A database of lattices using the G 6 representation of the Niggli-reduced cell as the search key provides a more robust and complete search than older techniques. Searching is implemented by finding the distance from the probe cell to other cells using a topological embedding of the Niggli reduction in G 6.
Keywords: Niggli reduction, Bravais lattice determination, computer programs
Abstract
A database of lattices using the G 6 representation of the Niggli-reduced cell as the search key provides a more robust and complete search than older techniques. Searching is implemented by finding the distance from the probe cell to other cells using a topological embedding of the Niggli reduction in G 6, so that all cells representing similar lattices will be found. The embedding provides the first fully linear measure of distances between unit cells. Comparison of results with those from older cell-based search algorithms suggests significant value in the new approach.
1. Introduction
Andrews & Bernstein (2012 ▶, 2014 ▶) introduced a topological embedding of the Niggli ‘cone’ of reduced cells with the goal of calculating a meaningful distance between unit cells. In the latter article by Andrews & Bernstein (2014 ▶), the embedding was used to determine likely Bravais lattices for a unit cell. Here we apply the embedding to searching within a database for lattices ‘close’ to the lattice of a given probe cell.
A crystallographic cell is a representation of a lattice, but each lattice can be represented just as well by any of an infinite number of such unit cells. Searching for matches to an experimentally determined crystallographic unit cell in a large collection of previously determined unit cells is a useful verification step in synchrotron data collection and can be a screen for ‘similar’ substances (Ramraj et al., 2011 ▶; Mighell, 2002 ▶), but it is more useful to search for a match to the lattice represented by the experimentally determined cell, which may involve many more cells. For identification of substances with small cells, a unit-cell match may be sufficient for unique identification (Mighell, 2001 ▶).
As a result of experimental error and the occurrence of multiple cells representing the same lattice and differing choices of lattice centering, simple searches based on raw cell edges and angles can miss similarities. A database of lattices using the 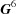 representation of the Niggli-reduced cell as the search key provides a more robust and complete search. Searching is implemented by finding the distances from the probe cell to other cells using a topological embedding of the cone of Niggli-reduced cells in
representation of the Niggli-reduced cell as the search key provides a more robust and complete search. Searching is implemented by finding the distances from the probe cell to other cells using a topological embedding of the cone of Niggli-reduced cells in 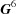 . Comparison of results with those from older cell-based search algorithms suggests significant value in the new approach.
. Comparison of results with those from older cell-based search algorithms suggests significant value in the new approach.
2. History
Tabulation of data for the identification of minerals dates to the 18th and 19th centuries. Data collected included interfacial angles of crystals (clearly related to unit-cell parameters) and optical effects [see the historical review by Burchard (1998 ▶)]. With the discovery of X-ray diffraction, those tables were supplanted by new collections. Early compilations that included unit-cell parameters arranged for material identification were ‘Crystal Structures’ (Wyckoff, 1931 ▶), ‘Crystal Data Determinative Tables’ (Donnay, 1943 ▶) and Handbook of Lattice Spacings and Structures of Metals and Alloys (Pearson, 1958 ▶). Early computerized searches were created by JCPDS in the mid-1960s (Johnson, 2013 ▶) and the Cambridge Structural Database and its search programs (Allen et al., 1973 ▶).
Those first searches were sensitive to the issues of differing equivalent presentations of the same lattice. The first effective algorithm for resolving that issue was reported by Andrews et al. (1980 ▶) using the V7 algorithm (NIH/EPA, 1980 ▶). Subsequently, other programs using the V7 algorithm have been described (see Table 1 ▶). The V7 algorithm has the advantage over simple Niggli-reduction-based cell searches of being stable under experimental error. However, sensitivity to a change in an angle is reduced as that angle nears 90°.
Table 1. Programs designed to perform effective searches in a unit-cell database.
3. Background
An effective search method must find ways to search for unit cells that represent ‘similar’ lattices, even when the cells are to be tabulated in ways that make the cells seem to be very different. A trivial example is
Clearly, these unit cells are almost identical, but simple tabulations might separate them. A somewhat more complex example includes the following primitive cells:
Here the relationship is not as obvious. The embedding of Andrews & Bernstein (2012 ▶, 2014 ▶) can be used to show that the distance between these two cells is quite small in 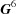 (0.004 Å2 in
(0.004 Å2 in 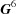 ).
).
