Abstract
We have identified the single Caenorhabditis elegans focal adhesion kinase (FAK) homolog KIN-32, which has the signature FAK structure including an N-terminal Four.1-Ezrin-Radixin-Moesin (FERM) domain followed by a tyrosine kinase domain and a C-terminal domain with weak homology to the focal adhesion targeting domain. The functional requirements for KIN-32 were examined using RNA interference depletion experiments and analysis of a deletion allele, kin-32(ok166), in which a large segment of the FERM domain is missing. Our results show that reduced levels of expression or absence of the FERM domain do not affect viability, fertility, or anatomy in C. elegans. Expression of an analogous FERM deletion in mouse FAK showed kinase activity in vitro and supported normal focal adhesion localization in cell culture. Thus, the FERM domain of KIN-32, and possibly KIN-32 activity in general, appears to be dispensable for normal C. elegans physiology.
Keywords: FERM, kinase, nematode, RNAi, FAK
INTRODUCTION
Interactions between cells and their extracellular environment play essential roles in controlling tissue architecture, cell survival, and cell migration. Cell–extracellular matrix (ECM) interactions are mediated by integrins, a large family of αβ heterodimeric transmembrane proteins (Hynes, 2002). Connections between the ECM and the actin cytoskeleton occur through integrin receptor clustering at focal adhesions, and mediate cell adhesion and changes in cell morphology. Integrins; cytoskeletal adaptor proteins such as vinculin, talin, and integrin linked kinase (ILK); and signaling molecules such as focal adhesion kinase (FAK) and Src are major constituents of these structures (Lo, 2006).
Dense bodies are the nematode equivalent of the vertebrate focal adhesion and contain many homologous proteins (Cox and Hardin, 2004). Loss of dense body proteins such as pat-2/α-integrin, pat-3/β-integrin, deb-1/vinculin, pat-4/ILK, unc-112/Mig2, unc-97/PINCH, or pat-6/actopaxin results in embryonic lethality (Williams and Waterston, 1994; Gettner et al., 1995; Benian et al., 1996; Riddle et al., 1997; Norman and Moerman, 2000; Mackinnon et al., 2002; Lin et al., 2003). In Caenorhabditis elegans, integrins are essential for embryonic development, muscle cell adhesion and contraction, axon outgrowth, and migration of gonadal distal tip cells (DTCs; Gettner et al., 1995; Baum and Garriga, 1997; Lee et al., 2001).
FAK is a cytoplasmic protein tyrosine kinase that is rapidly activated by integrin-ligand interactions and plays a central role in integrin-mediated signaling in mammalian cells (Mitra et al., 2005). The importance of FAK in mediating signals from ECM is illustrated by FAK−/− mice, which exhibit a null phenotype similar to fibronectin−/− animals (Ilic et al., 1995). These mice die during the early stages of embryogenesis due to defects in mesodermal development, and cells isolated from FAK−/− embryos exhibit migration defects in cell culture. FAK family proteins are composed of three domains: the N-terminal erythrocyte band Four.1-Ezrin-Radixin-Moesin (FERM) domain that mediates interactions with transmembrane proteins, a tyrosine kinase domain, and the C-terminal Focal Adhesion Targeting (FAT) domain (Mitra et al., 2005).
In this study, we have characterized the C. elegans FAK homolog KIN-32 using RNA interference (RNAi) depletion and analysis of a deletion allele, kin-32(ok166), which lacks a large part of the FERM domain. Our results show that kin-32 is not an essential gene in C. elegans.
RESULTS
KIN-32 Is the C. elegans FAK Homolog
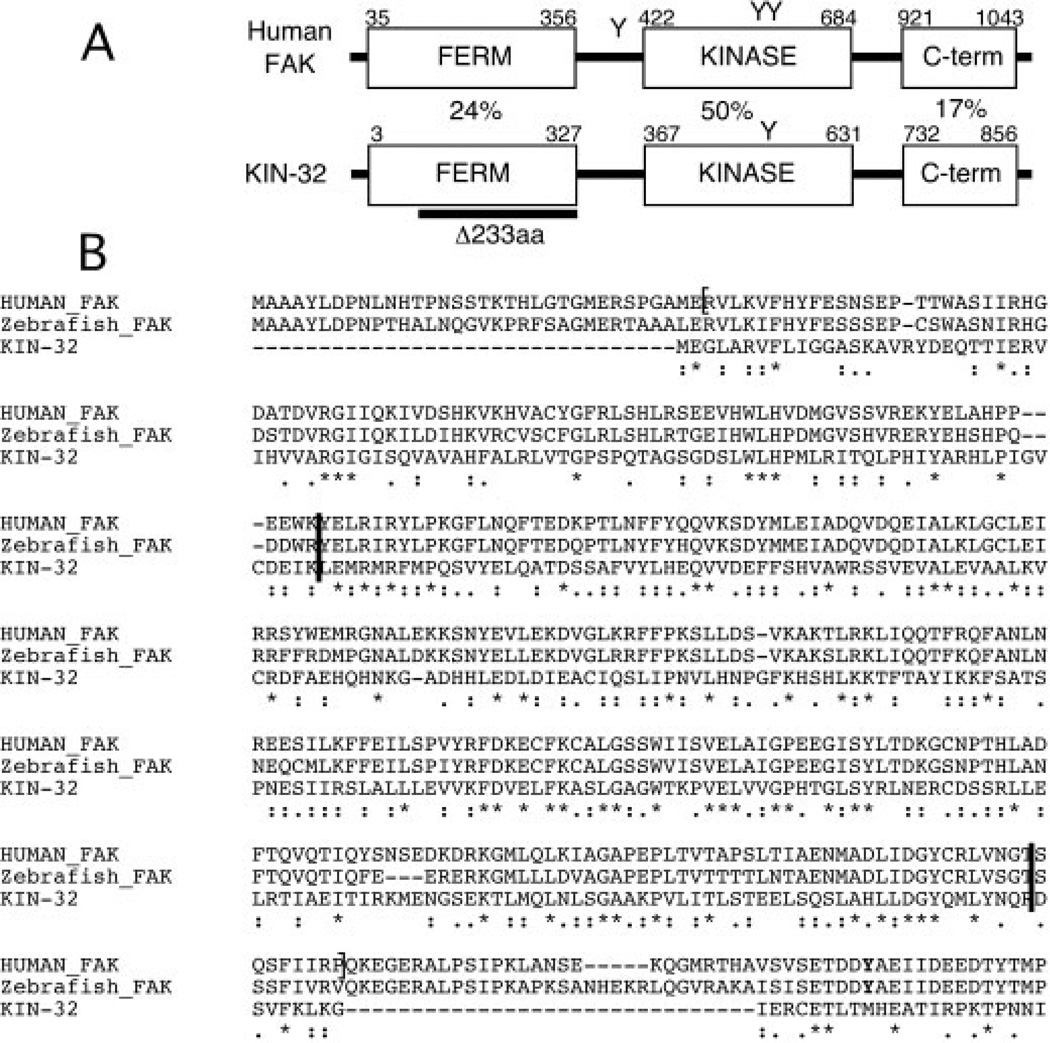
Using BLAST homology searches, we identified KIN-32 as the most similar protein to human FAK in the nematode C. elegans. The C. elegans homolog of c-Abl, ABL-1, is the next most similar protein, but has significant homology only in the kinase domain. The kin-32 gene has an overall sequence identity of 33% and 53% overall similarity to human FAK. KIN-32 has the conserved domain structure common to FAK-related proteins: an N-terminal FERM domain, a central tyrosine kinase domain and a C-terminal domain with similarity to the FAT domain (Fig. 1A).
