Abstract
Objective
Idiopathic osteoarthritis is the most common form of osteoarthritis (OA) world-wide and remains the leading cause of disability and the associated socio-economic burden in an increasing aging population. Traditionally, OA has been viewed as a degenerative joint disease characterized by progressive destruction of the articular cartilage and changes in the subchondral bone culminating in joint failure. However, the etiology of OA is multifactorial involving genetic, mechanical and environmental factors. Treatment modalities include analgesia, joint injection with steroids or hyaluronic acid, oral supplements including glucosamine and chondroitin sulfate, as well as physiotherapy. Thus, there is significant interest in the discovery of disease modifying agents. One such agent, glucosamine (GlcN) is commonly prescribed even though the therapeutic efficacy and mechanism of action remain controversial. Inflammatory cytokines, including IL-1β, and proteinases such as MMP-13 have been implicated in the pathogenesis and progression of OA together with an associated CpG demethylation in their promoters. We have investigated the potential of GlcN to modulate NF-kB activity and cytokine-induced abnormal gene expression in articular chondrocytes and, critically, whether this is associated with an epigenetic process.
Method
Human chondrocytes were isolated from the articular cartilage of femoral heads, obtained with ethical permission, following fractured neck of femur surgery. Chondrocytes were cultured for 5 weeks in six separate groups; (i) control culture, (ii) cultured with a mixture of 2.5 ng/ml IL-1β and 2.5 ng/ml oncostatin M (OSM), (iii) cultured with 2 mM N-acetyl GlcN (Sigma–Aldrich), (iv) cultured with a mixture of 2.5 ng/ml IL-1β, 2.5 ng/ml OSM and 2 mM GlcN, (v) cultured with 1.0 μM BAY 11-7082 (BAY; NF-kB inhibitor: Calbiochem, Darmstadt, Germany) and, (vi) cultured with a mixture of 2.5 ng/ml IL-1β, 2.5 ng/ml OSM and 1.0 μM BAY. The levels of IL1B and MMP13 mRNA were examined using qRT-PCR. The percentage DNA methylation in the CpG sites of the IL1β and MMP13 proximal promoter were quantified by pyrosequencing.
Result
IL1β expression was enhanced over 580-fold in articular chondrocytes treated with IL-1β and OSM. GlcN dramatically ameliorated the cytokine-induced expression by 4-fold. BAY alone increased IL1β expression by 3-fold. In the presence of BAY, IL-1β induced IL1B mRNA levels were decreased by 6-fold. The observed average percentage methylation of the −256 CpG site in the IL1β promoter was 65% in control cultures and decreased to 36% in the presence of IL-1β/OSM. GlcN and BAY alone had a negligible effect on the methylation status of the IL1B promoter. The cytokine-induced loss of methylation status in the IL1B promoter was ameliorated by both GlcN and BAY to 44% and 53%, respectively. IL-1β/OSM treatment increased MMP13 mRNA levels independently of either GlcN or BAY and no change in the methylation status of the MMP13 promoter was observed.
Conclusion
We demonstrate for the first time that GlcN and BAY can prevent cytokine-induced demethylation of a specific CpG site in the IL1β promoter and this was associated with decreased expression of IL1β. These studies provide a potential mechanism of action for OA disease modifying agents via NF-kB and, critically, demonstrate the need for further studies to elucidate the role that NF-kB may play in DNA demethylation in human chondrocytes.
Keywords: Osteoarthritis, Chondrocytes, Glucosamine, NF-kB, IL1B
1. Introduction
Idiopathic osteoarthritis (OA) is a late-onset, complex disease of the joint, characterized by progressive failure of the extracellular cartilage matrix, along with changes in the synovium and subchondral bone. OA is estimated to affect one in three individuals over the age of 60 and the primary etiology remains unclear. To date clinical intervention to slow or reverse OA progression is limited and, except for joint replacement, there remains no cure.