4. Implementation: 1 – distance
The program SAUC is structured to allow use of several alternative metrics for searching among cells in an attempt to identify cells representing similar lattices. To simplify comparisons between results with the different metrics, all have been linearized and normalized, i.e. converted to ångström units and scaled to be commensurate with the 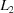 norm given below:
norm given below:
(a) A simple 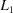 or
or 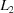 norm based on
norm based on
with the distance scaled by 6−1/2 in the case of the 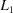 norm and unscaled in the case of the
norm and unscaled in the case of the 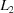 norm. The angles are assumed to be in radians and the edges in ångströms. The angles are converted to ångströms by multiplying by the average of the relevant edge lengths. This scaling of
norm. The angles are assumed to be in radians and the edges in ångströms. The angles are converted to ångströms by multiplying by the average of the relevant edge lengths. This scaling of 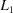 is suggested by the fact that in general
is suggested by the fact that in general 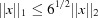 .
.
(b) The square root of the BGAOL (Andrews & Bernstein, 2014 ▶) Niggli cone embedding distance NCDist based on
Before taking the square root, the distances are scaled by 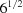 divided by the average length of the cell edges. The overall square root linearizes the metric to ångström units. The complex relationship between the NCDist distance and simpler norms such as
divided by the average length of the cell edges. The overall square root linearizes the metric to ångström units. The complex relationship between the NCDist distance and simpler norms such as 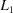 and
and 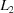 does not admit a single scaling that would align all distances. If it did, we could just use the
does not admit a single scaling that would align all distances. If it did, we could just use the 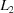 norm. However, as seen in Table 2 ▶ this scaling provides a rough approximation to the
norm. However, as seen in Table 2 ▶ this scaling provides a rough approximation to the 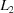 distance in the 1–2 Å range.
distance in the 1–2 Å range.
Table 2. Comparison of search results for cell (80.36, 80.36, 99.44, 90, 90, 120) from entry 1u4j in space group R3 (see Le Trong Stenkamp, 2007 ▶).
In each case the PDB code (Bernstein etal., 1977 ▶; Berman etal., 2000 ▶) found is shown with the distance metric for that method. In the case of Nearest-Cell (Ramraj etal., 2011 ▶), a second column with the square root of the metric is provided as well. The results are sorted by the NCDist distance. Results have been cut off at 3.5 in the NCDist metric. The three alternative cells cited by Le Trong Stenkamp (2007 ▶) are in bold and marked with an asterisk (*). The Nearest-Cell (‘N-C’) results are from the http://www.strubi.ox.ac.uk/nearest-cell/nearest-cell.cgi web site. The V7, NCDist (‘NCD’), 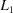 and
and 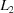 results are from SAUC.
results are from SAUC.
| PDB ID | N-C | 3(N-C)1/2 | NCD | V7 | L 1 | L 2 | Molecule | EC code |
|---|---|---|---|---|---|---|---|---|
| 1u4j* | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | Phospholipase A2 isoform 2 | 3.1.1.4 |
| 1g0z | 0 | 0 | 0 | 0 | 0 | 0 | Phospholipase A2 | 3.1.1.4 |
| 1g2x* | 0.1 | 1.0 | 0.9 | 0.2 | 0.4 | 0.5 | Phospholipase A2 | 3.1.1.4 |
| 2osn | 0.9 | 0.2 | 0.4 | 0.5 | Phospholipase A2 isoform 3 | 3.1.1.4 | ||