Fig. 1.
Domain structure and alignment of Caenorhabditis elegans KIN-32. A: Schematic diagram of human focal adhesion kinase (FAK) and C. elegans KIN-32, indicating the conserved Four.1-Ezrin-Radixin-Moesin (FERM), kinase, and Focal Adhesion Targeting (FAT) domain structure. Tyrosines 397, 576, and 577 are indicated (Y). Only Y577 is conserved in KIN-32. Percentage amino acid identity in each of the three domains is indicated between the diagrams. The 233 amino acid region deleted in kin-32(ok166) is indicated (dark bar). B: Alignment of the predicted protein sequence of the N-terminus of FAK from human, zebrafish and C. elegans KIN-32. The FERM domain is bracketed on the human sequence (Ceccarelli et al., 2006). Asterisks indicate identity, and colons and periods similarity of amino acids. The region between the two dark bars is deleted in kin-32(ok166). Tyrosine 397 is indicated in bold near the end of the alignment.
We used the programs ClustalW (Chenna et al., 2003) and MotifScan (Sigrist et al., 2002) to investigate the degree of homology between vertebrate and C. elegans FAKs. The ClustalW alignment of the entire FERM domains of human, zebrafish, and C. elegans proteins is depicted in Figure 1B. Our analysis shows amino acid sequence conservation with significant similarity between KIN-32 and the human FAK FERM domain (E value = 2.4e-37) and between protein tyrosine kinase domains (E value = 5.9e-113). Lower sequence homology was detected between KIN-32 and FAK (E value = 0.0022) FAT domains.
In addition to conserved domain organization, KIN-32 also has the conserved tyrosine Y577, which is a site for phosphorylation by Src kinase (Mitra et al., 2005; Fig. 1A). However, KIN-32 lacks some other features, including a critical amino acid for FAK function in vertebrates, Y397 (bolded in Fig. 1B), and the proline-rich region that follows the kinase domain in vertebrate FAK. Even so, sequence similarity between KIN-32 and FAK suggests that KIN-32 might play a similar role in integrin signaling, cell adhesion, or cell migration in the nematode.
kin-32 Gene Expression Pattern
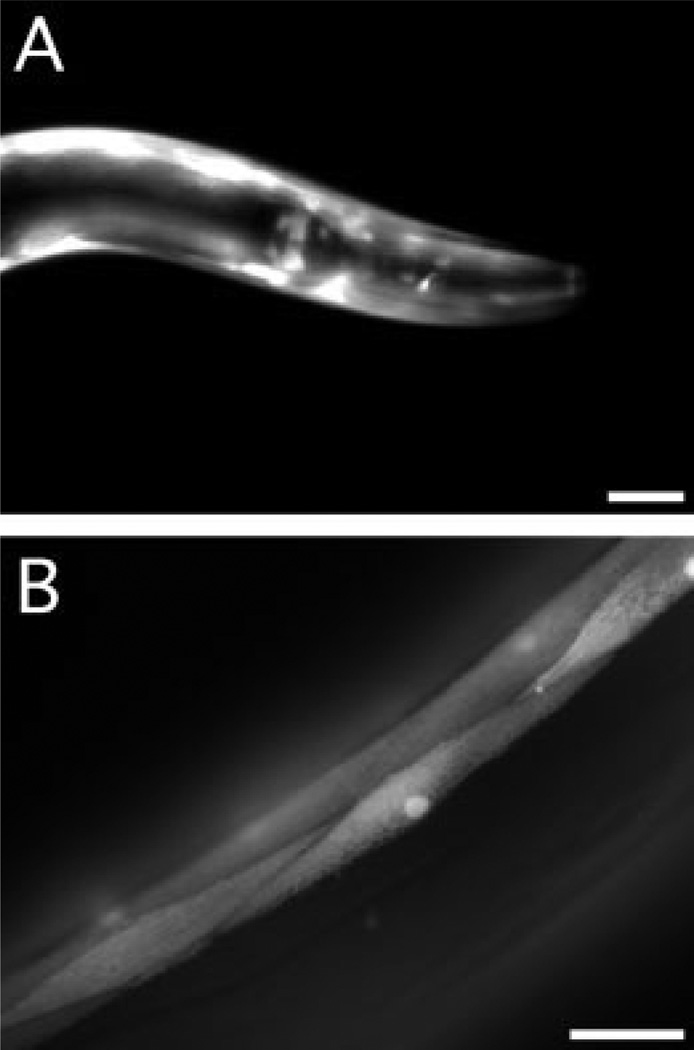
To determine the tissue distribution of KIN-32, we analyzed a nematode strain (BC10568) that expresses green fluorescent protein (GFP) under the control of 2,842 bp of the kin-32 promoter (kin-32::GFP). Fluorescence microscopy of transgenic animals showed that GFP was expressed in all stages starting with late embryos (data not shown). GFP was expressed in the body wall and sex muscles, head mesodermal cell, and various unidentified neurons in the head (Fig. 2A). Body wall muscle of larvae and adults strongly expressed GFP (Fig. 2B). Notably, we saw no GFP expression in the DTC, even in dissected gonad arms (data not shown). These results suggest that KIN-32 is appropriately distributed to interact with INA-1/PAT-3 or PAT-2/PAT-3 integrin heterodimers, which are expressed in the neurons and muscles, respectively, in embryos, larvae, and adults (Gettner et al., 1995; Baum and Garriga, 1997).
Fig. 2.
Expression pattern of the kin-32 promoter in adult hermaphrodites. The green fluorescent protein (GFP) expression pattern was examined by fluorescence microscopy of the kin-32::GFP (BC10568) transcriptional reporter line. A: GFP is seen in the body wall muscles and in unidentified neurons in the head. B: Con-focal microscopy shows GFP expression in the body wall muscles. Scale bars = 20 µm.