Proposed risk factors for the development of OA include heredity, obesity, reproductive factors, hypermobility, diabetes, hypertension, hyperuricemia, previous trauma to the joint, developmental variation in joint architecture and repetitive occupational or leisure activities, as well as factors which appear to be negatively associated with OA such as smoking and osteoporosis [1]. While current medical therapies for OA comprise pain relief, corticosteroids, non-steroidal anti-inflammatory drugs (NSAIDs), as well as oral supplements such as glucosamine and chondroitin sulfate. Thus, there is significant interest in the development of disease modifying agents. One such identified agent is glucosamine (GlcN) [2], administered as GlcN hydrochloride or sulfate form although the efficacy and mechanism of action of GlcN remains far form clear [3]. Originally advocated as a supplement for the extracellular matrix (ECM), recent studies suggest a different role of GlcN in the prevention of IL-1β mediated activation of nuclear factor kappa B (NF-kB) [4] and cytokine inducible gene expression in chondrocytes [5]. Zou et al. [6] showed that GlcN could attenuate LPS-induced NF-kB activation via O-linked N-acetylglucosa-mine (O-GlcNAc) resulting in a decreased systemic inflammatory reaction. If a similar mechanism exists for chondrocytes, GlcN may play a similar role as the NF-kB inhibitor and thus attenuate the response of chondrocytes to inflammatory cytokines. Epigenetics, defined as the heritable modification in gene function without changes in the DNA sequence, including DNA methylation, histone modification and changes in chromatin structure, are important in the maintenance of the phenotype of the normal adult chondrocyte [7]. We and others have shown that chondrocytes in OA cartilage undergo a phenotypic change acquiring a gene expression repertoire characterized by the aberrant expression of a number of catabolic genes including IL1β [8–11]. In addition, DNA demethylation at specific CpG sites accounts for the aberrant expression of matrix metalloproteinases (MMP) 3, 9 and 13, ADAMTS4 and IL1β in human articular chondrocytes [10–13]. Interestingly, the two CpG sites in the IL1β promoter (–299 and –256) shown previously to be differentially methylated between OA and normal chondrocytes bracket a binding site for NF-kB [14]. In recent studies, Kirillov and coworkers have suggested that NF-kB may be important in DNA demethylation [15], raising the prospect that IL-1β-induced IL1B mRNA expression could be prevented by the NF-kB inhibitor and an opportunity to examine whether there is an epigenetic association. In the current study, we have set out to investigate whether cytokine-induced expression of catabolic genes can be prevented by GlcN and an inhibitor that blocks NF-kB, and whether this is associated with epigenetic silencing.
2. Materials and methods
2.1. Histological assessment
Human articular cartilage was obtained after hemiarthroplasty following femoral neck fracture (#NOF) or total hip arthroplasty for OA respectively, with full patient consent and approval from the local ethics committee. Articular cartilage (~4 × 10 mm) was dissected from femoral heads within 6 h of surgery. Samples were fixed in freshly prepared paraformaldehyde overnight and processed into paraffin wax. The following primary antibodies were used for immunocytochemistry; rabbit anti-human MMP-13 (AHP751; Serotec, Oxford, UK), goat anti-human IL-1β (AN-201-NA: R&D Systems). After overnight incubation, binding of the primary antibodies was visualized with the aid of the appropriate biotinylated secondary antibody, followed by treatment with avidinperoxidase and 3-amino-9-ethyl-carbazole. Sections were counter-stained with 1% Alcian blue, viewed with a Zeiss Universal light microscope (Zeiss, Welwyn Garden City, UK), and images were captured with a digital camera.
2.2. Chondrocyte isolation
Human chondrocytes were isolated from the articular cartilage of #NOF patient samples following hemiarthroplasty (control) or the OA patients after total hip replacements (OA). Cartilage was dissected within 6 h of surgery.
For control cultures, we used only non-OA chondrocytes from the deep zone of tissues from patients with femoral neck fracture, as in previous studies [16]. For OA samples, only macroscopically degraded cartilage pieces adjacent to the weight bearing area were dissected, as these contained the typical OA chondrocytes with the altered ‘degradative’ phenotype [13]. To liberate the cells, cartilage pieces were cut into small fragments and digested with 10% trypsin (Lonza, Wokingham, UK) in phosphate buffered saline (PBS) for 30 min, in 1 mg/ml of hyaluronidase (Sigma–Aldrich, Gillingham, UK) in PBS for 15 min, and in 10 mg/ml of collagenase B (Roche, Lewes, UK) in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12; Invitrogen, Paisley, UK) for 12–15 h at 37 °C.
