Abstract
Objective
To investigate whether the abnormal expression of inducible nitric oxide synthase (iNOS) by osteoarthritic (OA) human chondrocytes is associated with changes in the DNA methylation status in the promoter and/or enhancer elements of iNOS.
Methods
Expression of iNOS was quantified by quantitative reverse transcriptase–polymerase chain reaction. The DNA methylation status of the iNOS promoter and enhancer regions was determined by bisulfite sequencing or pyrosequencing. The effect of CpG methylation on iNOS promoter and enhancer activities was determined using a CpG-free luciferase vector and a CpG methyltransferase. Cotransfections with expression vectors encoding NF-κB subunits were carried out to analyze iNOS promoter and enhancer activities in response to changes in methylation status.
Results
The 1,000-bp iNOS promoter has only 7 CpG sites, 6 of which were highly methylated in both control and OA samples. The CpG site at −289 and the sites in the starting coding region were largely unmethylated in both groups. The NF-κB enhancer region at −5.8 kb was significantly demethylated in OA samples compared with control samples. This enhancer element was transactivated by cotransfection with the NF-κB subunit p65, alone or together with p50. Critically, methylation treatment of the iNOS enhancer element significantly decreased its activity in a reporter assay.
Conclusion
These findings demonstrate the association between demethylation of specific NF-κB–responsive enhancer elements and the activation of iNOS transactivation in human OA chondrocytes, consistent with the differences in methylation status observed in vivo in normal and human OA cartilage and, importantly, show association with the OA process.
Idiopathic osteoarthritis (OA) is a late-onset, complex disease of the joint, characterized by progressive failure of the extracellular cartilage matrix, along with changes in the synovium and subchondral bone. A key feature of OA is the catabolic degradation of the cartilage matrix, which is mediated by aggrecanases and collagenases. The expression of these degradative proteases is elevated in human OA articular chondrocytes as compared to healthy chondrocytes, and enzyme activation results in matrix degradation, beginning with those located in the superficial zone and progressing to chondrocytes in the deep zone as the severity of OA increases. We have previously shown that this abnormal expression is associated with an epigenetic “unsilencing” of gene transcription (1–4), a finding that opens new avenues for future treatment of this debilitating disease (5).
Epigenetic alterations strongly affect gene expression without changing the DNA sequence. However, epigenetic mechanisms do not determine the short-term up- or down-regulation of a gene that is normally expressed in a specific cell, but involve silencing of all genes that do not belong to the repertoire of that cell type. The predominant epigenetic mechanisms are DNA methylation at the cytosine base of CpG dinucleotides and various histone modifications, as well as changes in higher-order chromatin structure.
Nitric oxide (NO) is a short-lived, multifunctional molecule involved in a myriad of key biologic processes. In chondrocytes, the most direct effect of NO appears to be the suppression of energy metabolism (6). The pathogenic involvement of NO in OA was first demonstrated when levels of nitrites, the stable end products of NO metabolism, were shown to be elevated in serum and synovial fluid samples from OA patients. Although the origin of the NO is unknown, articular chondrocytes are likely to be the major intraarticular source. Activated articular chondrocytes produce as much, if not more, NO than any other cell in the body (7). Unlike chondrocytes recovered from normal human articular cartilage, OA chondrocytes produce NO spontaneously (8) and express NO synthase (NOS) (9). Upon stimulation in culture, articular chondrocytes express the inducible isoform of NOS (iNOS) (10). Endogenously produced NO suppresses the synthesis of the cartilaginous matrix (11). Furthermore, chondrocytes in the superficial area of cartilage produce higher levels of NO as compared with cells in other layers (12).
Although iNOS is readily inducible in almost all rodent cell types in culture, normal human cells (except hepatocytes) are recalcitrant to induction, even with multiple cytokines. This lack of responsiveness has been shown to be due to epigenetic silencing by DNA methylation (13) and histone 3 lysine 9 (H3K9) methylation (14). At present, it is unclear whether the activation of iNOS in OA chondrocytes may also be attributed to epigenetic “unsilencing.” The iNOS promoter has a regulatory region around the TATA box, which is highly methylated in endothelial cells, but not hepatocytes (14). In addition, there are cytokine-responsive enhancers at −3665 to −5574 bp and −8093 to −8296 bp (15), although no difference in DNA methylation was found between endothelial cells and hepatocytes in the region between −5033 and −6199 bp, which contains numerous NF-κB binding sites (14). However, not all CpG sites in the 1-kb 5′-flanking regions or in the enhancers have been examined.
In the present study, we demonstrate that iNOS gene expression of human chondrocytes is regulated at the level of transcription and, critically, that this expression is mediated by the binding of NF-κB to the upstream sites of iNOS enhancer elements. Furthermore, we show for the first time the involvement of demethylation in specific NF-κB enhancer elements in the activation of iNOS in human OA chondrocytes. These data provide evidence linking the activation of iNOS and induction of the OA process.
MATERIALS AND METHODS
Cartilage dissection and chondrocyte isolation
Human articular cartilage was obtained from 15 patients who underwent hemiarthroplasty following femoral neck fracture (3 men and 12 women with a mean ± SD age of 76.8 ± 16.5 years) and from 13 OA patients who underwent total hip arthroplasty (10 men and 3 women with a mean ± SD age of 71.6 ± 8.2 years and an OA Research Society International–modified Mankin score [16] of 3–5). Informed consent was obtained from all patients, and the study was approved by the local ethics committee. Cartilage obtained from patients with femoral neck fracture is widely used as a suitable non-OA control. Cartilage was dissected within 6 hours after surgery. Only chondrocytes from the superficial layer of OA cartilage or the deep zone of cartilage from patients with femoral neck fracture were isolated. Small cartilage fragments were digested as previously described (1). Additional slices of cartilage were fixed overnight in freshly prepared paraformaldehyde and processed into paraffin wax for further immunohistochemical analysis.
DNA and RNA extraction
Total RNA and genomic DNA were extracted simultaneously from digested samples using an AllPrep DNA/RNA Mini kit (Qiagen), according to the manufacturer’s instructions. RNA was immediately reverse-transcribed with avian myeloblastosis virus reverse transcriptase and both oligo(dT)15 and random primers (17).