kin-32(ok166) Is a Deletion Allele
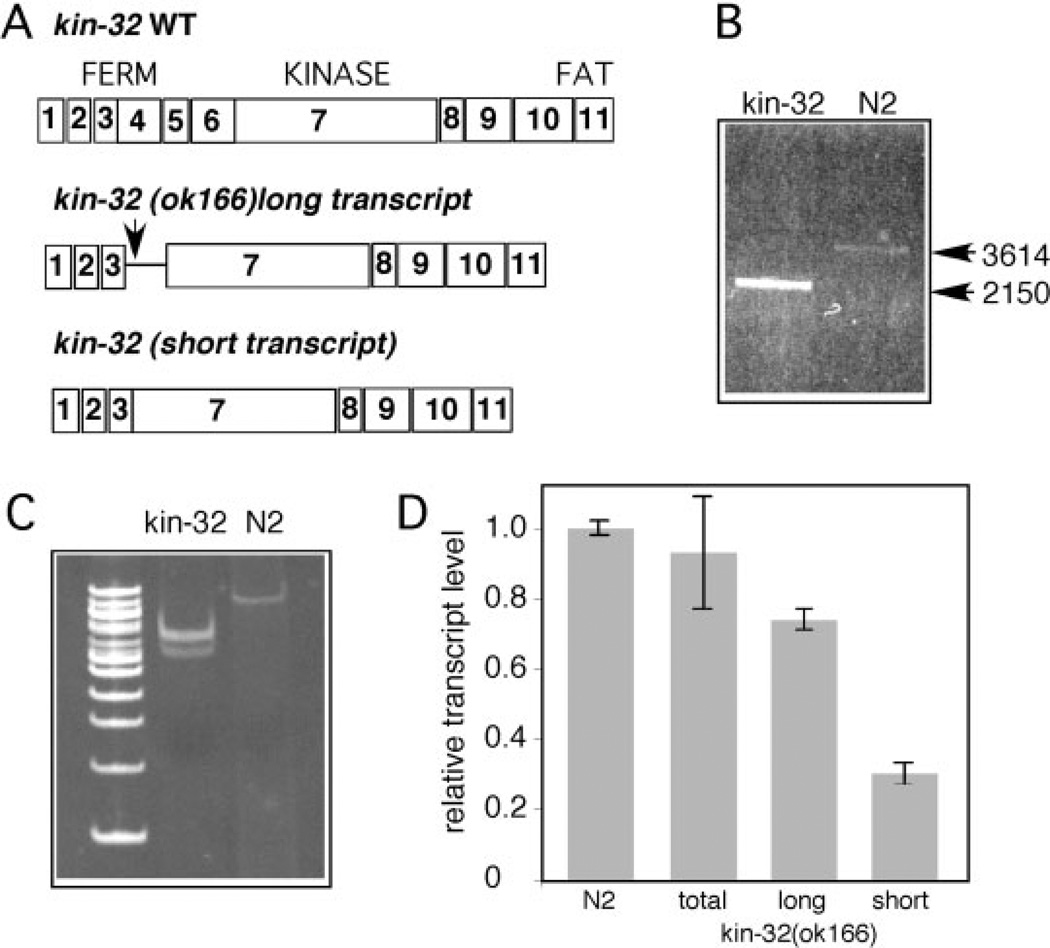
To investigate the role of KIN-32 in nematode development, we obtained a deletion allele, kin-32(ok166). PCR products from kin-32(ok166) genomic DNA revealed a 1,464-bp deletion that removes coding sequence for exons 4, 5, 6, and part of exon 7 (Fig. 3A,B). RT-PCR analysis of mRNA products from mixed-stage populations of N2 and kin-32(ok166) animals revealed two transcripts that differed in the region of the deletion (Fig. 3C). The sequence of the longer transcript encoded a truncated 93 amino acid peptide with sequence from exons 1–3 and terminated in intron 3 with multiple in-frame stop codons (Fig. 3A). The shorter transcript encoded a 634-amino acid protein missing 233 residues from the FERM domain (Figs. 1, 3A). Quantitative reverse transcriptase-polymerase chain reaction (RTPCR) experiments using primers specific to each kin-32(ok166) variant showed somewhat more of the long transcript. Using primers in the first two exons of KIN-32, which are present in mutant and wild-type animals, we found that the overall level of expression of kin-32 mRNA was similar in N2 and kin-32(ok166) animals (Fig. 3D). This analysis suggests that kin-32(ok166) animals produce a KIN-32 protein missing the majority of the FERM domain and probably expressed at a reduced level compared with wild-type KIN-32.
Fig. 3.
Analysis of kin-32(ok166) deletion mutant. A: Schematic of KIN-32, depicting protein domains and exon organization (numbered boxes) of the wild-type (top) and deletion transcripts inferred from cDNA sequencing. Arrow indicates position of a stop codon in the long transcript resulting from inclusion of a portion of intron 3 (thin line) in the kin-32(ok166) message. The short cDNA is predicted to encode exons 1–3 and 7–11. B: Polymerase chain reaction (PCR) of genomic DNA from kin-32(ok166) and N2 wild-type using primers flanking the deleted sequence shows a large deletion. Sizes of amplicons are indicated on the right. C: Reverse transcriptase-PCR (RT-PCR) analysis of total RNA extracted from kin-32(ok166) (lane 2) and N2 (lane 3) animals reveals two shorter messages in kin-32(ok166). Size markers indicate 100-bp intervals (100-bp ladder Invitrogen). D: Quantitative RT-PCR analysis of total RNA extracted from kin-32(ok166) and N2 animals was used to determine average mRNA levels (± SD). Values are normalized to internal controls and wild-type message level in N2 was set to 1. Overall levels of kin-32 mRNA in kin-32(ok166) (“total”) and N2 animals was similar. The long transcript from kin-32(ok166) was present at a higher level than the short mRNA. Data are obtained from three independent experiments.
kin-32(ok166) and kin-32 RNAi Animals Are Essentially Normal
Somewhat to our surprise, the kin-32(ok166) animals are viable, fertile, and exhibit no obvious morphological or behavioral defects. Because kin-32(ok166) is probably not a null allele, we depleted kin-32 by RNAi and looked for loss-of-function phenotypes. We determined by quantitative RT-PCR that RNAi treatment reduced kin-32 transcript level to 59 ± 13% of controls. Similar levels of depletion of talin or integrins result in strong phenotypes (Cram et al., 2003). Using an RNAi-sensitive strain, rrf-3(pk1426) for bacterially mediated RNAi (Simmer et al., 2002), or injection of in vitro synthesized dsRNA, a method for producing stronger RNAi phenotypes (Timmons, 2006), also resulted in no discernable differences from control animals. The kin-32(ok166) animals treated with kin-32 RNAi also showed no obvious phenotypes (data not shown). Neither kin-32(ok166) nor kin-32 RNAi animals had observable anatomical differences from wild-type N2 animals, and no significant differences in fertility, longevity, or heat shock sensitivity were detected (Table 1, Table 2).
TABLE 1.
Comparison of kin-32(ok166) and N2 Phenotypes
| Phenotype scoreda | N2 wild-type (N) | kin-32(ok166) (N) | Other (N) |
|---|---|---|---|
| No. of progeny | 173 ± 69 | 159 ± 30 | |
| Heat shock survival | 71% (100) | 83% (117) | |
| Chemotactic index | 0.50 (497) | 0.58 (412) | 0.10 (465)b |
| Body length, L4 | 0.91 ± 0.18 mm (67) | 0.94 ± 0.17 mm (64) | |
| Body length, young adult | 1.15 ± 0.24 mm (94) | 1.20 ± 0.16 mm (100) | |
| DTC migration defects | 5% (256) | 5% (304) | 7% (688)c |
Methods used to score phenotypes are described in the Experimental Procedures section.
Chemotactic index for eat-4(ky5) control nematodes.
Defective gonadal distal tip cells (DTCs) migration in N2 hermaphrodites treated with kin-32 RNAi.
TABLE 2.
Qualitative Analyses of kin-32(ok166) and Wild-Type Nematodes
| Phenotype scoreda | ok166 | RNA interference |
|---|---|---|
| Longevity | ✓ | |
| Fitnessb | ✓ | |
| Muscle filament and dense body organizationc | ✓ | ✓ |
| Movement/coordinationd | ✓ | ✓ |
| Overall phosphotyrosine levels | ✓ |
Checks indicate no difference in phenotype between mutant or RNA interference–treated nematodes and wild-type N2 animals. No check mark = not determined. Methods for scoring are described in the Experimental Procedures section.
Fitness was assessed by competition assay as described in the text.
N2, kin-32(ok166), and kin-32 RNA interference–treated animals was visualized by fluorescence microscopy of stained young adult animals. See also Figure 4.
No obvious defects in coordination and movement were observed during any of the experiments on kin-32(ok166) or kin-32 RNA interference–treated animals.
To detect a more subtle defect in fitness of kin-32(ok166) animals, kin-32(ok166) hermaphrodites were crossed to N2 males and 100 heterozygous F1 cross progeny picked. Approximately 1,000 animals were transferred to new plates every 3 days for 12 generations, after which PCR analysis across the kin-32 genomic region was performed on 24 randomly selected animals. In that group, 13 had the wild-type gene and 11 had the kin-32(ok166) deletion, not significantly different from equal proportions by χ2 analysis. We conclude that kin-32(ok166) animals show no difference in fitness from wild-type (Table 2). These results indicate that the FERM domain of KIN-32 is not needed for viability or fertility of C. elegans under laboratory conditions.