2.3. Chondrocyte culture
Before treatment, chondrocytes were cultured for 48 h at a density of 2–4 × 105 cells/25 cm2 flask in 5 ml of DMEM/F12 supplemented with 5% fetal calf serum (FCS; Invitrogen, Paisley, UK), 1% insulin-transferrin-selenium (Sigma–Aldrich), 100 units/ml of penicillin and 100 μg/ml of streptomycin (Lonza), and 100 μg/ml of ascorbic acid (Sigma–Aldrich) in 5% CO2 at 37 °C. Chondrocytes were divided into six groups: (i) control culture, (ii) cultured with a mixture of 2.5 ng/ml IL-1β and 2.5 ng/ml oncostatin M (OSM), (iii) cultured with 2 mM N-acetyl GlcN (Sigma–Aldrich), (iv) cultured with a mixture of 2.5 ng/ml IL-1β, 2.5 ng/ml OSM and 2 mM GlcN, (v) cultured with 1.0 μM BAY 11-7082 (BAY; NF-kB inhibitor: Calbiochem, Darmstadt, Germany) and, (vi) cultured with a mixture of 2.5 ng/ml IL-1β, 2.5 ng/ml OSM and 1.0 μM BAY. Media and reagents were changed twice weekly. The primary cultures were maintained for 5 weeks until confluence.
2.4. DNA and RNA extraction
Total RNA and genomic DNA were extracted simultaneously from each sample with the use of an AllPrep DNA/RNA Mini Kit (Qiagen, Crawley, UK) according to the manufacturer's instructions. RNA was immediately reverse-transcribed with avian myeloblastosis virus reverse transcriptase and both oligo(dT)15 and random primers [17].
2.5. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Relative quantification of gene expression was performed with an ABI Prism 7500 detection system (Applied Biosystems, Warring-ton, UK). Reactions were performed in triplicate, with GAPDH as the internal control. Primer Express 3.0 software (Applied Biosystems) was used to design primers across exon-exon boundaries. Messenger RNA expression was quantified according to the 2–ΔΔCt method. The 20 μl reaction mixture was used contained 1 μl of complementary DNA, 10 μl of Power SYBR Green PCR Master Mix (Applied Biosystems), and 250 nM of each primer. Thermal cycler condition consisted of an initial activation step at 95 °C for 10 min, followed by a 2-step PCR program of 95 °C for 15 s and 60 °C for 60 s for 40 cycles. A dissociation curve was obtained for each quantitative PCR run. The primers used for RT-PCR are shown in Table 1a.
Table 1.
(a) Primer pairs used for qRT-PCR*; (b) Primer pairs and sequencing primers used for pyrosequencing*.
| Amplicon ID (amplicon length, bp) | Primer (sequence 5′ to 3′) |
|---|---|
| (a) | |
| GAPDH (108) | F (CCAGGTGGTCTCCTCTGACTTC) R (TCATACCAGGAAATGAGCTTGACA) |
| IL1B (126) | F (TGGCAATGAGGATGACTTGTTC) R (CTGTAGTGGTGGTCGGAGATT) |
| MMP13 (71) | F (TTAAGGAGCATGGCGACTTCT) R (CCCAGGAGGAAAAGCATGAG) |
| DNMT1 (64) | F (CAGGCCCAATGAGACTGACA) R (GTGGGTGTTCTCAGGCCTGTAG) |
| (b) | |
| IL1B_1 (157) | F (ATGGAAGGGTAAGGAGTAGTAA) R (CCCACATATACTAAATTTAAACATTCTT) S (ATACTAAATTTAAACATTCTTCTA) |
| IL1B_2 (129) | F (ATGAAGATTGGTTGAAGAGAATTTTAGA) R (ATTTCTCAACCTCCTACTTCTACTTTTAA) S (ATTTTAGAGTAGTTTGTTGTG) |
| MMP13_1 (216) | F (AATTAGTATTAAGTTTTTTTTTATGGAAGT) R (TTCAACAAAATCTCAAAACCCATCTAA) S1 (AAATTTTTTTTTTTTTACCTTCTAT) S2 (CTCAAAACCCATCTAAC) |
| MMP13_2 (246) | F (ATGGGTTTTGAGATTTTG) R (ACCCCTAAATACATCTTAAATA) S1 (CAATCACTTAAAAATAAACATACTT) S2 (AATAATACCTAAAAACTATTATC) |
The primers for IL1B qRT-PCR were designed by and purchased from PrimerDesign Ltd, Southampton, UK.
F = forward; R = reverse; S = sequencing.