| 2cmp | 0.3 | 1.7 | 1.5 | 0.7 | 1.3 | 1.5 | Terminase small subunit | |
| 3kp8 | 0.43 | 0.66 | 1.1 | 1.7 | 1.5 | 1.8 | VKORC1/thioredoxin domain protein | |
| 3mij | 1.7 | 1.0 | 0.8 | 0.8 | RNA [5-R(*UP*AP*GP*GP*GP*UP*UP*AP*GP*GP*GP*U)-3] | |||
| 3e56 | 0.4 | 1.9 | 1.9 | 1.6 | 1.5 | 1.5 | Putative uncharacterized protein | |
| 1csq | 0.5 | 2.1 | 2.0 | 1.8 | 1.2 | 1.6 | Cold shock protein B (CSPB) | |
| 4den | 2.1 | 2.0 | 2.0 | 2.0 | Actinohivin | |||
| 3svi | 0.5 | 2.2 | 2.1 | 1.9 | 1.5 | 1.8 | Type III effector HopAB2 | |
| 1fkf | 0.8 | 2.7 | 2.4 | 2.7 | 2.4 | 2.9 | FK506 binding protein | 5.2.1.8 |
| 1fkj | 0.8 | 2.7 | 2.4 | 2.7 | 2.4 | 2.9 | FK506 binding protein | 5.2.1.8 |
| 1fkd | 0.9 | 2.8 | 2.5 | 2.8 | 2.5 | 3.0 | FK506 binding protein | 5.2.1.8 |
| 1bkf | 0.9 | 2.9 | 2.5 | 2.8 | 3.1 | 3.4 | Subtilisin Carlsberg | 3.4.21.62 |
| 2fke | 0.9 | 2.9 | 2.6 | 3.0 | 2.5 | 3.1 | FK506 binding protein | 5.2.1.8 |
| 3tjy | 0.9 | 2.8 | 2.6 | 3.0 | 2.5 | 2.9 | Effector protein HopAB3 | |
| 2i5l | 1.1 | 3.1 | 2.7 | 3.7 | 4.3 | 4.4 | Cold shock protein CSPB | |
| 3p63 | 1.3 | 3.4 | 2.9 | 4.0 | 4.4 | 4.9 | Ferredoxin | |
| 2wce | 1.2 | 3.3 | 3.0 | 3.5 | 5.9 | 6.4 | Protein S100-A12 | |
| 4sga | 1.4 | 3.5 | 3.1 | 4.8 | 5.7 | 5.8 | Proteinase A (SGPA) | 3.4.21.80 |
| 2cxd | 1.4 | 3.5 | 3.1 | 4.8 | 4.7 | 5.4 | Conserved hypothetical protein, TTHA0068 | |
| 5sga | 1.4 | 3.5 | 3.1 | 4.9 | 5.8 | 5.9 | Proteinase A (SGPA) | 3.4.21.80 |
| 2sga | 1.4 | 3.6 | 3.1 | 4.9 | 5.8 | 5.9 | Proteinase A | 3.4.21.80 |
| 3sga | 1.4 | 3.6 | 3.1 | 5.0 | 5.9 | 6.0 | Proteinase A (SGPA) | 3.4.21.80 |
| 1f9p | 1.4 | 3.5 | 3.1 | 4.7 | 4.1 | 4.8 | Connective tissue activating peptide III | |
| 2yzu | 3.1 | 4.8 | 6.3 | 6.7 | Thioredoxin | |||
| 1sgc | 1.5 | 3.7 | 3.2 | 5.2 | 6.2 | 6.3 | Proteinase A | 3.4.21.80 |
| 1pkr | 1.4 | 3.6 | 3.2 | 4.7 | 3.9 | 4.9 | Plasminogen | 3.4.21.7 |
| 1gus | 1.2 | 3.2 | 3.2 | 0.3 | 3.4 | 4.6 | Molybdate binding protein II | |
| 2vri | 1.5 | 3.7 | 3.2 | 5.2 | 5.0 | 5.7 | Non-structural protein 3 | 3.4.19.12 |
| 2cvk | 3.2 | 5.2 | 7.0 | 7.3 | Thioredoxin | |||
| 2c9q | 1.5 | 3.6 | 3.3 | 4.8 | 4.6 | 4.9 | Copper resistance protein C | |
| 1fe5* | 1.2 | 3.3 | 3.3 | 2.3 | 6.0 | 7.0 | Phospholipase A2 | 3.1.1.4 |
| 1dpy | 1.2 | 3.3 | 3.3 | 2.3 | 6.0 | 7.0 | Phospholipase A2 | 3.1.1.4 |
| 2he2 | 1.5 | 3.6 | 3.3 | 4.8 | 4.5 | 5.1 | Discs large homolog 2 | |
| 3su6 | 1.5 | 3.7 | 3.3 | 5.0 | 4.5 | 5.4 | NS3 protease, NS4A protein | |
| 1cdc | 3.3 | 4.9 | 4.3 | 5.4 | CD2 | |||
| 1gut | 1.2 | 3.3 | 3.3 | 0.1 | 3.4 | 4.8 | Molybdate binding protein II | |
| 2it5 | 1.6 | 3.8 | 3.3 | 5.6 | 5.7 | 6.4 | CD209 antigen, DCSIGN-CRD | |
| 3su5 | 1.6 | 3.8 | 3.3 | 5.1 | 4.8 | 5.7 | NS3 protease, NS4A protein | |
| 3su1 | 1.6 | 3.8 | 3.3 | 5.2 | 4.7 | 5.7 | Genome polyprotein | |
| 3su2 | 1.6 | 3.8 | 3.3 | 5.2 | 4.7 | 5.6 | Genome polyprotein | |
| 3cyo | 3.4 | 5.6 | 7.6 | 7.9 | Transmembrane protein | |||
| 1sl4 | 1.7 | 3.9 | 3.4 | 5.8 | 6.2 | 6.8 | mDC-SIGN1B type I isoform | |
| 3su3 | 1.6 | 3.8 | 3.4 | 5.3 | 4.9 | 5.9 | NS3 protease, NS4A protein | |
| 2it6 | 1.7 | 3.9 | 3.4 | 6.0 | 6.4 | 7.0 | CD209 antigen | |
| 3sv7 | 1.7 | 3.9 | 3.5 | 5.6 | 5.5 | 6.4 | NS3 protease, NS4A protein |
(c) The V7 distances based on individual components linearized to ångström units,
and scaled by 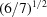 to adjust for the change in dimensionality. V is the volume. As with the NCDist scaling, as seen in Table 2 ▶ this scaling provides a rough approximation to the
to adjust for the change in dimensionality. V is the volume. As with the NCDist scaling, as seen in Table 2 ▶ this scaling provides a rough approximation to the 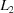 distance in the 1–2 Å range.
distance in the 1–2 Å range.