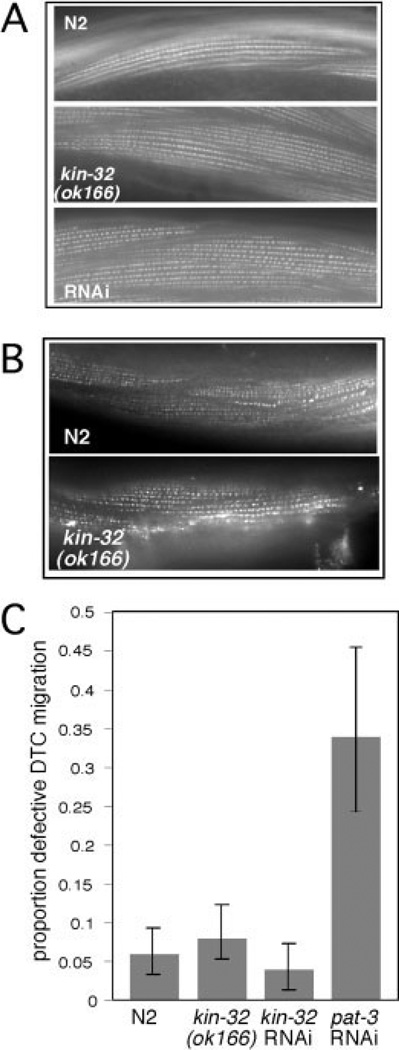
Inhibition of integrin function in larval and adult nematodes results in defects in muscle function, DTC migration, and malformation of the gonad arms (Baum and Garriga, 1997; Lee et al., 2001). Because FAK is a known mediator of integrin signaling in various organisms (Mitra et al., 2005), we tested the mutant and RNAi-treated animals for integrin related defects. We found that the kin-32(ok166) and kin-32 RNAi animals had no detectable defects in muscle cytoskeletal structure as assessed by phalloidin staining of actin filaments (Fig. 4A) and staining with antibodies to muscle filament components α-actinin (Fig. 4B) and UNC-89 (not shown). DTC migration and gonad formation were also normal (Fig. 4C). In addition, kin-32(ok166) and RNAi animals had wild-type organization of muscle dense bodies as assessed by PAT-3/β integrin and ATN-1/α-actinin staining (Table 2). Distribution of the M-line component UNC-89 was also found to be normal in kin-32(ok166) animals (data not shown). Preliminary examination of kin-32(ok166) and kin-32 RNAi animals showed no uncoordinated movement in reversal assays. We also depleted other proteins involved in integrin signaling including pat-3/β integrin, src-1/Src, talin, and pat-4/ILK by bacterially mediated RNAi in N2 and kin-32(ok166) animals. After 48 hr of RNAi treatment, DTC migration was assessed in the RNAi-treated animals. No significant differences between N2 and RNAi-treated kin-32(ok166) animals were found in any of the experiments (Table 1).
Fig. 4.
kin-32(ok166) and kin-32 RNA interference (RNAi) -treated muscles and gonads are not significantly different from N2. A: Rhodamine-conjugated phalloidin was used to stain actin filaments in body wall muscle cells of N2 animals (top), kin-32(ok166) animals (middle), and kin-32 RNAi-treated animals (bottom). B: Anti-α-actinin (MH35) antibodies were used to stain dense body structures in the muscle cells of N2 animals (top) or kin-32(ok166) animals (bottom). C: kin-32(ok166), kin-32 RNAi-treated, and N2 hermaphrodites were examined for defects in gonad formation. Feeding RNAi for pat-3/β-integrin was used as a positive control. Number of gonad arms assayed (N): N2, N = 314; kin-32(ok166) N = 216; kin-32 RNAi N = 196; pat-3 RNAi N=82. Percentage of animals with misshapen gonad arms is indicated by the shaded bars, and the error bars indicate the 95% confidence interval.
Activity of Mouse FAK Does Not Depend on the FERM Domain
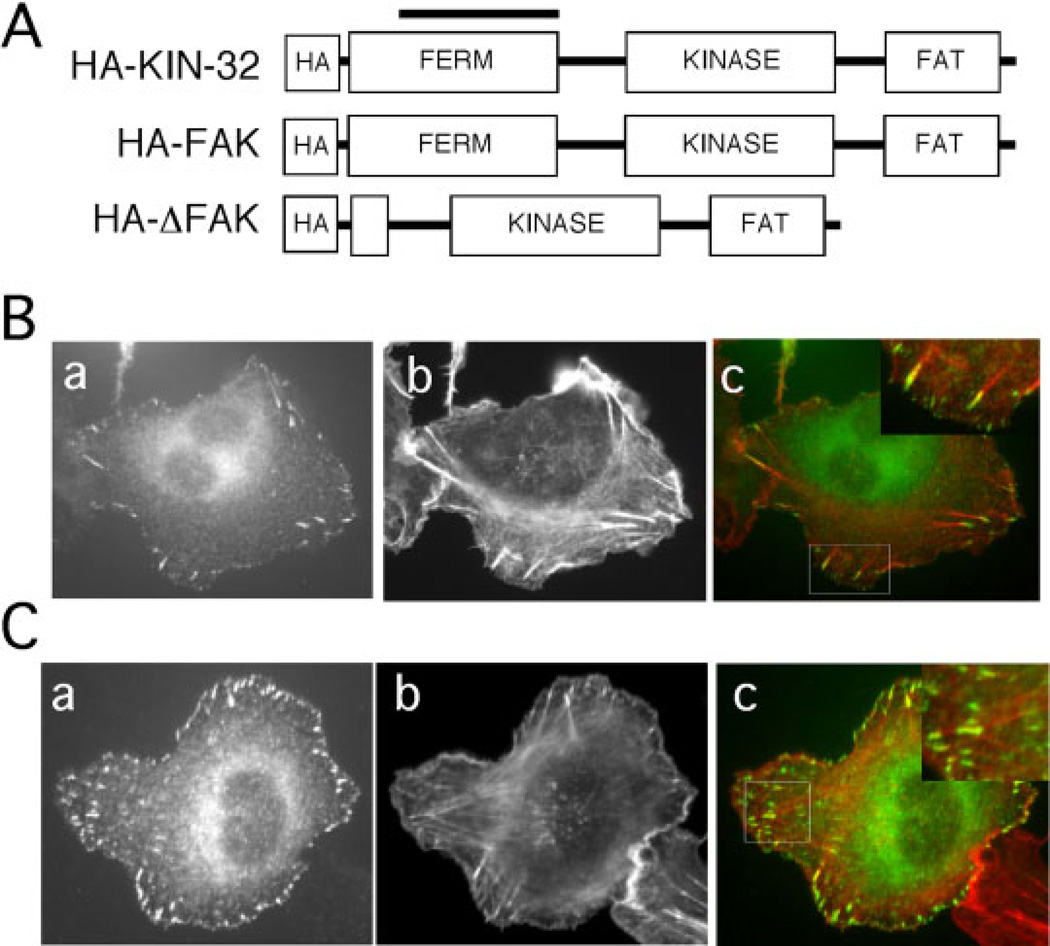
Deletion of 233 amino acids of the FERM domain of KIN-32 in the kin-32(ok166) animals resulted in no measurable behavioral or anatomical defects. To investigate the cell biological function of this region of the protein, and to test whether the deleted region is necessary for proper FAK localization and function in mammalian cells, we constructed the analogous deletion (residues 122–355) in a mouse FAK expression construct (HA-ΔFAK, Fig. 5A). HT1080 fibrosarcoma cell transfectants expressing HA-ΔFAK or a control full-length HA-FAK construct were allowed to spread on fibronectin-coated coverslips for 90 min, then fixed and stained with an anti-HA antibody. Staining was seen in a punctate pattern at points of cell contact with the substratum (Fig. 5B,C), and termini of actin filaments colocalized with anti-HA staining, as is typical of focal adhesions (Fig. 5, merged images). These results demonstrate localization of both HA-FAK and HA-ΔFAK to focal adhesions and show that the FERM domain is not required for focal adhesion localization.