2.6. Analysis of DNA methylation
Bisulfite treatment was performed for the genomic DNA (500 ng) of each sample using the EZ DNA Methylation-Gold™ Kit (Zymo Research Corporation, CA 92867, USA) according to the manufacturer's instructions. After bisulfite treatment, a 40 μl PCR was carried out in 3.2 μl bisulfite treated DNA (30 ng), 36 μl Platinum® PCR Supermix High Fidelity (Invitrogen), and 200 nM of each primer. Thermal cycler conditions consisted of an initial activation step at 95 °C for 5 min, followed by a 3-step PCR program of 95 °C for 15 s, 55 °C for 30 s (55 °C for IL1B_1, 2, and MMP13_1; 50 °C for MMP13_2) and 72 °C for 30 s for 50 cycles. Following PCR, gel electrophoresis to determine purity was performed using 10 μl of each PCR product. Percent DNA methylation in the IL1β and the MMP13 promoter were quantified using PyroMark™ MD (Qiagen) according to the manufacturer's instructions. The primers used for pyrosequencing (see Table 1b) were designed with Pyrosequencing™ Assay Design Software Ver 2.0 (Qiagen). Two primer sets were designed for both IL1β and the MMP13 promoter. The –299 and –256 CpG sites of IL1B promoter were covered by IL1B_1 primer set and the –20 and +13 CpG sites were covered by IL1B_2 primer set. MMP13_1 primer set covered –344, –324 and –225 CpG sites of MMP13 promoter, sequencing primer S1 was used for –344 and –324 CpGs, and S2 for –225 CpG. MMP13_2 primer set covered –135, –115, –110 and –14 CpG sites of MMP13 promoter, in addition, sequencing primer S1 was used to assess –135, –115 and –110 CpGs, and S2 for –14 CpG.
2.7. Features of the promoter regions
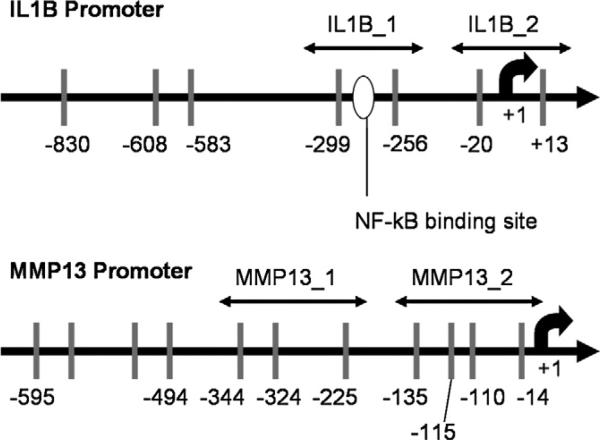
The 5′-flanking regions which contain the promoters of IL1B (GenBank accession No. NG008851) and MMP13 (GenBank accession No. NG021404) are shown in Fig. 1. Both IL1B and MMP13 promoter regions have “sparse” CpG distribution. Notably, the –299 and –256 CpG sites of IL1B promoter bracket a NF-kB binding site.
Fig. 1.
IL1B and MMP13 promoters. Vertical bars represent CpG sites. IL1B has 7 CpG sites in 900 bp and MMP13 has 11 CpG sites in 600 bp. The –299 and –256 CpG sites of the IL-1β promoter bracket a potential binding site for NF-kB. The locations of polymerase chain reaction primers are indicated.
2.8. Enzyme-linked immunosorbent assay (ELISA) for IL-1β in culture media
At the end of culture period, the culture medium containing exogenous cytokines was removed, and after thorough washing with PBS, 5 ml of fresh medium with 1% FCS was added. After 48 h, the culture supernatants were harvested and stored at –80 °C until analysis with the human IL-1β/IL-1F2 Quantikine ELISA Kit (R&D Systems, Abingdon, UK).
2.9. Statistical analysis
The data for gene expression and methylation analysis were analyzed in Microsoft Excel (Microsoft, Redmond, WA) using Wilcoxon's signed rank test. P values less than 0.05 were considered significant. Other data are expressed as the mean ± SEM.
