of the TK transgene. As a result, a pCAG-attBrP2ATK plasmid integration vector was generated (Figure 2A). We also added a rox and loxP flanked multiple cloning site (MCS) to introduce the genes of interest (GOI) and then replaced the IRES-AcGFP-Nuc cassette with an EGFP-expressing cassette driven by an EF1a promoter. To determine the expression efficiency of the newly generated integration vector, we transiently transfected the HEK293 cells with the TK constructs driven by either a CMV promoter or a CAGG promoter. Western blot assay showed that the TK product was robustly expressed under CAGGS promoter (Supplementary Fig. S2A). The cells were observed by fluorescence microscopy at 48 h after transfection. pCAG-attBrP2ATK-transfected HEK293 cells displayed a strong fluorescent GFP signal (Supplementary Fig. S2B). This result suggested the appropriate function of the EF1a promoter. Therefore, pCAG-attBrP2ATK, which possibly resulted in strong transgene expression in primary isolated cells, was used in further experiments.
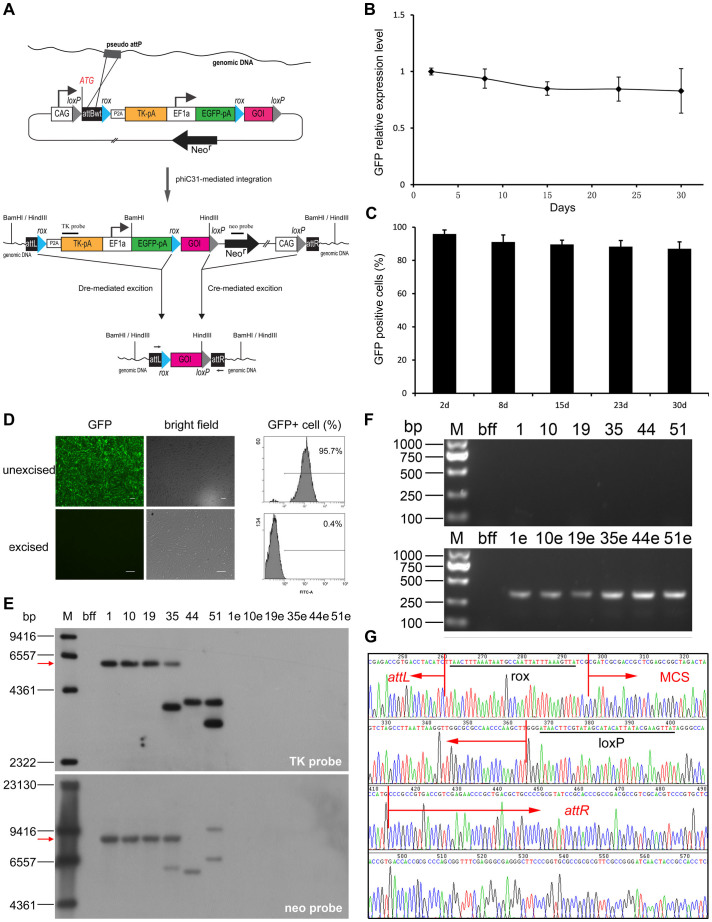
Figure 2. Site-specific recombinase-based integration and excision in primary isolated bovine fetal fibroblasts.
