Abstract
Many individually rare missense substitutions are encountered during deep resequencing of candidate susceptibility genes and clinical mutation screening of known susceptibility genes. BRCA1 and BRCA2 are among the most resequenced of all genes, and clinical mutation screening of these genes provides an extensive data set for analysis of rare missense substitutions. Align-GVGD is a mathematically simple missense substitution analysis algorithm, based on the Grantham difference, which has already contributed to classification of missense substitutions in BRCA1, BRCA2, and CHEK2. However, the distribution of genetic risk as a function of Align-GVGD's output variables Grantham variation (GV) and Grantham deviation (GD) has not been well characterized. Here, we used data from the Myriad Genetic Laboratories database of nearly 70,000 full-sequence tests plus two risk estimates, one approximating the odds ratio and the other reflecting strength of selection, to display the distribution of risk in the GV-GD plane as a series of surfaces. We abstracted contours from the surfaces and used the contours to define a sequence of missense substitution grades ordered from greatest risk to least risk. The grades were validated internally using a third, personal and family history-based, measure of risk. The Align-GVGD grades defined here are applicable to both the genetic epidemiology problem of classifying rare missense substitutions observed in known susceptibility genes and the molecular epidemiology problem of analyzing rare missense substitutions observed during case-control mutation screening studies of candidate susceptibility genes.
Keywords: BRCA1, BRCA2, Align-GVGD, unclassified variant, missense substitution, protein multiple sequence alignment
INTRODUCTION
The human gene pool harbors a vast number of rare missense substitutions, and approximately 70% of these are at least mildly deleterious [Kryukov et al., 2007]. In the fields of genetic and molecular epidemiology, large numbers of individually rare missense substitutions are most often encountered in two research settings: analysis of the unclassified variants (UVs) observed during clinical mutation screening of known susceptibility genes [Chan et al., 2007; Easton et al., 2007], and case-control mutation screening of candidate intermediate-risk susceptibility genes [Cohen et al., 2004; Landi et al., 2005; Rahman et al., 2006; Renwick et al., 2006; Seal et al., 2006]. In silico assessment of the expected effects of individual rare missense substitutions could contribute to both of these research areas if the assessment methods achieve a sufficient signal-to-noise ratio.
In the clinical mutation screening setting, we know by definition that at least some sequence variants that damage function of the susceptibility gene of interest are pathogenic for the disease of interest. When a patient is found to carry a specific missense UV, the research goal is to assess either the probability that, or the degree to which, that specific UV is pathogenic. For analysis of missense UVs in this setting, the current state of the art is to compile a multicomponent likelihood ratio that integrates assessment methods ranging from segregation analysis to immunohistochemical analysis [Goldgar et al., 2004, 2008; Easton et al., 2007; Spurdle et al., 2008]. By its nature, the multicomponent likelihood approach will benefit from either improvements to the methods already integrated or addition of new methods that summarize data sources independent to those already integrated.
On the other hand, in an archetypal case-control mutation screening study, we do not know whether damaging sequence variants in the gene of interest are pathogenic with respect to the disease of interest. In this setting, the research goal is to combine statistical evidence from the distribution of rare missense substitutions observed in cases and controls with statistical evidence from the distributions of more easily recognized damaging sequence variants, such as protein-truncating variants and clear splice-junction variants, to make an overall assessment of the status of the candidate gene. No systematic method for analysis of rare missense substitutions in this context has been described. The challenges here are twofold: to provide a method of assigning a risk estimate to each individual substitution that in aggregate has a good enough signal-to-noise ratio to actually contribute to the statistical analysis, and to provide those risk estimates in a format that allows them to be assessed along with other types of sequence variation.
Over the last several years, we and others have devised algorithms for in silico assessment of missense substitutions (for overview, see Tavtigian et al. [2008]). Most of these rely, at least in part, on the evolutionary information inherent in protein multiple sequence alignments and thus bear relatively directly on the question of whether a substitution is deleterious to evolutionary fitness but less directly on the probability to damage protein function or the probability of pathogenicity [Kryukov et al., 2007]. Derivations and/or optimizations of the classifiers tied to these algorithms have sometimes been based on measures of sensitivity and specificity against either in vitro, phage, bacterial, or yeast functional assays [Ng and Henikoff, 2001; Stone and Sidow, 2005; Karchin et al., 2007]; i.e., measurements of damage to protein function, an indirect surrogate for pathogenicity. In other cases, the sensitivity and specificity for “correct” classification of “known” neutral and pathogenic variants was used to optimize classifiers [Sunyaev et al., 2001; Yue et al., 2005; Ferrer-Costa et al., 2004; Capriotti et al., 2008], an approach that depends on the accuracy of classification of variants in the database from which sequence variants were extracted.
An alternative approach is to start with well-characterized susceptibility genes for which a rich data set is available, bypass the mapping between in silico assessment of sequence variation and either damage to protein function or results with “classified” variants, and instead investigate the relationship between in silico assessment of sequence variation and evidence of genetic risk in patients. Ideally, such an approach would lead to in silico prediction of a relative odds ratio of disease for each sequence variant, with only an overall scale factor to be fitted in a candidate gene study. Applied to candidate gene data, regressing the subjects’ case/control status on the predicted odds ratio of the missense variants that they carry would enable us to fit the scaling factor and test for the candidate gene's effect on the phenotype.
We describe in four steps an approach to the desired in silico predictions.
Select a dataset that contains a large number of missense substitutions and observational data that are linked to genetic risk. For this we use a data freeze from Myriad Genetics Laboratory's (Salt Lake, Utah) clinical BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185) mutation screening data (BRACAnalyis®).
Adopt a summary measure thought to predict missense substitution severity that can be applied to all of the missense substitutions in the dataset. Here we use a pair of variables, the Grantham variation (GV) and Grantham deviation (GD) [Tavtigian et al., 2006], which are based on the commonly used Grantham difference [Grantham, 1974].
Adopt an estimate of genetic risk that can be measured from pools of missense substitutions present in the data set used. For this we combine two measures: the ascertainment ratio (AR) [Tavtigian et al., 2006] and the enrichment ratio for single-nucleotide substitutions (ERS; derived here). The AR is a measure of genetic risk closely related to an odds ratio, but designed to be applied to aggregated pools of rare sequence variants. The ERS is closely related to the standard population genetics measure dN/dS (ω), which is a codon-based measure of evolutionary selective pressure [Yang, 1998]. As applied here, the ERS reflects the contribution of genetic risk to selection of patients for BRCA1 and BRCA2 testing.
Calibrate the relationship between the measures of missense substitution severity, GV and GD, and the measures of genetic risk AR and ERS. The outcome of this process, which is the major concern of this work, is to stratify an initial set of undifferentiated rare missense substitutions into a sequence of grades ordered by genetic risk.
Finally, we use an independent assessment of the proportion of sequence variants within each grade that are estimated to be pathogenic to validate the calibration described in step 4. Specifically, we use the family history likelihood ratio (FamHx-LR) [Goldgar et al., 2004; Easton et al., 2007], which is a measure of the relative likelihood that a given variant (or proportion of pooled variants) is pathogenic compared to neutral with respect to genetic risk, as an internal validation.
We describe each of these steps in more detail below and in the Supplementary Appendix (available online at http://www.interscience.wiley.com/jpages/1059-7794/suppmat).
METHODS
Step 1. BRCA1 and BRCA2 Data Set
BRCA1 and BRCA2 mutation screening was carried out at Myriad Genetic Laboratories as described previously [Tavtigian et al., 1997, 2006]. The analyses described here are based on results of full sequence tests of both genes from 68,000 BRACAnalysis® subjects of whom 4,867 were reported to carry a high-risk BRCA1 variant and 3,561 were reported to carry a high-risk BRCA2 variant. For a test to have been performed, a test request form must have been completed by the ordering health care provider, and the form must have been signed by an appropriate individual indicating that “informed consent has been signed and is on file.” The mutation screening data are arranged by sequence variant rather than by subject. For missense substitutions, the data include nucleotide and amino acid identity, total number of heterozygous and homozygous observations, number of observations in subjects who also carried a clearly pathogenic mutation in the same gene, and number of observations in subjects who also carried a clearly deleterious mutation in the opposite gene. For silent substitutions and nonsense substitutions, our data are limited to nucleotide and amino acid identities. We also know the total number of clearly deleterious mutations reported from each gene and the number of subjects who carried clearly deleterious mutations in both genes. These are all of the data required to calculate the AR and ERS. Analyses of the personal and family history of tested probands to calculate FamHx-LRs derive from a virtually identical series of subjects, as described previously [Easton et al., 2007]. We refer to these data as the B1&2 68K set.