3. Results
3.1. Localization of MMP-13 and IL-1β producing chondrocytes
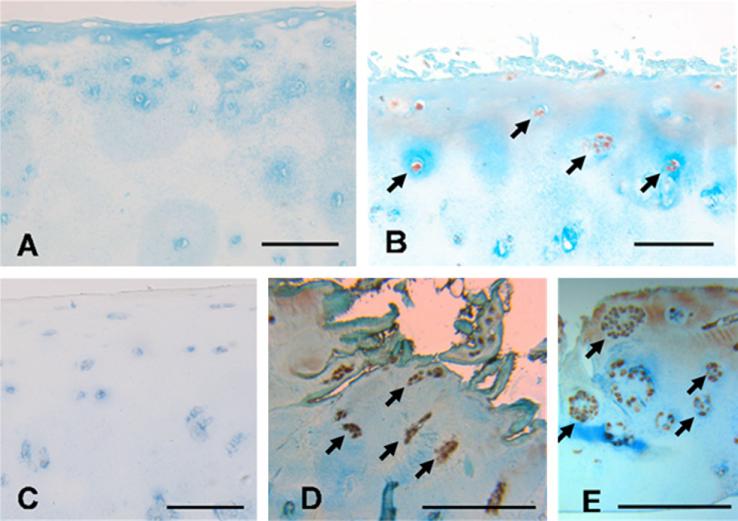
Immunocytochemical analysis demonstrated that the majority of articular chondrocytes from #NOF patients (Mankin score 1–2) do not produce IL-1β (Fig. 2A) or MMP-13 (Fig. 2C). In contrast, chondrocytes in the superficial zone of cartilage from OA patients (Mankin score 4–8) showed positive staining of IL-1β (Fig. 2B) and MMP-13 (Fig. 2D) indicating the presence of chondrocytes in the superficial zone of OA with a “degradative” phenotype. As illustrated in Fig. 2E, these “degradative” chondrocytes formed clusters of clonal chondrocytes and were observed to produce MMP-13. No clonal chondrocyte populations were observed in #NOF samples.
Fig. 2.
Immunocytochemical analysis of articular chondrocytes from #NOF patients (Mankin score 1–2) stained negatively for IL-1β (A) and MMP-13 (C). Chondrocytes in the cartilage from OA patients (Mankin score 4–8) stained positively for IL-1β (B) (arrows) and MMP-13 (D) (arrows). Clonal chondrocytes producing MMP-13 can be observed in E. Scale bar is 100 μm.
3.2. Glucosamine and BAY inhibit the cytokine induced expression of IL1B
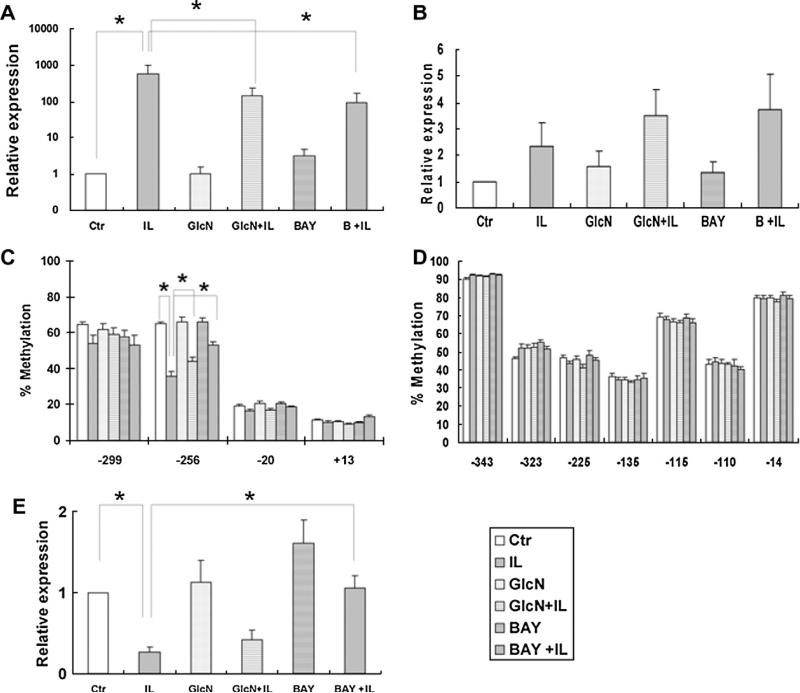
Examination of gene expression encoding IL-1β showed a dramatic and significant expression (approximately 600-fold increase) of IL1B mRNA levels by primary chondrocytes in the presence of IL-1β/OSM (Fig. 3A). GlcN alone had no effect on IL1B mRNA; however, GlcN, ameliorated IL-1β induced expression of IL1B by 4 fold. The NF-kB inhibitor, BAY, produced a modest increase in IL1B expression (3-fold) and reduced the IL-1β-induced IL1B expression by 6-fold.
Fig. 3.