(A) Schematic of the pCAG-attBrP2ATK construct and intermediates in pseudo-attP-site integration and Cre/Dre-mediated excision. AttBrP2ATK fusion gene is ubiquitously expressed under the control of the CAGGS promoter until phiC31-mediated integration. EF1a-EGFP is used for visual fluorescence monitoring of the stably-transfected primary cells before Dre-mediated excision and indicates the unexcised transgenic cells after excision. “GOI” represents multiple cloning sites where any DNA sequence can be inserted. TK probe and neo-probe are used for Southern blot analysis to detect Dre- and Cre- mediated excision, respectively. Two arrows that point to opposite directions represent primer pairs used to verify the excision of both selectable makers and plasmid backbone. (B) Relative real-time RT-PCR analysis of EGFP mRNA expression was performed in stably transfected bovine fetal fibroblast cells. The decline of mRNA from 2 to 30 days was not significant (P > 0.05). Error bars denote SEM. (B) Flow cytometry analysis of the percentage of GFP-positive cells was performed in stably transfected bovine fetal fibroblast cells. The decline of percentage of GFP+ cells from 2 to 30 days was not significant (P > 0.05). Error bars denote SEM. (D) Fluorescence phenotype of transgenic cells before and after excision. Scale bar = 20 μm. Approximately 95% of unexcised cells were GFP+ and almost all of the excised cells were GFP negative after FACS sorting. (E) Southern blot analysis of transgenic cell lines before and after Cre/Dre-mediated excision. The single-copy safe harbor integrated clones (#1, #10, and #19) showed a single band of 5.9 kb by using a TK probe or a single band of 8.5 kb by using neo-probe, as depicted by arrow. The unexcised clones (#1, #10, #19, #35, #44, and # 51) carried at least one copy of TK gene or neo-gene, which was no longer detectable after excision (#1e, #10e, #19e, #35e, #44e, and # 51e). Full-length blots are presented in Supplementary Figure S9. (F) Cre- and Dre-mediated excision was further demonstrated by PCR. The genomic DNA of untransfected bovine fetal fibroblast cells was used as a negative control. Only the excised cells showed the expected band (334 bp). The gels have been run under the same experimental conditions and full-length gels are presented in Supplementary Figure S10. (G) Panel F, sequencing result of the specific band. The selectable markers and plasmid bacterial backbone were completely removed from the excised cells; rox- and loxP-flanked MCS was retained.
Bovine fetal fibroblasts isolated from the skin of a Holstein female fetus (aged 50 d to 60 d) were transfected with pCAG-attBrP2ATK and functional phiC31 integrase mRNA to investigate whether or nor the newly generated vector can be used to generate stable cell lines in primary cells. Using G418/GCV dual selection, we obtained 58 individual cell colonies at 15 d post-transfection and these colonies displayed a strong fluorescent signal. By contrast, not a single clone was obtained from bovine fetal fibroblasts transfected in the absence of functional integrase. To screen single-copy integration in an evaluated safe harbor20, we performed a two-step nested PCR. Among the 58 clones, 9 showed an appropriately sized PCR product (Supplementary Fig. S3A). Absolute quantity PCR was then applied to determine single-copy integration among these safe-harbor-integrated clones (Supplementary Figs. S3B and S3C). In addition, random integration events were not detected in the 58 clones by using PCR for non recombined attB site (Fig. S3D). Thus we speculated that the remaining clones were result of an integration in a different pseudo-attP site. Half-nested inverse PCR was performed for detection of integration sites and a few of different pseudo-attP sites were identified (Supplementary Fig. S4). Before Cre/Dre-mediated excision was performed, single-copy transgenic bovine fetal fibroblasts were cultured for four weeks. Relative EGFP expression level and the percentage of GFP-positive cells were measured during this period. The results showed no significant decrease in EGFP expression level (Figure 2B) and the percentage of GFP-positive cells (Figure 2C).
Three clones (#1, #10, and #19) carrying a single integration of pCAG-attBrP2ATK in the pre-defined safe harbor were selected for excision assay, and three other identified clones (#35, #44, and #51) were used as controls. To remove the selectable marker and plasmid backbone, we transiently exposed the selected cell clones to cell-permeant Cre and Dre proteins (Supplementary Fig. S5). At 5 d post-transduction, these cells were prepared for FACS. The rox-flanked sequence-excised clones were easily detected and sorted for expansion by visually tracking the loss of EGFP expression. In general, approximately 50% of the stably transfected cells exhibited a loss of EGFP expression (Supplementary Fig. S6). The loss of GFP fluorescent signal was observed in the sorted cells under a fluorescence microscope (Figure 2D). Excision was verified by Southern blot (Figure 2E, upper panel). All of the analyzed cells also revealed that the loxP-flanked sequences were excised (Figure 2E, lower panel). Cre- and Dre-mediated excision was further demonstrated by PCR using primers that flanked the integration site. Only the excised cells showed the expected band, which was purified by gel extraction and sequenced (Figure 2F). The sequencing result showed that the selectable markers and plasmid bacterial backbone were completely removed, but rox- and loxP-flanked GOI (MCS) were retained (Figure 2G). To investigate the effect of phiC31 mediated integration and Cre/Dre-mediated excision on transgene expression, we inserted a luciferase gene under the control of CMV promoter into the MCS of pCAG-attBrP2ATK and generated transgenic bovine fetal fibroblast clones with a single insertion at the pre-defined safe harbor. Furthermore, we performed the luciferase assay, demonstrating that no significant decline of luciferase activity was observed during multiple passages or even after excision of selection markers (Supplementary Fig. S7A). Consistently, western blot analysis also revealed robust and stable expression of luciferase gene after site-specific integration and excision of the selection system (Supplementary Fig. S7B).