These metrics are applied to reduced primitive cells 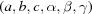 and, when the reciprocal cell
and, when the reciprocal cell 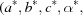
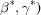 is needed for the V7 metric, that cell is also reduced.
is needed for the V7 metric, that cell is also reduced.
In order to facilitate comparisons with older searches that just consider simple ranges in 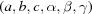 , an option for performing such searches is also included in SAUC.
, an option for performing such searches is also included in SAUC.
4.1. Validity of using the square root
The use of the square root on a metric preserves the triangle inequality, which is important in order to conform to the rules of metric spaces (Fréchet, 1906 ▶). This allows us to work with more general distance functions, such as NCDist, rather than just those that can be expressed as simple Euclidean distances. The triangle inequality states that, for any triangle, the sum of the lengths of any two sides is greater than the length of the third side. In metric space terms, the metric 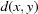 of a metric space M satisfies
of a metric space M satisfies 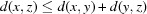 ,
, 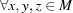 . Suppose a function f satisfies the following conditions:
. Suppose a function f satisfies the following conditions:
Then, if 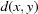 satisfies the triangle inequality,
satisfies the triangle inequality, 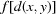 will also satisfy the triangle inequality:
will also satisfy the triangle inequality:
The square root satisfies the stated requirements. It is monotone, and
which is clearly true.
5. Implementation: 2 – searching
Range searching in a mapped embedding needs to be done using a nearest-neighbor algorithm (or ‘post-office problem’ algorithm; Knuth, 1973 ▶). Exact matches are unlikely since most unit cells representing lattices in a database are experimental, and probe cells are also likely to have been calculated from experimental data. Several efficient nearest-neighbor algorithms are available; we have used an implementation of NearTree (Andrews, 2001 ▶; http://sf.net/projects/neartree).
In order to insert new data into the database, each new cell must be examined and compared with some appropriate subset of the already inserted cells in order to place the new cell in the right place. In other words, there is a search of the database to do for each insertion. If there are N cells in the database, the typical time for a search of a tree-based database is proportional to the logarithm of N, so we say the search time is 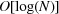 , read as ‘big Oh of log N’, and the time to load the entire database by building the entire tree is
, read as ‘big Oh of log N’, and the time to load the entire database by building the entire tree is 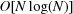 , read as ‘big Oh of N log N’. The raw unit cell data are loaded into the tree once and serialized to a dump file on disk; subsequent searches do not need to wait for the
, read as ‘big Oh of N log N’. The raw unit cell data are loaded into the tree once and serialized to a dump file on disk; subsequent searches do not need to wait for the 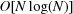 build time of NearTree, which for the 70 000+ cells from the Protein Data Bank (PDB; Bernstein et al., 1977 ▶; Berman et al., 2000 ▶) can take half an hour in the BGAOL NCDist metric. The linearization makes the search space more compact and reduces the tree depth, thereby speeding searches. Because the PDB unit-cell database contains many identical cells, we modified NearTree to handle the duplicates in auxiliary lists, thereby further reducing the tree depth and speeding up searches.
build time of NearTree, which for the 70 000+ cells from the Protein Data Bank (PDB; Bernstein et al., 1977 ▶; Berman et al., 2000 ▶) can take half an hour in the BGAOL NCDist metric. The linearization makes the search space more compact and reduces the tree depth, thereby speeding searches. Because the PDB unit-cell database contains many identical cells, we modified NearTree to handle the duplicates in auxiliary lists, thereby further reducing the tree depth and speeding up searches.
6. Comparison of search methods
The simplest approach to lattice searching is a straightforward box search on ranges in unit-cell edge lengths a, b and c and possibly on unit-cell angles α, β and γ, as for example in the ‘cell dimensions’ option in the RCSB advanced search at http://www.rcsb.org/pdb/search/advSearch.do for the PDB. In the following examples, we will call that type of search ‘range’. For the reasons discussed above, such simple searches can fail to find unit cells with very different angles that actually represent similar lattices. Such searches are best characterized as cell searches rather than as lattice searches.