The gene expression and DNA methylation in cultured primary chondrocytes culture. Each group represents chondrocytes without treatment (control), chondrocytes incubated with 2.5 ng/ml IL-1β and 2.5 ng/ml OSM, with 2 mM GlcN, with IL-1β/ OSM and GlcN, with 1.0 μM BAY and with IL-1β/OSM and BAY, respectively. * P < 0.05. (A) IL1B mRNA levels; (B) MMP13 mRNA levels. (C) The methylation status of 4 CpG sites in the IL1B proximal promoter. The% methylation of the –256 CpG site was decreased from 65% in control cultures to 36% in cells treated with IL-1β/OSM. GlcN and BAY alone had negligible effects on methylation status. The cytokine-induced loss of methylation was recovered by GlcN and BAY to 44% and 53%, respectively. The altered expression of IL1B was associated inversely with the % methylation at the –256 CpG site. The% methylation of –20 and +13 CpG sites was independent of treatment. (D) The % methylation of any of the 7 CpG sites of MMP13 promoter was largely unaffected by treatment. E. DNMT1 expression was reduced by treatment with IL-1β/OSM. BAY was able to maintain DNMT1 expression at the control level. GlcN did not prevent the cytokine-induced DNMT1 down-regulation, and the DNMT1 expression in cells treated with GlcN + IL-1β/OSM was 2.5-fold lower compared to the control cultures. The data for qRT-PCR were run in triplicate and the mean of three different trials shown. The data for pyrosequencing were run in duplicate. Data from N = 6, mean ± SEM.
3.3. Glucosamine and BAY do not modulate MMP13 expression in OA chondrocytes
Primary OA chondrocytes treated with IL-1β/OSM displayed increased levels of MMP13 mRNA, independent of culture in the presence or absence of GlcN or BAY. There was no statistical significance between any of the treatment groups (Fig. 3B).
3.4. Cytokine induced loss of methylation in IL1B promoter was ameliorated by glucosamine and BAY
To examine the possible mechanism of cytokine-induced IL1B expression in primary OA chondrocytes, the percentage methylation at the –299, –256, –20 and +13 CpG sites in the IL1B promoter was analyzed (Fig. 3C).
The percentage methylation at the –256 CpG site was 65% in control cultures. The percentage methylation significantly decreased to 36% after treatment of the cells with IL-1β/OSM. GlcN and BAY alone had a negligible effect on the percentage methylation status of the IL1B promoter. In contrast, the cytokine-induced loss of methylation was significantly recovered by GlcN and BAY to 44% and 53%, respectively. The methylation status of CpG sites at –299, –20 and +13 did not show any significant difference between any groups. In the presence of GlcN and BAY, IL-1β induced IL1B expression was alleviated and this was associated with hypermethylation.
3.5. Methylation status of the MMP13 promoter is unaffected by cytokine treatment
The methylation status of the 7 CpG sites of MMP13 proximal promoter was examined following culture under control conditions and after treatment with IL-1β/OSM in the absence or presence GlcN or BAY. The methylation status remained unaltered irrespective of culture condition with percentage methylation recorded at each CpG site as follows; 92% at –344, 52% at –324, 45% at –225, 35% at –135, 67% at –110, 43% at –110, and 80% at –14 (Fig. 3D).
3.6. Increased IL1B expression correlated with increased production of IL-1β protein
Primary OA chondrocytes cultured in the presence of IL-1β/OSM produced significant amounts of IL-1β protein (0.094–0.134 pg of IL-1β per μg of DNA per hour). In contrast, no IL-1β could be detected in cultures of primary OA chondrocytes maintained in control medium or treated with GlcN or BAY. Critically, no IL-1β was detected after incubation with GlcN + IL-1β/OSM or BAY + IL-1β/ OSM, further confirming the ability of both agents to inhibit IL-1β induced cytokine expression.
3.7. BAY inhibits cytokine induced down-regulation of DNMT1 expression
IL-1β/OSM reduced DNMT1 expression by 0.3-fold (Fig. 3E). This down regulation of DNMT1 expression was inhibited by BAY with expression levels maintained at control levels. In contrast, GlcN did not alleviate the cytokine-induced DNMT1 down-regulation.