Step 2. Sequences, Alignments, and Measurements of Substitution Severity
Analysis of missense substitutions using sequence alignment-based missense analysis software requires appropriately informative protein multiple sequence alignments [Greenblatt et al., 2003; Ng and Henikoff, 2003; Tavtigian et al., 2008]. BRCA1 analyses are based on a full-length alignment of nine mammalian BRCA1 sequences plus chicken, frog, and puffer fish. Experimentally determined sequences from the sea urchin BRCA1 ortholog were added to the RING (human residues 1–102) and BRCT domain (human residues 1641–1863) segments of the alignment. BRCA2 analyses are based on a full-length alignment of seven mammalian BRCA2 sequences plus chicken, frog, and puffer fish. Experimentally determined sequences from the sea urchin BRCA2 ortholog were added to the DNA binding domain (DBD domain) (human residues 2401–3110) segment of the alignment. Alignments were made using the M-Coffee tool suite (www.igs.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi) [Wallace et al., 2006] followed by minor hand realignment. Accession numbers are given in Table 1, and the alignments (or updated versions thereof) are available online at http://agvgd.iarc.fr/alignments.php as a public resource for assessment of missense substitutions in these breast cancer susceptibility genes. Based on a Fitch-type 3 rate constant model modified from Fitch and Markowitz [1970] and Abkevich et al. [2004], approximately 45% of positions in the concatenated RING-BRCT-DBD alignment appear to be under functional constraint, and the alignment has a maximum likelihood estimate of 3.3 substitutions per position. Because the remainder of these proteins has much greater cross-species sequence variability, corresponding full-length alignments would contain more than five substitutions per position.
TABLE 1.
BRCA1 and BRCA2 Sequences Used in the Protein Multiple Sequence Alignments (PMSAs)
| Species | BRCA1 | BRCA2 |
|---|---|---|
| Human | NP_009225.1 | AAB07223.1 |
| Chimpanzee | Q9GKK8.2 | XP_509619.2 |
| Gorilla | Q6J6I8.1 | — |
| Orangutan | Q6J6J0.1 | — |
| Rhesus monkey | Q6J6I9.1 | XP_001118184.1 |
| Mouse | NP_033894.2 | — |
| Rat | — | AAB71378.1 |
| Dog | NP_001013434.1 | NP_001006654.2 |
| Cow | NP_848668.1 | XP_583622.2 |
| Opossum | AAX92675.1 | ABP48762.1 |
| Chicken | NP_989500.1 | AAL89470.1 |
| Frog | ||
| X. laevis | AAL13037.1 | — |
| X. tropicali | — | ABP48763.1 |
| Puffer fish | AAR89523.1 | ABQ42581.1 |
| Sea urchin | ABL86143.1 | ABP57025.1 |
All of the missense substitutions that can arise via a single-nucleotide substitution to the canonical BRCA1 and BRCA2 coding sequences were then scored using the missense analysis program Align-GVGD (http://agvgd.iarc.fr/agvgd_input.php). This online algorithm calculates two variables that, together, are related to missense substitution severity: GV and GD. Both variables are extensions of the original Grantham difference [Grantham, 1974]; GV is a quantitative measure of the observed range of variation at a position in a protein multiple sequence alignment (PMSA), and GD is a quantitative measure of the distance between a missense substitution and the range of variation observed at its position in the alignment [Tavtigian et al., 2006]. GV = 0 corresponds to a residue that is invariant in the alignment, GV = 60–65 is the upper limit of conservative variation across species, and GV > 100 is indicative of positions that are under little functional constraint. GD = 0 corresponds to a missense substitution that is within the cross-species range of variation at its position in the protein; at invariant positions (GV = 0); GD = 60–65 is the upper limit of a conservative missense substitution; and GD > 100 is indicative of radical substitution. GV and GD were calculated at three depths of alignment: through frog, through puffer fish, and through sea urchin.
Step 3. Risk Estimations
Missense substitution data set
Starting with all of the missense substitutions observed during full-sequence mutation screening of BRCA1 and BRCA2 from a series of 68,000 subjects, we focused on those observed in the BRCA1 RING, BRCA1 BRCT repeat, and BRCA2 DBD domains because these are the only domains of these proteins where some missense substitutions have already been proven to confer relatively high risk of breast cancer due to missense-induced effects per se [Goldgar et al., 2004; Easton et al., 2007]. We excluded the substitutions observed at M1 (because they are likely pathogenic due to interference with translation initiation independent of any missense effect per se) and those observed at the nine canonical C3HC4 RING residues (because substitutions at cysteine have very high Grantham differences, inclusion of which could bias our results toward very high values of GD in a way that might only be characteristic of the minority of proteins that are functionally dependent on multi-cysteine motifs). After these exclusions, our data set contained a total of 453 missense substitutions. Only one of these, the known neutral variant BRCA1 M1652I, had a carrier frequency above 1% (2.8%); 225 of the substitutions were observed exactly once each.
Ascertainment ratio
Consider those individuals in the B1&2 68K test series who carry no high-risk variants in BRCA1 but who do carry BRCA1 missense variants in some potentially deleterious set M. Let the number of these who have a clearly high-risk mutation in BRCA2 be a1 and those who do not be b1. Under the rationale that the high-risk BRCA2 variants carried by the a1 subjects largely explains their presence in the sample series, the allele frequencies for deleterious BRCA1 variants in these subjects should be closer to population allele frequencies than the allele frequencies for deleterious BRCA1 variants in the b1 subjects will be. Thus the a1 subjects can be thought of as pseudocontrols, the b1 subjects can be thought of as pseudocases, and the ratio b1/a1 is an (nonnormalized) estimate of the odds for breast cancer for a carrier of a missense substitution in M. Comparing this ratio with the analogous quantity d1/c1 for a clearly benign set of sequence variants, the reference set, gives us the AR for BRCA1, which is an estimate of the odds ratio for carriers of BRCA1 missense substitutions in M [Tavtigian et al., 2006]. For calculation of the AR for BRCA2, similarly define a2, b2, c2, and d2.
| (1) |
The reference set of clearly benign BRCA1 and BRCA2 missense substitutions is defined in the Supplementary Appendix.
ERS nucleotide substitutions
For each nucleotide in a canonical DNA sequence, there are three possible single-nucleotide substitutions. However, these substitutions are not equally likely to occur because of differences in the underlying substitution rate constants. Using the dinucleotide substitution rate constants given by Lunter and Hein [2004], averaging sense and antisense orientations, we can calculate a relative substitution rate for every possible single-nucleotide substitution to a DNA sequence, ri. In a protein coding sequence, each of the possible single-nucleotide substitutions can also be classified as silent, nonsense, or missense. The probability that a new mutation (i.e., a new germline sequence variant at the moment that it comes into existence) will fall into a particular class C is given by:
| (2) |
Hence, under the null hypothesis of no selection, we can obtain from the total number of variants observed in a mutation screening study, oT, the number expected in any class, eC = pCoT, and compare this to the actual number observed, oC. Thus, we define the ERS for any class of substitutions C as the observed/expected ratio for that class normalized by the same ratio for silent substitutions:
| (3) |
Step 4. Distribution of Genetic Risk Within the GV-GD Plane and Risk Surface Calculations
While there are only a finite number of points in the GV-GD plane that can be occupied depending on the specific amino acids seen in the alignment and the set of observed and possible substitutions, it is useful to envision the AR and ERS as smooth two dimensional risk surfaces. To do this:
Gaussian smoothing was used to load the discretely distributed data underlying the AR and ERS onto a regular two-dimensional (2D) grid. This was done at each of three depths of alignment: frog, puffer fish, and sea urchin.
The AR and the ERS were calculated across the grid, yielding smooth 2D risk surfaces that could be displayed as heat maps. These were merged to a weighted geometric mean (ARBRCA10.11) × (ARBRCA20.22) × (ERS0.67), the joint risk estimate, which could also be displayed as a heat map.
