To the editor:
GATA2 encodes a transcription factor that regulates hematopoiesis and vascular development, and germline mutations cause MonoMAC syndrome,1,2 Emberger syndrome,3 and some cases of mild chronic neutropenia.4 Patients with each of these disorders are predisposed to myelodysplastic syndrome (MDS) and acute myeloid leukemia that is frequently associated with somatic acquisition of monosomy 7.5,6
Juvenile myelomonocytic leukemia (JMML) is an aggressive childhood malignancy with overlapping features of an MDS and myeloproliferative neoplasm (MPN) that is caused by mutations that aberrantly activate Ras/MAPK signaling. Mutations in NF1, NRAS, KRAS, PTPN11, and CBL are found in 85% to 90% of newly diagnosed patients.7,8 Given the frequent occurrence of somatic monosomy 7 in JMML7 as well as the fact that there are several germline syndromes that predispose children to developing both transient and aggressive forms of JMML, we hypothesized that mutations in GATA2 play a role in its development.
Specimens from 57 patients with JMML were screened for GATA2 mutations. Patient samples and clinical data were collected from the Children’s Oncology Group trial AAML0122.9 DNA was extracted from peripheral blood or bone marrow as per previous protocols and whole genome amplified using Qiagen REPLI-g kits. We performed bidirectional Sanger sequencing (Beckman Coulter Genomics, Danvers, MA) of the entire coding region of GATA2 (NM_001145661.1) and aligned the sequences using CLC software (CLC Bio, Aarhus, Denmark). Only missense, splice site, or nonsense mutations were evaluated using SIFT (Sorting Tolerant from Intolerant)10 to predict the impact of identified mutations on the structure and function in the resultant protein.
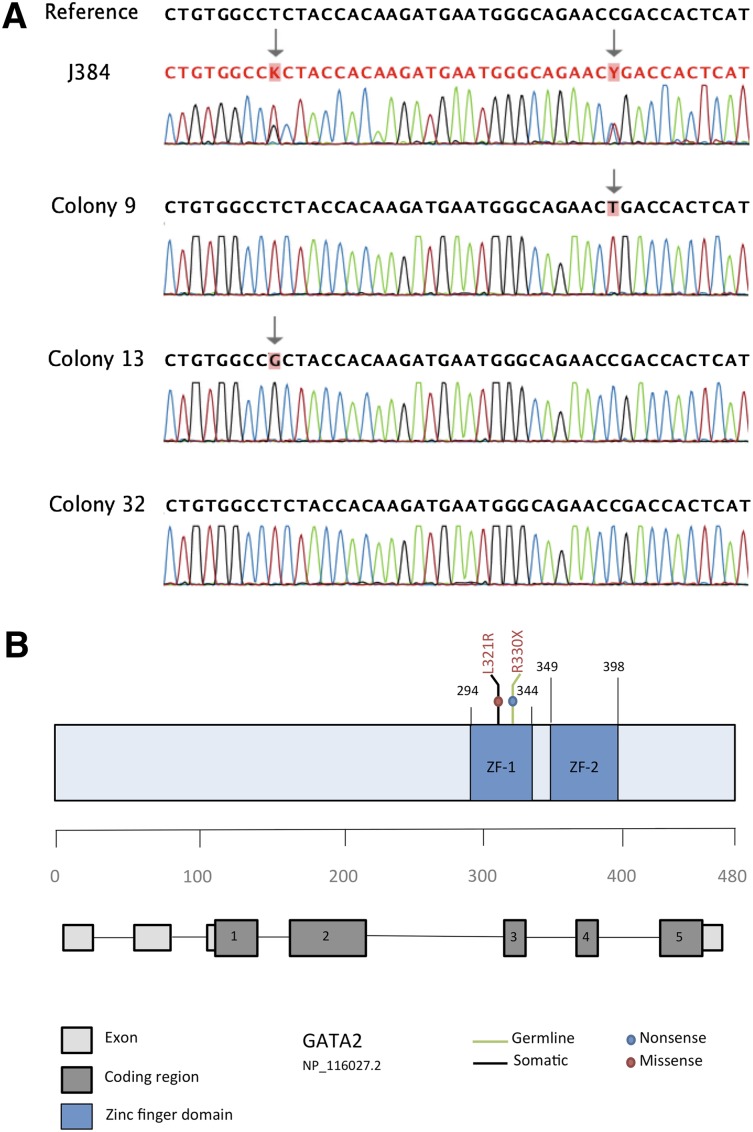
GATA2 was mutated in only 1 of 57 JMML specimens. Peripheral blood from patient J384 contained a nonsense point mutation (Figure 1) at c.988C>T (R330X), which has been reported as a germline mutation in patients with mild chronic neutropenia.4 Patient J384 also carried a second missense point mutation at c.962T>G (L321R), which was predicted to be damaging by SIFT, PolyPhen 2.0, and Mutation Assessor. Subcloning of the mutant amplicon using a TA cloning kit with pCR 2.1 vector (Invitrogen) followed by directed sequencing of individually transformed colonies revealed that the 2 sequence variants only occurred in a trans configuration. Out of 40 amplicons sequenced, 20 displayed the c.988C>T transition, 16 had the c.962T>G variant, and 4 were wild-type. We therefore hypothesize that c.988C>T was inherited as a germline event, whereas c.962T>G represents a somatic acquisition. Unfortunately, granulocyte-macrophage progenitor colonies at diagnosis were not available for analysis, nor was parental DNA. Previous studies of patient J384 revealed a subclonal KRAS G12D mutation (c.35G>A) as well as monosomy 7 in his JMML bone marrow.
Figure 1.