Fig. 5.
Localization of HA-ΔFAK. A: Schematic diagram of mammalian focal adhesion kinase (FAK) and Caenorhabditis elegans KIN-32, indicating the conserved domain structure and the N-terminal HA tag. HA-KIN-32 is the C. elegans cDNA with an HA tag for expression in mammalian cells under the control of the CMV promoter. The line above HA-KIN-32 indicates the location of the ok166 deletion. HA-FAK is full-length mouse FAK, and HA-ΔFAK is a deletion construct analogous to kin-32(ok166) (see Fig. 1B for deleted residues). B,C: HT1080 cells expressing HA-FAK (B) or HA-ΔFAK (C) were plated on fibronectin-coated coverslips and then stained with anti-HA antibodies (a) and rhodamine-phalloidin (b). Merged images are shown in c. Higher magnifications of boxed areas are presented in insets and show termini of actin filaments colocalized with anti-HA staining.
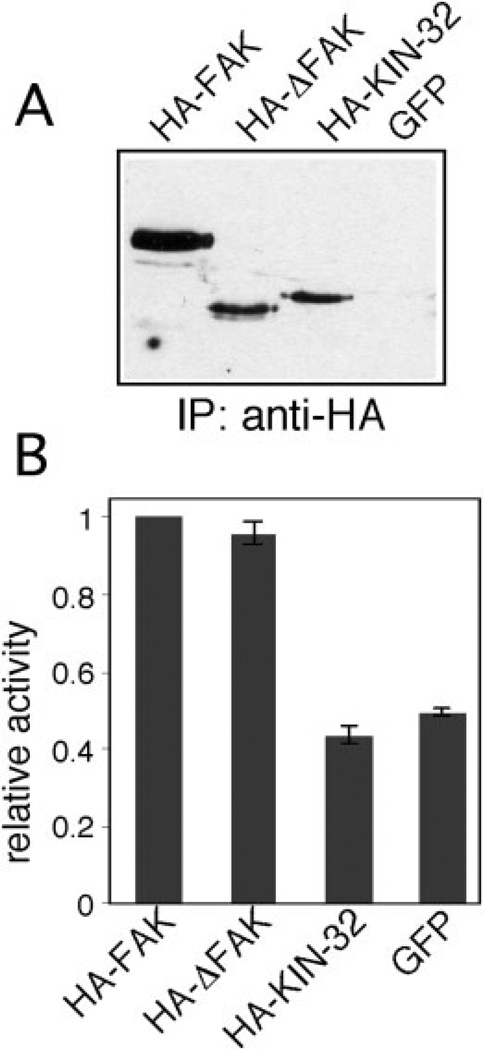
FAK kinase activity is required for many of its functions in vivo (Mitra et al., 2005). To investigate the enzymatic properties of ΔFAK and KIN-32, we immunoprecipitated HA-FAK, HA-ΔFAK, or HA-KIN-32 (see Fig. 5A) from lysates of 3T3 transfectants after plating on fibronectin-coated substrates (Fig. 6A). GFP-transfected cells were used as a negative control. Parallel immunoprecipitates were tested for in vitro kinase activity against a general tyrosine kinase substrate, polyglutamine-tyrosine. No measurable activity was detected in the HA-KIN-32 sample suggesting that KIN-32 is not an active kinase (Fig. 6B). In contrast, we found that HA-FAK and HA-ΔFAK kinase activities were readily detectable and significantly above the background in GFP transfectants. These results demonstrate that the FERM domain deletion in kin-32(ok166) is dispensable for kinase activity in mammalian FAK.
Fig. 6.
Analysis of kinase activity. NIH3T3 cells transfected with HA-focal adhesion kinase (FAK), HA-ΔFAK, or HA-KIN-32 constructs were plated on fibronectin-coated cell culture plates for 30 min, lysed, and immunoprecipitated with anti-HA antibodies. A: Half of the immunoprecipitate was immunoblotted with anti-HA antibodies. The immunoprecipitate in the green fluorescent protein (GFP) lane is a negative control. B: The other half of the immunoprecipitate was used for in vitro phosphorylation of the tyrosine kinase substrate poly(Glu-Tyr) with γ32P-ATP. Kinase activity relative to HA-FAK is indicated by the shaded bars (± SD).
DISCUSSION
This work represents the first study of the biological properties and biochemical function of the C. elegans FAK homolog KIN-32. Our KIN-32 depletion experiments by bacterially mediated or injection RNAi strongly suggest that KIN-32 is not an essential protein in C. elegans. In addition, we performed a battery of experiments on kin-32(ok166), a deletion allele that lacks more than three-quarters of the FERM domain. We show that this domain is not necessary for normal physiology of C. elegans under laboratory conditions. In addition, we also demonstrate that a similar deletion within the FERM domain of mouse FAK did not affect its ability to localize to the focal adhesions or to exhibit in vitro kinase activity. Our results are consistent with in vitro studies showing that the FERM domain is dispensable for kinase activity in mammalian FAK. Deletions encompassing the entire FERM domain or parts of it gave functional proteins that localized to focal adhesions in cultured cells and exhibited kinase activity (Hildebrand et al., 1993; Cooper et al., 2003). Our analysis of kin-32(ok166) provides in vivo evidence that the FERM domain is not essential for FAK function.
FERM domains are multifunctional protein and lipid-binding motifs found in proteins associated with the cytoskeleton, such as ezrin, radixin, and moesin (Chishti et al., 1998). FERM domains mediate the interaction of talin with the cytoplasmic domains of integrins (Calderwood, 2004). Interaction between the FERM domains of ezrin and FAK facilitate FAK activation by integrins (Poullet et al., 2001) and the guanine exchange factor Trio can increase FAK activation by binding the FAK FERM domain (Medley et al., 2003). The FERM domain has also been implicated in FAK regulation of receptor tyrosine kinase signaling and G-protein coupled receptor signaling (Sieg et al., 2000; Streblow et al., 2003). For example, in FAK null mouse embryo fibroblasts, FAK constructs lacking the FERM domain do not rescue cell migration in response to EGF stimulation (Sieg et al., 2000). Despite conservation of the FAK FERM domain in KIN-32 and presence of potential FAK FERM interacting proteins Trio (unc-73) and ezrin (erm-1) in C. elegans, we have found that the FERM domain of C. elegans KIN-32 is dispensable for nematode growth and development. In addition, an internal deletion of 233 amino acids of the FERM domain of mouse FAK yielded an active kinase that localized to focal adhesions in HT1080 and 3T3 cells. This deletion construct was also able to localize to focal adhesions in FAK−/− mouse embryo fibroblasts (unpublished observations). These results are consistent with previous analyses in which deletion of the entire FAK FERM domain results in increased phosphorylation and constitutive activation of FAK (Cooper et al., 2003). This inhibition could be intramolecular (Cooper et al., 2003; Lietha et al., 2007) or intermolecular (Dunty et al., 2004) with FERM domain interactions serving to keep the kinase in an inactive state until activated by recruitment to focal adhesions.