To place portable gradations of genetic risk on the heat maps, we abstracted visible contour of the maps into simple equations, which we will refer to as contour curves. From inspection of the heat maps, we concluded that fractional monomials of the form GD = GD0+tan(a) GVb provide a suitable family of equations to do this. Owing to the definition of GD with respect to GV, we further decided that the contour curves should originate from a series of fixed values of GD0 (GD at GV = 0): GD0 = 65, 55, 45, 35, 25, and 15. Individual contour curves were selected by systematic grid search over the free fractional monomial variables a and b.
A bootstrapping approach was used to estimate confidence intervals for the grid points of the risk surfaces, along the contour curves, and over the grades of missense substitutions defined by the intervals between the contour curves.
The AR, ERS, Gaussian smoothing, and risk surface manipulations are described in greater detail in the Supplementary Appendix.
Grade Risk Estimates
The contour curves serve as gradations, defining grades of sequence variants that should confer similar within-grade genetic risks. We took three different approaches to calculating risk for various grades.
Direct joint risk estimate. All of the missense substitutions with GV-GD coordinates falling between two specified contour curves were pooled and used to calculate the BRCA1 AR, BRCA2 AR, overall ERS, and joint risk estimate. Such joint risk estimates were generally calculated at all three depths of alignment of interest.
Smoothed joint risk estimate. We lay down a grid of exactly 50 equally-spaced points with GV-GD coordinates that fall between two specified contour curves. Joint risk estimates for each of the 50 points within such a grade were calculated using Gaussian weighting exactly as was done for grid points on the heat maps. These 50 values were then averaged to get a smoothed joint risk estimate for the grade. Such joint risk estimates were calculated at all three depths of alignment of interest.
Summary family history. All of the summary personal/family histories of subjects who carried missense substitutions with GV-GD coordinates falling within a grade defined between two specified contour curves (and no other clearly deleterious, high-risk variant) were used to estimate the proportion, α, of the variants within the grade that were deleterious using the heterogeneity likelihood defined in Easton et al. [2007]. Approximate 95% confidence intervals for the heterogeneity proportion were obtained by finding the values αL and αU for which the overall likelihood differed from that at α by an amount equivalent to a likelihood ratio test significant at the 0.05 level [Easton et al., 2007].
RESULTS
The distribution of genetic risk in the GV-GD plane has not yet been systematically explored. To visualize the distribution of evidence of genetic risk among BRCA1 and BRCA2 missense substitutions, we calculated GV and GD for each substitution at each of three depths of alignment: frog, puffer fish, and sea urchin. We then created a grid of points in the GV-GD plane and calculated two measurements of genetic risk, the AR and the ERS, at each point on the grid. These risk estimates used a log-distance Gaussian weighting approach to load the required observational data (number of observations in pseudocases, number of observations in pseudocontrols, underlying dinucleotide substitution rate constants) onto each grid point before the AR and ERS for that grid point were calculated. Point risk estimates were then displayed as heat maps.
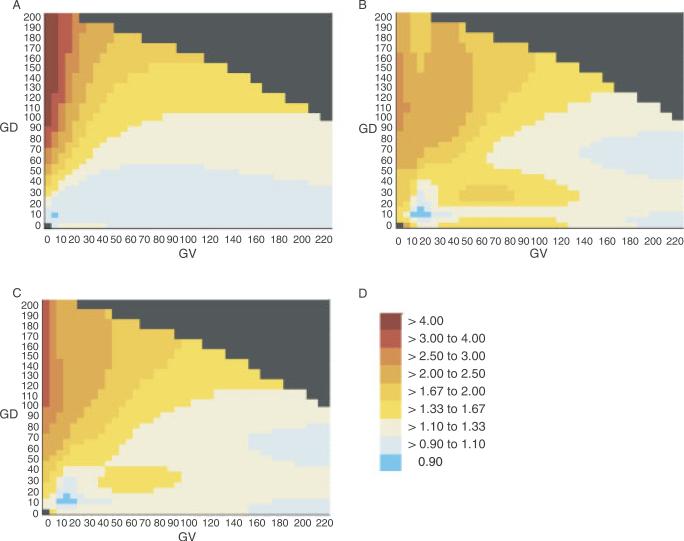
AR and ERS heat maps for the complete alignment through sea urchin are displayed in Figure 1. Although the observational data used to calculate the AR and ERS are somewhat independent, there is some commonality in the distribution of genetic risk in the GV-GD plane that they detect: highest risk at GV = 0 (positions that are invariant in the multiple sequence alignment) and GD > 90 (nonconservative to radical substitutions), low risk at high values of GV and low values of GD, and in the higher risk portions of the heat maps, gradations in risk that bring to mind contours. The AR heat map appears more regular than the ERS map, but this is partly an artifact limited to the example that we have chosen (alignment through sea urchin) and also partly attributable to the need for a larger standard deviation in the Gaussian weighting of the AR data than the ERS data in order to smooth away inappropriately high risk-estimates at some points on the grid. The AR and ERS were merged into a single joint risk estimate and joint heat map (Fig. 1C), and all subsequent analyses are based on these or similar joint risk estimates.
FIGURE 1.
AR and ERS heat maps. A: AR heat map using the sequence alignment through sea urchin. B: ERS heat map using the sequence alignment through sea urchin. C: Joint risk estimate heat map using the sequence alignment through sea urchin. D: Risk estimate color lookup table. Note: This table is used for all of the heat maps in Figures 1 and 2.
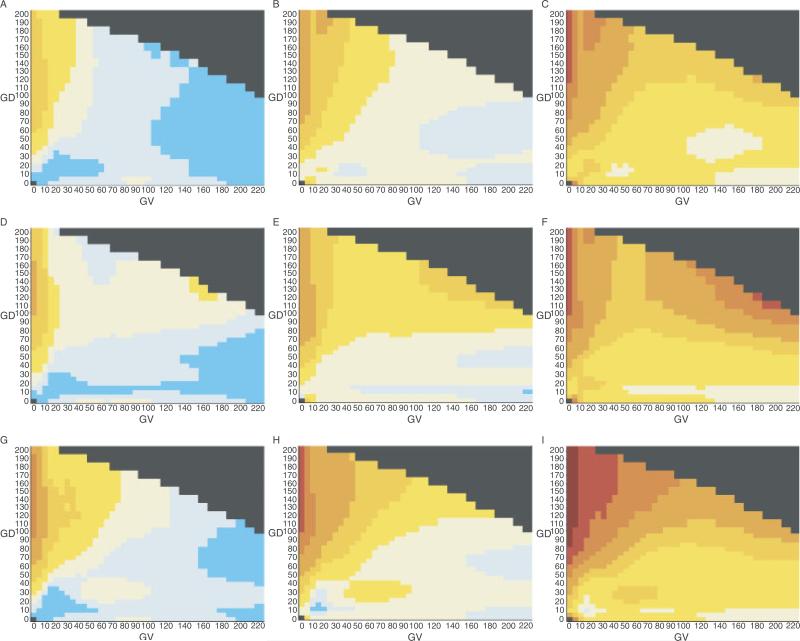
To evaluate the stability of the pattern of risk distribution in the GV-GD plane, we made bootstrap estimates of the confidence intervals for each grid point of the heat maps. These are displayed in Figure 2, where the first column of heat maps are the 5% lower bound, the central column are the actual data, and the third column are the 95% upper bound. All but one of the maps evince the general pattern described above (highest risk at GV = 0 and GD > 90, low risk at high values of GV and low values of GD, and gradations of risk that descend from the upper left regions of the maps. The main exception is the 95th centile bootstrap of the data for the alignment through puffer fish, which shows a noticeable area of high risk in the upper right area of the map. As displayed in Figure 3, the upper right region of the GV-GD plane is very sparsely populated with missense substitutions. The difference in this area between the heat maps through frog, puffer fish, and sea urchin can be explained in terms of the effects of two individual sequence variants. Analyzed with the alignment through frog, the upper right region of the map contains 34 very rare substitutions plus one more common substitution. The more common substitution, BRCA2 A2466V, has a frequency of about 0.7% and an AR point estimate of 0.8. In the bootstrap sampling, the data from this variant contributes to keeping the AR estimates of the upper right quadrant of the heat map fairly stable. Analyzed with the alignment through sea urchin, the upper right region of the map contains 41 very rare substitutions plus one more common substitution. The more common substitution, BRCA2 I2490T, has a frequency of about 0.7% and an AR point estimate of 0.6. In the bootstrap sampling, the data from this variant keeps the AR estimates of the upper right quadrant of the heat map fairly stable. On the other hand, analyzed with the alignment through puffer fish, the upper right region of the map contains 39 variants, all of which are extremely rare. Some of the AR samplings choose just one or even zero pseudocontrols, resulting in unstable and sometimes very high AR estimates that in turn contribute to high joint risk estimates. For calculation of the ERS, the equivalent of controls are the dinucleotide substitution rate constants of all possible substitutions. These provide a rich, dispersed data set from which more stable estimates can be obtained.