4. Discussion
There is a significant unmet need for therapies to address OA in an increasing aging population. Approaches to treatment have been compounded by the observation that the etiology of OA is multifactorial, involving a combination of genetic and environmental influences. This current study demonstrates that an inhibitor of NF-kB and the disease modifying agent, glucosamine, significantly ameliorates the IL-1β-induced increase of IL1B expression in human chondrocytes and, that this was associated with the alleviation of demethylation in the specific CpG site in the IL1B promoter. In contrast, we observed that GlcN and BAY had no effect on the modulation of cytokine-induced MMP13 expression and, furthermore, there was no alteration in the methylation status of the MMP13 promoter following GlcN and BAY treatment; possibly relating to the promoter sequence. In support of our observations, ELISA analysis demonstrated the ability of GLcN and BAY to attenuate IL-1β/OSM-induced IL-1β protein synthesis.
It is known that BAY11-7082 is an inhibitor of NF-kB activation through blockade of IkB-a phosphorylation. Also, GlcN inhibits the IL-1β-induced activation of NF-kB [4,18]. We have previously reported an alteration in the methylation status of the –256 CpG site in the IL1B promoter which is located adjacent to a binding site for NF-kB [12]. On the other hand, there is no NF-kB binding site close to CpG sites in the MMP13 proximal promoter. We have previously reported that long-term aberrant expression of IL1B in OA is a consequence of DNA demethylation after treatment of cells with inflammatory cytokines [12]. Furthermore in OA cartilage, the acquired degradative heritable phenotype is transmitted to the daughter cells in the cluster of clonal chondrocytes by epigenetic mechanisms [10,13]. Based on these findings, we assume that GlcN and BAY can modulate the gene expression patterns by altering DNA methylation status. Of course inflammatory cytokines including IL-1β, OSM and TNFα act not only via the NF-kB pathway but also through number of other signaling pathways (JNK, p38, and ERK), and complete suppression of cytokine/protease activity would require coordinated inhibition of more than one pathway [11,19,20]. Thus, although the current studies cannot advocate the use of BAY in OA, the observations indicate the importance and need to further dissect the mechanisms, efficacy and safety of NF-kB inhibition, and the epigenetic pathways involved. It is important to note that there are a number of procedures to quantify the methylation of CpG sites, including methylation-sensitive restriction enzymes (MSRE) and real-time PCR [17] as in this study. However, this method is limited to CpG sites for which appropriate MSRE are available. Thus, although we have previously reported using bisulfite sequencing that the −256 CpG site of the IL1B promoter is differentially methylated [12], in the absence of a relevant MSRE, this could not be examined; however, pyrosequencing was used to determine the percentage methylation of the site.
In the current study, DNMT1 expression was reduced by cytokine treatment and while GlcN had no discernible effect, BAY treatment retained DNMT1 expression at comparable levels to control cultures. The modulation of DNMT1 expression may account for the enhanced effect of BAY to attenuate cytokine-induced demethylation at the –256 CpG site of IL1B promoter.
In conclusion, we demonstrate for the first time, that both GlcN and BAY alleviate cytokine-induced demethylation of a specific CpG site in the IL1B promoter, which is associated with decreased gene and protein expression. This study provides further evidence and support that the long-term cytokine-stimulated induction of IL1B in human articular chondrocytes in vitro involves the loss of DNA methylation and indicates a role for NF-kB in DNA methylation and potential of strategies to intervene in the process. The results support the need for further studies in this area to address the role of epigenetics in OA and the potential therein for innovative therapeutic strategies for OA in an aging population.
Acknowledgments
The authors thank the orthopedic surgeons at Southampton General Hospital for provision of femoral heads from patients undergoing total hip replacement surgery. Grant support from National Institutes of Health grants R21-AR054887 (MBG and HIR) and R01-AG022021 (MBG), Wessex Medical Research (M19; HIR and RO) and BBSRC G006970/1 (RO) is gratefully acknowledged. This paper is dedicated to the memory of Dr. Helmtrud (Trudy) Roach.