The single-copy safe harbor integrated clean transgenic cells free of selectable markers and excised vector backbone were used as nuclei donors to produce transgenic SCNT embryos. In addition, normal bovine fetal fibroblasts derived from the same fetus and with a similar passage number were used as a control sample to investigate the effect of transgenic procedures on the developmental potential of SCNT embryos. No significant differences in cleavage and blastocyst formation rates were observed when excised cells were used as nuclei donors compared with untransfected bovine fetal fibroblast cells (77.4 ± 2.6% vs. 78.6 ± 4.3%, P = 0.7 for cleavage rate; 32.6 ± 2.9% vs. 36.3 ± 4.0%, P = 0.3 for blastocyst formation rate). After the embryo was transferred, similar pregnancy rates were also observed compared with the control group (25.9 ± 2.4% vs. 32.3 ± 4.7%, P = 0.1).
Discussion
We presented a new strategy that can be used to select phiC31-mediated pseudo-site-specific integration from random integration and produce clean transgenic cells free of selectable markers and vector backbone. We used phiC31 integrase to achieve site-specific genomic integration at pseudo sites. We also used attB-TK fusion proteins as negative selection markers to exclude random integration. Clean transgenic cells were obtained by fluorescence-activated cell sorting after Cre- and Dre-recombinase treatment.
PhiC31 system provides a more efficient and safer gene delivery approach than a traditional random integration-based method25. However, random integration events are observed in phiC31-mediated transfection13,26. Extensive studies have been conducted to improve the efficiency of the phiC31 system12,27,28,29,30, which contribute to enhancing and stabilizing transgene expression without preventing random integration. To address this issue, scholars developed two-step integration strategies19,31,32. An attP-containing docking site cassette is pre-inserted by random or targeted approaches into cell lines; a second round of phiC31-mediated integration is then performed accompanied with an appropriate drug selection strategy to identify site-specific integration events. The two-step selection strategies are based on a silent selection marker activated by an incoming plasmid-delivered promoter or vice versa; hence, a highly reliable integration system is developed and random integration is reduced. Nevertheless, multiple transfections and drug selections are tedious processes and can cause significant cell death. Thus, two-step strategies may be acceptable for a robust cell line but undesirable for an isolated primary cell, which is the main resource of donor cell for ex vivo gene therapy and generation of a large transgenic animal.
In our study, HSV-1-TK or attB-TK fusion proteins were used as negative selectable markers to exclude random integration in phiC31-mediated integration. Our strategy is based on the principle that these cells, in which random integration has occurred, likely express a wild-type or fusion TK protein ubiquitously; thus, such cells are specifically eliminated by the antiviral agent GCV. In this method, genomic pseudo-attP-site-integrated clones are enriched. Our original scheme used 285 bp full-length attB sites. We constructed a series of full-length attB-TK fusion proteins without a linker, or with (Gly4Ser)3 linker33, or P2A “self-cleaving” peptide34 between these proteins. Western blot assay of HEK293 cell lysates showed TK-specific bands with the predicted size; however, some proteolytic breakdown products were also found as revealed in a previous study35. Expression levels were qualitatively higher in the cells transfected with attBrP2ATK constructs than those transfected with attBrTK or attBG4STK. Nevertheless, no difference in the subcellular localization of different fusion proteins and wild-type TK protein was observed. Therefore, the differences in GCV sensitivity of different TK-construct-transfected cells were mainly affected by the expression level of functional TK protein. Also, we investigated whether or not minimal attB sizes of 35 bp can support pseudo-attP integration in our system. Our results showed that either of full-length or 35 bp attB sites was competent for the site-specific integration mediated by phiC31 integrase. However, full-length attB showed a slight increase in colony formation compared with 35 bp attB. This increase presumably indicated a favorable sequence context of the full-length attB site compared with the minimal attB site. Under stringent reaction conditions, the reduced attB site may function less effectively than the full-length site36.