FIGURE 2.
Joint risk estimate heat maps and confidence intervals. Row 1: Heat maps based on the sequence alignment through frog; Row 2: heat maps based on the sequence alignment through puffer fish; Row 3: heat maps based on the sequence alignment through sea urchin; Column 1: 5th centile heat maps; Column 2: actual data heat maps; Column 3: 95th centile heat maps.
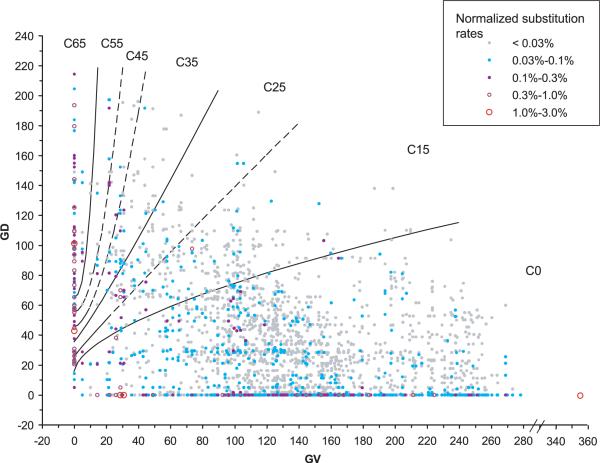
FIGURE 3.
Align-GVGD contour curves displayed with distributions of substitution probability. Each point or circle on the graph represents a coordinate in the GV-GD plane that is occupied by one or more of the possible missense substitutions that can result from a single-nucleotide substitution in the BRCA1 RING domain, BRCT domain, or BRCA2 DNA binding domain. Beyond values of GV and GD, each possible single-nucleotide substitution is associated with an underlying dinucleotide substitution rate constant [Lunter and Hein, 2004]. The color and size of the point or circle represent the rate constant. If two or more possible substitutions have exactly the same GV and GD coordinates, then their underlying dinucleotide substitution rate constants are added together to get the total rate constant for that coordinate.The color/size intensities of the substitution rate constant representations are normalized so that the total for the figure is 100%. Visual integration of the color/size intensities for substitution within a grade gives the proportion of all possible missense substitutions that are grouped in that grade.This particular graph displays the missense distribution obtained from the sequence alignment through sea urchin. The equations of the contour curves are: GD = 65+tan(10°) × (GV2.5); GD = 55+tan(10°) × (GV2.0); GD = 45+tan(15°) × (GV1.7); GD = 35+tan(50°) × (GV1.1); GD = 25+tan(55°) × (GV0.95); GD = 15+tan(75°) × (GV0.6).
Contours
Ideally, we would like to reduce the output of Align-GVGD from a pair of continuous variables to a single continuous variable that is analogous to the original Grantham difference and has genetic risk estimates attached to its magnitude. To the extent that the heat maps of the GV-GD plane evince quasicontinuous contour, we can take a step in that direction by collapsing the Align-GVGD output to a single-graded variable by following joint risk estimate contours from positions anchored at selected values of GD and GV = 0 to positions at some (usually higher) value of GD and GV >0. On visual inspection, most of the contours originating from the GD axis with joint risk estimates above ~1.2 resemble curves from the family of fractional monomials GD = GD0+tan(a) GVb. Therefore, we should be able to approximate the contours with equations from that family; the selected equations then serve as easily calculated gradations. Working from fixed values of GD0, this family of equations has only two free variables and is therefore easily scanned by grid search to find the individual equations that have the least mean deviation from their joint risk estimate at GD0.
To begin, we picked two reference values of GD0: 65 and 15. GD0 = 65 was chosen because the border between what molecular biologists and biochemists would generally consider the upper limit of conservative substitution and nonconservative substitution falls between a Grantham difference of 60 and 65. Therefore, a contour anchored to GD0 = 65, GV = 0 corresponds to the lower bound on risk conferred by a nonconservative substitution at an invariant position. On the other hand, the pairwise Grantham differences between the three large nonpolar/nonaromatic amino acids leucine, isoleucine, and methionine are all <15, whereas the Grantham differences between amino acid pairs that differ by at least a methyl group are above 20. Therefore, a contour anchored to GD0 = 15, GV = 0 corresponds to the upper bound on risk conferred by essentially structurally isomeric substitution at an invariant position.
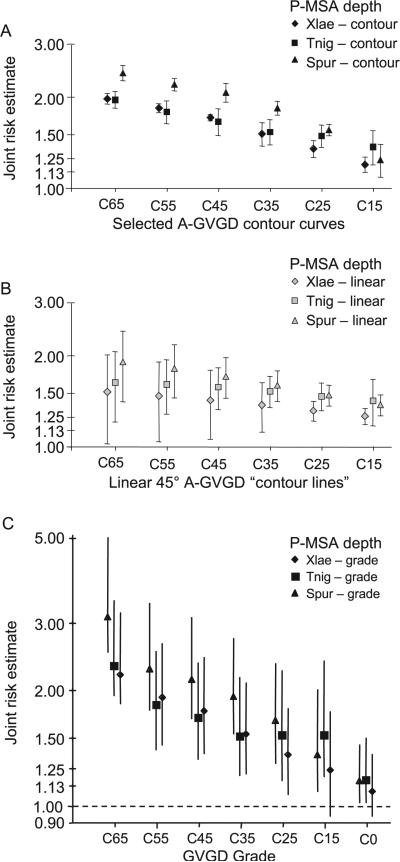
After selecting contour curves anchored at GD0 = 15 and GD0 = 65, we selected contour curves anchored at GD0 = 25, 35, 45, and 55. Together, the six contour curves serve as gradations in the GV-GD plane extending from the genetic risk equivalent of the border between nonconservative and conservative substitution at an invariant position to the genetic risk equivalent of the border between single methyl substitution and isomeric substitution at an invariant position. The mean joint risk estimates for each of the six contour curves at each of the three depths of alignment are displayed in Figure 4A. At each depth of alignment, the six risk estimates form the expected ordered sequence with highest risk at the contour curve with GD0 = 65 and least risk at the contour curve with GD0 = 15. Along the higher-risk contour curves, with GD0 from 65 to 35, joint risk estimates determined from the alignment through sea urchin are repeatedly greater than those determined for the alignments through frog or puffer fish. On the other hand, at the contour curve with GD0 = 15, all three joint risk estimates fall within one standard deviation of 1.25.
FIGURE 4.
Contour curve and grade risk estimates. A: Joint risk estimates averaged from 20 points spaced evenly along each selected contour curve at each of the three depths of alignment. Closed error bars give ±1 standard deviation from the joint risk estimate at the GD0 intercept. B: Joint risk estimates averaged from 20 evenly points spaced evenly along each linear 45° reference “contour line.” Closed error bars give ±1 standard deviation from the joint risk estimate at the GD0 intercept. C: Joint risk estimates averaged from 50 evenly spaced points within each grade. Open error bars give the 95th percentile confidence intervals, determined from 1,000 bootstrap samplings over the underlying data.