Searching on primitive reduced cells greatly improves the reliability of a search, as for example in Nearest-Cell (Ramraj et al., 2011 ▶), which uses a metric based on the reduced cell and all permutations of axes. While an improvement over simple range searches as discussed above, such searches can also miss similar lattices if the number of alternative lattice presentations considered is not complete. One way to reduce such gaps in searches is to use only parameters that do not depend on the choice of reduced presentation. The approach of Andrews et al. (1980 ▶) using seven parameters (three reduced cell edges, three reduced reciprocal cell edges and the volume), ‘V7’, helps, but has difficulty distinguishing cells with angles near 90°. The NCDist approach used here, derived from the work of Andrews & Bernstein (2012 ▶, 2014 ▶), both fills in the gaps and handles angles near 90°.
Consider, for example, the unit cells of phospholipase A2 discussed by Le Trong & Stenkamp (2007 ▶). They present three alternative cells from three different PDB entries that are actually for the same structure: (57.98, 57.98, 57.98, 92.02, 92.02, 92.02) from entry 1fe5 (Singh et al., 2001 ▶) in space group R32, (80.36, 80.36, 99.44, 90, 90, 120) from entry 1u4j (Singh et al., 2005b
▶) in space group R3 and (80.949, 80.572, 57.098, 90.0, 90.35, 90.0) from entry 1g2x (Singh et al., 2005a
▶) in space group C2. No simple range search can bring these three cells together. For example, if we use the PDB advanced cell dimensions search around the cell from 1u4j with edge ranges of 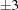 Å and angle ranges of
Å and angle ranges of 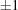 °, we get 28 hits: 1cg5, 1cnv, 1fw2, 1g0z, 1gs7, 1gs8, 1hau, 1ild, 1ilz, 1im0, 1lr0, 1ndt, 1oe1, 1oe2, 1oe3, 1qd5, 1u4j, 2bm3, 2bo0, 2h8a, 2hz5, 2ohg, 2rew, 2wce, 3i06, 3kku, 3q98, 3rp2, of which only three actually have cells close to the target using the linearized NCDist metric: 2wce at 2.96 Å, 1g0z at 0 Å and 1u4j, the target itself. The remaining cells are, as we will see, rejected under the Nearest-Cell and the V7 metric. The simple range searches are not appropriate to this problem.
°, we get 28 hits: 1cg5, 1cnv, 1fw2, 1g0z, 1gs7, 1gs8, 1hau, 1ild, 1ilz, 1im0, 1lr0, 1ndt, 1oe1, 1oe2, 1oe3, 1qd5, 1u4j, 2bm3, 2bo0, 2h8a, 2hz5, 2ohg, 2rew, 2wce, 3i06, 3kku, 3q98, 3rp2, of which only three actually have cells close to the target using the linearized NCDist metric: 2wce at 2.96 Å, 1g0z at 0 Å and 1u4j, the target itself. The remaining cells are, as we will see, rejected under the Nearest-Cell and the V7 metric. The simple range searches are not appropriate to this problem.
Table 2 ▶ shows partial results from a lattice search using Nearest-Cell compared to results from NCDist, V7, 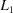 and
and 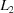 searches using SAUC. The searches were first done in May 2013 and then redone in October 2013, because Nearest-Cell had been improved. The results reported here are from October 2013. We have restricted the searches to NCDist distances
searches using SAUC. The searches were first done in May 2013 and then redone in October 2013, because Nearest-Cell had been improved. The results reported here are from October 2013. We have restricted the searches to NCDist distances 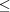 3.5 Å. The Nearest-Cell metric appears to be in Å2. An extra column with three times the square root of the Nearest-Cell metric has been introduced to facilitate comparison with the linearized SAUC V7 and NCDist metrics. The searches showed consistent behavior: the three cells noted by Le Trong & Stenkamp (2007 ▶) are found in the same relative positions by all the searches. All cells found by Nearest-Cell are also found by all the SAUC searches. Of the 48 structures found by all three metrics within 3.5 Å under the NCDist metric, four (1g0z, 1g2x, 1dpy and 1fe5) are EC class 3.1.1.4 phospholipase A2 structures, and three (1pkr, 1sgc and 1vri) are other hydrolases (EC classes 3.4.21.7, 3.4.21.80 and 3.4.19.2, respectively). However, six cells found by NCDist, V7,
3.5 Å. The Nearest-Cell metric appears to be in Å2. An extra column with three times the square root of the Nearest-Cell metric has been introduced to facilitate comparison with the linearized SAUC V7 and NCDist metrics. The searches showed consistent behavior: the three cells noted by Le Trong & Stenkamp (2007 ▶) are found in the same relative positions by all the searches. All cells found by Nearest-Cell are also found by all the SAUC searches. Of the 48 structures found by all three metrics within 3.5 Å under the NCDist metric, four (1g0z, 1g2x, 1dpy and 1fe5) are EC class 3.1.1.4 phospholipase A2 structures, and three (1pkr, 1sgc and 1vri) are other hydrolases (EC classes 3.4.21.7, 3.4.21.80 and 3.4.19.2, respectively). However, six cells found by NCDist, V7, 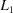 and