AcGFP-Nuc protein expressed from IRES as part of the bicistronic transcript was used to sort TK-fusion-gene-expressing stably transfected HEK293 cells by FACS for cytotoxicity assay. The IRES-AcGFP-Nuc cassette was also used as a fluorescent reporter to monitor random integration events when phiC31-mediated site-specific integration in HEK293 cells was evaluated. After G418 selection was performed, cell colonies showing GFP-fluorescent signal indicated that the attB site between the CMV promoter and the TK-IRES-AcGFP-Nuc cassette was not cleaved; this result was probably caused by non-site-specific integration. As indicated by the GFP fluorescent signal, at least 16.6% of colonies were generated in the presence of phiC31 integrase involved in random integration; almost all of the stably transfected clones generated without phiC31 integrase were GFP positive. Cells transfected with phiC31 integrase showed 3.6-fold to 4.6-fold increase in colony-forming ability compared with that without phiC31. The fold increase in this study was slightly lower than that in previous studies5,37. We speculated that the DNA/RNA co-transfection method/parameters used in our study may have contributed to the difference in fold increase. The delivery efficiency or stability of phiC31 mRNA was less than that of the attB-containing donor plasmid; this condition may result in a high proportion (15/38, in our previous study20) of unwanted random integration of donor plasmid during co-transfection. However, the exact mechanisms remain unknown. In addition, EGFP expression induced by an EF1a promoter was conducted to monitor stably transfected primary cells by visual fluorescence before excision; this procedure was also performed to determine the unexcised transgenic cells after excision. To ensure a normal fluorescent indicator function, we determined the EGFP expression level and GFP-positive percentage of the selected bovine fetal fibroblast clones for a time course of four weeks. The stability of EGFP expression and no loss in percentage of GFP-positive cells were considered as the result of phiC31-mediated integration.
Once transgenic cells are obtained, the selectable marker is no longer necessary but raises public concerns on the biological safety of ex vivo gene therapy or generation of large transgenic animals. The presence of CpG nucleotides and bacterial sequences in the donor plasmid backbone can also lead to post-integrative silencing of the transgene38. A recent study attempted to address post-integrative gene silencing by reducing the number of CpG dinucleotides in the plasmid backbone29. We could excise donor plasmid backbone and selectable markers after transgenic cells were obtained. In this study, cell-permeant Cre and Dre proteins were used to excise the cell clones carrying a single copy of transgene integrated in a previously evaluated safe harbor, where the expression of neighboring genes is unaffected by transgene integration and excision20. Cre recombination resulted in the excision of neoR gene and vector backbone. Furthermore, Dre recombination resulted in the excision of TK gene and EGFP-expressing cassette. The EGFP-expressing cassette may indicate that rox-flanked sequences were not excised. After protein transduction and FACS were conducted, the cells that lost GFP fluorescence were subjected to Southern blot assay and PCR test. The results showed that rox-flanked and loxP-flanked sequences were removed completely in each tested cell clone. Considering that no crossover recombination occurs between Cre-rox or Dre-loxP39, we found that the MCS sequence used to introduce GOI was not deleted in any of the sorted cells after protein transduction. Furthermore, when luciferase gene was introduced in the pre-defined safe harbor in bovine genome, robust and stable expression of luciferase was observed during multiple passages or even after excision of selection system, which provided a potential application to generate selectable marker- and vector-backbone-free cells without disturbing transgene expression. In addition, the development competency of SCNT embryos was not compromised when these clean cells were used as nuclear donors compared with untransfected cells.