Identification of 2 distinct GATA2 mutations in patient J384. (A) Bidirectional sequencing of patient sample J384 revealed 2 distinct sequence variants in both the forward (shown here) and reverse strands. Sequencing of 40 individual colony picks after subcloning revealed that each sequence variant occurred in a trans configuration (CP9 and CP13 are shown here as examples). In addition, 10% of colony picks (ie, CP32) revealed a wild-type sequence, indicating that at least 1 of the 2 variants was a somatic event. (B) Schematic representation of the GATA2 gene highlighting the proposed germline and somatic mutations identified in patient J384. Both mutations are present in coding region 3, which translates to the zinc finger 1 region of the protein.
Patient J384 met the World Health Organization diagnostic criteria for JMML based on having splenomegaly, an absolute monocyte count >1000 (1 × 109/μL), <20% bone marrow blasts, absence of t(9;22) or BCR/ABL, and presence of myeloid precursors in peripheral blood with a white blood cell count >10 000 (10 × 109/μL).9 However, he also possessed features less common in JMML, including older age at diagnosis (4 years and 10 months) and, in particular, a normal response in myeloid progenitor cells to the cytokine granulocyte-macrophage colony-stimulating factor in methylcellulose culture, which is rarely seen in JMML.11
We speculate that a somatic “second hit” in the normal GATA2 allele accelerated the onset of MDS in patient J384, with monosomy 7 and the oncogenic KRAS mutation cooperating to drive clonal outgrowth resulting a JMML-like MDS/MPN disease. Patient J384 died within 3 months of his initial diagnosis without undergoing transplant. Cryopreserved bone marrow was not available to define the acquisition of pathogenic sequence variants in this patient.
This case is the first description of GATA2 mutations in a patient suspected of having JMML but who, on retrospective review, did not manifest all of the classic features. In addition, this is the first report of biallelic GATA2 mutations in a hematologic malignancy. Finally, this is also the first reported case of a concurrent KRAS mutation and monosomy 7 in a patient with a presumed germline GATA2 mutation. Although GATA2 mutations are rare in JMML, this work expands the spectrum of hematologic cancers associated with germline GATA2 mutations and supports sequencing this gene in patients with atypical features of JMML, particularly those with monosomy 7.
Authorship
Acknowledgments: The authors thank the patients and their families for participating in the Children’s Oncology Group trial AAML0122, without which this research would not be possible. This work was supported by the Leukemia and Lymphoma Society, grant 6059-09 (M.L.L.); the National Institutes of Health, National Cancer Institute, grant T32 CA128583 (E.S.), grant R37 CA72614 (K.M.S.), and grant R01 CA095621 (P.D.E.); the Frank A. Campini Foundation (E.S., M.L.L., K.M.S.); and the Team Conner Foundation (M.L.L.). K.S. is an American Cancer Society Research Professor.
Contribution: E.S., K.M.S., and M.L.L. designed the experiment and analyzed the data; E.S. performed the experiments and wrote the paper; K.M.S. and M.L.L. edited the paper; and Y.L.L., P.D.E., R.P.C., and T.M.C. provided patient samples and clinical data. All authors reviewed the literature, contributed to specific sections, and reviewed the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mignon Loh, Department of Pediatrics, Benioff Children’s Hospital/Helen Diller Family Comprehensive Cancer Center, 513 Parnassus Ave, HSE-302, Box 0519, University of California, San Francisco, San Francisco, CA 94143; e-mail: lohm@peds.ucsf.edu.
References
- 1.Hsu AP, Sampaio EP, Khan J, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118(10):2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hahn CN, Chong C-E, Carmichael CL, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43(10):1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostergaard P, Simpson MA, Connell FC, et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet. 2011;43(10):929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- 4.Pasquet M, Bellanné-Chantelot C, Tavitian S, et al. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood. 2013;121(5):822–829. doi: 10.1182/blood-2012-08-447367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalra R, Dale D, Freedman M, et al. Monosomy 7 and activating RAS mutations accompany malignant transformation in patients with congenital neutropenia. Blood. 1995;86(12):4579–4586. [PubMed] [Google Scholar]
- 6.Calvo KR, Vinh DC, Maric I, et al. Myelodysplasia in autosomal dominant and sporadic monocytopenia immunodeficiency syndrome: diagnostic features and clinical implications. Haematologica. 2011;96(8):1221–1225. doi: 10.3324/haematol.2011.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergstraesser E, Hasle H, Rogge T, et al. Non-hematopoietic stem cell transplantation treatment of juvenile myelomonocytic leukemia: a retrospective analysis and definition of response criteria. Pediatr Blood Cancer. 2007;49(5):629–633. doi: 10.1002/pbc.21038. [DOI] [PubMed] [Google Scholar]
- 8.Loh ML. Recent advances in the pathogenesis and treatment of juvenile myelomonocytic leukaemia. Br J Haematol. 2011;152(6):677–687. doi: 10.1111/j.1365-2141.2010.08525.x. [DOI] [PubMed] [Google Scholar]
- 9.Castleberry RP, Loh ML, Jayaprakash N, et al. Phase II window study of the farnesyltransferase inhibitor R115777 (Zarnestra®) in untreated juvenile myelomonocytic leukemia (JMML): a Children’s Oncology Group study. Blood. 2005;106(11):2587. [Google Scholar]
- 10.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 11.Liu YL, Castleberry RP, Emanuel PD. PTEN deficiency is a common defect in juvenile myelomonocytic leukemia. Leuk Res. 2009;33(5):671–677. doi: 10.1016/j.leukres.2008.09.036. [DOI] [PubMed] [Google Scholar]