and 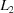 in SAUC were not found by Nearest-Cell (2osn, 3mij, 4den, 2yzu, 1cdc and 2cvk). Of those six, one (2osn) is an EC class 3.1.1.4 phospholipase A2 structure. In the early Nearest-Cell search in May 2013, prior to the release of SAUC, Nearest-Cell also failed to find five additional cells (2cmp, 2sga, 3sga, 4sga and 5sga). Of those five, four (2sga, 3sga, 4sga and 5sga) are hydrolases, specifically EC class 3.4.21.80 proteinase A. Two of the still missing six (2yzu and 2cvk) are thioredoxin, for which the ProMOL (Craig et al., 2013 ▶) motif finder shows significant active site homologies to multiple hydrolase motifs (2yzu has site homologies to 132l, 135l and 1lz1 in EC class 3.2.1.17 and to 4hoh in EC class 3.1.27.3, 2cvk to 1amy in EC class 3.2.1.1, to 1bf2 in EC class 3.2.1.68, to 1eyi in class 3.2.3.11 etc.). For 1cdc, a ‘metastable structure of CD2’, ProMOL shows an active site homology to 1alk of EC class 3.1.3.1, another hydrolase.
in SAUC were not found by Nearest-Cell (2osn, 3mij, 4den, 2yzu, 1cdc and 2cvk). Of those six, one (2osn) is an EC class 3.1.1.4 phospholipase A2 structure. In the early Nearest-Cell search in May 2013, prior to the release of SAUC, Nearest-Cell also failed to find five additional cells (2cmp, 2sga, 3sga, 4sga and 5sga). Of those five, four (2sga, 3sga, 4sga and 5sga) are hydrolases, specifically EC class 3.4.21.80 proteinase A. Two of the still missing six (2yzu and 2cvk) are thioredoxin, for which the ProMOL (Craig et al., 2013 ▶) motif finder shows significant active site homologies to multiple hydrolase motifs (2yzu has site homologies to 132l, 135l and 1lz1 in EC class 3.2.1.17 and to 4hoh in EC class 3.1.27.3, 2cvk to 1amy in EC class 3.2.1.1, to 1bf2 in EC class 3.2.1.68, to 1eyi in class 3.2.3.11 etc.). For 1cdc, a ‘metastable structure of CD2’, ProMOL shows an active site homology to 1alk of EC class 3.1.3.1, another hydrolase.
The significant gaps in the Nearest-Cell search do not appear to be a result of the distance for the Nearest-Cell search having been cut off at too small a value. For the common hits between the square root of the Nearest-Cell metric and the linearized NCDist metric, a linear fit is excellent, with 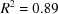 , and no points are very far from the line. The agreement of the linearized V7 with the other two metrics is much noisier because of loss of sensitivity of the V7 metric for angles near 90° and the inherent difficulty the V7 metric has in discriminating between the
, and no points are very far from the line. The agreement of the linearized V7 with the other two metrics is much noisier because of loss of sensitivity of the V7 metric for angles near 90° and the inherent difficulty the V7 metric has in discriminating between the 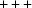 and
and  parts of the Niggli cone. For example, 1gut (Schüttelkopf et al., 2002 ▶) is at a distance of 3.3 from 1uj4 in both the Nearest-Cell and linearized NCDist metrics, respectively, but only 0.1 in the V7 metric. The 1gut cell is (78.961, 82.328, 57.031, 90.00, 93.44, 90.00) in C121, Z = 24, with a primitive cell (57.031, 57.0367, 57.0367, 92.3918, 92.3804, 92.3804), which corresponds to a
parts of the Niggli cone. For example, 1gut (Schüttelkopf et al., 2002 ▶) is at a distance of 3.3 from 1uj4 in both the Nearest-Cell and linearized NCDist metrics, respectively, but only 0.1 in the V7 metric. The 1gut cell is (78.961, 82.328, 57.031, 90.00, 93.44, 90.00) in C121, Z = 24, with a primitive cell (57.031, 57.0367, 57.0367, 92.3918, 92.3804, 92.3804), which corresponds to a  vector (3252.53, 3253.18, 3253.18, −271.53, −270.208, −270.208) and a linearized V7 vector (52.8004, 52.8057, 52.8057, 52.7101, 52.7101, 52.7053, 52.7569).The 1u4j cell is (80.36, 80.36, 99.44, 90, 90, 120) in R3, Z = 18, with a primitive cell (57.02, 57.02, 57.02, 89.605, 89.605, 89.605), which corresponds to a
vector (3252.53, 3253.18, 3253.18, −271.53, −270.208, −270.208) and a linearized V7 vector (52.8004, 52.8057, 52.8057, 52.7101, 52.7101, 52.7053, 52.7569).The 1u4j cell is (80.36, 80.36, 99.44, 90, 90, 120) in R3, Z = 18, with a primitive cell (57.02, 57.02, 57.02, 89.605, 89.605, 89.605), which corresponds to a 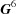 vector (3251.28, 3251.28, 3251.28, 44.8265, 44.8265, 44.8265) and a linearized V7 vector (52.7902, 52.7902, 52.7902, 52.7878, 52.7878, 52.7878, 52.789). This is almost identical to the 1gut V7 vector, even though the corresponding primitive cells and
vector (3251.28, 3251.28, 3251.28, 44.8265, 44.8265, 44.8265) and a linearized V7 vector (52.7902, 52.7902, 52.7902, 52.7878, 52.7878, 52.7878, 52.789). This is almost identical to the 1gut V7 vector, even though the corresponding primitive cells and 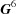 cells differ significantly.