In conclusion, a simple phiC31-mediated strategy was presented in this study to integrate a transgene in a safe genomic harbor by preventing random integration and combining cell-permeant protein transduction and fluorescence-activated cell sorting to remove the selection system and the vector backbone. This non-viral approach provides a simple and safe alternative to produce clean transgenic cells free of selectable markers and plasmid bacterial backbone. Therefore, our strategy is a valuable tool in gene therapy and animal transgenesis.
Methods
Ethics statement
The experimental procedure was approved by the Animal Care Commission of the College of Veterinary Medicine, Northwest A&F University (China). Bovine ovaries from slaughtered mature cows were collected from Tumen Abattoir (Xi'An, China). A newborn female Holstein calf was used to obtain nuclear donor cell cultures and beef-breed Angus cows were used as recipient animals (Yangling Keyuan Cloning Co., Ltd., P. R. China).
Plasmid construct
Multiple TK fusion constructs were generated as described in SUPPLEMENTARY MATERIALS AND METHODS (Figure 1A). The primer sequences, restriction sites, and templates used in the study are shown in Table S1.
Cell culture and transfection
HEK293 cells were grown in Dulbecco's modified Eagle medium (DMEM; GIBCO, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; GIBCO) at 37°C in a humidified atmosphere with 5% CO2. Bovine fetal fibroblasts were isolated from a Holstein female fetus (aged 50 d to 60 d; Yangling Keyuan Biotechnology Inc., China) by disaggregating the body without the head and viscera; the fibroblasts were then culture in DMEM supplemented with 10% FBS at 38.5°C in a humidified atmosphere with 5% CO2. Bovine fetal fibroblasts at confluency were collected by trypsinization and subjected to passaging or cryopreservation.
Cell transfection was performed as described previously20. In brief, HEK293 cells or bovine fetal fibroblasts were re-suspended in 0.2 mL of diluted electroporation working buffer containing a donor plasmid only (5 μg) or a donor plasmid (5 μg) with phiC31 mRNA (1 μg). Electroporation was performed using BTX ECM2001 (BTX, San Diego, CA) set to a single pulse of 2 ms at 510 V. At 24 h after electroporation, the cells were diluted in a 10 cm dish containing fresh medium supplemented with G418 (400 μg/mL to 600 μg/mL) or G418/GCV (400 μg/mL to 600 μg/mL and 1 μg/mL to 10 μg/mL) to select stably transfected cells. Selection was performed for 12 d to 15 d until individual colonies were obtained and counted.
Western blot
The expression of thymidine kinase protein was detected by western blot. Cell lysates were prepared from HEK293 cells transfected with an empty vector control (pIRES2-AcGFP1-Nuc) or different TK constructs (attB35TK, attBrTK, attBrG4STK, attBrP2ATK, and wt-TK). At 48 h post-transfection, the cells were harvested and re-suspended in phosphate buffered saline (PBS). Total protein (10 μg) was loaded per lane in a 12% SDS-PAGE gel. The gel was transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA) and subjected to western blot analysis according to a standard protocol. A goat polyclonal antibody against HSV-1 thymidine kinase (1:1000; Santa Cruz Biotechnology Inc., Santa Cruz, CA) and HRP-labeled Donkey Anti-Goat IgG (H + L; 1:1000; Beyotime, Jiangsu, China) were used to detect TK products. A rabbit polyclonal antibody against GAPDH (1:1000, Sigma) and HRP-labeled goat anti-rabbit IgG (H + L; 1:1000; Beyotime) were used to detect GAPDH, which was used as an internal control.
Cytotoxicity assay
HEK293 cells were transfected with different TK constructs in the absence of phiC31 integrase. Transfected cells were determined by GFP expression and sorted by FACS performed at 48 h after transfection. The sorted GFP-positive cells were seeded in 96-well plates at a cell density of 5.0 × 103 cells/well. GCV concentrations (0, 0.01, 0.1, 1, 10, and 20 μg/mL) were added to the transfected cells. After 4 d, the killing effect of GCV was measured using a WST-1 cell proliferation and cytotoxicity assay kit (Beyotime) according to the manufacturer's instructions.