Quantitative reverse transcription–polymerase chain reaction (qRT-PCR)
Relative quantification of gene expression was performed with an ABI Prism 7500 detection system (Applied Biosystems). Primer Express 3.0 software (Applied Biosystems) was used to design primers. The 20-μl reaction mixture was prepared in triplicate, containing 1 μl of complementary DNA, 10 μl of Power SYBR Green PCR Master Mix (Applied Biosystems), and 250 nM of each primer. Thermal cycler conditions included an initial activation step at 95°C for 10 minutes, followed by a 2-step PCR program of 95°C for 15 seconds and 60°C for 60 seconds for 40 cycles. The 2−ΔΔCt method was used for relative quantification of gene expression, and the data were normalized to GAPDH expression. Primer information is available upon request from the corresponding author.
Immunohistochemistry
Formalin-fixed paraffin-embedded cartilage sections were deparaffinized and permeabilized in 0.5% Triton X-100 (Sigma) for 1 hour. Any endogenous peroxidase activity was blocked by incubating the slides in 3% H2O2 for 5 minutes. The specimens were incubated overnight at 4°C in anti-iNOS (BML-SA200; Enzo Life Sciences International) diluted 1:250 with 1% bovine serum albumin–phosphate buffered saline. The antibody was visualized using the appropriate biotinylated secondary antibody, followed by treatment with avidin–peroxidase and 3-amino-9-ethyl-carbazole. Sections were counterstained with 1% Alcian blue, and viewed with a Zeiss Universal light microscope, followed by image capture with a digital camera.
Analysis of DNA methylation
Bisulfite modification
Bisulfite modification was performed with 500 ng of each genomic DNA using an EZ DNA Methylation-Gold kit (Zymo Research Corporation) according to the manufacturer’s instructions.
Bisulfite modified sequencing
The MethPrimer website (http://www.urogene.org/methprimer) was used to design primers without CpG sites. Thirteen CpG sites located between −1173 and +117 bp were analyzed with 2 pairs of nested PCR primers. Primer information is available upon request from the corresponding author.
Thermocycler conditions comprised an activation step at 94°C for 2 minutes, followed by a 3-step PCR program, which consisted of 94°C for 30 seconds, 55°C for 60 seconds, 72°C for 60 seconds for 35 cycles, and a final extension at 72°C for 3 minutes. The PCR products were diluted 50×, and inner reactions were run. The PCR products were cloned with a TOPO TA cloning kit using One Shot TOP 10 Chemically Competent Escherichia coli (Invitrogen), followed by purification of the plasmids with a PureLink Quick Plasmid MiniPrep kit (Invitrogen). Five plasmids were sequenced for each sample by Eurofins MWG Operon.
Pyrosequencing
After bisulfite modification, a 40-μl PCR was carried out in 3.2 μl bisulfite-modified DNA (30 ng), 36 μl Platinum PCR Supermix High Fidelity (Invitrogen), and 200 nM of each primer. Thermal cycling conditions consisted of an initial activation step at 95°C for 5 minutes, followed by a 3-step PCR program of 95°C for 15 seconds, annealing for 30 seconds (58°C for iNOS1, iNOS2, and −5.2 kb enhancer, 52°C for −5.4 kb enhancer, and 56°C for −5.8 kb enhancer), and 72°C for 30 seconds for 40–50 cycles. PCR products were checked by 1% agarose gel electrophoresis using 10 μl of each product. The percent DNA methylation in the human iNOS enhancer and promoter were quantified using PyroMark MD (Qiagen) according to the manufacturer’s instructions. Five primer sets were designed to cover all 13 CpG sites within 1,290 bp of the iNOS promoter and 16 CpG sites within ~900 bp of the NF-κB enhancer elements (GenBank accession no. AF017634). The primers used for pyrosequencing were designed with Pyrosequencing Assay Design Software version 2.0 (Qiagen). Primer information is available upon request from the corresponding author.
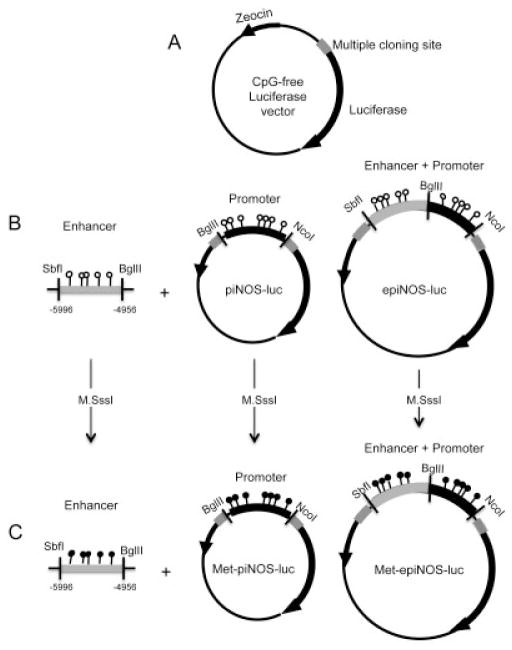
Plasmids, stable transfections, and reporter gene assay
The iNOS promoter (piNOS) and enhancer plus promoter (epiNOS) constructs were generated by PCR amplification using Platinum PCR Supermix High Fidelity (Invitrogen), and genomic DNA from human articular chondrocytes was used as a template. Information about primers used for cloning is available upon request from the corresponding author. The resulting PCR products were cloned in the CpG-free vector. The vector has no CpG sites in the whole vector sequence and was generated as described by Hashimoto et al (unpublished observations) according to the report by Klug and collaborators (18). The piNOS-luc construct was generated by using the FastDigest restriction enzymes Bgl II and Nco I (Fermentas) to cut the DNA from −1144 to +25 bp. Another construct with the enhancer was generated by using the FastDigest restriction enzymes Sbf I and Bgl II (Fermentas) to cut the DNA from −5996 to −4906 bp, and subsequently, the epiNOS construct was generated by ligation of both constructs. The authenticity of the DNA insert was confirmed by sequence analysis. E coli GT115 competent cells (InvivoGen) were used for the transformation.