KIN-32 lacked kinase activity when tested in vitro with a general tyrosine kinase substrate. It is possible that KIN-32 is a kinase in C. elegans, but was not activated by adhesion when expressed in 3T3 cells. However, it seems more likely that differences in critical functional residues between mammalian FAK and KIN-32 explain the lack of kinase activity. In fact, KIN-32 lacks many of the tyrosine residues critical for FAK function in vertebrates, including Y397, the major site of autophosphorylation and Src binding (Mitra et al., 2005). In addition, in the analogous positions, KIN-32 has a valine and a tyrosine rather than two tyrosines (Y576, Y577), a variation that might impair kinase activity, especially in combination with the lack of Y397. Y576 and Y577 are both major sites of Src family kinase phosphorylation in vivo. Mutation of either Y576 or Y577 to F moderately decreased kinase activity by approximately 30% of normal levels (Calalb et al., 1995). Mutation of both Y576 and Y577 to F resulted in a 50% decrease in kinase activity, similar to Y397F FAK activity in the same assay. The absence of all three tyrosines (Y397, 576, 577F) decreased activity by 80%. Therefore, it seems probable that KIN-32, which lacks two of these three tyrosines, does not have significant kinase activity in C. elegans. Instead KIN-32 may perform an adaptor function recruiting proteins such as SRC-1, DEB-1/vinculin, and/or talin to integrin cytoplasmic domains in muscle and other adherent cells.
FAK is essential in vertebrates where it is required for mesoderm development (Ilic et al., 1995). FAK−/− cells in culture exhibit reduced migration and increased numbers of focal adhesions, suggesting that FAK may be required for disassembly of integrin-based adhesions (Ren et al., 2000; Webb et al., 2004). This idea is further supported by the increase in cell motility with overexpression of FAK in CHO cells (Cary et al., 1996). In C. elegans, KIN-32 may not be required for the normal dynamics of relatively stable adhesion structures such as body wall muscle dense bodies. C. elegans nematodes have little mesenchymal tissue and limited numbers of migrating cells; therefore, KIN-32 may not have the prominent role observed during mesodermal cell migrations in vertebrate embryogenesis. Our results suggest that the truncated KIN-32 protein in kin-32(ok166) animals or residual protein left following kin-32 RNAi are sufficient to fulfill all necessary functions of KIN-32. Alternatively, it is possible that KIN-32 is completely dispensable for C. elegans viability. In Drosophila, the FAK homolog, Fak56, is not required for development or viability. However, Fak56 is lethal when overexpressed, apparently due to excessive down-regulation of integrin function (Grabbe et al., 2004). We have been unable to generate nematode lines overexpressing KIN-32, suggesting that its overexpression is deleterious to C. elegans viability (E.J. Cram, unpublished observations). We expect KIN-32 may modulate integrin function in response to stresses encountered by C. elegans in the wild that we were unable to duplicate in the laboratory.
EXPERIMENTAL PROCEDURES
Strains
All Caenorhabditis elegans nematodes were grown under standard conditions (Brenner, 1974). N2 and the deletion allele RB776 kin-32(ok166) were obtained from Caenorhabditis Genetics Center. The strain BC10568; [dpy-5(+), kin-32::GFP] was generously provided by David Baillie, Simon Fraser University, British Columbia.
Characterization of kin-32(ok166)
To characterize the deletion in kin-32(ok166) animals, genomic DNA was isolated, and primers flanking the predicted deletion site were designed (numbers reflect position on genomic sequence relative to the ATG). kin-32 F1549, 5′ GACAAGTTTGTTCTGT-CCC 3′; kin-32 R5163, 5′ GTCATGT-TCCTATATGCTC 3′.
PCR amplification from N2 and kin-32(ok166) genomic DNA yielded products of the expected sizes, and sequencing of these PCR products confirmed the 1,464 bp deletion. RNA from mixed-stage kin-32(ok166) and N2 nematodes was isolated and 1µg was used to synthesize cDNA with Superscript reverse transcriptase (Invitrogen, Carlsbad, CA) as previously described (Cram et al., 2003). For PCR amplification, 1 µl of cDNA reaction product was used in each reaction with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) in a 96-well plate. Primer design and concentrations were optimized according to the manufacturer’s recommendations. Amplification of the invariant C. elegans gene ama-1 was used as a loading and amplification control with primers: ama-1 F3462, 5′ACGTT-GAAAAAGGTAACATGCAATACA 3′; ama-1 R3598, 5′GAAGCCATGGAG-AGGTACGCGATAGAT 3′.
All assays were amplified and evaluated in real-time using the Applied Biosystems Prism 7900HT Fast Real-Time PCR detection system and the levels of kin-32 transcripts were calculated by comparison to a serial dilution standard. To visualize the two transcripts, cDNA was amplified by PCR using primers: kin-32 F14, 5′CACGTGATTCCTGATTGGTG 3′; kin-32 R591, 5′ TTGACTCTTGAAG-GATGAGTTG 3′.
Total levels of kin-32 transcripts in kin-32(ok166) and N2 nematodes were monitored by amplifying cDNA with the following sets of primers: kin-32 F9, 5′ GTCTAGCACGTGTATTCC 3′; kin-32 R116, 5′ GTGAGATTCCGAT-TCCTCG 3′.
Levels of the longer kin-32(ok166) transcript containing exon 3 were specifically amplified using: kin-32 splice primer F, 5′ GCGATGAAATTAAG-GATAGTGTG 3′; kin-32 R1035, 5′ AGCTGCAGAGTTTTCGGTCGAATC-GTTGC 3′. Levels of the shorter kin-32(ok166) transcript were amplified using kin-32 R1035 plus: kin-32 F144, 5′ AGGCTAGCTCGACTGGTCACG-GGTCC 3′.
Primer numbering scheme in all cases is based on the wild-type cDNA sequence with respect to the ATG.
RNAi and analysis of DTC migration
The RNAi feeding protocol was essentially as described (Timmons et al., 2001). Briefly, HT115(DE3) bacteria were cultured overnight in LB supplemented with 40 µg/ml ampicillin and seeded onto NGM agar supplemented with carbenicillin (25 µg/ml) and IPTG (1 mM). Double-stranded RNA expression was induced overnight at room temperature on the IPTG plates. Eggs were released from gravid hermaphrodites using alkaline hypochlorite solution (Hope, 1999), washed with M9 buffer and transferred to seeded plates. RNAi phenotypes were monitored in young adults following 48 hr incubation at 23°C. To analyze gonad morphology, hermaphrodites were mounted in a drop of M9 buffer containing 0.1 M sodium azide on a cover slip coated with 2% agarose and examined using a Nikon Eclipse TE 2000-U microscope with DIC optics. Defects such as inappropriate or extra turns, migration in the wrong direction, or aberrant stops were scored as DTC migration abnormalities.
To construct the kin-32 RNAi clone, primers at base positions 375 and 1035 relative to the ATG in the predicted cDNA sequence were used for RT-PCR amplification with N2 total RNA as the template. Primers included NheI and HindIII sites at the 5′ and 3′ ends, respectively, and these sites were used to clone the PCR product into pPD129.36 RNAi feeding vector (Fraser et al., 2000). The pat-3 RNAi clone is previously described (Cram et al., 2003). Primers used were kin-32 R1035 and kin-32 F375 5′AG-GCTAGCGAATTCTTTTCTCACGTT-GCC 3′.