Flow cytometric analysis
At 2 or 5 d after transfection, 2 × 107 cells from each culture transfected with TK fusion constructs only or TK fusion constructs and phiC31 mRNA were harvested by trypsinization and re-suspended in PBS containing 2% FBS. The cells were then analyzed by flow cytometry to determine GFP expression. Flow cytometric analysis was performed using BD FACSAria (BD Biosciences, San Jose, CA).
Preparation of recombinant cell-permeable protein and protein transduction
pET-28a(+)-His-TAT-Dre-NLS plasmids were constructed by cloning the TAT-Dre-NLS sequence into His6 expression vector pET-28a(+). The expression vector pET-28a(+)-His-NLS-TAT-Cre20 and pET-28a(+)-His-TAT-Dre-NLS were used to transform Escherichia coli strain BL21 (DE3) separately for IPTG-inducible expression of His-tagged Cre and Dre proteins. The expression and purification of His-tagged Cre and Dre proteins were performed according to the detailed protocol described previously20. Protein concentrations were measured using a BCA protein assay kit (Beyotime). For protein transduction experiments, 2 × 106 cells were seeded on a 24-well plate and grown for 24 h. Cell-permeant proteins were sterilized by filtration using a 0.22 μm filter (Millipore). The cells were incubated for 4 h in a medium containing 100 μg/mL of His-NLS-TAT-Cre and 100 μg/mL of His-TAT-Dre-NLS proteins. After transduction, the cells were washed with PBS and cultured for another 72 h in a normal medium before flow cytometric analysis was conducted.
Southern blot
Approximately 10 μg to 20 μg of genomic DNA from untransfected cells or stably transfected cell colonies was digested with BamHI and HindIII (New England Biolabs, Beijing, China) overnight and resolved by agarose gel electrophoresis. The DNA was transferred, subjected to UV cross linking on a Hybond N+ nylon membrane (Roche, South San Francisco, CA), and hybridized with a TK probe or a neo-probe generated using a DIG High Prime Labeling and Detection Starter Kit II (Roche). The primer sequences were listed as follows: for TK probe, 5′-CAGCAAGAAGCCACGGAAGT-3′ (forward) and 5′-GCCCGAAACAGGGTAAATAACG-3′ (reverse); for neo-probe, 5′-GGATTGCACGCAGGTTCTCCG-3′ (forward) and 5′-CGCCGCCAAGCTCTTCAGCAA-3′ (reverse).
Statistical analysis
Each experiment was performed at least 3 times. Data were analyzed using SPSS 20.0 (IBM Corporation, Somers, NY, USA) with one-way ANOVA and least-significant difference tests. Data were reported as mean ± SEM. P < 0.05 was considered significant.
Author Contributions
Y.Y. and Y.Z. conceived and designed the experiments; Y.Y., Q.T., Z.X.L., J.H.T., Y.Z.W. and Y.S.W. performed the experiments; Y.Y., F.S., J.L. and Y.Z. analyzed the data; Y.Y. and Y.Z. wrote the paper. All authors reviewed the manuscript.
Supplementary Material
Supplementary information
Supplementary Figure S8
Supplementary Figure S9
Supplementary Figure S10
Acknowledgments
We thank Prof. Zekun Guo and Dr. Yongyan Wu for their technical assistance. We also thank Mr. Younan Wang for facilitating the transport of the Holstein cow ovaries used in this study. This work was supported by the National Major Project for Production of Transgenic Breeding (No. 2013ZX08007-004) and the National High Technology Research and Development Program of China (863 Program) (No. 2011AA100303).