In vitro DNA methylation and transient transfection
The methylated plasmids (Met-piNOS-luc and Met-epiNOS-luc) were generated by incubating 1 μg of plasmid DNA with 4 units/μl of CpG methyltransferase (M.SssI; New England Biolabs) in reaction buffer and 1,600 μM S-adenosyl methionine according to the manufacturer’s instructions. Complete methylation was verified by plasmid DNA bisulfite modification and pyrosequencing using iNOS-2 primers. The methylated plasmid DNA was purified using a GeneJET PCR purification kit (Fermentas) and transfected (0.5 μg) into C-28/I2 chondrocyte cells in parallel with the unmethylated piNOS-luc and epiNOS-luc, along with control Renilla vector using Turbofect reagent (Fermentas). In cotransfections, the expression vectors for NF-κB (p50 [Addgene plasmid 21965], p65 [Addgene plasmid 21966], or p50/p65) were used (0.06 μg). Blank expression vector pCMV4 (generously provided by Dr. David Russell, University of Texas) was used as a control. Total DNA was normalized with empty vectors in the transfection mixture. In all of the transient transfections, the internal control reporter (pRLV Renilla luciferase) was present at 1 ng/well. The luciferase assay was conducted using a 96-well luminometer with the dual luciferase substrate system (Promega) 24 hours after transfection. Relative luciferase activity was normalized to the internal control Renilla luciferase activity. Each experiment was repeated at least 3 times, and each data point was calculated as the mean ± SD of 3 wells per experiment.
Statistical analysis
Statistical analysis was performed using SPSS software version 17.0. Except where indicated otherwise, data are the mean ± SD of at least 3 multiple independent experiments. Significance was determined by analysis of variance with post hoc t-test and by Mann-Whitney U test. P values less than 0.05 were considered significant.
RESULTS
Differential expression profile of iNOS in articular chondrocytes
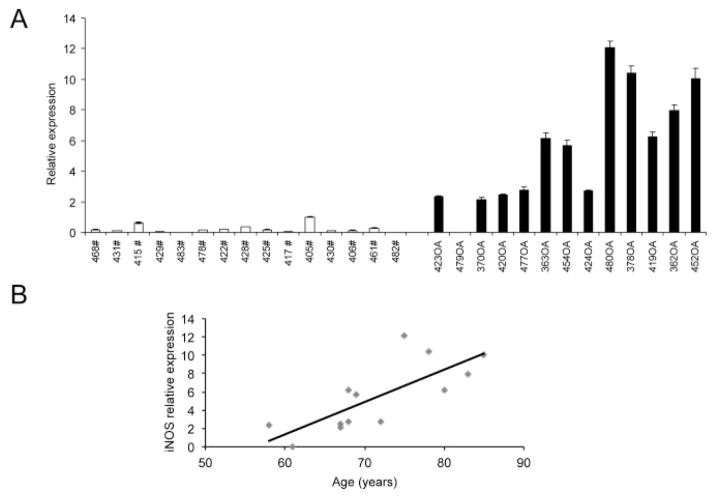
We first examined iNOS expression in articular chondrocytes from healthy controls (with femoral neck fracture) and OA patients. Differential expression of iNOS was determined by qRT-PCR analysis. A significant increase in relative iNOS expression was observed in OA chondrocytes compared to control (mean ± SD 0.22 ± 0.27 in controls versus 5.46 ± 3.77 in OA samples) (Figure 1A). Furthermore, a significant correlation between donor age and iNOS expression in OA chondrocytes was observed (Figure 1B). However, we observed no difference in iNOS expression between men and women.
Figure 1.
Inducible nitric oxide synthase (iNOS) expression in primary human chondrocytes obtained from control subjects with femoral neck fracture and patients with osteoarthritis (OA). A, Differential expression of iNOS in primary human chondrocytes obtained from control subjects with femoral neck fracture (#; n = 15) and those obtained from OA patients (n = 13), analyzed by quantitative reverse transcription–polymerase chain reaction. Values are the mean ± SD of triplicate determinations per sample. Results for each group are shown in order of increasing patient age from left to right. B, Correlation between donor age and iNOS expression in OA chondrocytes, as determined by Spearman’s rank order correlation (R2 = 0.85, P < 0.0005).
Localization of iNOS-producing chondrocytes
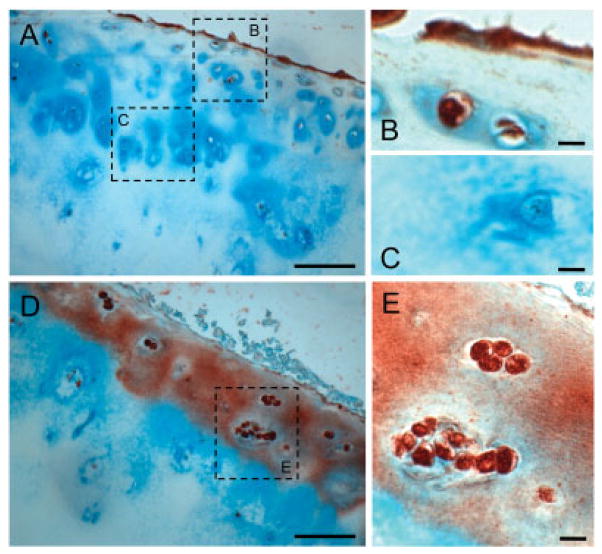
Immunohistochemical analysis demonstrated that the majority of articular chondrocytes from patients with femoral neck fracture (Mankin score 0–2) produced no detectable iNOS (Figure 2A), with only a few immune-positive cells in the superficial layer (magnified in Figure 2B) and negative-staining chondrocytes in the deep layer (magnified in Figure 2C). In contrast, cartilage from patients with late-stage OA showed intensely stained iNOS-positive cells distributed throughout the cartilage (Figure 2D). Furthermore, all cells within the clusters of chondrocytes were iNOS-positive in patients with severe OA (Mankin score 14) (magnified in Figure 2E).
Figure 2.
Localization of inducible nitric oxide synthase (iNOS) in human articular cartilage by immunohistochemistry. A, Cartilage from a patient with femoral neck fracture (Mankin score 0–2). B and C, Higher-magnification views of the boxed areas in A. Only a few cells in the superficial layer were immunopositive for iNOS (B), whereas the bulk of chondrocytes were still negative (C). D, Cartilage from a patient with late-stage osteoarthritis (Mankin score 14). E, Higher-magnification view of the boxed area in D. All of the chondrocytes in the cluster were immunopositive for iNOS. Bars in A and D = 100 μm; bars in B, C, and E = 10 μm.