cells differ significantly.
The results for the 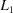 and
and 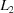 norms are problematic for database use. Notice the large discontinuity in distances for both the
norms are problematic for database use. Notice the large discontinuity in distances for both the 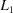 and
and 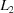 distances between 3tjy and 2i5l. There are 13 ‘misplaced’ cells in the gap in the
distances between 3tjy and 2i5l. There are 13 ‘misplaced’ cells in the gap in the 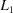 distance ordering and eight ‘misplaced’ cells in the gap in the
distance ordering and eight ‘misplaced’ cells in the gap in the 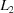 ordering. These cells are misplaced in the sense that, because these distance functions do not consider the reduction ambiguities in database searches, these searches are inserting a large number of false positives among the true positives, making search results much harder to use effectively.
ordering. These cells are misplaced in the sense that, because these distance functions do not consider the reduction ambiguities in database searches, these searches are inserting a large number of false positives among the true positives, making search results much harder to use effectively.
7. SAUC program availability
SAUC is an open-source program released under the GPL and LGPL on Sourceforge in the iterate project at http://sf.net/projects/iterate/.
Specifically, a recent release is available at http://downloads.sf.net/iterate/sauc-0.6.4.tar.gz.
A web site on which searches may be done and from which the latest release may be retrieved is located at http://iterate.sf.net/sauc.
Snapshots of this web site are shown in Figs. 1 ▶ and 2 ▶.
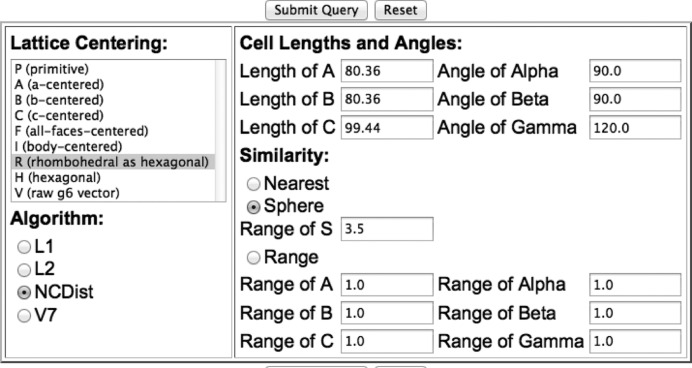
Figure 1.
Query box from the SAUC web site at http://iterate.sf.net/sauc.
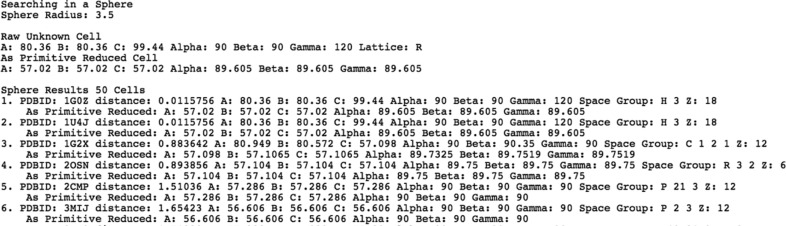
Figure 2.
Partial results from a SAUC web site query.