Microscopy of nematodes
Fluorescence microscopy was performed with a Nikon Eclipse TE 2000-U microscope equipped for epifluorescence. Images were captured with a Cooke SensiCam High Performance camera using IP Lab software (Scanalytics, BD Biosciences, Rockville, MD). Fluorescence in BC10568 kin-32::GFP animals was visualized by mounting young adult animals in a drop of M9 buffer containing 0.1 M sodium azide on a coverslip coated with 2% agarose. To visualize muscle structures, nematodes were frozen on poly-L-lysine coated slides, fixed with methanol for 5 min, permeabilized with 0.1% Nonidet P40, and stained essentially as described (Lee et al., 2001). Primary antibodies (MH35 for ATN-1 and MH42 for UNC-89) were kindly provided by Dr. Michelle Hresko (Washington University School of Medicine, St. Louis, MO) and were used at 1:100 dilution in phosphate buffered saline (PBS) with 1% goat serum. Secondary fluorescein isothiocyanate (FITC) -conjugated goat anti-mouse antibodies were diluted 1:400 in PBS with 1% goat serum. For phalloidin staining, nematodes were fixed with methanol and acetone and stained with 0.4 U/ml of rhodamine-conjugated phalloidin (Molecular Probes, Invitrogen) for 2 hr at room temperature.
Phenotypic characterization of kin-32(ok166)
Fertility was determined by transferring L4 kin-32(ok166) or N2 individuals to fresh plates and counting larval progeny after 48 hr. For lifespan assays, 40 young adult kin-32(ok166) and N2 animals (10 animals per plate) were transferred to fresh NGM plates seeded with OP50 and maintained in a 20°C incubator. Animals were transferred to fresh plates every 2 days, at which time nematodes were scored as dead if they failed to respond to light touch with a platinum wire. N2 and kin-32(ok166) survival curves were indistinguishable over the course of the 25-day experiment. L2 kin-32(ok166) and N2 larvae were subjected to the following heat shock regimen: 33°C 1 hr, 22°C 4 hr, 37°C 2 hr, 20°C overnight. Controls were incubated at 20°C. Survivors after 24 hr were counted. Significance was determined by calculating the binomial 95% confidence interval for the proportion of living animals. For chemotaxis assays, well-fed L4 kin-32(ok166), eat-4(ky5), and N2 larvae were transferred in a drop of distilled water to the center of chemotaxis plates. eat-4(ky5) animals are defective in olfactory chemotaxis. Preference for isoamyl alcohol versus the ethanol control was assessed by calculating the chemotactic index: no. of animals at attractant – no. of animals at control)/total (Bargmann et al., 1993). A higher number indicates a greater preference for the attractant. Body lengths were measured on images of synchronized populations of kin-32(ok166) and N2 animals using Spot-Advanced software. Overall phosphotyrosine levels in 50 solubilized N2 or kin-32(ok166) animals were analyzed by sodium dodecyl sulfate-polyacryl-amide gel electorphoresis (SDS-PAGE) and immunoblotting with anti-phosphotyrosine monoclonal antibodies: Tyr-100 (Cell Signaling Technology) and PT66 (Sigma).
Mammalian expression and immunofluorescence
The plasmid pRcCMV-wtFAKHA, which is triple HA tagged, was a gift of Dr. Steven Hanks (Vanderbilt University, Nashville, TN) and is referred to here as HA-FAK. The HA-ΔFAK plasmid was constructed by deleting 233 amino acids from the FERM domain in HA-FAK. Two fragments of HA-FAK flanking the deletion were amplified by PCR, each with a KpnI restriction site to facilitate re-ligation of the fragments. An EcoRV site in the pRc-CMV vector and a naturally occurring ClaI site in FAK were used to clone in the resulting deleted fragment. To construct HA-KIN-32, kin-32 cDNA was amplified by RT-PCR from RNA isolated from mixed stage wild-type nematodes (numbers on primer sequences refer to position on cDNA) and cloned into T-vector (Promega, Madison, WI). HA was amplified by PCR from the HA-FAK vector with ClaI ends and cloned 5′ to the kin-32 ATG using the T-vector ClaI site, then HA-KIN-32 was inserted in-frame between the KpnI and NotI sites of pcDNA3. Vectors were confirmed by restriction digest and sequencing.
Primers used in the construction of HA-ΔFAK were as follows: FAK F935, 5′ CTGGAATTCTGCAGATATCCATC 3′; FAK R1389, 5′ CAAGGTACCTT-TCCACTCCTCTGG 3′; FAK F2110, 5′ GGAGGTACCCAATCTTTCATCATC 3′; and FAK R2475, 5′ GTGAGGATG-GTCAAACTGACGCATTG 3′.
Primers used in the construction of HA-KIN-32 were as follows: kin-32 F1, 5′ ATGGAAGGTCTAGCACGTG-TATTC 3′; kin-32 R2590, 5′ GCTC-CCGGGCTAACAATTAGATAA 3′; HA-F, 5′ GGTACCGGCCGCATGTTT-TACCC 3′; and HA-R, 5′ ATCGAT-CACTGAGCACGTAATC 3′.
HT1080 fibrosarcoma cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen) with 10% fetal bovine serum (Hyclone). Cells were transfected with HA-FAK or HA-ΔFAK constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations. The pcGFP5 construct, provided by Don Winkelmann (Robert Wood Johnson Medical School, Piscataway, NJ) was used as a transfection reporter. Diffuse GFP expression did not interfere with visualization of focal adhesions using fluorescein-labeled antibodies. Transfected cells were selected in medium plus 1 mg/ml G418 (Invitrogen).
For focal adhesion analysis, cells were plated on coverslips coated with a 10 µg/ml solution of plasma fibronectin and then blocked with 1% BSA in PBS. The 2 × 105 transfectants were plated for 90 min at 37°C, fixed with 3.7% formaldehyde in PBS, and permeabilized with 0.5% NP40. Fixed cells were stained with anti-HA antibodies (Covance at 1:500), Texas Red phalloidin (1:500), and fluorescein-goat anti-mouse antibodies (1:400). Images were collected as described above.
Immunoblotting and kinase assay
NIH3T3 cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone Laboratories, Logan, UT) and 50 U/ml penicillin and 50 µg/ml streptomycin. For transfection, fibroblasts were seeded at a density of 1 × 105 cells per well of a six-well culture dish and transfected with 1 µg of total DNA and LipofectAMINE (Invitrogen) according to the manufacturer’s instructions. The pcGFP5 construct, provided by Don Winkelmann (Robert Wood Johnson Medical School, Piscataway, NJ) was used as a transfection reporter. Assays were conducted 48 hr after transfection.
NIH 3T3 cells transfected with HA-FAK, HA-ΔFAK, HA-KIN-32, or GFP were trypsinized and replated in serum-free medium on 10 µg/ml human fibronectin coated plates for 30 min. We confirmed this protocol activates FAK by immunoblotting with anti-phosphotyrosine 397 antibodies (Biosource International). Immunoprecipitation of HA constructs was conducted essentially as described (Zhang et al., 1999). Cells were lysed in ice-cold NP40 immunoprecipitation buffer 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM ethyleneglycoltetraacetic acid, 0.1% NP-40, 50 mM NaF, 1 mM Na3VO4, 0.2 M phenyl-methyl sulfonyl fluoride, 10 µg/ml aprotinin, and 10 µg/ml leupeptin. Approximately 500 µg protein were immunoprecipitated with a 1:150 dilution of HA.11 monoclonal antibody (Covance, Princeton, NJ) and Protein A Sepharose beads for 3 hr at 4°C. After rinsing with NP-40 immunoprecipitation buffer, the samples were evenly divided for the Western blot and kinase assay. For the Western blot, proteins were separated using SDS-PAGE and transferred to a nitrocellulose membrane. Proteins were detected using the 12CA5 HA monoclonal antibody (1:100), horseradish peroxidase-conjugated IgG (Pierce, Rockford, IL) and the ECL detection system (GE Healthcare [Amersham Biosciences], Piscataway, NJ). The other half of the sample was used for the kinase assay. Immunoprecipitates were rinsed with kinase buffer (20 mM HEPES, pH 7.4, 10% glycerol, 10 mM MgCl2, 10 mM MnCl2, and 150 mM NaCl). Excess buffer was removed to leave a 30-µl reaction volume, to which was added 25 µCi of γ32P ATP (3,000 µCi/mmol), 50 µg poly (Glu-Tyr) substrate (Sigma Aldrich, St. Louis, MO) and 20 µM ATP. After 30 min at 32°C, the reaction was quenched with gel loading buffer, samples were separated using SDS-PAGE, fixed, dried, and quantified using a STORM 860 PhosphorImager with Image-QuaNT software (Molecular Dynamics, Inc., Sunnyvale, CA).