Identification of differentially methylated CpG sites in the iNOS promoter
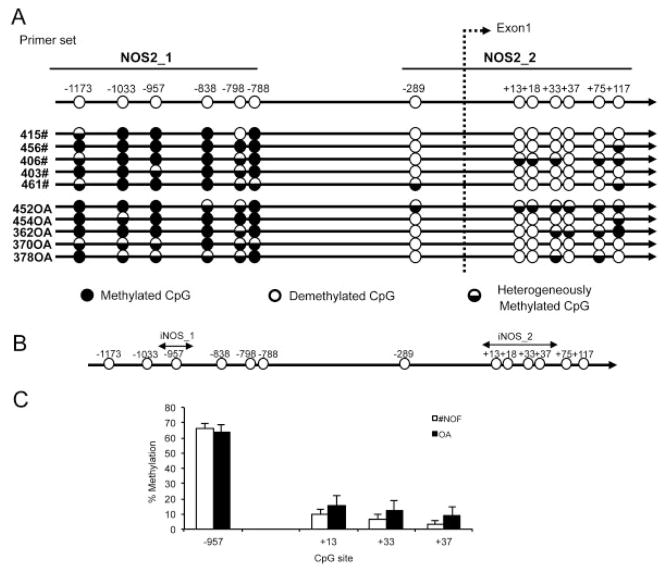
Of 13 CpG sites in the 1.3-kb sequence (7 CpG sites in the promoter and 6 in exon 1), 6 CpG sites in the promoter were predominantly methylated and the remaining 7 sites were predominantly demethylated in both samples from patients with femoral neck fracture and samples from patients with OA (Figure 3A). These results indicate that epigenetic regulation of iNOS in human chondrocytes does not involve the region of the promoter between −1400 and +117 bp. Subsequently, pyrosequencing analysis was used to confirm these results and to quantify the percentage methylation of the CpG sites. Pyrosequencing for CpG sites at −957 (iNOS_1 primer) and +13 to +37 (iNOS_2 primer) confirmed the bisulfite sequencing results (Figures 3B and C).
Figure 3.
CpG methylation in the inducible nitric oxide synthase (iNOS) proximal promoter. A, Bisulfite sequencing analysis of the iNOS promoter in chondrocytes obtained from patients with femoral neck fracture (#NOF) and patients with osteoarthritis (OA) (n = 5 patients per group). Each circle represents the average result of 5 sequence clones. B, Pyrosequencing analysis of the iNOS promoter. Circles indicate the locations of CpG sites in the iNOS proximal promoter. Arrows indicate the sequences used for PCR primers for pyrosequencing. C, Percentage methylation at each specific CpG site in the iNOS proximal promoter in human articular chondrocytes from patients with femoral neck fracture and patients with OA. Values are the mean ± SD (n = 11 patients per group).
Identification of differentially methylated CpG sites in NF-κB enhancer elements of the iNOS gene
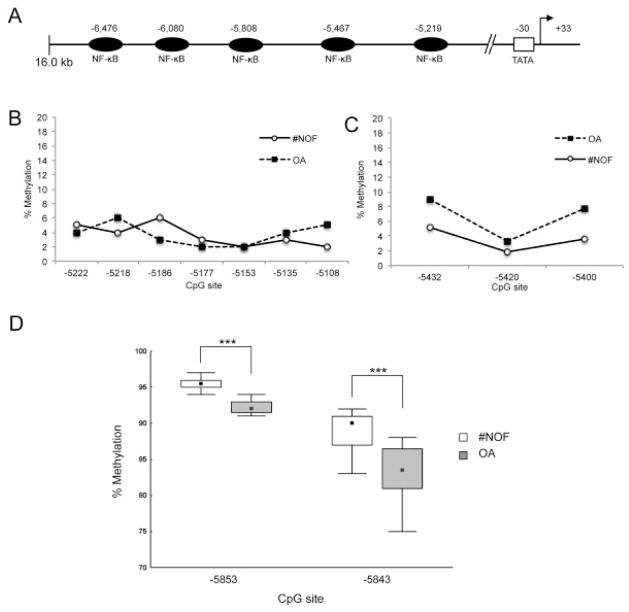
It is known that the human iNOS gene possesses multiple functional NF-κB binding sites that confer cytokine inducibility (19) with the functional promoter elements upstream of −4.7 kb. The promoter elements consist of a novel enhancer region of ~800 bp, which contains 4 functional NF-κB binding sites (Figure 4A). For pyrosequencing analysis, 3 primer sets were designed to analyze the CpG sites closest to the NF-κB binding sites localized at −5.8 kb, −5.4 kb, and −5.2 kb. All CpG sites analyzed for the enhancer elements at −5.4 kb and −5.2 kb were found to be completely demethylated in samples from both patients with femoral neck fracture and patients with OA (Figures 4B and C). However, 2 CpG sites, at −5853 and −5843, were significantly demethylated in OA samples in comparison to samples from patients with femoral neck fracture (mean ± SD 96 ± 1.03% for femoral neck fracture versus 92 ± 1.88% for OA at −5853 and 89 ± 2.77% for femoral neck fracture versus 83 ± 3.77% for OA at −5843) (P < 0.001) (Figure 4D).
Figure 4.
The 5′-flanking region of the human inducible nitric oxide synthase (iNOS) gene is a target for the transcription factor NF-κB. A, Schematic diagram of NF-κB binding sites in the iNOS promoter. B–D, Pyrosequencing analysis of the DNA methylation status of CpG sites in the NF-κB binding element at −5.2 kb (B), −5.4 kb (C), and −5.8 kb (D) in the iNOS gene in chondrocytes from patients with femoral neck fracture (#NOF) and patients with osteoarthritis (OA). Values in B and C are the mean (n = 12 patients per group). Data in D are shown as box plots, where the boxes represent the 25th to 75th percentiles, the squares within the boxes represent the median, and the error bars represent the 10th and 90th percentiles. *** = P < 0.01 by Mann-Whitney U test.
Expression of the iNOS gene in human chondrocytes is mediated by the p65 subunit of NF-κB
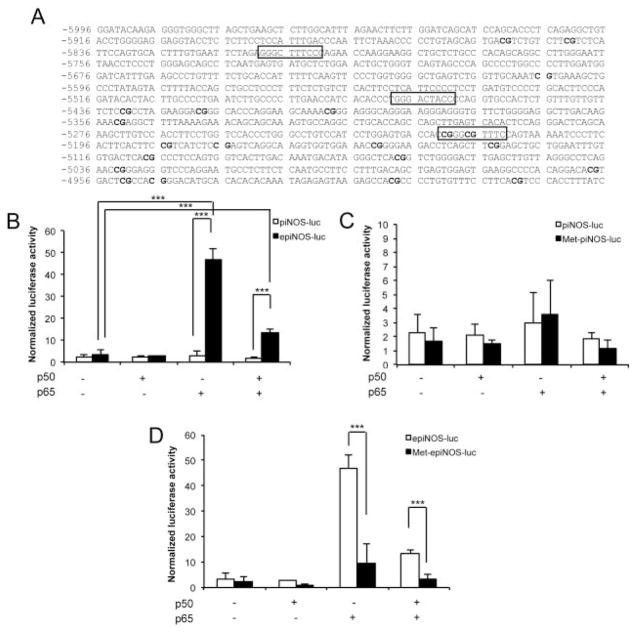
The effect of transcription factor NF-κB on the regulation of the human iNOS gene in chondrocytes was determined using a CpG-free reporter assay. The iNOS promoter (piNOS) and the combination of enhancer and promoter (epiNOS) constructs were cloned into the CpG-free vector (Figures 5A–C). The epiNOS construct contains the 5′-flanking region of the human iNOS gene from −5996 to −4956, containing 3 NF-κB binding sites and 21 CpG sites, most of which have previously been analyzed by pyrosequencing (Figure 6A). The reporter vectors were then transfected into C-28/I2 cells, a human chondrocyte cell line, with or without cotransfection of expression vectors coding p50 (pCMV4-p50) and p65 (pCMV4-p65), alone and in combination, to analyze the effect of NF-κB on iNOS activity.