References
- 1.Dennison EM, Hindmarsh PC, Kellingray S, Fall CH, Cooper C. Growth hormone predicts bone density in elderly women. Bone. 2003;32:434–440. doi: 10.1016/s8756-3282(03)00035-8. [DOI] [PubMed] [Google Scholar]
- 2.Bohne W. Glucosamines in the conservative treatment of arthrosis. Med. Welt. 1969;30:1668–1671. [PubMed] [Google Scholar]
- 3.Black C, Clar C, Henderson R, MacEachern C, McNamee P, Quayyum Z, Royle P, Thomas S. The clinical effectiveness of glucosamine and chondroitin supplements in slowing or arresting progression of osteoarthritis of the knee: a systematic review and economic evaluation. Health Technol. Assess. 2009;13:1–148. doi: 10.3310/hta13520. [DOI] [PubMed] [Google Scholar]
- 4.Largo R, varez-Soria MA, ez-Ortego I, Calvo E, Sanchez-Pernaute O, Egido J, Herrero-Beaumont G. Glucosamine inhibits IL-1beta-induced NFkappaB activation in human osteoarthritic chondrocytes. Osteoarthritis Cartilage. 2003;11:290–298. doi: 10.1016/s1063-4584(03)00028-1. [DOI] [PubMed] [Google Scholar]
- 5.Shikhman AR, Kuhn K, Alaaeddine N, Lotz M. N-acetylglucosamine prevents IL-1 beta-mediated activation of human chondrocytes. J. Immunol. 2001;166:5155–5160. doi: 10.4049/jimmunol.166.8.5155. [DOI] [PubMed] [Google Scholar]
- 6.Zou L, Yang S, Champattanachai V, Hu S, Chaudry IH, Marchase RB, Chatham JC. Glucosamine improves cardiac function following trauma-hemorrhage by increased protein O-GlcNAcylation and attenuation of NF-{kappa}B signaling. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H515–H523. doi: 10.1152/ajpheart.01025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roach HI, Aigner T. DNA methylation in osteoarthritic chondrocytes: a new molecular target. Osteoarthritis Cartilage. 2007;15:128–137. doi: 10.1016/j.joca.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Aida Y, Maeno M, Suzuki N, Namba A, Motohashi M, Matsumoto M, Makimura M, Matsumura H. The effect of IL-1beta on the expression of inflammatory cytokines and their receptors in human hondrocytes. Life Sci. 2006;79:764–771. doi: 10.1016/j.lfs.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 9.Goldring SR, Goldring MB. The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin. Orthop. Relat. Res. 2004;6:S27–S36. doi: 10.1097/01.blo.0000144854.66565.8f. [DOI] [PubMed] [Google Scholar]
- 10.Cheung KS, Hashimoto K, Yamada N, Roach HI. Expression of ADAMTS-4 by chondrocytes in the surface zone of human osteoarthritic cartilage is regulated by epigenetic DNA de-methylation. Rheumatol. Int. 2009;29:525–534. doi: 10.1007/s00296-008-0744-z. [DOI] [PubMed] [Google Scholar]
- 11.Roach HI. Potential directions for drug development for osteoarthritis. Expert Opin. Drug Discovery. 2008;3:1–12. doi: 10.1517/17460441.3.5.475. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto K, Oreffo RO, Gibson MB, Goldring MB, Roach HI. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 2009;60:3303–3313. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, Kokubun S, Bronner F. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 14.Stylianou E, Saklatvala J. Interleukin-1. Int. J. Biochem. Cell Biol. 1998;30:1075–1079. doi: 10.1016/s1357-2725(98)00081-8. [DOI] [PubMed] [Google Scholar]
- 15.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Igkappa locus. Nat. Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 16.da Silva MA, Yamada N, Clarke NM, Roach HI. Cellular and epigenetic features of a young healthy and a young osteoarthritic cartilage compared with aged control and OA cartilage. J. Orthop. Res. 2009;27:593–601. doi: 10.1002/jor.20799. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto K, Kokubun S, Itoi E, Roach HI. Improved quantification of DNA methylation using methylation-sensitive restriction enzymes and real-time PCR. Epigenetics. 2007;2:86–91. doi: 10.4161/epi.2.2.4203. [DOI] [PubMed] [Google Scholar]
- 18.Wu S, Flint JK, Rezvani G, De LF. Nuclear factor-kappaB p65 facilitates longitudinal bone growth by inducing growth plate chondrocyte proliferation and differentiation and by preventing apoptosis. J. Biol. Chem. 2007;282:33698–33706. doi: 10.1074/jbc.M702991200. [DOI] [PubMed] [Google Scholar]
- 19.Fan Z, Soder S, Oehler S, Fundel K, Aigner T. Activation of interleukin-1 signaling cascades in normal and osteoarthritic articular cartilage. Am. J. Pathol. 2007;171:938–946. doi: 10.2353/ajpath.2007.061083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roman-Blas JA, Jimenez SA. NF-kappaB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthritis Cartilage. 2006;14:839–848. doi: 10.1016/j.joca.2006.04.008. [DOI] [PubMed] [Google Scholar]