How well do the contour curves approach being real contours? To address this question, we first asked “how variable are the joint risk estimates for a series of GV-GD points along each contour curve, and what is the probability that a randomly chosen set of points will have equal or less variability?” The joint risk estimate for each contour curve was measured by averaging the joint risk estimates of 20 GV-GD points spaced equally along each curve. As determined from the mean risk estimates and standard deviations for 10,000 sets of 20 randomly selected points in the GV-GD plane, fewer than two random selections per thousand have a standard deviation of the mean risk estimate that is less than or equal to that measured for any of the six contour curves at any of the three depths of alignment. Thus the P values against the null hypothesis that the contour curves are unrelated to contours are each <0.002, and most (15/18) are <0.001 (data not shown). As a second approach to the question, we compared performance of the six selected contour curves to the corresponding family of six straight lines given by
| (4) |
on the grounds that one could consider this family of straight lines of slope 45° as a sort of reasonable null hypothesis/starting point for candidate contour curves. The mean joint risk estimates and standard deviations about the joint risk estimate at GD0 for the six selected contour curves are given in Figure 4A. The corresponding data for each of the six straight line candidate contours are given in Figure 4B. From these data, the selected contour curves can be seen to outperform the family of 45° straight lines in two ways: 1) The slopes for the mean joint risk estimate data sequence are greater for the selected contour curves than for the 45° lines; for the alignment through frog, the ratio of slopes across the selected contours vs. across the 45° lines is 3.1:1 and for the alignment through sea urchin this ratio is 2.1:1. Thus the selected contour curves generate a steeper risk gradient than do the 45° lines. 2) The standard deviations are tighter for the selected curves than for the 45° lines. Perhaps most tellingly, the one standard deviation interval around the joint risk estimates for the selected contour curves at GD0 = 65 excludes all of the other point estimates in the same depth of alignment sequence, whereas the one standard deviation interval around the joint risk estimates for the 45° straight lines at GD0 = 65 includes almost all of the other point estimates in the same depth of alignment sequence.
In Figure 3, the six contour curves are displayed over a representation of the probability distribution, in the GV-GD plane, of all possible missense substitutions that can result from a single-nucleotide substitution to the RING and BRCT domains of BRCA1 and the DNA binding domain of BRCA2. That is, summation over a portion of the GV-GD plane displayed in this figure will give the proportion of all newly occurring missense substitutions expected to fall in that portion of the plane. Note that the ratio of observed substitutions to possible substitutions is the fundamental idea underlying the ERS. Therefore, division of the observed distribution of missense substitutions in the B1&2 68K set by the expected distribution given in Figure 3 would result essentially in the heat map of Figure 1B, with some scaling provided by the corresponding silent substitution data.
The six contour curves define seven grades of variants. We name the grades after the contour curves that provide their lower bound. Thus the highest risk grade is contour 65 (C65), and the lowest risk grade is C0. These two grades are also in some sense special because they contain a long segment of one of the axes and, as is visible from Figure 3, a disproportionate fraction of the probability density of all possible missense substitutions. C65 contains a long segment of the GV = 0 axis and therefore all possible substitutions at invariant positions that have GD≥65. At the same time, there are only two types of variable positions, Ile-Leu (GV = 4.9) and Ile-Met (GV = 10.1), at which some missense substitutions can fall into C65. Because of these characteristics, the vast majority of missense substitutions belonging to the grade C65 will fall at invariant positions, and the logic behind alignment-based classification algorithms suggests that this grade should be highly enriched for damaging substitutions. In contrast, the grade C0 contains the entire GD = 0 axis. As we have previously demonstrated, most substitutions with GD = 0 are either neutral or very nearly so [Tavtigian et al., 2006]. As the GD = 0 variants actually make up a large fraction of the observed variants in this grade, we expect the joint risk estimate for this grade to be quite low.
We took three different approaches to measuring genetic risk conferred by the pools of variants in these grades. The first was pooling of all sequence variants in each grade and direct measurement of the joint risk estimate. The advantage of direct measurement is offset by the disadvantage that direct joint risk estimate measurements do not allow a direct confidence interval calculation, and the small number of pseudocontrols in some of the grades prevents estimating confidence intervals by a bootstrap approach. The second approach was to lay down a grid of equally-spaced points within each grade and then use the Gaussian weighting approach to measure mean joint risk estimates. While this approach is less direct, it captures some data from all of the sequence variants in the data set, allowing estimation of confidence intervals by a bootstrap approach. The third approach is to use the personal and family histories of probands who carried the sequence variants in each grade to calculate FamHx-LRs. This method is direct and allows estimation of confidence intervals, but the nature of the variable calculated differs from the odds-ratio–like joint risk estimates. However, the grades C55, C45, and C35 are sparsely populated, and there are simply not enough data at this time to make direct measurements on these grades, either joint risk estimate or FamHx-LR, with any confidence. Consequently, for direct measurement of risk in the intermediate grades, we pooled from two to five grades. As the contour curve at GD0 = 35 most evenly divides the missense substitutions between the contours GD0 = C65 and GD0 = C15 into two groups, we pooled grades C55, C45, and C35 (C35–C55) and grades C25 and C15 (C15–C25) for these measurements.
Results from these pooled measurements are summarized in Table 2. Whether the genetic risk estimate of the grade C65 is measured directly or by Gaussian smoothing, its risk estimate is greater than that of the contour curve with GD0 = C65. Similarly, whether measured directly or by Gaussian smoothing, the risk estimates for the grade combination C35–C55 lie between those of the C35 lower boundary and the C55 upper boundary. The risk estimates for the grade combination C15–C25 lie between those of the C15 lower boundary and C35 upper boundary, and the risk estimates for the grade C0 lie below that of the C15 lower boundary.
TABLE 2.
Risk Estimates for BRCA1 and BRCA2 A-GVGD Grades in a Four-Grade System
| PMSA through frog |
Through puffer fish |
Through sea urchin |
||||
|---|---|---|---|---|---|---|
| Risk estimate | (95% CI) | Risk estimate | (95% CI) | Risk estimate | (95%CI) | |
| A. Directly calculated point estimates of the joint risk estimatea | ||||||
| C65 | 2.80 | 3.52 | 3.84 | |||
| C35-C55 | 1.85 | 1 .87 | 2.25 | |||
| C15-C25 | 1.44 | 1 .51 | 1.61 | |||
| C0 | 1.13 | 1.14 | 1.13 | |||
|
B. Point estimates of the joint risk estimate calculated by Gaussian smoothing | ||||||
| C65b | 2.20 | (1.85-3.19) | 2.32 | (1.94-3.34) | 3.12 | (2.52-5.02) |
| C35-C55 | 1.72 | (1.39-2.27) | 1.66 | (1.34-2.22) | 2.10 | (1.68-2.88) |
| C15-C25 | 1.28 | (1.02-1.70) | 1.52 | (1.21-2.21) | 1.46 | (1.20-2.00) |
| C0 | 1.09 | (0.94-1.36) | 1.17 | (1.02-1.50) | 1.16 | (1.02-1.44) |
|
C. Estimated proportions of pathogenic variants within each grade based on summary family history likelihood ratiosa | ||||||
| C65 | 0.60 | (0.42-0.76) | 0.78 | (0.55-0.93) | 0.81 | (0.61-0.95) |
| C35-C55 | 0.46 | (0.21-0.73) | 0.72 | (0.41-0.96) | 0.66 | (0.34-0.93) |
| C15-C25 | 0.21 | (0.03-0.48) | 0.19 | (0.04-0.42) | 0.29 | (0.09-0.56) |
| C0 | 0.01 | (0.00-0.07)c | 0.01 | (0.00-0.06)c | 0.01 | (0.00-0.05)c |
By direct calculation, the joint risk estimate for nonsense substitutions in BRCA1 and BRCA2 (excluding BRCA2 K3326X and any downstream nonsense substitutions) is 7.20. For the FamHx-LR the proportion of pathogenic truncating variants is 1.00, by definition.
Careful examination of the set of all possible BRCA1/2 RING/BRCT/DBD missense substitutions revealed that > 96% of all possible substitutions in grade C65 fall at GV = 0 and most of the rest fall at GV = 4.9. Accordingly, for Gaussian smoothing measurements of this grade, we used a set of 50 points in which 48 were equally spaced along the relevant portion of the GV = 0 axis and the remaining 2 were equally spaced along GV = 5.
In fact, the FamHx-LR point estimates for Class C0 are 0.00 at all three depths of P-MSA. However, these point estimates will serve as prior probabilities in analyses of unclassified missense substitutions in future studies. Because it would impossible to modify a prior probability of 0.00, we report these point estimates as 0.01.
The estimated proportions of high-risk missense substitutions contained within each grade are shown in Table 2. It is important to remember that these estimates are constructed under the assumption that each given missense substitution is either high-risk (in the same sense as a truncating mutation) or neutral with respect to risk. The behavior of these estimates in the situation where all missense substitutions within a pool are of intermediate risk (or mixed-risk) is unknown. Nevertheless, results from the FamHx-LR followed a pattern very similar to that detected by the joint risk estimate. The trend of grade C65 having the highest relative odds of a high-risk variant and grade C0 having the lowest odds was present at every depth of alignment. In 5 out of 9 pairwise comparisons, the 95% confidence interval of one grade, measured by a linkage analysis–type heterogeneity estimate, excluded the point estimate of the adjacent grade. The 95% confidence intervals of one grade always excluded the point estimate for a grade from which it was separated by an intervening grade. In addition, the estimated proportions of pathogenic variants based on FamHx-LRs for the grades C65, C35–C55, and C15–C25 were significantly greater than 0.0 at all three depths of alignment.