Figure 5.
Various inducible nitric oxide synthase (iNOS) constructs generated using the CpG-free vector. A, Map of the CpG-free reporter vector. The vector, including its multiple cloning site, is completely free of CpG dinucleotides. It contains a Zeocin resistance gene. B, The iNOS promoter (piNOS) and enhancer plus promoter (epiNOS) constructs before methylation. C, The iNOS promoter (Met-piNOS) and enhancer plus promoter (Met-epiNOS) constructs after CpG methylation treatment with M.SssI.
Figure 6.
NF-κB–mediated inducible nitric oxide synthase (iNOS) transactivation is affected by the CpG methylation of its enhancer region in human chondrocytes. A, Sequence of the 5′-flanking region of the human iNOS gene from −5996 to −4956. The 3 putative NF-κB binding sites in the construct are highlighted. B, Transcriptional activation of the iNOS promoter (piNOS) and enhancer plus promoter (epiNOS) elements by cotransfection with the p65 NF-κB subunit. C, Lack of effect of CpG methylation on the iNOS promoter construct (Met-piNOS). D, Significant reduction in the p65 overexpression–induced enhanced iNOS activity in the enhancer plus promoter construct (Met-epiNOS) after CpG methylation. Values are the mean ± SD of 3–5 independent experiments, each performed in triplicate. *** = P < 0.01 by analysis of variance with post hoc t-test.
No difference was found in relative iNOS activity between the reporter containing the promoter (piNOS-luc) and the reporter with combination of enhancer and promoter elements (epiNOS-luc) (mean ± SD 0.9 ± 0.6 versus 0.9 ± 0.5). Methylation of either construct reduced iNOS activity, although the reduction was not statistically significant (0.89 ± 0.55 versus 0.54 ± 0.40 for piNOS-luc and 0.91 ± 0.52 versus 0.52 ± 0.36 for epiNOS-luc; data not shown).
In contrast, when the epiNOS construct was cotransfected with pCMV4-p65, alone or in combination with pCMV4-p50, iNOS-driven reporter activities were significantly elevated. Interestingly, the enhanced activity was higher with overexpression of p65 alone (3.4 ± 2.2% versus 46.7 ± 5.4%) than with overexpression of p50 and p65 combined (3.4 ± 2.2% versus 13.5 ± 1.5%) (Figure 6B). Furthermore, cotransfection of NF-κB subunits did not alter the activity of the iNOS proximal promoter (piNOS) (Figure 6B).
NF-κB–mediated iNOS transactivation is affected by the CpG methylation of its enhancer region in human chondrocytes
The iNOS promoter construct (piNOS) and the iNOS enhancer plus promoter construct (epiNOS) were methylated in vitro using CpG methyltransferase (M.SssI) to analyze the effect of CpG methylation on iNOS activity. The suppressive effect of CpG methylation on the iNOS proximal promoter (Met-piNOS) was not significant (2.3 ± 1.3% for piNOS versus 1.7 ± 0.9% for Met-piNOS) (Figure 6C). Similar results were observed when subunits of NF-κB were overexpressed by cotransfection. In contrast, with the epiNOS construct, the enhanced iNOS activity induced by p65 overexpression was significantly reduced after methylation treatment (Met-epiNOS) (46.7 ± 5.4% for epiNOS versus 9.6 ± 7.7% for Met-epiNOS). This suppressive effect of methylation treatment also occurred with the p65/p50-driven transactivated epiNOS construct (13.5 ± 1.5% for epiNOS versus 3.4 ± 1.8% for Met-epiNOS) (Figure 6D).
DISCUSSION
In the present study, we examined the role and importance of methylation status in the expression of iNOS, the major NOS responsible for NO production in OA, a late-onset polygenic disease. Methylation of genomic DNA represents a significant mechanism for regulating tissue-specific gene expression (4), and epigenetic dysregulation, in combination with genetic predisposition, is thought to be involved in several complex, non-Mendelian diseases, such as Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, and hyperparathyroidism (20). NO synthesis is enhanced in OA cartilage (21). The increased expression of iNOS and cyclooxygenase 2 (COX-2) in OA chondrocytes is, in part, a consequence of enhanced stress-induced signaling that can be stimulated by mechanical signals (22) or proinflammatory cytokines, particularly interleukin-1β (IL-1β), which acts in an autocrine/paracrine manner to perpetuate a catabolic state resulting in progressive destruction of articular cartilage (23). Consistent with these findings, iNOS expression was observed in samples from OA patients in this study and, furthermore, iNOS expression was found to correlate with patient age and disease severity.
We have previously shown an association between loss of DNA methylation in the promoter and abnormal expression of matrix metalloproteinase 3 (MMP-3), MMP-9, MMP-13, and ADAMTS-4 by OA chondrocytes (4). However, in contrast to our observations for catabolic enzymes (3,4) and the proinflammatory cytokine IL-1β (2), loss of DNA methylation at specific CpG sites in the proximal promoter of the iNOS gene could not account for its aberrant expression. The methylation status of the iNOS proximal promoter was analyzed by bisulfite sequencing and pyrosequencing. Identical results were observed: 6 CpG sites localized between −1173 and −788 bp were predominantly methylated in healthy and OA chondrocytes. In contrast, one CpG site at −289, the CpG site closest to the transcription start site, and 6 CpG sites localized in the coding region near the transcription start site of the iNOS gene were predominantly demethylated in all samples analyzed.