References
- Thorpe H. M. & Smith M. C. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. U. S. A. 95, 5505–5510 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J., Foster S. G. & Tobek I. Physical and genetic analysis of IS110, a transposable element of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 200, 235–239 (1985). [DOI] [PubMed] [Google Scholar]
- Kudriavtseva E. A., Yun Yang L., Voeikova T. A. & Lomovskaia N. D. Use of a plasmid with integrative function of phage phiC31 for transfer of cloned genes into Streptomyces strains. Genetika 30, 886–897 (1994). [PubMed] [Google Scholar]
- Sclimenti C. R., Thyagarajan B. & Calos M. P. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 29, 5044–5051 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B., Olivares E. C., Hollis R. P., Ginsburg D. S. & Calos M. P. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol. Cell. Biol. 21, 3926–3934, 10.1128/MCB.21.12.3926-3934.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalberg T. W. et al. Integration specificity of phage phiC31 integrase in the human genome. J. Mol. Biol. 357, 28–48, 10.1016/j.jmb.2005.11.098 (2006). [DOI] [PubMed] [Google Scholar]
- Ginsburg D. S. & Calos M. P. Site-specific integration with phiC31 integrase for prolonged expression of therapeutic genes. Adv. Genet. 54, 179–187, 10.1016/S0065-2660(05)54008-2 (2005). [DOI] [PubMed] [Google Scholar]
- Aneja M. K., Imker R. & Rudolph C. Phage phiC31 integrase-mediated genomic integration and long-term gene expression in the lung after nonviral gene delivery. J. Gene Med. 9, 967–975, 10.1002/jgm.1090 (2007). [DOI] [PubMed] [Google Scholar]
- Karow M. & Calos M. P. The therapeutic potential of PhiC31 integrase as a gene therapy system. Expert Opin. Biol. Ther. 11, 1287–1296, 10.1517/14712598.2011.601293 (2011). [DOI] [PubMed] [Google Scholar]
- Hollis R. P. et al. Phage integrases for the construction and manipulation of transgenic mammals. Reprod. Biol. Endocrinol. 1, 79, 10.1186/1477-7827-1-79 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. W. et al. Identification of pseudo attP sites for phage phiC31 integrase in bovine genome. Biochem. Biophys. Res. Commun. 345, 984–988, 10.1016/j.bbrc.2006.04.145 (2006). [DOI] [PubMed] [Google Scholar]
- Nishiumi F. et al. Simultaneous single cell stable expression of 2-4 cDNAs in HeLaS3 using psiC31 integrase system. Cell Struct. Funct. 34, 47–59 (2009). [DOI] [PubMed] [Google Scholar]
- Thyagarajan B. et al. Creation of engineered human embryonic stem cell lines using phiC31 integrase. Stem Cells 26, 119–126, 10.1634/stemcells.2007-0283 (2008). [DOI] [PubMed] [Google Scholar]
- Woodard L. E., Hillman R. T., Keravala A., Lee S. & Calos M. P. Effect of nuclear localization and hydrodynamic delivery-induced cell division on phi C31 integrase activity. Gene Ther. 17, 217–226, 10.1038/gt.2009.136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L. et al. Generation of induced pluripotent stem cells using site-specific integration with phage integrase. Proc. Natl. Acad. Sci. U. S. A. 107, 19467–19472, 10.1073/pnas.1012677107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Skjorringe T., Gjetting T. & Jensen T. G. PhiC31 integrase induces a DNA damage response and chromosomal rearrangements in human adult fibroblasts. BMC Biotechnol. 9, 8, 3110.1186/1472-6750-9-31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jeppesen I., Nielsen K. & Jensen T. G. Phi c31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 13, 1188–1190, 10.1038/sj.gt.3302789 (2006). [DOI] [PubMed] [Google Scholar]
- Fish M. P., Groth A. C., Calos M. P. & Nusse R. Creating transgenic Drosophila by microinjecting the site-specific phiC31 integrase mRNA and a transgene-containing donor plasmid. Nat. Protoc. 2, 2325–2331, 10.1038/nprot.2007.328 (2007). [DOI] [PubMed] [Google Scholar]
- Monetti C. et al. PhiC31 integrase facilitates genetic approaches combining multiple recombinases. Methods 53, 380–385, 10.1016/j.ymeth.2010.12.023 (2011). [DOI] [PubMed] [Google Scholar]