Acknowledgments
The authors acknowledge the invaluable assistance of Frances C. Bernstein. The work by HJB, KJM, MA and MTK has been supported in part by NIH NIGMS grant GM078077. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agency. LCA would like to thank Frances and Herbert Bernstein for hosting him during hurricane Sandy and its aftermath. Elizabeth Kincaid has contributed significant support in many ways. Our thanks to Ronald E. Stenkamp for pointing us to the highly relevant work of Le Trong & Stenkamp (2007 ▶). We wish to express our deepest gratitude to the editors and referees for very helpful suggestions, consultations and cooperation.
References
- Allen, F. H., Kennard, O., Motherwell, W. D. S., Town, W. G. & Watson, D. G. (1973). J. Chem. Doc. 13, 119–123.
- Andrews, L. (2001). C/C++ Users J. 19, 40–49.
- Andrews, L. C. & Bernstein, H. J. (2012). arXiv: 1203.5146. http://arxiv.org/abs/1203.5146.
- Andrews, L. C. & Bernstein, H. J. (2014). J. Appl. Cryst. 47, 346–359. [DOI] [PMC free article] [PubMed]
- Andrews, L. C., Bernstein, H. J. & Pelletier, G. A. (1980). Acta Cryst. A36, 248–252.
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Bernstein, F. C., Koetzle, T. F., Williams, G. J., Meyer, E. F., Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T. & Tasumi, M. (1977). J. Mol. Biol. 112, 535–542. [DOI] [PubMed]
- Burchard, U. (1998). Mineral. Rec. 29, 517–583.
- Craig, P. A., Hanson, B., Westin, C., Rosa, M., Bernstein, H. J., Grier, A., Osipovitch, M., MacDonald, M., Dodge, G., Boli, P. M., Corwin, C. W. & Kessler, H. (2013). BMC Bioinformatics. Submitted. [DOI] [PMC free article] [PubMed]
- Donnay, J. D. H. (1943). Am. Mineral. 28, 313–327.
- Fréchet, M. M. (1906). Rend. Circ. Mater. Palermo, 22, 1–72.
- Johnson, G. G. (2013). Personal communication.
- Knuth, D. E. (1973). Sorting and Searching. The Art of Computer Programming, Vol. 3. Reading: Addison Wesley.
- Le Trong, I. & Stenkamp, R. E. (2007). Acta Cryst. D63, 548–549. [DOI] [PubMed]
- Mighell, A. D. (2001). J. Res. Natl Inst. Stand. Technol. 106, 983–996. [DOI] [PMC free article] [PubMed]
- Mighell, A. D. (2002). J. Res. Natl Inst. Stand. Technol. 107, 425–430. [DOI] [PMC free article] [PubMed]
- NIH/EPA (1980). User’s Manual NIH-EPA Chemical Information System, ch. User’s Guide to Cryst The X-ray Crystallographic Search System National Institutes of Health, Environmental Protection Agency, Washington, DC, USA.
- Pearson, W. B. (1958). Handbook of Lattice Spacings and Structures of Metals and Alloys. International Series of Monographs on Metal and Physics and Physical Metallurgy, edited by G. V. Raynor. Oxford: Pergamon Press.
- Ramraj, V., Esnouf, R. & Diprose, J. (2011). Nearest-Cell. A Fast and Easy Tool for Locating Crystal Matches in the PDB. Technical Report, Division of Structural Biology, University of Oxford, UK. http://www.strubi.ox.ac.uk/nearest-cell/nearest-cell.cgi. [DOI] [PMC free article] [PubMed]
- Schüttelkopf, A. W., Harrison, J. A., Boxer, D. H. & Hunter, W. N. (2002). J. Biol. Chem. 277, 15013–15020. [DOI] [PubMed]
- Singh, G., Gourinath, S., Saravanan, K., Sharma, S., Bhanumathi, S., Betzel, C., Srinivasan, A. & Singh, T. P. (2005a). Acta Cryst. F61, 8–13. [DOI] [PMC free article] [PubMed]
- Singh, G., Gourinath, S., Sarvanan, K., Sharma, S., Bhanumathi, S., Betzel, Ch., Yadav, S., Srinivasan, A. & Singh, T. P. (2005b). J. Struct. Biol. 149, 264–272. [DOI] [PubMed]
- Singh, G., Gourinath, S., Sharma, S., Paramasivam, M., Srinivasan, A. & Singh, T. P. (2001). J. Mol. Biol. 307, 1049–1059. [DOI] [PubMed]
- Thomas, I. R., Bruno, I. J., Cole, J. C., Macrae, C. F., Pidcock, E. & Wood, P. A. (2010). J. Appl. Cryst. 43, 362–366. [DOI] [PMC free article] [PubMed]
- Toby, B. (1994). Personal communication.
- Wyckoff, R. W. G. (1931). The Structure of Crystals, No. 19. New York: The Chemical Catalog Company.