Expression of iNOS is primarily regulated by transcriptional and posttranscriptional mechanisms (24) with iNOS expression typically requiring exposure to inflammatory stimuli (25–27). Taylor and coworkers have reported that NF-κB plays a crucial role in human iNOS gene regulation (19) and identified NF-κB response elements in the human iNOS promoter. It is known that NF-κB regulation is dependent on induction of ESE1 transcription through NF-κB binding sites on iNOS (28) and COX-2 (29) promoters, although, in this case, it is important to note that the ESE1 binding sites are distant from CpG sites in the iNOS promoter. Unlike the murine iNOS promoter, the sequences within the first 1.0 kb of the 5′-flanking region of the human iNOS gene are not sufficient for iNOS induction (13), but rather inducible NF-κB binding elements located at more than −4.7 kb upstream of the transcription start site are known to be required for cytokine activation of the promoter. Specifically, a unique upstream cytokine-responsive enhancer region from −5.2 to −6.1 kb in the human iNOS gene contains 4 cis-acting NF-κB binding elements (19).
CpG analysis of enhancer elements by pyrosequencing indicated that CpG sites located close to the −5.8 kb (but not −5.5 or −5.2 kb) enhancer element displayed a significant demethylation in OA chondrocytes, which could account for the increased iNOS expression in OA cartilage. These results are consistent with the findings of Taylor et al (19) since the authors showed that within the context of the 7.2-kb construct, the site at −5.8 kb is essential for iNOS promoter activity. However, sites at −5.2, −5.5, and −6.1 kb are also functionally important and regulate iNOS gene expression, as already indicated (19).
Interestingly, the NF-κB elements from −5.2 to −6.1 kb in the human iNOS gene are spaced in approximate multiples of nucleosome units (200 bp), which may contribute to the 3-dimensional structure necessary for efficient iNOS transcription (19). The configuration of the enhancer region, particularly sequence similarity between species, its position from the transcription start site, and the pattern of conserved transcription factor binding sites (30) are likely to be important in iNOS regulation, not only in humans, but also across species (19).
Promoter regions without CpG islands are variably methylated in normal cells, typically cell specific, reflecting the transcriptional activity of the gene (13). To assess the influence of the transcription factor NF-κB and methylation in iNOS expression in human chondrocytes, we used a CpG-free vector (18) system providing simple and robust analysis of CpG methylation effects on the iNOS gene promoter within a transfection assay (18). NF-κB complexes comprise a number of different subunits, namely, p50, p52, RelA (p65), RelB, and c-Rel/Rel, which associate in various homo- or heterodimers through an amino acid sequence known as the Rel homology domain (31). Of these, the prototypical NF-κB complex (canonical pathway) is a heterodimer composed of p65 and p50 subunits. The p50/p65 heterodimers and p65 homodimers represent transcriptionally active forms of NF-κB.
The activity of the iNOS enhancer plus promoter construct was augmented by the overexpression of p65 or combination of p65/p50. However, significant activity was not detected following overexpression of p50, and the relative activity was higher with overexpression of p65 alone than with overexpression of p50/p65. This observation could be a consequence of p50/p50 homodimers acting as repressors of NF-κB–dependent genes by inhibiting transactivation of the p50/p65 heterodimers (32,33). This inhibitory function can be enhanced when p50/p50 homodimers interact with the transcription corepressor histone deacetylase (HDAC) (34). Guan et al have reported that constitutive DNA binding of p50/p50 is an important mechanism used by immune cells to suppress expression of NF-κB target genes (35). Furthermore, p65/p65 homodimers are more uniquely associated with tumor necrosis factor receptor–associated factor 6 and IL-1/lipopolysaccharide (LPS) signaling, consistent with IκBβ-mediated activation and resistance to IκBζ inhibition, both of which are triggered by either IL-1 or LPS (36).
NF-κB activation has been widely implicated in inflammatory diseases, and significant attention has been focused on the development of antiinflammatory drugs targeting NF-κB (37). Homodimers of the p50 subunit lacking transactivation domains have been shown to repress expression of NF-κB targets genes including tumor necrosis factor α (38). Interestingly, in some cases p50/p50 homodimers can also activate gene expression when complexed with coactivators, such as Bcl-3 (39,40). This cofactor-mediated transactivating effect of p50/p50 on one gene is not believed to undermine the suppressive function of the homodimer on other genes (35). Given that NF-κB p65 plays a major role in the expression of key inflammatory cytokines involved in the pathogenesis of arthritis, it is likely that NF-κB p65–specific small interfering RNA could be developed as a powerful approach to prevent induction of the degeneration of cartilage in OA in vitro (41). NF-κB signaling molecules orchestrate a number of proinflammatory and stress-like processes and control aspects of cellular differentiation, further substantiating their potential as OA therapeutic targets (42).
Finally, we also observed that the p65 or p65/p50-driven iNOS transactivation was abolished by CpG methylation treatment. DNA methylation can repress gene transcription either by directly inhibiting the interaction of transcription factors with their regulatory sequences, or through attraction of methylated DNA binding proteins that in turn recruit HDACs and histone methyltransferases, resulting in an inactive chromatin structure (43). Thus, changes in DNA methylation status at key CpG sites may be dependent on binding of NF-κB family members to initiate demethylation (44,45). We have recently shown that an inhibitor of NF-κB and the disease-modifying agent glucosamine significantly ameliorate the IL-1β–induced IL1B expression in human chondrocytes. This event is associated with demethylation in the specific CpG site in the IL1B promoter (1).
NO can activate DNA methyltransferases through a posttranscriptional mechanism to induce DNA methylation and repress the expression of several genes, including FMR1 and HPRT (46). It is possible that methylation of the iNOS promoter presents a feedback inhibition by which the end product NO can silence iNOS itself and hence its own production, thus reducing host cell damage (13). Surprisingly, even though it is difficult to induce iNOS expression in vitro, there are important human disease processes in which iNOS pathophysiology plays a prominent role, particularly chronic inflammatory conditions such as rheumatoid arthritis and tuberculosis (47,48), indicating the importance of understanding the epigenetic regulation of iNOS in the pathogenesis of iNOS expression in various diseases (14). Recently, Chan et al have shown that the human iNOS gene is epigenetically silenced/repressed not only by DNA methylation but also by H3K9 methylation (14). HDAC inhibitors suppress IL-1–induced NO and prostaglandin E2 synthesis, iNOS and COX-2 expression, as well as proteoglycan degradation. The suppressive effects of HDAC inhibitors are not due to impaired DNA binding activity of NF-κB (49). H3K4 methylation by Set-1A contributes to the induction of iNOS and COX2 expression by IL-1 and thus HDAC and Set-1A may be novel therapeutic targets for OA and other conditions (50).