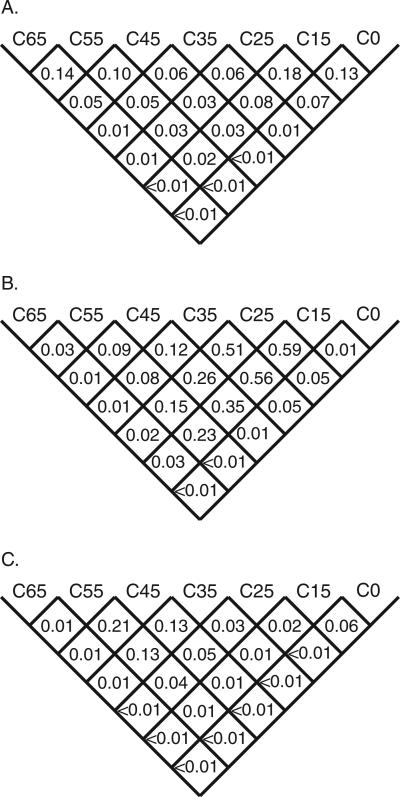
Measured by the Gaussian weighting approach, the question of how well the joint risk estimates for two grades are resolved from each other is subtly different from measurement of their confidence intervals. In addition to the normal difference in interpretation between the meanings of P values and confidence intervals estimated by bootstrapping over observational data, an additional consequence of the Gaussian smoothing is that the risk estimates for pairs of grades will be somewhat correlated within a single bootstrap cycle. Therefore, in Figure 5 we present a series of pairwise P values that summarize the degree to which the grades, in the full seven-grade system, are distinct from each other. In 5 out of 18 comparisons between adjacent grades, the higher grade has a higher joint risk estimate in more than 95% of the bootstrap measurements. Comparing grades separated by one to five intervening grades, this test of pairwise resolution rises to 5 out of 15, 9 out of 12, 8 out of 9, 6 out of 6, and 3 out of 3 pairs, respectively. Using the PMSA through sea urchin, in 16 out of 21 grade-grade comparisons, the higher grade has a higher joint risk estimate in more than 95% of the bootstrap measurements, and 13 out of 21 comparisons meet or exceed a 99th centile criterion. In this respect, the alignment through frog is intermediate, with 11 out of 21 comparisons exceeding a 95th centile criterion. The alignment through puffer fish has the weakest performance, with 9 out of 21 comparisons exceeding a 95th centile criterion.
FIGURE 5.
Grade cross-comparisons.The value given in each cell is the probability, from bootstrap sampling, that the average joint risk estimate for missense substitutions falling into the lower numbered grade of the pair-wise comparison is greater than the average joint risk estimate for missense substitutions falling into the higher numbered grade. A: Using the alignment through frog. B: Using the alignment through puffer fish. C: Using the alignment through sea urchin.
DISCUSSION
The Grantham difference has been a popular source of data that contributes assessments of the pathogenicity of missense substitutions. While the Grantham difference does have some utility in this regard, we have demonstrated previously that extending the Grantham difference to a two-variable system (GV and GD) that incorporates information from PMSAs of genes of interest improves the ability to distinguish between substitutions that are likely to be pathogenic and those that are not [Tavtigian et al., 2006]. Here we have: 1) used missense substitution data from BRCA1 and BRCA2 to display the distribution of genetic risk in the GV-GD plane; 2) abstracted contours of genetic risk in the GV-GD plane into a sequence of six contour curves; and 3) used the contour curves to collapse the continuous two-variable GV-GD system into a 1D sequence of seven grades that are ordered by empirical measurements of genetic risk.
Notably, our analyses neither depended on existing classifications of BRCA1 and BRCA2 missense substitutions into risk categories such as pathogenic or neutral nor depended on results from functional assays. Instead, our analyses depended on using observational data inherent in a large BRCA1 and BRCA2 mutation screening series, the B1&2 68K set, to calculate three measurements of genetic risk: the AR, the ERS, and the FamHx-LR. In turn, the AR and ERS contributed to the display of genetic risk in the GV-GD plane, the abstraction of contours into contour curves, and initial measurements of genetic risk in the ordered missense substitution grades defined by the contour curves. The FamHx-LR, which is discussed at length elsewhere [Easton et al., 2007], was then used to validate the ordering of the missense substitution grades.
As the process used to select contour curves, characterize them, and characterize the grades that they define relied heavily on the AR, ERS, and Gaussian weighting, these merit further discussion. While the AR is a relatively direct estimate of the ratio of odds of disease between carriers and noncarriers of sets of substitutions, interpretation of the ERS is more complex. Point estimates of the ERS correspond to the standard population genetics measure dN/dS (ω), corrected for dinucleotide substitution rate constants. ERS point estimates also correspond to the NA/NS ratios recently described by Kryukov et al. [2007], except that the formula for ERS specifies how the dinucleotide substitution rate constant correction is incorporated. Thus the ERS is an accepted measure for strength of selection and, applied to an unbiased population sample, would measure the degree to which a specific grade of substitutions is subject to natural selection. However, in our application—where 1) loss of function mutations in the genes of interest (BRCA1 and BRCA2) are known to confer high risk of the trait of interest (risk of breast or ovarian cancer) and 2) the subjects are strongly biased toward personal and/or family history of breast or ovarian cancer—ascertainment selection for rare variants that increase risk of the trait of interest may outweigh other factors. Thus we expect a positive association with pathogenic sequence variants. Under this assumption then, the ERS will also be positively correlated to the disease odds ratio. However, to the extent that purifying selection must tend to limit the frequency of deleterious substitutions in BRCA1 and BRCA2, the ERS should tend to underestimate true odds ratios and the constant of proportionality between the disease odds ratio and the ERS cannot be estimated a priori.
The analysis required to associate AR, ERS, or joint risk estimates to specific missense mutations is not self-evident due to the rarity of individual mutations. For example, standard logistic regression models of the form
| (5) |
where XJ is the number of copies of the jth UV and exp(βJ) is the associated odds ratio, cannot be fitted at all unless there is at least one observation of each UV in each of the case and control sets; the estimates will not be reliable unless several observations fall in each set. However in datasets such as the present B1&2 68K set or the 36-gene dataset analyzed in Kryukov et al. [2007], approximately 50% of the individual sequence variants are observed exactly once each. Therefore, we are forced to some form of smoothing, pooling, or both. The Gaussian weighting that we used provided a device to load all of the observational data from the sequence variants in our dataset onto a grid or geometrically-defined set of points and then calculate the variables of interest, culminating in a joint risk estimate, at those points. This approach allowed us to: 1) smooth away very local irregularities in the data; 2) generalize risk estimates across geometrically-defined regions of the GV-GD plane; and 3) see long-distance trends in the data.
The resulting heat maps provide a visual representation of the distribution of the joint risk estimate in the GV-GD plane. Three consistent trends were evident from these maps. 1) Highest genetic risk is in the upper left of the GV-GD plane, at GV = 0, e.g., positions that are invariant across species, and GD >65, e.g., substitutions that are nonconservative to radical. 2) Risk decreases with increasing GV, decreasing GD, or both. 3) In the consistently high-risk area of the GV-GD plane, the risk increases with increasing depth of PMSA. In addition to these consistent trends, there was inconsistent evidence, observed mostly using the sequence alignment through puffer fish, of elevated risk in the upper right region of the GV-GD plane. We note, however, that this area of the GV-GD plane is very thinly populated with sequence variants. Thus, to the extent that sequence variants mapping to this region confer increased risk, their pooled attributable fraction will be small because their summed frequency will usually be very small.