ACKNOWLEDGMENTS
C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources, National Institutes of Health. We thank R. Daniel Slone, Nicolette Wangler, Reem Hussein, Horia Negulescu, and Xuyan Feng for help with experiments. J.E.S. was funded by the National Institutes of Health, and E.J.C. was funded by the Damon Runyon Cancer Research Fund and the Robert Black Charitable Foundation.
Grant sponsor: NIH; Grant number: R01 GM059383; Grant sponsor: Damon Runyon Cancer Research Fund; Grant number: 1706-02.
REFERENCES
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Baum PD, Garriga G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron. 1997;19:51–62. doi: 10.1016/s0896-6273(00)80347-5. [DOI] [PubMed] [Google Scholar]
- Benian GM, Tinley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calalb MB, Polte TR, Hanks SK. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol Cell Biol. 1995;15:954–963. doi: 10.1128/mcb.15.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA. Talin controls integrin activation. Biochem Soc Trans. 2004;32:434–437. doi: 10.1042/BST0320434. [DOI] [PubMed] [Google Scholar]
- Cary LA, Chang JF, Guan JL. Stimulation of cell migration by overexpression of focal adhesion kinase and its association with Src and Fyn. J Cell Sci. 1996;109:1787–1794. doi: 10.1242/jcs.109.7.1787. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Song HK, Poy F, Schaller MD, Eck MJ. Crystal structure of the FERM domain of focal adhesion kinase. J Biol Chem. 2006;281:252–259. doi: 10.1074/jbc.M509188200. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishti AH, Kim AC, Marfatia SM, Lutch-man M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, Chasis JA, Conboy JG, Gascard P, Takakuwa Y, Huang SC, Benz EJ, Jr, Bretscher A, Fehon RG, Gusella JF, Ramesh V, Solomon F, Marchesi VT, Tsukita S, Tsukita S, Hoover KB, et al. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci. 1998;23:281–282. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- Cooper LA, Shen TL, Guan JL. Regulation of focal adhesion kinase by its amino-terminal domain through an autoinhibitory interaction. Mol Cell Biol. 2003;23:8030–8041. doi: 10.1128/MCB.23.22.8030-8041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox EA, Hardin J. Sticky worms: adhesion complexes in C. elegans. J Cell Sci. 2004;117:1885–1897. doi: 10.1242/jcs.01176. [DOI] [PubMed] [Google Scholar]
- Cram EJ, Clark SG, Schwarzbauer JE. Talin loss-of-function uncovers roles in cell contractility and migration in C. elegans. J Cell Sci. 2003;116:3871–3878. doi: 10.1242/jcs.00705. [DOI] [PubMed] [Google Scholar]
- Dunty JM, Gabarra-Niecko V, King ML, Ceccarelli DF, Eck MJ, Schaller MD. FERM domain interaction promotes FAK signaling. Mol Cell Biol. 2004;24:5353–5368. doi: 10.1128/MCB.24.12.5353-5368.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gettner SN, Kenyon C, Reichardt LF. Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabbe C, Zervas CG, Hunter T, Brown NH, Palmer RH. Focal adhesion kinase is not required for integrin function or viability in Drosophila. Development. 2004;131:5795–5805. doi: 10.1242/dev.01462. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD, Schaller MD, Parsons JT. Identification of sequences required for the efficient localization of the focal adhesion kinase, pp125FAK, to cellular focal adhesions. J Cell Biol. 1993;123:993–1005. doi: 10.1083/jcb.123.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA, editor. C. elegans, a practical approach. Oxford: Oxford University Press; 1999. [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- Lee M, Cram EJ, Shen B, Schwarzbauer JE. Roles for beta(pat-3) integrins in development and function of Caenorhabditis elegans muscles and gonads. J Biol Chem. 2001;276:36404–36410. doi: 10.1074/jbc.M105795200. [DOI] [PubMed] [Google Scholar]
- Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, Eck MJ. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–1187. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Qadota H, Moerman DG, Williams BD. C. elegans PAT-6/actopaxin plays a critical role in the assembly of integrin adhesion complexes in vivo. Curr Biol. 2003;13:922–932. doi: 10.1016/s0960-9822(03)00372-5. [DOI] [PubMed] [Google Scholar]
- Lo SH. Focal adhesions: what’s new inside. Dev Biol. 2006;294:280–291. doi: 10.1016/j.ydbio.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- Medley QG, Buchbinder EG, Tachibana K, Ngo H, Serra-Pages C, Streuli M. Signaling between focal adhesion kinase and trio. J Biol Chem. 2003;278:13265–13270. doi: 10.1074/jbc.M300277200. [DOI] [PubMed] [Google Scholar]
- Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- Norman KR, Moerman DG. The let-268 locus of Caenorhabditis elegans encodes a procollagen lysyl hydroxylase that is essential for type IV collagen secretion. Dev Biol. 2000;227:690–705. doi: 10.1006/dbio.2000.9897. [DOI] [PubMed] [Google Scholar]
- Poullet P, Gautreau A, Kadare G, Girault JA, Louvard D, Arpin M. Ezrin interacts with focal adhesion kinase and induces its activation independently of cell-matrix adhesion. J Biol Chem. 2001;276:37686–37691. doi: 10.1074/jbc.M106175200. [DOI] [PubMed] [Google Scholar]
- Ren XD, Kiosses WB, Sieg DJ, Otey CA, Schlaepfer DD, Schwartz MA. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J Cell Sci. 2000;113:3673–3678. doi: 10.1242/jcs.113.20.3673. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. p. 1222. [Google Scholar]
- Sieg DJ, Hauck CR, Ilic D, Klingbeil CK, Schaefer E, Damsky CH, Schlaepfer DD. FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol. 2000;2:249–256. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Streblow DN, Vomaske J, Smith P, Melnychuk R, Hall L, Pancheva D, Smit M, Casarosa P, Schlaepfer DD, Nelson JA. Human cytomegalovirus chemokine receptor US28-induced smooth muscle cell migration is mediated by focal adhesion kinase and Src. J Biol Chem. 2003;278:50456–50465. doi: 10.1074/jbc.M307936200. [DOI] [PubMed] [Google Scholar]
- Timmons L. Delivery methods for RNA interference in C. elegans. Methods Mol Biol. 2006;351:119–125. doi: 10.1385/1-59745-151-7:119. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chattopadhyay A, Ji QS, Owen JD, Ruest PJ, Carpenter G, Hanks SK. Focal adhesion kinase promotes phospholipase C-gamma1 activity. Proc Natl Acad Sci U S A. 1999;96:9021–9026. doi: 10.1073/pnas.96.16.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]