In summary, to our knowledge, this is the first study to show the involvement of demethylation in specific NF-κB enhancer elements in the activation of iNOS in human OA chondrocytes. These data provide evidence linking the activation of iNOS, demethylation and, critically, induction of the OA process, indicating that this pathway could be a potential target for pharmacologic intervention in the treatment of OA and other arthritic diseases.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Oreffo had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. De Andrés, Imagawa, Hashimoto, Roach, Goldring, Oreffo.
Acquisition of data. De Andrés, Imagawa, Hashimoto.
Analysis and interpretation of data. De Andrés, Imagawa, Hashimoto, Gonzalez, Roach, Goldring, Oreffo.
References
- 1.Imagawa K, de Andres MC, Hashimoto K, Pitt D, Itoi E, Goldring MB, et al. The epigenetic effect of glucosamine and a nuclear factor-κB (NF-kB) inhibitor on primary human chondrocytes—implications for osteoarthritis. Biochem Biophys Res Commun. 2011;405:362–7. doi: 10.1016/j.bbrc.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashimoto K, Oreffo RO, Gibson MB, Goldring MB, Roach HI. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 2009;60:3303–13. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung KS, Hashimoto K, Yamada N, Roach HI. Expression of ADAMTS-4 by chondrocytes in the surface zone of human osteoarthritic cartilage is regulated by epigenetic DNA demethylation. Rheumatol Int. 2009;29:525–34. doi: 10.1007/s00296-008-0744-z. [DOI] [PubMed] [Google Scholar]
- 4.Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110–24. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 5.Roach HI, Aigner T. DNA methylation in osteoarthritic chondrocytes: a new molecular target. Osteoarthritis Cartilage. 2007;15:128–37. doi: 10.1016/j.joca.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Maneiro E, Lopez-Armada MJ, de Andres MC, Carames B, Martin MA, Bonilla A, et al. Effect of nitric oxide on mitochondrial respiratory activity of human articular chondrocytes. Ann Rheum Dis. 2005;64:388–95. doi: 10.1136/ard.2004.022152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadler J, Stefanovic-Racic M, Billiar TR, Curran RD, McIntyre LA, Georgescu HI, et al. Articular chondrocytes synthesize nitric oxide in response to cytokines and lipopolysaccharide. J Immunol. 1991;147:3915–20. [PubMed] [Google Scholar]
- 8.Amin AR, Attur M, Patel RN, Thakker GD, Marshall PJ, Rediske J, et al. Superinduction of cyclooxygenase-2 activity in human osteoarthritis-affected cartilage: influence of nitric oxide. J Clin Invest. 1997;99:1231–7. doi: 10.1172/JCI119280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amin AR, Di Cesare PE, Vyas P, Attur M, Tzeng E, Billiar TR, et al. The expression and regulation of nitric oxide synthase in human osteoarthritis-affected chondrocytes: evidence for upregulated neuronal nitric oxide synthase. J Exp Med. 1995;182:2097–102. doi: 10.1084/jem.182.6.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charles IG, Palmer RM, Hickery MS, Bayliss MT, Chubb AP, Hall VS, et al. Cloning, characterization, and expression of a cDNA encoding an inducible nitric oxide synthase from the human chondrocyte. Proc Natl Acad Sci U S A. 1993;90:11419–23. doi: 10.1073/pnas.90.23.11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taskiran D, Stefanovic-Racic M, Georgescu H, Evans C. Nitric oxide mediates suppression of cartilage proteoglycan synthesis by interleukin-1. Biochem Biophys Res Commun. 1994;200:142–8. doi: 10.1006/bbrc.1994.1426. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi T, Abe E, Yamate T, Taguchi Y, Jasin HE. Nitric oxide production by superficial and deep articular chondrocytes. Arthritis Rheum. 1997;40:261–9. doi: 10.1002/art.1780400210. [DOI] [PubMed] [Google Scholar]
- 13.Yu Z, Kone BC. Hypermethylation of the inducible nitric-oxide synthase gene promoter inhibits its transcription. J Biol Chem. 2004;279:46954–61. doi: 10.1074/jbc.M407192200. [DOI] [PubMed] [Google Scholar]
- 14.Chan GC, Fish JE, Mawji IA, Leung DD, Rachlis AC, Marsden PA. Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J Immunol. 2005;175:3846–61. doi: 10.4049/jimmunol.175.6.3846. [DOI] [PubMed] [Google Scholar]
- 15.Chu SC, Marks-Konczalik J, Wu HP, Banks TC, Moss J. Analysis of the cytokine-stimulated human inducible nitric oxide synthase (iNOS) gene: characterization of differences between human and mouse iNOS promoters. Biochem Biophys Res Commun. 1998;248:871–8. doi: 10.1006/bbrc.1998.9062. [DOI] [PubMed] [Google Scholar]
- 16.Pritzker KP, Gay S, Jimenez SA, Ostergaard K, Pelletier JP, Revell PA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14:13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto K, Kokubun S, Itoi E, Roach HI. Improved quantification of DNA methylation using methylation-sensitive restriction enzymes and real-time PCR. Epigenetics. 2007;2:86–91. doi: 10.4161/epi.2.2.4203. [DOI] [PubMed] [Google Scholar]
- 18.Klug M, Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1:127–30. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 19.Taylor BS, de Vera ME, Ganster RW, Wang Q, Shapiro RA, Morris SM, Jr, et al. Multiple NF-κB enhancer elements regulate cytokine induction of the human inducible nitric oxide synthase gene. J Biol Chem. 1998;273:15148–56. doi: 10.1074/jbc.273.24.15148. [DOI] [PubMed] [Google Scholar]
- 20.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–7. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 21.Amin AR, Dave M, Attur M, Abramson SB. COX-2, NO, and cartilage damage and repair. Curr Rheumatol Rep. 2000;2:447–53. doi: 10.1007/s11926-000-0019-5. [DOI] [PubMed] [Google Scholar]
- 22.Nam J, Aguda BD, Rath B, Agarwal S. Biomechanical thresholds regulate inflammation through the NF-κB pathway: experiments and modeling. PLoS One. 2009;4:e5262. doi: 10.1371/journal.pone.0005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abramson SB, Attur M, Amin AR, Clancy R. Nitric oxide and inflammatory mediators in the perpetuation of osteoarthritis. Curr Rheumatol Rep. 2001;3:535–41. doi: 10.1007/s11926-001-0069-3. [DOI] [PubMed] [Google Scholar]