These results contradict our original GV-GD classifiers [Tavtigian et al., 2006], and predictions from the popular online classifier “sorting intolerant from tolerant,” or SIFT [Ng and Henikoff, 2003] in one important way. Our original classifier placed all substitutions at invariant positions in the most likely deleterious class (ED1) (for a visual comparison of the old vs. new Align-GVGD classifiers superposed on one of the heat maps (see Supplementary Figs. S1 and S2) as does SIFT (score = 0.00). Indeed, using PMSAs that include orthologous sequences from frog, puffer fish, and sea urchin, there can be little doubt but that the alignments capture sufficient evolutionary time that essentially all of the invariant positions must be functionally constrained and that purifying selection must be acting against any missense substitution at these positions. Reinterpreted in a classifier that attempts the binary classification pathogenic–not pathogenic, the evolutionary argument that all of the substitutions at invariant positions must be deleterious results in their classification as pathogenic. Yet one consistent trend in the heat maps is that, taken as a pool, substitutions at invariant positions that nonetheless have low values of GD do not have extremely high joint risk estimates. This observation is also easily rationalized: even at an extremely functionally constrained position, a very conservative substitution will sometimes be less damaging to protein function than a nonconservative substitution. Reinterpreted from a perspective of genetic risk as a continuous variable, and the goal of a graded classifier that will group variants of similar risk together, substitutions that have GV = 0 and low GD should not be categorized with substitutions that have GV = 0 and high GD because, on average, we measure a large difference in joint risk estimate between them.
The seven-tiered classifier that we have constructed begins with a grade, C65, in which the vast majority of included variants should be deleterious, the joint risk estimate measures to detects markedly elevated genetic risk, and the FamHx-LR measures consist largely of pathogenic sequence variants. The classifier ends with a grade, C0, that has the opposite characteristics: the majority of included variants should be at most slightly deleterious, the joint risk estimate detects only slightly elevated risk, and the FamHx-LR measures consist almost entirely of substitutions that are either neutral or of little clinical significance. Interposed are five grades across which most effects should increase or decrease in an ordered fashion.
These seven grades are based on six contour curves, which serve as portable abstractions from, and good approximations to, the real heat map contours. Abstract because the real contours have many small irregularities that trace the real data whereas the contour curves are smooth functions, and also because there are actually three real contours at each anchor point, one for each depth of alignment, whereas the contour curves are a compromise across the three depths of alignment. Portable because the contour curves are simple equations; given GV and GD, one can calculate in a few moments the grade into which a substitution falls. The Grantham difference framework also provides a physical interpretation of missense substitution severity. The contour curves and grades were constructed so that a missense substitution of grade X that falls at a position that is variable in the underlying PMSA should have similar effects to a missense substitution with Grantham difference X falling at an invariant position in the underlying PMSA. This classifier system should prove useful in both genetic epidemiology and molecular epidemiology applications.
One approach that has proved effective for clinically relevant analysis of unclassified variants in BRCA1 and BRCA2 has been a multicomponent likelihood ratio based integration of assessment methods that range from segregation analysis to immunohistochemical analysis [Goldgar et al., 2004, 2008; Chenevix-Trench et al., 2006; Lovelock et al., 2006; Tavtigian et al., 2006]. The original likelihood ratio–based approach incorporated Grantham differences and sequence conservation as independent components, with the sequence conservation element estimated via a Fitch Covarion theory approach [Abkevich et al., 2004; Goldgar et al., 2004]. Recently, Easton et al. [2007] updated this approach by using the FamHx-LR to estimate the proportion of variants predicted to confer increased cancer risk for sets of missense substitutions and in-frame indels defined by position in the gene and sequence conservation at that position. The analysis output posterior probabilities potentially suitable for use as prior probabilities in downstream analyses of sequence variants falling into the same position and conservation classes. However, the results as presented were descriptive rather than utilitarian because the key table (Table 5 of Easton et al. [2007]) described overlapping classes by domain and sequence conservation such that a single variant could fall into two classes with distinctly different results. In Table 3 we give the correspondence between two key data lines from Table 5 of Easton et al. [2007] and the results described here. The vast majority of all possible missense substitutions in BRCA1 and BRCA2 fall uniquely into one of the categories of position and/or A-GVGD grade defined in the right half of our Table 3. Thus the output is a set of utilitarian posterior probabilities intended for use as prior probabilities in future studies of missense substitutions in these genes.
TABLE 3.
Correspondence Between Key Results From Easton et al. [2007] and This Analysis
| From Easton et al. [2007], Table 5 | Reparsing based on this work | ||||||
|---|---|---|---|---|---|---|---|
| Missense or in-frame indel, by domain | n | α (95% CI) | Missense substitutions, by domain | Class | n | Joint risk estimatea | α (95% CI) |
| BRCA1 BRCTor BRCA2 DBD | BRCA1 RINGb or BRCTor BRCA2 DBD | C65 | 98 | 3.12 | 0.81 (0.61-0.95) | ||
| C55 | 15 | 2.28 | 0.66 (0.34-0.93) | ||||
| C45 | 12 | 2.14 | |||||
| 323 | 0.35 (0.26-0.45)c | C35 | 28 | 1.93 | |||
| C25 | 28 | 1.67 | 0.29 (0.09-0.56) | ||||
| C15 | 30 | 1.36 | |||||
| C0 | 242 | 1.16 | 0.01 (0.00-0.06)d | ||||
| Not BRCT nor DBD | 854 | 0.00 (0.00-0.04)c | Not BRCA1 RING, BRCT | GD = 0 | 554 | 1.04 | 0.01 (0.00-0.04)d per Easton etal. [2007] |
| Not BRCA2 DBDe | GD > 0f | 677 | 1.05 | ||||
Calculated by Gaussian smoothing.
Excluding substitutions at M1 and the eight canonical C3HC4 residues.
Please note that the heterogeneity analysis calculations inTable 5 of Easton et al. [2007] included some in-frame insertion deletion variants and also included data from segregation analyses. Recalculation for RING, BRCT, and DBD missense substitutions (and excluding substitutions at M1 and the eight canonical C3HC4 residues) using the FamHx-LR alone yielded a heterogeneity point estimate of 0.32. The point estimate for the set of missense substitutions lying outside of the RING, BRCT, and DBD domains remained 0.00.
In fact, the FamHx-LR heterogeneity analysis point estimate for this Class is 0.00. However, these point estimates will serve as prior probabilities in future analyses of unclassified BRCA1 and BRCA2 missense substitutions. Because it would be impossible to modify a prior probability of 0.00, we report these point estimates as 0.01.
And also excluding substitutions at BRCA2 M1 and substitutions that fall within 2 bp of a splice junction.
A detailed study of the potential genetic risk due to missense substitutions in the remaining small well-conserved BRCA1 and BRCA2 sequence elements will be the subject of a future study.
Case-control mutation screening has emerged as an approach to assess whether sequence variation in candidate intermediate risk susceptibility genes actually contribute to disease risk [Cohen et al., 2004; Landi et al., 2005; Rahman et al., 2006; Renwick et al., 2006; Seal et al., 2006]. Such studies are typically powered to detect the pooled effect of many rare protein truncating variants (pooled frequency >1%) and average odds ratios of greater than 2.0 per allele. The resulting sample sizes are around 1,000 cases and 1,000 controls. This design does not yield sufficient power to estimate the effect of individual missense mutations for three reasons:
Most missense mutations are observed only once. It is therefore impossible to compare frequencies between cases and controls.
Power decreases with variant frequency and will be insufficient even where several copies are observed.
The large number of distinct missense mutations exacerbates the multiple comparison problem and requires an even more stringent significance threshold to control the rate of false positives.
The results presented here can resolve all three issues. By pooling variants with similar predicted risk, we increase the effective frequency of observation and hence avoid the first two problems. Simultaneously, we reduce the number of comparisons (or degrees of freedom). Several specific options are available to exploit the methods we describe, providing different balances between the strength of assumptions and the potential increases in power. In each case, we augment the seven defined grades of missense mutations with two others. A low-risk grade 0 containing only wild-type alleles and common variants; and a high-risk grade 8, containing protein truncating and splice-junction modifying (PT+SJM) variants.
Approach 1
A test of any heterogeneity between the nine grades is applied. This test will have eight degrees of freedom, but makes no assumption of the relative magnitude of effect found within each grade. The test itself could be a Pearson's chi-squared test to address significance only, or logistic regression combined with a likelihood ratio test to additionally estimate an effect size for each grade. If age of onset data are available, there may be cases where these could be replaced by the log-rank test or a survival analysis regression method, respectively.
Approach 2
The 7+2 grades are treated as ordered categories, and a one-degree of freedom test for linear trend is applied, with the same choice of analyses. The reduction in degrees of freedom will provide additional power providing the ordering obtained from our analysis of BRCA1 and BRCA2 missense substitutions extrapolates reliably to other genes.