- Yu Y. et al. A site-specific recombinase-based method to produce antibiotic selectable marker free transgenic cattle. PloS ONE 8, e62457, 10.1371/journal.pone.0062457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. X. et al. TAT-phiC31 integrase mediates DNA recombination in mammalian cells. J. Biotechnol. 142, 107–113, 10.1016/j.jbiotec.2009.03.018 (2009). [DOI] [PubMed] [Google Scholar]
- Sharma N., Moldt B., Dalsgaard T., Jensen T. G. & Mikkelsen J. G. Regulated gene insertion by steroid-induced PhiC31 integrase. Nucleic Acids Res. 36, e67, 10.1093/nar/gkn298 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maucksch C. et al. Cell type differences in activity of the Streptomyces bacteriophage phiC31 integrase. Nucleic Acids Res. 36, 5462–5471, 10.1093/nar/gkn532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Z. et al. DAXX interacts with phage PhiC31 integrase and inhibits recombination. Nucleic Acids Res. 34, 6298–6304, 10.1093/nar/gkl890 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. & Bullock C. Site-specific gene targeting for gene expression in eukaryotes. Curr. Opin. Biotechnol. 11, 455–460 (2000). [DOI] [PubMed] [Google Scholar]
- Liu Y. et al. Generation of platform human embryonic stem cell lines that allow efficient targeting at a predetermined genomic location. Stem Cells Dev. 18, 1459–1472, 10.1089/scd.2009.0047 (2009). [DOI] [PubMed] [Google Scholar]
- Raymond C. S. & Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PloS ONE 2, e162, 10.1371/journal.pone.0000162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreas S., Schwenk F., Kuter-Luks B., Faust N. & Kuhn R. Enhanced efficiency through nuclear localization signal fusion on phage PhiC31-integrase: activity comparison with Cre and FLPe recombinase in mammalian cells. Nucleic Acids Res. 30, 2299–2306 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneja M. K. et al. Optimization of Streptomyces bacteriophage phi C31 integrase system to prevent post integrative gene silencing in pulmonary type II cells. Exp. Mol. Med. 41, 919–934, 10.3858/emm.2009.41.12.098 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Nakamura S., Sakurai T., Akasaka K. & Sato M. Improvement of a phiC31 integrase-based gene delivery system that confers high and continuous transgene expression. New Biotechnol. 28, 312–319, 10.1016/j.nbt.2010.11.001 (2011). [DOI] [PubMed] [Google Scholar]
- Michael I. P. et al. Highly efficient site-specific transgenesis in cancer cell lines. Mol. Cancer 11, 89, 10.1186/1476-4598-11-89 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. M., Krohn J., Bhattacharya S. & Davies B. A Comparison of Exogenous Promoter Activity at the ROSA26 Locus Using a PhiC31 Integrase Mediated Cassette Exchange Approach in Mouse ES Cells. Plos ONE 6, e23376, 10.1371/journal.pone.0023376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston J. S. et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 85, 5879–5883 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H. et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS ONE 6, e18556, 10.1371/journal.pone.0018556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulski K. R., Kim J. H., Kim S. H. & Freytag S. O. Glioma cells transduced with an Escherichia coli CD/HSV-1 TK fusion gene exhibit enhanced metabolic suicide and radiosensitivity. Hum. Gene Ther. 8, 73–85, 10.1089/hum.1997.8.1-73 (1997). [DOI] [PubMed] [Google Scholar]
- Groth A. C., Olivares E. C., Thyagarajan B. & Calos M. P. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. U. S. A. 97, 5995–6000, 10.1073/pnas.090527097 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt A., Engler J. A., Xu H., Cherry A. M. & Kay M. A. Molecular analysis of chromosomal rearrangements in mammalian cells after phiC31-mediated integration. Hum. Gene Ther. 17, 1077–1094, 10.1089/hum.2006.17.1077 (2006). [DOI] [PubMed] [Google Scholar]
- Hodges B. L., Taylor K. M., Joseph M. F., Bourgeois S. A. & Scheule R. K. Long-term transgene expression from plasmid DNA gene therapy vectors is negatively affected by CpG dinucleotides. Mol. Ther. 10, 269–278, 10.1016/j.ymthe.2004.04.018 (2004). [DOI] [PubMed] [Google Scholar]
- Anastassiadis K. et al. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model. Mech. 2, 508–515, 10.1242/dmm.003087 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary Figure S8
Supplementary Figure S9
Supplementary Figure S10