- 24.Kleinert H, Schwarz PM, Forstermann U. Regulation of the expression of inducible nitric oxide synthase. Biol Chem. 2003;384:1343–64. doi: 10.1515/BC.2003.152. [DOI] [PubMed] [Google Scholar]
- 25.Nathan C, Xie QW. Regulation of biosynthesis of nitric oxide. J Biol Chem. 1994;269:13725–8. [PubMed] [Google Scholar]
- 26.Geller DA, de Vera ME, Russell DA, Shapiro RA, Nussler AK, Simmons RL, et al. A central role for IL-1β in the in vitro and in vivo regulation of hepatic inducible nitric oxide synthase. IL-1β induces hepatic nitric oxide synthesis. J Immunol. 1995;155:4890–8. [PubMed] [Google Scholar]
- 27.Geller DA, Lowenstein CJ, Shapiro RA, Nussler AK, Di Silvio M, Wang SC, et al. Molecular cloning and expression of inducible nitric oxide synthase from human hepatocytes. Proc Natl Acad Sci U S A. 1993;90:3491–5. doi: 10.1073/pnas.90.8.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudders S, Gaspar J, Madore R, Voland C, Grall F, Patel A, et al. ESE-1 is a novel transcriptional mediator of inflammation that interacts with NF-κB to regulate the inducible nitric-oxide synthase gene. J Biol Chem. 2001;276:3302–9. doi: 10.1074/jbc.M006507200. [DOI] [PubMed] [Google Scholar]
- 29.Grall FT, Prall WC, Wei W, Gu X, Cho JY, Choy BK, et al. The Ets transcription factor ESE-1 mediates induction of the COX-2 gene by LPS in monocytes. FEBS J. 2005;272:1676–87. doi: 10.1111/j.1742-4658.2005.04592.x. [DOI] [PubMed] [Google Scholar]
- 30.Rico D, Vaquerizas JM, Dopazo H, Bosca L. Identification of conserved domains in the promoter regions of nitric oxide synthase 2: implications for the species-specific transcription and evolutionary differences. BMC Genomics. 2007;8:271. doi: 10.1186/1471-2164-8-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 32.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–36. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 33.Sanjabi S, Hoffmann A, Liou HC, Baltimore D, Smale ST. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc Natl Acad Sci U S A. 2000;97:12705–10. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vermeulen L, De Wilde G, Van Damme P, Vanden Berghe W, Haegeman G. Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1) EMBO J. 2003;22:1313–24. doi: 10.1093/emboj/cdg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan H, Hou S, Ricciardi RP. DNA binding of repressor nuclear factor-κB p50/p50 depends on phosphorylation of Ser337 by the protein kinase A catalytic subunit. J Biol Chem. 2005;280:9957–62. doi: 10.1074/jbc.M412180200. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida Y, Kumar A, Koyama Y, Peng H, Arman A, Boch JA, et al. Interleukin 1 activates STAT3/nuclear factor-κB cross-talk via a unique TRAF6- and p65-dependent mechanism. J Biol Chem. 2004;279:1768–76. doi: 10.1074/jbc.M311498200. [DOI] [PubMed] [Google Scholar]
- 37.Karin M, Yamamoto Y, Wang QM. The IKK NF-κB system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 38.Kastenbauer S, Ziegler-Heitbrock HW. NF-κB1 (p50) is upregulated in lipopolysaccharide tolerance and can block tumor necrosis factor gene expression. Infect Immun. 1999;67:1553–9. doi: 10.1128/iai.67.4.1553-1559.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thornburg NJ, Pathmanathan R, Raab-Traub N. Activation of nuclear factor-κB p50 homodimer/Bcl-3 complexes in nasopharyngeal carcinoma. Cancer Res. 2003;63:8293–301. [PubMed] [Google Scholar]
- 40.Budunova IV, Perez P, Vaden VR, Spiegelman VS, Slaga TJ, Jorcano JL. Increased expression of p50-NF-κB and constitutive activation of NF-κB transcription factors during mouse skin carcinogenesis. Oncogene. 1999;18:7423–31. doi: 10.1038/sj.onc.1203104. [DOI] [PubMed] [Google Scholar]
- 41.Lianxu C, Hongti J, Changlong Y. NF-κBp65-specific siRNA inhibits expression of genes of COX-2, NOS-2 and MMP-9 in rat IL-1β-induced and TNF-α-induced chondrocytes. Osteoarthritis Cartilage. 2006;14:367–76. doi: 10.1016/j.joca.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Marcu KB, Otero M, Olivotto E, Borzi RM, Goldring MB. NF-κB signaling: multiple angles to target OA. Curr Drug Targets. 2010;11:599–613. doi: 10.2174/138945010791011938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation—how does it all fit together? J Cell Biochem. 2002;87:117–25. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- 44.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-κB, Lin28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-κB in B-cell-specific demethylation of the Igκ locus. Nat Genet. 1996;13:435–41. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 46.Hmadcha A, Bedoya FJ, Sobrino F, Pintado E. Methylation-dependent gene silencing induced by interleukin 1β via nitric oxide production. J Exp Med. 1999;190:1595–604. doi: 10.1084/jem.190.11.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St Clair EW, Wilkinson WE, Lang T, Sanders L, Misukonis MA, Gilkeson GS, et al. Increased expression of blood mononuclear cell nitric oxide synthase type 2 in rheumatoid arthritis patients. J Exp Med. 1996;184:1173–8. doi: 10.1084/jem.184.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholson S, Bonecini-Almeida MD, Lapa e Silva JR, Nathan C, Xie QW, Mumford R, et al. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J Exp Med. 1996;183:2293–302. doi: 10.1084/jem.183.5.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chabane N, Zayed N, Afif H, Mfuna-Endam L, Benderdour M, Boileau C, et al. Histone deacetylase inhibitors suppress interleukin-1β-induced nitric oxide and prostaglandin E2 production in human chondrocytes. Osteoarthritis Cartilage. 2008;16:1267–74. doi: 10.1016/j.joca.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 50.El Mansouri FE, Chabane N, Zayed N, Kapoor M, Benderdour M, Martel-Pelletier J, et al. Contribution of H3K4 methylation by SET-1A to interleukin-1–induced cyclooxygenase 2 and inducible nitric oxide synthase expression in human osteoarthritis chondrocytes. Arthritis Rheum. 2011;63:168–79. doi: 10.1002/art.27762. [DOI] [PubMed] [Google Scholar]