Approach 3
An intermediate approach is to consider that the PT+SJM variants are qualitatively different to the missense variants, implying that their effect should be estimated independently. Hence a two-degree of freedom test is applied using a logistic regression model in which grade 0 plus the seven missense grades are entered as a continuous variable, together with an indicator of membership in the PT+SJM grade 8. The hypothesis that modifications to the gene affect disease outcome is provided by the likelihood ratio test of this two-degrees of freedom model against the null model (risk independent of all variants).
Finally, the relative contribution of each specific grade of missense substitutions, and the PT+SJM grade 8, can be calculated as an attributable fraction, using the observed frequency in controls and the odds ratios calculated from any one of the three approaches.
In conclusion, the mutation screening data analysis methods that we propose allow powerful tests of association between a gene, with all its observed missense, truncating, and splice-modifying mutations, and a specific disease via case-control data. Of course, these approaches add the burden of building good-quality PMSAs for each gene of interest to provide background data required for the analysis. However, we have been able to build ATM and CHEK2 alignments to the phylogenetic depth used here for analysis of BRCA1 and BRCA2 (http://agvgd.iarc.fr/alignments.php) and believe that the cost and effort involved in case-control mutation screening warrant the implied extra work to achieve a more complete data analysis. Beyond identifying new genes implicated in specific diseases, the Align-GVGD grades defined here have application to the classification of variants within genes of known pathogenic potential. For BRCA1 and BRCA2 they have been assigned empirically determined posterior probabilities that the missense substitutions they contain are pathogenic. These posterior probabilities are specifically intended for use as prior probabilities in future studies of unclassified missense substitutions in these two genes. The approach through which these probabilities were determined may be adapted to analyses of unclassified missense substitutions in other high-risk cancer susceptibility genes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Amie Deffenbaugh and Lynne Ann Burbidge for compiling the BRCA1 and BRCA2 missense substitution dataset used in this analysis. We thank Paul B. Samollow and R. Andrew Cameron for providing opossum and sea urchin tissue samples, respectively. We thank Davit Babikyan, Laure Barjhoux, Yuan Chen, David Frank, Stephanie Monnier, and Andre Zharkikh for contributing to model organism BRCA1 and BRCA2 sequencing efforts. We thank Andre Zharkikh and Diana Iliev for calculating substitution rate constants from the alignments. We thank James McKay, Jon Wakefield, and Rayjean Hung for helpful discussions and critical reading of the manuscript. Finally, we also thank the entire Breast Information Core (BIC) database steering committee for their encouragement of our efforts in BRCA variant classification.
Grant sponsors: Canadian Institutes of Health Research (CIHR) INHERIT BRCAs Research Program; Mayo Clinic Breast Cancer SPORE; Grant number: P50 CA116201.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Abkevich V, Zharkikh A, Deffenbaugh A, Frank D, Chen Y, Shattuck D, Skolnick MH, Gutin A, Tavtigian SV. Analysis of missense variation in human BRCA1 in the context of interspecific sequence variation. J Med Gen. 2004;41:492–507. doi: 10.1136/jmg.2003.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Arbiza L, Casadio R, Dopazo J, Dopazo H, Marti-Renom MA. Use of estimated evolutionary strength at the codon level improves the prediction of disease-related protein mutations in humans. Hum Mutat. 2008;29:198–204. doi: 10.1002/humu.20628. [DOI] [PubMed] [Google Scholar]
- Chan PA, Duraisamy S, Miller PJ, Newell JA, McBride C, Bond JP, Raevaara T, Ollila S, Nystrom M, Grimm AJ, Christodoulou J, Oetting WS, Greenblatt MS. Interpreting missense variants: comparing computational methods in human disease genes CDKN2A, MLH1, MSH2, MECP2, and tyrosinase (TYR). Hum Mutat. 2007;28:683–693. doi: 10.1002/humu.20492. [DOI] [PubMed] [Google Scholar]
- Chenevix-Trench G, Healey S, Lakhani S, Waring P, Cummings M, Brinkworth R, Deffenbaugh AM, Burbidge LA, Pruss D, Judkins T, Scholl T, Bekessy A, Marsh A, Lovelock P, Wong M, Tesoriero A, Renard H, Southey M, Hopper JL, Yannoukakos K, Brown M, Easton D, Tavtigian SV, Goldgar D, Spurdle AB. Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res. 2006;66:2019–2027. doi: 10.1158/0008-5472.CAN-05-3546. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, Tavtigian SV, Monteiro AN, Iversen ES, Couch FJ, Goldgar DE. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Costa C, Orozco M, de la Cruz X. Sequence-based prediction of pathological mutations. Proteins. 2004;57:811–819. doi: 10.1002/prot.20252. [DOI] [PubMed] [Google Scholar]
- Fitch WM, Markowitz E. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem Genet. 1970;4:579–593. doi: 10.1007/BF00486096. [DOI] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Deffenbaugh AM, Monteiro AN, Tavtigian SV, Couch FJ. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet. 2004;75:535–544. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Byrnes GB, Spurdle AB, Iversen ES, Greenblatt MS, IARC Unclassified Genetic Variants Working Group Integration of various data sources for classifying uncertain variants into a single model. Hum Mutat. 2008;29:1265–1272. doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Greenblatt MS, Beaudet JG, Gump JR, Godin KS, Trombley L, Koh J, Bond JP. Detailed computational study of p53 and p16: using evolutionary sequence analysis and disease-associated mutations to predict the functional consequences of allelic variants. Oncogene. 2003;22:1150–1163. doi: 10.1038/sj.onc.1206101. [DOI] [PubMed] [Google Scholar]
- Karchin R, Monteiro AN, Tavtigian SV, Carvalho MA, Sali A. Functional impact of missense variants in BRCA1 predicted by supervised learning. PLoS Comput Biol. 2007;3:e26. doi: 10.1371/journal.pcbi.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Kanetsky PA, Tsang S, Gold B, Munroe D, Rebbeck T, Swoyer J, Ter-Minassian M, Hedayati M, Grossman L, Goldstein AM, Calista D, Pfeiffer RM. MC1R, ASIP, and DNA repair in sporadic and familial melanoma in a Mediterranean population. J Natl Cancer Inst. 2005;97:998–1007. doi: 10.1093/jnci/dji176. [DOI] [PubMed] [Google Scholar]
- Lovelock PK, Healey S, Au W, Sum EY, Tesoriero A, Wong EM, Hinson S, Brinkworth R, Bekessy A, Diez O, Izatt L, Solomon E, Jenkins M, Renard H, Hopper J, Waring P, Tavtigian SV, Goldgar D, Lindeman GJ, Visvader JE, Couch FJ, Henderson BR, Southey M, Chenevix-Trench G, Spurdle AB, Brown MA. Genetic, functional, and histopathological evaluation of two C-terminal BRCA1 missense variants. J Med Genet. 2006;43:74–83. doi: 10.1136/jmg.2005.033258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Hein J. A nucleotide substitution model with nearest-neighbour interactions. Bioinformatics. 2004;20(Suppl 1):I216–I223. doi: 10.1093/bioinformatics/bth901. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2006;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- Spurdle AB, Lakhani SR, Healey S, Parry S, Da Silva LM, Brinkworth R, Hopper JL, Brown MA, Babikyan D, Chenevix-Trench G, Tavtigian SV, Goldgar DE. Clinical classification of BRCA1 and BRCA2 DNA sequence variants: the value of cytokeratin profiles and evolutionary analysis--a report from the kConFab Investigators. J Clin Oncol. 2008;26:1657–26. 1663. doi: 10.1200/JCO.2007.13.2779. [DOI] [PubMed] [Google Scholar]
- Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe Wr, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Oliphant A, Shattuck-Eidens D, Bartel PL, Thomas A, Frank TS, Pruss D, Skolnick MH. Genomic organization, functional analysis, and mutation screening of BRCA1 and BRCA2. In: Fortner JG, Sharp PA, editors. Accomplishments in cancer research, 1996. Lippincott-Raven; New York: 1997. pp. 189–204. [Google Scholar]
- Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Greenblatt MS, Lesueur F, Byrnes GB, IARC Unclassified Genetic Variants Working Group In silico analysis of missense substitutions using sequence-alignment based methods. Hum Mutat. 2008;29:1327–1336. doi: 10.1002/humu.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.